Abstract
Background and study aims Capsule endoscopy (CE) is the preferred method for small bowel (SB) exploration. With a mean number of 50,000 SB frames per video, SBCE reading is time-consuming and tedious (30 to 60 minutes per video). We describe a large, multicenter database named CAD-CAP (Computer-Assisted Diagnosis for CAPsule Endoscopy, CAD-CAP). This database aims to serve the development of CAD tools for CE reading.
Materials and methods Twelve French endoscopy centers were involved. All available third-generation SB-CE videos (Pillcam, Medtronic) were retrospectively selected from these centers and deidentified. Any pathological frame was extracted and included in the database. Manual segmentation of findings within these frames was performed by two pre-med students trained and supervised by an expert reader. All frames were then classified by type and clinical relevance by a panel of three expert readers. An automated extraction process was also developed to create a dataset of normal, proofread, control images from normal, complete, SB-CE videos.
Results Four-thousand-one-hundred-and-seventy-four SB-CE were included. Of them, 1,480 videos (35 %) containing at least one pathological finding were selected. Findings from 5,184 frames (with their short video sequences) were extracted and delimited: 718 frames with fresh blood, 3,097 frames with vascular lesions, and 1,369 frames with inflammatory and ulcerative lesions. Twenty-thousand normal frames were extracted from 206 SB-CE normal videos. CAD-CAP has already been used for development of automated tools for angiectasia detection and also for two international challenges on medical computerized analysis.
Introduction
Capsule Endoscopy (CE) has rapidly become the standard minimally invasive method for visualization of the small bowel (SB). CE is considered the first-line method of investigation of SB diseases 1 and may become the leading diagnostic tool for the entire gastrointestinal tract. With a mean number of 50,000 SB frames per video, SB-CE reading is time-consuming and tedious (30 to 60 minutes per video) 2 . This entails an inherent risk of missed lesions during the reading process by physicians.
Computer-aided diagnosis (CAD) algorithms are increasingly used to assist physicians in reading and interpreting medical images. The field of CAD has rapidly expanded from radiology 3 4 to many other settings, such as pathology, dermatology, cardiology, and ophthalmology 5 6 7 . In August 2018, the U.S. Food and Drug Administration approved the first medical device to use artificial intelligence (AI) for screening diabetic retinopathy 8 . Development of CAD has also emerged in endoscopy, mostly in the setting of colon polyp detection and characterization 5 9 10 . We and others 11 12 13 , believe that CAD will meet the challenge of automated lesion detection in SB-CE, thus improving diagnostic accuracy and relieving the burden of SB-CE reading 14 .
CAD algorithms dedicated to SB-CE combine elements of AI and computer vision. This development is mostly based on machine learning (ML) algorithms for automated detection of relevant patterns in CE still frames and videos 15 16 . To be efficient, ML algorithms must be trained on large databases. Any data (still frame or video) should be labeled, and any finding within frames/videos can be annotated or delimitated to better guide and supervise the ML process. Only a limited number of CE images databases is available 13 . Herein, we describe a large, multicenter database named CAD-CAP (computer-assisted diagnosis for capsule endoscopy). This database aims to serve development of CAD tools, from the initial steps (ML) to the last preclinical steps (assessment and comparison of performances).
Materials and methods
CAD-CAP is a national multicenter database approved by the French Data Protection Authority. Twelve French endoscopic units participated to the database ( Table 1 and Table 2 ). A research group specialized in the computerized analysis of medical images was also involved.
Table 1. Characteristics of the CAD-CAP database.
| Characteristic | |
| Total number of SB3-CE (n) | 4174 |
| Total number of SB3-CE (n) videos with at least one abnormal finding | 1480 |
| Total number of frames with abnormal findings (n) | 5124 |
| Total number of normal frames (n) | 20,000 |
| Gender ratio M/F (%) | 59/41 % |
| Mean ± SD age (years) | 64 ± 15 |
CAD-CAP, computer-assisted diagnosis for capsule endoscopy; SB3-CE, third-generation small bowel capsule endoscopy; SD, standard deviation
Table 2. CAD-CAP database details and contribution of the centers.
| Centers | Number of SB3-CE provided |
| Brest Hospital | 340 |
| Henri Mondor Hospital, Créteil | 358 |
| Lomme Hospital | 173 |
| Lille Hospital | 445 |
| Edouard Herriot Hospital, Lyon | 450 |
| Nantes Hospital | 242 |
| Nice Hospital | 471 |
| Cochin Hospital, Paris | 466 |
| HEGP, Paris | 395 |
| Saint Antoine Hospital, Paris | 541 |
| Tenon Hospital, Paris | 58 |
| Strasbourg Hospital | 235 |
SB3-CE: third-generation small bowel capsule endoscopy; HEGP: Hôpital Européen Georges Pompidou .
Data collection and preparation of locally read SB-CE
In September 2016, all third-generation SB-CE videos (Pillcam SB3 system, Medtronic, Minnesota, United States) registered in the 12 participating endoscopy units were retrospectively collected, and de-identified. Clinical data were noted (age, gender, indication for SB-CE). Any SB icon selected by the local reader was considered as a frame of interest. Each frame of interest was extracted and included in the CAD-CAP database, together with a short adjacent video sequence that included 25 frames upstream and downstream the index frame. Two pre-med students were trained, and supervised by an expert reader (XD), to select and to delimitate any lesion found into the selected frames of interest. The delimitation process used Adobe Photoshop CS6 (Adobe Systems, United States) and GIMP softwares (GNOME Foundation, United States) with a Wacom (Wacom Co., Ltd, Japan) pen tablet connected to a laptop.
Central reading of selected frames
During several face-to-face meetings, three expert SB-CE readers (XD, SL, JPLM) screened all selected frames of interest (and associated 9-second video sequence including 25 frames upstream and 25 downstream the index still frames), and all delimitations of abnormal findings within each selected frames. Blurred or unanalyzable frames were excluded. Findings' delimitations were also reviewed, and remade when necessary.
Abnormal findings were first sorted into three different categories: (a) fresh blood and clots; (b) vascular findings; (c) ulcerative/inflammatory findings. Then, in these categories, all abnormal findings were sorted again according to their relevance. Relevance was defined as followed by a group of four SB-CE experts (FC, XD, GR, JCS). Fresh blood and clots 17 , typical angiectasia 18 and ulcerated lesions 19 were considered "highly relevant" findings. Non-ulcerated but inflammatory findings (for instance erythema, edema, denudation) 20 were considered "moderately relevant" findings. Subtle vascular lesions (for instance erythematous patches, red spots/dots, phlebectasia, and so-called "diminutive angiectasia") were considered "poorly relevant" findings 18 .
Control frames
A set of 206 normal and complete SB-CE videos was used to create a control dataset. Still frames were automatically extracted from these latter videos, every 1 % of the SB sequence (between 1 % and 100 % of the SB sequence). Thus, 100 normal frames were extracted per video. Again, for any normal extracted still frame, a short video sequence was also captured (with 25 frames upstream and downstream the index frame). All supposedly normal frames were reviewed by two readers (XD, RL). Doubtful, blurred or irrelevant frames were excluded.
Database splitting into two datasets
All the frames (and short video sequences) were randomly grouped through a computer-generated algorithm into two different, independent subsets of still frames, of equal sizes for each category (considering the presence and type of abnormal finding and the category of relevance). One dataset was dedicated to the development and training of algorithms (machine learning). The other dataset was dedicated to the testing and validation of algorithms.
Results
We collected 4,174 third-generation SB-CE from the 12 participating centers ( Table 1 ). We included in the database 1,480 videos (35 %) containing at least one frame of interest (with abnormal findings), and 206 normal videos (as controls). Clinical characteristics from these 1480 SB-CE were collected (59 % men, 41 % female, mean age 64). SB-CE were mainly indicated for obscure gastrointestinal bleeding (67 %), suspected Crohn’s disease (CD) (12 %), and sometimes (21 %) for others various indications such as coeliac disease and Peutz-Jeghers polyposis. In total we extracted 6,013 still frames (with their adjacent short video sequences). Abnormal findings were delimitated by the pre-med students within 5,124 frames, and then reviewed and sorted by the experts ( Fig. 1 ). Of the frames, 3103 contained images of vascular abnormalities (817 frames with highly relevant images of angiectasia, 2286 frames with other, poorly relevant, images of vascular lesions), 1370 frames contained images of ulcerative/inflammatory findings (1057 frames with highly relevant images of ulcerated lesions, 313 frames with moderately relevant images of inflammatory but not ulcerated lesions). Highly relevant images of fresh blood and clots were contained in 651 frames ( Fig. 2 ). Three-hundred-and-ninety-one frames with images of miscellaneous lesions (including tumors and polyps) were not delimitated and not included in this first version of the database, because this number was considered too low for machine learning). Twenty-thousand normal frames (controls, with their short video sequences) were extracted from the 206 SB-CE normal and complete videos. Two subsets of 12,562 still frames (2562 with abnormal findings with an associated, delimitated frame, and 10,000 normal, control, frames) were selected.
Fig. 1.
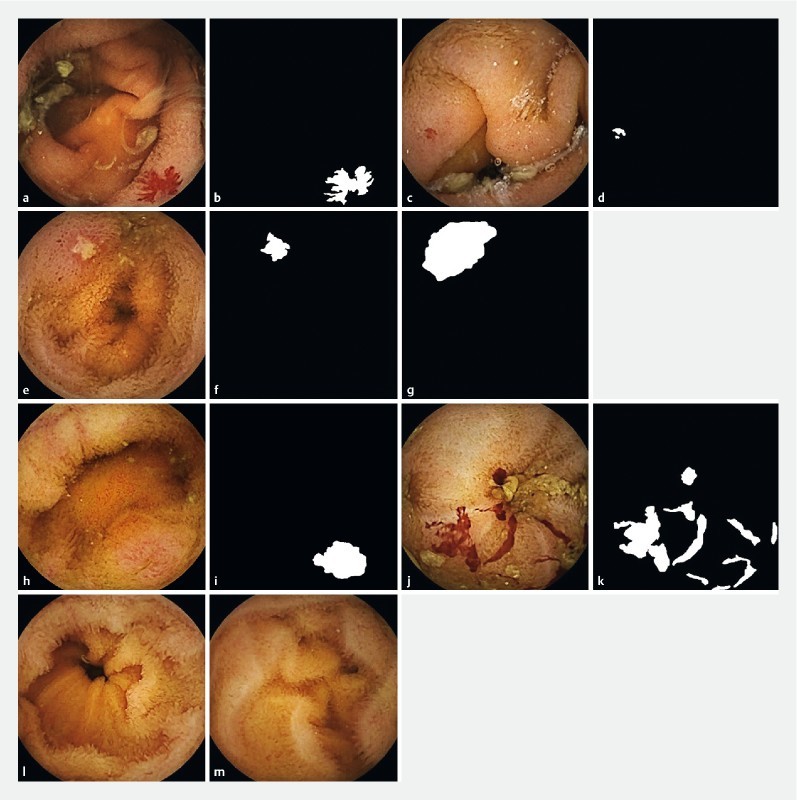
Examples of native small bowel capsule endoscopy frames, with their corresponding delimitations. a Native deidentified still frame showing a vascular lesion considered "highly relevant.” b Manual delimitation of the vascular lesion within frame 1a. c Native deidentified still frame showing a vascular lesion considered " poorly relevant.” d Manual delimitation of the vascular lesion within frame 1c. e Native deidentified still frame showing an ulcerative lesion (considered "highly relevant"). f Manual delimitation of the ulcerative lesion within frame 1e g Manual delimitation of the ulcerative lesion including mucosal inflammatory changes within frame 1e. h Native deidentified still frame showing an ulcero-inflammatory lesion (considered "moderately relevant"). i Manual delimitation of the ulcero-inflammatory lesion within frame 1 hour. j Native deidentified still frame showing fresh blood (considered "highly relevant"). k Manual delimitation of the fresh blood within frame 1j. l , m Two normal control frames (no delimitation needed)
Fig. 2.
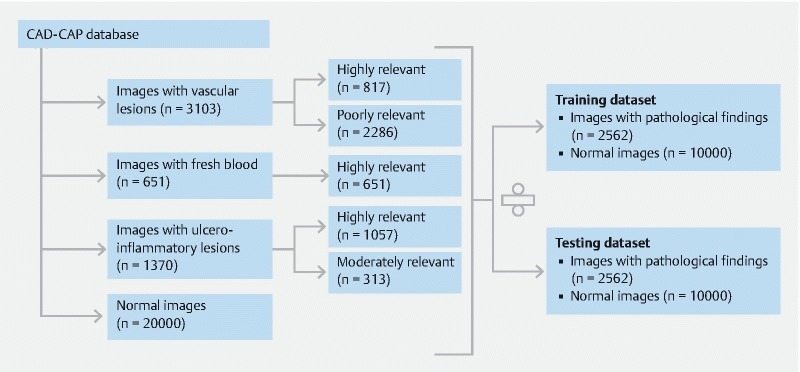
CAD-CAP database distribution.
Discussion
The multicenter CAD-CAP database was created to promote the development of AI applications in the field of SB-CE. This database has several strengths, the first being the volume of collected data. Data collection was multicenter, thus providing over 4,000 SB-CE capsules videos and reports. Six thousand frames with abnormal findings and 20,000 normal frames, were provided. This makes CAD-CAP the largest published SB3-CE database so far. The second strength is the quality of the collected data. Only third-generation SB-CE were considered, which provides homogenous frames and videos (in terms of resolution, luminosity, and overall quality). CAD-CAP is composed of supervised and validated, delimitated findings from SB-CE. Three levels of pertinency (relevance) of abnormal findings were defined before frames were sorted by a group of expert readers. Future CAD software could therefore be challenged at different levels (diagnosis performance for not/poorly/moderately/highly relevant lesions) by using the CAD-CAP database. It is unknown whether private CE or AI companies have such databases. Since 2017, the KID project by Koulaouzidis A et al. 13 has aimed to provide an Internet-based digital video atlas of capsule endoscopy for research purposes.
The CAD-CAP database has some limitations. First, some computer vision experts may consider the number of frames collected insufficient. For comparison, 128,175 retinal images frames were used by Gulshan et al. 7 to develop a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Another 129,450 skin images were used by Esteva et al. 6 for development of an automated tool for classification of malignant vs benign skin lesions. However, the quality of collected data (with delimitated abnormal findings within each frame of interest), and the number of normal frames used as controls may balance out this limitation. Second, the CAD-CAP database only used data from the SB-CE PillCam SB3 system, thus restraining potential development of CAD algorithms with other SB-CE systems that have different image processing inner characteristics. Third, SB preparation quality was not considered in this database because of the lack of fully validated cleansing score for SB-CE 21 . Poor preparation quality should be taken into consideration as it may generate false-positive findings. Fourth, and most importantly, polyps/tumors are certainly "highly relevant findings," but such frames were not included in this first version of the CAD-CAP database because of their limited number. We plan to collect a significant number of frames with polypoid lesions from ongoing studies and include them in the next version of the database. Fifth, the clinical relevance of many SB findings is not determined yet in the literature. To overcome this limitation, our group is currently conducting a study based on an illustrated script questionnaire, aiming to better determine the clinical relevance of SB-CE findings. This study includes 576 frames with various types and numbers of findings, and different clinical settings. It should provide a strong asset for a better classification of SB lesions seen in CE, thus allowing updating a second version of CAD-CAP with a more accurate findings’ classification. Sixth, there was no sample size calculation. We tried to collect as many CE as possible, and to accumulate a high number of high-quality images (i. e., with a clear delimitation of any finding within frames).
AI and machine learning processes are increasingly used for medical and research purposes. This shift provides a large variety of perspectives concerning image analysis and automated diagnostic tools. Because of the exponential growth in healthcare data, new processing methods are needed for analysis of large data sets. That will completely transform our current clinical practice. Image recognition software can already make predictions as effectively as, or even better than, physicians 10 22 . Machine learning directly from medical data could prevent clinical errors due to human cognitive biases, positively impacting patient care 23 .
In medicine, AI is still in its nascent stage, but it should not be long before it becomes a part of everyday life in daily practice. For example, the first FDA-approved AI tool in medicine has already shown high sensitivity (87 %–90 %) and specificity (98 %) for detecting moderately severe retinopathy and macular edema, compared with 54 ophthalmologists and senior residents 7 . Similarly, another application has demonstrated the efficacy of an automated tool for classifying appearance of skin lesions 6 . Many others are under development.
Unsurprisingly, the AI revolution has also started in the field of endoscopy. Developed algorithms are now able to differentiate diminutive adenomas from hyperplastic polyps during colonoscopy in real time with high accuracy 5 . Other studies have shown that an automated detection tool for Barrett’s esophagus neoplasia can assist endoscopists in detecting early neoplasia on volumetric laser endomicroscopy 24 . Many more tools and studies are to come, as the field of AI is expanding exponentially 25 .
Indeed, AI applications are also emerging in the field of CE. In the next few years, automated software should be able to facilitate more efficient reporting. Many studies have been conducted to establish a cleansing score for SB-CE quality preparation. Different approaches have focused on the abundance of bubbles 26 , red over green color 27 28 , and quality of bowel preparation in colon capsule endoscopy 29 . Algorithms based on machine learning approaches have also been developed to detect SB lesions 12 28 30 . Aoki et al. 16 developed a CAD algorithm for detection of erosions and ulcerations in SB-CE based on a deep convolutional neural network with sensitivity, specificity, and accuracy of 88.2 %, 90.9 % and 90.8 %, respectively. Although AI in endoscopy remains very promising, it still requires further research and clinical trials to be validated in daily clinical practice.
The CAD-CAP database has already been used for international challenges in medical image computing. During the 2017 and 2018 Medical Image Computing and Computer-Assisted Intervention (MICCAI) meetings, 14 and 20 research teams, respectively, were challenged to develop CAD tools dedicated to SB-CE 31 . We recently reported of an automated, highly sensitive, highly specific algorithm for diagnosis of SB angiectasia 12 , which was developed using a subset of frames from the CAD-CAP database. This tool reached high performance with 100 % sensitivity and negative predictive value and a 96 % specificity and positive predictive value. Patent application and industrial partnership are also under consideration.
Conclusion
In conclusion, the CAD-CAP database aims to promote development of CAD reading in SB-CE. It represents an opportunity to design fully-automated software for detection of all SB lesions to facilitate and improve the future of CE reading, reviewing and reporting.
Acknowledgements
The CAD-CP database was developed with the support of the Société Nationale Française de Gastroentérologie (SNFGE, grant FARE) and with the support of MSD France.
Footnotes
Competing interests Dr. Saurin is a consultant for Capsovision, Medtronic, and Intromedic. Dr. Rahmi is a consultant for Medtronic. Dr. Leenhardt is cofounder and shareholder of Augmented Endoscopy, and has given lectures for Abbvie. Dr. Dray is cofounder and shareholder of Augmented Endoscopy and has acted as a consultant for Alfasigma, Bouchara Recordati, Boston Scientific, Fujifilm, Medtronic, and Pentax. Dr. Huvelin is a consultant for Medtronic.
References
- 1.Iddan G, Meron G, Glukhovsky A et al. Wireless capsule endoscopy. Nature. 2000;405:417. doi: 10.1038/35013140. [DOI] [PubMed] [Google Scholar]
- 2.McAlindon M E, Ching H-L, Yung D et al. Capsule endoscopy of the small bowel. Ann Transl Med. 2016;4:369. doi: 10.21037/atm.2016.09.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajpurkar P, Irvin J, Zhu Ket al. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learningarXiv preprint arXiv:1711.05225 (2017)
- 4.Chilamkurthy S, Ghosh R, Tanamala S et al. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. The Lancet. 2018;392:2388–2396. doi: 10.1016/S0140-6736(18)31645-3. [DOI] [PubMed] [Google Scholar]
- 5.Byrne M F, Chapados N, Soudan F et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94–100. doi: 10.1136/gutjnl-2017-314547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteva A, Kuprel B, Novoa R A et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulshan V, Peng L, Coram M et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 8.Commissioner O of the Press Announcements - FDA permits marketing of artificial intelligence-based device to detect certain diabetes-related eye problems https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm604357.htm[Apr 2018]
- 9.Byrne M F, Shahidi N, Rex D K. Will Computer-Aided Detection and Diagnosis Revolutionize Colonoscopy? Gastroenterology. 2017;153:1460–14640. doi: 10.1053/j.gastro.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Chen P-J, Lin M-C, Lai M-J et al. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568–575. doi: 10.1053/j.gastro.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Iakovidis D K, Koulaouzidis A. Automatic lesion detection in capsule endoscopy based on color saliency: closer to an essential adjunct for reviewing software. Gastrointest Endosc. 2014;80:877–883. doi: 10.1016/j.gie.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Leenhardt R, Vasseur P, Li C et al. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest Endosc. 2019;89:189–194. doi: 10.1016/j.gie.2018.06.036. [DOI] [PubMed] [Google Scholar]
- 13.Koulaouzidis A, Iakovidis D K, Yung D E et al. KID Project: an internet-based digital video atlas of capsule endoscopy for research purposes. Endosc Int Open. 2017;5:E477–E483. doi: 10.1055/s-0043-105488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iakovidis D K, Koulaouzidis A. Software for enhanced video capsule endoscopy: challenges for essential progress. Nat Rev Gastroenterol Hepatol. 2015;12:172–186. doi: 10.1038/nrgastro.2015.13. [DOI] [PubMed] [Google Scholar]
- 15.Lee J-G, Jun S, Cho Y-W et al. Deep Learning in Medical Imaging: General Overview. Korean J Radiol. 2017;18:570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoki T, Yamada A, Aoyama K et al. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc. 2019;89:357–36300. doi: 10.1016/j.gie.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 17.Saurin J-C, Delvaux M, Gaudin J-L et al. Diagnostic value of endoscopic capsule in patients with obscure digestive bleeding: blinded comparison with video push-enteroscopy. Endoscopy. 2003;35:576–584. doi: 10.1055/s-2003-40244. [DOI] [PubMed] [Google Scholar]
- 18.Leenhardt R, Li C, Koulaouzidis A Nomenclature and Semantic Description of Vascular Lesions in Small Bowel Capsule Endoscopy: an International Delphi Consensus Statement. Endosc Int Open. 2019. pp. E372–E379. [DOI] [PMC free article] [PubMed]
- 19.Buisson A, Filippi J, Amiot A et al. Su1229 Definitions of the Endoscopic Lesions in Crohnʼs Disease: Reproductibility Study and GETAID Expert Consensus. Gastroenterology. 2015;148:S445. [Google Scholar]
- 20.Gal E, Geller A, Fraser G et al. Assessment and validation of the new capsule endoscopy Crohn’s disease activity index (CECDAI) Dig Dis Sci. 2008;53:1933–1937. doi: 10.1007/s10620-007-0084-y. [DOI] [PubMed] [Google Scholar]
- 21.Yung D E, Rondonotti E, Sykes C et al. Systematic review and meta-analysis: is bowel preparation still necessary in small bowel capsule endoscopy? Expert Rev Gastroenterol Hepatol. 2017;11:979–993. doi: 10.1080/17474124.2017.1359540. [DOI] [PubMed] [Google Scholar]
- 22.Mori Y, Kudo S-E, Misawa M et al. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357–366. doi: 10.7326/M18-0249. [DOI] [PubMed] [Google Scholar]
- 23.Miller D D, Brown E W. Artificial Intelligence in Medical Practice: The Question to the Answer? Am J Med. 2018;131:129–133. doi: 10.1016/j.amjmed.2017.10.035. [DOI] [PubMed] [Google Scholar]
- 24.Swager A-F, van der Sommen F, Klomp S R et al. Computer-aided detection of early Barrett’s neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc. 2017;86:839–846. doi: 10.1016/j.gie.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Kashin S.Artificial intelligence: the rise of the machinesUnited European Gastroenterology Week. 20-24 oct 2018, Vienna, Austria.
- 26.Pietri O, Rezgui G, Histace A et al. Development and validation of an automated algorithm to evaluate the abundance of bubbles in small bowel capsule endoscopy. Endosc Int Open. 2018;6:E462–E469. doi: 10.1055/a-0573-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Weyenberg S JB, De Leest H TJI, Mulder C JJ. Description of a novel grading system to assess the quality of bowel preparation in video capsule endoscopy. Endoscopy. 2011;43:406–411. doi: 10.1055/s-0030-1256228. [DOI] [PubMed] [Google Scholar]
- 28.Abou Ali E, Histace A, Camus M et al. Development and validation of a computed assessment of cleansing score for evaluation of quality of small-bowel visualization in capsule endoscopy. Endosc Int Open. 2018;6:E646–E651. doi: 10.1055/a-0581-8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becq A, Histace A, Camus M et al. Development of a computed cleansing score to assess quality of bowel preparation in colon capsule endoscopy. Endosc Int Open. 2018;6:E844–E850. doi: 10.1055/a-0577-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan S, Xu L, Fan Y et al. Computer-aided detection of small intestinal ulcer and erosion in wireless capsule endoscopy images. Phys Med Biol. 2018;63:165001. doi: 10.1088/1361-6560/aad51c. [DOI] [PubMed] [Google Scholar]
- 31.Bernal J, Tajkbaksh N, Sanchez F J et al. Comparative Validation of Polyp Detection Methods in Video Colonoscopy: Results From the MICCAI 2015 Endoscopic Vision Challenge. IEEE Trans Med Imaging. 2017;36:1231–1249. doi: 10.1109/TMI.2017.2664042. [DOI] [PubMed] [Google Scholar]


