1. Introduction
The need continues for new prostate cancer (PCa) biomarkers for use in early detection, recurrence and metastatic disease diagnostics, therapeutic monitoring, and prediction of treatment benefit. As the urology, radiology and oncology disciplines continue to evaluate how to minimize the overdiagnosis and overtreatment associated with current prostate specific antigen (PSA) serum tests [1], there has been significant emphasis placed on multiparametric magnetic resonance imaging (mpMRI) based detection of clinically significant PCa. It is used for multiple indications including stratification of patients into low, intermediate, and high risk groups that guide treatment regimens from active surveillance to radical prostatectomy. Although an important step forward, inherent limitations with the use of MR in the detection of clinically significant cancer are still present and contribute to both false positives and false negatives [2]. Therefore optimizing tissue biomarker discovery that can be used as “ground truth” for further optimization of clinical imaging protocols represents a significant unmet need in the field.
As a potential alternative, genomic-based liquid biopsy targets of cell free DNA (cfDNA) and circulating tumor cells (CTC) have emerged [3]. A recent consensus report of clinical experts in circulating prostate cancer biomarkers highlights these needs, especially in regards to developing biomarkers for treatment decisions, efficacy and metastasis [4]. These opinions were all based on developing genomic-based cfDNA and CTC assays. Genomics-based classification and risk stratification have made great strides, but many limitations still exist due to the continued interpretation of complex genomic data, its relation to impact on clinical outcomes, and not every mutation may be clinically actionable. Therefore, employing complementary research strategies to better clarify the functional readout of the cancer genome, i.e, how pathways downstream of genome regulation enhance tumorigenesis, may lead to new approaches for diagnosis and treatment for advanced PCa, as well as new biomarker targets.
One well-known functional readout of the cancer genome is metabolism. Carbohydrate, or sugar, metabolism, specifically glucose, has been documented as a hallmark of tumorigenesis that enhances cell energetics (e.g. ATP synthesis) and biosynthetic processes (e.g. nucleotide and amino acid synthesis) [5]. This provides the basis of fluorodeoxyglucose positron emission tomographic (FDG-PET) imaging where tumor FDG uptake inversely correlates with survival in numerous cancers, including prostate cancer [6]. However, carbohydrates also can be incorporated into multiple types of complex carbohydrate structures that comprise the cell surface glycocalyx. These glycans are represented by N-linked and O-linked glycoproteins, glycosphingolipids, and glycosaminoglycans attached to proteoglycans and can interact with multiple extracellular matrix (ECM) proteins in the stroma, many of which are also glycosylated, as well various cell types including the immune system [7]. The role of the stroma in promoting and supporting cancer progression is a major area of cancer research, including PCa. Malignant transformation of the ECM disrupts the homeostatic tissue microenvironment, altering tissue processes of adhesion, cell death, migration, and proliferation to promote tumor growth [8]. Overall, targeting carbohydrate utilization as a functional end-product of metabolism, combined with targeted analysis of ECM constituents, could lead to novel diagnostic and therapeutic biomarkers of PCa across the clinical spectrum of the disease.
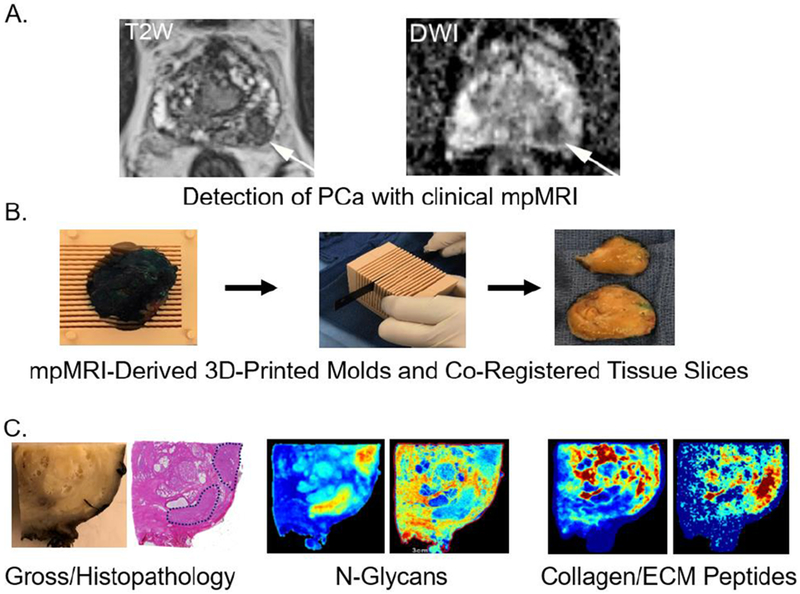
Herein, we propose a strategy for PCa biomarker discovery in tissues, that spatially registers tissue glycosylation and protein expression of tumor, ECM, and immune components by merging matrix assisted laser desorption ionization (MALDI) imaging mass spectrometry and clinical prostate multiparametric magnetic resonance imaging (mpMRI). Specifically, MRI scans that are performed prior to prostatectomy are used to generate histologic 3D printed molds that are used to section the prostate in the same orientation as the MRI scan [9]. This allows for the coregistration of MRI imaging biomarkers and histology. Once the tumor tissue is sectioned by the pathologist using these molds, it is formalin fixed and paraffin embedded. Molecular MALDI imaging mass spectrometry of glycans and ECM proteins from the same patient tissues are then performed on unstained slides that directly correspond to the clinical imaging slices (see Figure 1).
Figure 1. An integrated three dimensional multimodality work flow to evaluate new glycomic and extracellular matrix biomarker candidates for prostate cancer.
A. Representative pre-prostatectomy MRI images for T2W (T2 Weighted Image) and DWI (Diffusion-weighted image). B. A representative 3D-mold from mpMRI used for preparing prostatectomy tissue co-registered with mpMRI coordinates. C. Gross pathology and standard H&E stained tissue section, with two representative MALDI-IMS images of tumor and stroma localized N-glycans and collagenase derived ECM peptides in the same tissue slice.
2. Glycan and protein biomarkers of the PCa extracellular matrix.
Complex carbohydrates attached to proteins and lipids represent a major class of potential biomarkers because of their known correlation with the presence, pathology stage and metastatic potential of prostate cancers. It is well documented that malignant transformation and cancer progression result in fundamental changes in the glycosylation patterns of cell surface and secreted glycoproteins [7,10]. Historically, the majority of current FDA-approved tumor biomarkers are circulating glycoproteins (e.g.prostate specific antigen; PSA) or glycan antigens like sialyl Lewis A (e.g. CA 19-9 for hepatobiliary cancers). Of note, imaging of the prostate-specific membrane antigen (PSMA) glycoprotein with PET/CT is becoming increasingly popular in the detection of clinically significant PCa but is not yet FDA approved [6]. Biochemically, two major classes of carbohydrate antigens, di-fucosylated Lewis Y/F77 and high mannose structures, have been shown to be most consistently associated with advanced PCa [10]. One group has used several new methods based upon mass spectrometry imaging of prostate cancer FFPE tissues to further define the types of N-glycans detected in tumor, stroma and normal gland regions of the prostate [11]. This method allows for the co-localization of the glycans with various immunostains for the tumor microenvironment. Multiple tumor-associated glycans have been identified and current efforts are in progress to link these changes to prostate tumor progression. Further, defining these N-glycan changes could potentially lead to evaluation of other sugar-based clinical imaging probes, as well as correlates with existing glycoprotein imaging targets like PSMA.
Stromal organization is dependent on post-translational modifications of glycosylation, hydroxylation of lysines and prolines, and sulfation [12]. Recently, a novel MALDI imaging mass spectrometry approach to identify stromal and ECM proteins following digestion with collagenase was used to profile FFPE tumor tissues [12]. This method has the advantage of identifying many different types of collagen sub-types and multiple ECM proteins that interact with collagen. These molecules are difficult to identify using standard trypsin-based proteomic analysis workflows, as many ECM proteins, typified by collagens, are poor trypsin substrates. The collagenase imaging MS approach has recently been merged with the N-glycan imaging MS methods to perform the analyses on the same tissue slice. This allows co-localization and mapping of unique glycan and ECM features within the tumor microenvironment (see Figure 1). Of note, as the starting material for the experiments are FFPE tissue slides derived from large amounts of MRI-co-registered tissues, multiple sequential tissues can be analyzed to generate three dimensional molecular image maps. The workflow is also inclusive of any immunohistochemistry correlates of known PCa tissue markers, as well as immune or immunotherapy related targets.
3. ECM, glycans and tumor-immune interactions
Within the PCa tumor microenvironment, it is now established that there is significant crosstalk between active stroma that contributes to tumor progression and immune responses [13]. Stromal cells, which may originate from the normal adjacent tissue or from the tumor, promote connective tissue remodeling through ECM protein degradation and secretion [6]. Spatial analyses at a genetic level demonstrate that PCa progression involves molecular gradients that integrate a stromal response with inflammation [13], and PCa tumors with increased stromal density may be more immunosuppressive [14]. The interaction of the immune system with tumor and stroma is largely influenced by carbohydrate-binding lectins on immune cells and other carbohydrate binding proteins like galectins [15]. More studies are needed to understand how ECM stromal protein and carbohydrate composition influences the tumor-immune microenvironment.
4. Summary and future directions
The role of the prostate stroma in preventing or promoting effective immune responses to tumor is an active area of research, and it is certainly a contributor to the largely poor responses of prostate cancers to emerging immunotherapy strategies [13,15]. The imaging mass spectrometry approaches developed [11,12] yield unique spatially localized targets integrally linked with molecular changes in the prostate tumor microenvironment. The glycan and ECM biomolecules being identified have not been previously evaluated as biomarkers. The approach we describe represents a framework to identify new prostate cancer biomarkers, and certainly other methods could be used or incorporated. Overall, we hypothesize that our multi-modality approach of combining clinical imaging, molecular imaging mass spectrometry and multiplexed immunohistochemistry for three dimensional visualization and characterization of prostate tumors will identify multiple novel diagnostic and therapeutic biomarker targets. This approach can be fully integrated with existing clinical, histopathology and genomic assay workflows.
Acknowledgments
Funding
This work was supported by research grants from the U.S. National Institutes of Health and National Cancer Institute, R01 CA212409 and U54 MD010706.
Footnotes
Declaration of interest
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewers Disclosure
Peer reviewers on this manuscript have no relevant financial relationships or otherwise to disclose.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Fleshner K, Carlsson SV, Roobol MJ. The effect of the USPSTF PSA screening recommendation on prostate cancer incidence patterns in the USA. Nat Rev Urol. 2017; 14(1): 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mason BR, Eastham JA, Davis BJ, et al. Current Status of MRI and PET in the NCCN Guidelines for Prostate Cancer. J Natl Compr Canc Netw. 2019; 17(5): 506–513. [DOI] [PubMed] [Google Scholar]
- 3.Di Meo A, Bartlett J, Cheng Y, et al. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol Cancer. 2017; 16(1): 80. [DOI] [PMC free article] [PubMed] [Google Scholar]; (* An overview of genomic-based liquid biopsy targets)
- 4.Sumanasuriya S, Omlin A, Armstrong A, et al. Consensus Statement on Circulating Biomarkers for Advanced Prostate Cancer. Eur Urol Oncol. 2018; 1(2): 151–159. [DOI] [PubMed] [Google Scholar]; (* A clinical opinion summary of current needs and uses of circulating prostate cancer biomarkers)
- 5.Cutruzzolà F, Giardina G, Marani M, et al. Glucose Metabolism in the Progression of Prostate Cancer.Front Physiol. 2017; 8:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraum TJ, Ludwig DR, Kim EH, et al. Prostate cancer PET tracers: essentials for the urologist. Can J Urol. 2018; 25(4): 9371–9383. [PubMed] [Google Scholar]
- 7.Rodrigues JG, Balmaña M, Macedo JA, et al. Glycosylation in cancer: Selected roles in tumour progression, immune modulation and metastasis. Cell Immunol. 2018; 333: 46–57. [DOI] [PubMed] [Google Scholar]
- 8.Kalluri R The biology and function of fibroblasts in cancer. Nature Reviews Cancer 2016; 16(9): 582–598. [DOI] [PubMed] [Google Scholar]
- 9.Priester A, Natarajan S, Le JD, et al. A system for evaluating magnetic resonance imaging of prostate cancer using patient-specific 3D printed molds. Am J Clin Exp Urol. 2014; 2(2):127–135 [PMC free article] [PubMed] [Google Scholar]
- 10.Drake RR, Jones EE, Powers TW, Nyalwidhe JO. Altered glycosylation in prostate cancer. Adv Cancer Res. 2015; 126: 345–382. [DOI] [PubMed] [Google Scholar]; *A historic review of glycosylation research for prostate cancer
- 11.Drake RR, West CA, Mehta AS, Angel PM. MALDI Mass Spectrometry Imaging of N-Linked Glycans in Tissues. Adv Exp Med Biol. 2018; 1104:59–76. [DOI] [PubMed] [Google Scholar]; *An overview of the N-glycan imaging mass spectrometry methodology
- 12.Angel PM, Comte-Walters S, Ball LE, et al. Mapping Extracellular Matrix Proteins in Formalin-Fixed, Paraffin-Embedded Tissues by MALDI Imaging Mass Spectrometry. J Proteome Res. 2018; 17(1): 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]; **The first report of using collagenase and elastase in mass spectrometry imaging applications
- 13.Jansen CS, Prokhnevska N, Kissick HT. The requirement for immune infiltration and organization in the tumor microenvironment for successful immunotherapy in prostate cancer. Urol Oncol. 2018; S1078-1439(18): 30393–30394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berglund E, Maaskola J, Schultz N, et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nature communications 2018; 9(1): 2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez-Bosch N, Rodriguez-Vida A, Juanpere N, et al. Galectins in prostate and bladder cancer: tumorigenic roles and clinical opportunities. Nat Rev Urol. 2019; 16(7): 433–445. [DOI] [PubMed] [Google Scholar]