Abstract
The brain’s primary circadian pacemaker, the suprachiasmatic nucleus (SCN), is required to translate day-length and circadian rhythms into neuronal, hormonal, and behavioral rhythms. Here, we identify the homeodomain transcription factor Ventral anterior homeobox 1 (Vax1) as required for SCN development, vasoactive intestinal peptide expression, and SCN output. Previous work has shown that VAX1 is required for gonadotropin-releasing hormone (GnRH/LHRH) neuron development, a neuronal population controlling reproductive status. Surprisingly, the ectopic expression of a Gnrh-Cre-allele (Gnrhcre) in the SCN confirmed the requirement of both VAX1 (Vax1flox/flox:Gnrhcre, Vax1Gnrh-cre) and Sine Oculis Homeobox Protein 6 (Six6flox/flox:Gnrhcre, Six6Gnrh-cre) in SCN function in adulthood. To dissociate the role of Vax1 and Six6 in GnRH neuron and SCN function, we used another Gnrh-cre allele that targets GnRH neurons, but not the SCN (Lhrhcre). Both Six6Lhrh-cre and Vax1Lhrh-cre were infertile, and in contrast to Vax1Gnrh-cre and Six6Gnrh-cre mice, Six6Lhrh-cre and Vax1Lhrh-cre had normal circadian behavior. Unexpectedly, ~1/4 of the Six6Gnrh-cre mice were unable to entrain to light, showing that ectopic expression of Gnrhcre impaired function of the retino-hypothalamic tract that relays light information to the brain. This study identifies VAX1, and confirms SIX6, as transcription factors required for SCN development and function and demonstrates the importance of understanding how ectopic CRE expression can impact the results.
Keywords: Suprachiasmatic nucleus, gonadotropin-releasing hormone neuron, Ventral Anterior Homeobox 1, Sine Oculis Homeobox Protein 6, circadian rhythm, mouse model validation
Introduction
Correct brain development relies on the precise timing and dosage of a large number of transcription factors, where homeodomain-binding proteins are one of the largest groups of transcription factors required for hypothalamic development [1–4]. Homeodomain-binding transcription factors preferentially bind ATTA and related sites in target gene promoters allowing regulation of gene expression. In contrast to most transcription factor families, homeodomain binding transcription factor dosage is critical during embryogenesis, and their haploinsufficiency can impair development of specific neuronal populations causing infertility as well as other behavioral and physiological deregulations [5–11]. Specifically, haploinsufficiency of homeodomain transcription factors including Sine Oculis-related Homeobox 3 (Six3), Orthodenticle Homeobox (Otx2), and Ventral Anterior Homeobox 1 (Vax1) impact ventral forebrain and hypothalamic development thereby reducing fertility [12].
The incredible diversity of peptides and transcription factors expressed by hypothalamic neurons arises from a complex developmental program allowing specification of each neuronal subtype [13, 4, 14–16]. A small bilateral structure in the ventral hypothalamus called the suprachiasmatic nucleus (SCN) is considered to be the body’s master circadian pacemaker [17, 14]. The SCN translates light information received through the retinohypothalamic tract of the eye to the body, allowing neuronal and peripheral circadian rhythms to be aligned to the environmental light-dark cycle and season of the year, thus promoting optimal cellular and behavioral function of the organism [18, 19]. The critical role of the SCN as a circadian timekeeper is demonstrated in animal models with disrupted SCN function through ablations or genetic mutations, which produce impaired circadian rhythms, metabolic deregulation and poor fertility, among other physiological disruptions [20–25].
The important role of the SCN in fertility is demonstrated by the elimination of the luteinizing hormone (LH) surge driving ovulation after SCN ablation [26]. Although the specific mechanism of SCN timing of the LH surge is still unclear, both vasoactive intestinal polypeptide (VIP) and arginine vasopressin (AVP) neurons of the SCN directly project onto two hypothalamic neuronal populations required for fertility, the kisspeptin and gonadotropin-releasing hormone (GnRH) neurons [27–35]. Interestingly, SCN and GnRH neurons share some striking similarities. Both depend on homeodomain transcription factors to develop correctly and both possess endogenous rhythms [36, 12, 22, 37, 38]. Major differences between GnRH and SCN neuron development include their timing and migratory patterns. Generation of GnRH neurons occurs in the olfactory placode at ~embryonic day 11 (E11), followed by their migration to the anterior hypothalamus, where most GnRH neurons are located at E17 [39]. In contrast, the SCN arises from a rapid cell proliferation of the neuroepithelium caudal to the optic recess between ~E12–15 in the mouse [22, 40–42]. Numerous homeodomain transcription factors are highly expressed in the neuroepithelium and ventral forebrain between E12 and E16, including Six3, Six6, LIM homeobox 1 (Lhx1) [22], and Vax1 (this paper and [40, 41]). Thus far, it has been shown that Six3 and Six6 are required for both GnRH neuron and SCN development [43, 44, 22, 45], whereas Lhx1 is required for SCN development [17], and Vax1 is necessary for GnRH neuron development [9, 46]. Based on the Allen Brain Atlas (www.brain-map.org), Vax1 possesses a high expression pattern in the ventral forebrain from E10-E18, and at postnatal day 0 (P0) Vax1 knock-out (Vax1KO) mice lack midline crossing, are holoprosencephalic and die soon after birth due to severe brain development defects, in combination with an incapacity to suckle due to cleft lip and palate [40, 47]. In contrast, Six6KO mice survive but lack SCN morphology, SCN expression of AVP, and VIP, causing impaired circadian wheel-running activity [44].
Materials and Methods
Mouse breeding.
All animal procedures were performed according to protocols approved by University of California, San Diego Institutional Animal Care and Use Committee and the Institutional Animal Care and Use Committee of Michigan State University and conducted in accordance with the Guide for the Care and Use of Laboratory Animals. Vax1tm1Grl [41], here referred to as Vax1HET and Vax1KO mice, Vax1flox mice [46], Six6flox [45], and Bmalflox (The Jackson Laboratory #007668 [B6.129S4(Cg)-Arntltm1Weit/J]) mice were crossed with Lhrhcre [48] [Tg(Gnrh1-cre)1Dlc] or Gnrhcre [49] [Tg(Gnrh1-cre)35Awo] mice. All mice were kept on a C57BL/6J background. Mice were housed with lights ON from 6 AM till 6 PM and killed by CO2 or isoflurane (Vet One, Meridian, ID) overdose and decapitation.
Wheel-running behavior.
Female and male mice of 10–14 weeks of age at the start of the experiment were housed individually in cages with running wheels. Food and water were available ad libitum during the entire experiment. After 1-week acclimation to the polypropylene cages (17.8 × 25.4 × 15.2 cm) containing a metal running wheel (11.4 cm diameter), locomotor activity rhythms were monitored with a VitalView data collection system (Version 4.2, Minimitter, Bend OR) that compiled in 6 min bins the number of electrical closures triggered by half wheel rotations. Running wheel activity in Light 12h:Dark 12h (LD12:12) was monitored for 2 weeks. Subsequently, mice were placed for 4 weeks in constant darkness (DD). Cage changes were scheduled at 3-week intervals. Wheel-running activity was analyzed using ClockLab Analysis (ActiMetrics Software).
Visual evoked potentials.
Visual evoked potential measurements were taken after the wheel-running behavior tests. This protocol was adapted from published studies [50]. Mice were dark-adapted for 12 hours before the procedure. Animals were anesthetized, given an eye drop of 0.5% proparacaine as analgesic, followed by 1% tropicamide for dilation. The top of the mouse head was cleaned with an antiseptic solution. A scalpel was used to incise the scalp skin, and a metal electrode was inserted into the primary visual cortex through the skull, 0.8 mm deep from cranial surface, 2.3 mm lateral to the lambda. A platinum subdermal needle (Grass Telefactor) was inserted through the animal’s mouth as reference, and through the tail as ground. Flashes of light at 2 log cd*s/m2 were delivered through a full-field Ganzfeld bowl at 2 Hz. Signal was amplified, digitally processed by the software (Veris Instruments), then exported and peak-to-peak responses were analyzed in Excel (Microsoft). To isolate visual evoked potentials of the measured eye from the crossed signal originating in the contralateral eye, a black aluminum foil eyepatch was used to cover the eye not undergoing measurement. Following the readings, the animals were euthanized, their eyes collected and processed for immunohistochemistry (IHC) and image analysis.
Cell culture.
NIH3T3 (American Type Culture Collection) and COS-1 (American Type Culture Collection) cell lines were cultured in DMEM (Mediatech), containing 10% fetal bovine serum (Gemini Bio), and 1x penicillin-streptomycin (Life Technologies/Invitrogen) in a humidified 5% CO2 incubator at 37°C. For luciferase assays NIH3T3 cells were seeded into 24-well plates (Nunc) at 35,000 cells per well. Transfection of cells was performed 48 h after the cells were plated. COS-1 cells used for electrophoretic mobility shift assay (EMSA) were harvested at sub-confluency 48–56 h after seeding in 10 cm dishes (Nunc).
Transfections and luciferase assays.
Transient transfections for luciferase assays were performed using PolyJet™ (SignaGen Laboratories, Rockville, MD), whereas Fugene was used for plasmid overexpression for EMSA’s, following manufacturer’s recommendations. For luciferase assays, NIH3T3 cells were co-transfected as indicated in the figure legends, with 150 ng/well luciferase reporter plasmids and expression plasmids at the indicated concentration. 100 ng/well thymidine kinase-β-galactosidase reporter plasmid was added and served as an internal control. The plasmids used were previously published [51, 52]. Site directed mutagenesis of the homeodomain binding sites in the mouse ~1 Kb Vip-luciferase plasmid was performed using the NEB Q5 Site-Directed Mutagenesis Protocol (New England Biolabs Inc.), following manufacturer’s instructions. Primers for NEB Q5 site-directed mutagenesis were designed using the NEB Base Changer (Table 1). To equalize the amount of DNA transfected into cells, we systematically equalized plasmid concentrations by adding the corresponding plasmid backbone. For luciferase assays, cells were harvested 24 h after transfection in lysis buffer [100 mM potassium phosphate (pH 7.8) and 0.2% Triton X-100]. Luciferase and β-galactosidase assays were performed as previously described [53]. Luciferase values are normalized to β-galactosidase values to control for transfection efficiency. Values are normalized to pGL3 and are expressed as fold change as compared to control plasmid as indicated in the figure legends. Data represent the mean ± SEM of at least three independent experiments done in triplicate.
Table 1. Primers used for Site-Directed Mutagenesis in the 1 Kb Vip-luciferase plasmid.
Table of primers used to mutate ATTA sites within the 1 Kb mouse Vip-promoter (Vip-pGL3) plasmid and EMSA probes. Position refers to the number of base pairs upstream of the transcription start site. All primers were designed using NEBase Changer. Underlined bold sequences highlights the bases which were mutated.
| Mutant | Position | Genotype | Sequence |
|---|---|---|---|
| Vip −909 | −914 → −901 | Wildtype | TCCCAATTAAATGT |
| Vip −862 | −867 → −854 | Wildtype | AAAAAATTAAGCAT |
| Vip −656 | −661 → −648 | Wildtype | TGATCATTATTTAT |
| Vip −474 | −479 → −468 | Wildtype | AAGTGTAATAATAG |
| Vip −471 | −476 → −463 | Wildtype | TGTAATAATAGAAA |
| Vip −432 | −437 → −424 | Wildtype | TTTAAATTAGAGTA |
| Vip −408 | −413 → −400 | Wildtype | AACTTTAATTCAGA |
| Vip −365 | −370 → −357 | Wildtype | AAGGCATTAAAGCA |
| Vip −243 | −248 → −235 | Wildtype | ATAACATTATAATC |
| Vip −239 | −244 → −231 | Wildtype | CATTATAATCATCT |
| Vip −204 | −209 → −196 | Wildtype | TAAAGATTAAAGCA |
| Vip −94 | −99 → −86 | Wildtype | ACATTTAATAAGAA |
Cytoplasmic and nuclear extracts.
COS-1 cells were scraped in hypotonic buffer (20 mM Tris-HCl, pH 7.4, 10 mM NaCl, 1 mM MgCl2, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1x protease inhibitor cocktail; Sigma-Aldrich) and left on ice to swell. Cells were lysed and nuclei were collected by centrifugation (4°C, 1700 g, 4 min). Nuclear proteins were extracted on ice for 30 min in hypertonic buffer [20 mM HEPES, pH 7.9, 20% glycerol, 420 mM KCl, 2 mM MgCl2, 10 mM NaF, 0.1 mM EDTA, 0.1 mM EGTA, 1x protease inhibitor cocktail (Sigma-Aldrich), and 1 mM phenylmethylsulfonylfluoride]. Debris was eliminated by centrifugation (4°C, 20,000 g, 10 min), and supernatant was snap-frozen and stored at −80°C.
Determination of pubertal onset and fertility assessment.
As previously described [54], pubertal onset in male mice is observed by preputial separation, whereas vaginal opening is indicative of pubertal onset in female mice. Males and females were observed every day starting at approximately 23–25 days of age. At 11–14 weeks of age, virgin Vax1flox/flox (WT), Vax1flox/fllox:Lhrhcre (Vax1Lhrh-cre), Six6flox/flox (WT), and Six6flox/flox:Gnrhcre (Six6Gnrh-cre) were housed in pairs with the corresponding controls (Vax1flox/flox or Six6flox/flox) mice. The number of litters produced was recorded for 90 days.
Determination of estrous cyclicity.
To assess estrous cyclicity, vaginal smears were performed daily between 9–11 am on 3–5 month-old mice by vaginal lavage. Smears were collected on glass slides and counterstained with 0.1% of methylene blue (Spectrum, Gardena, CA). Cell type was observed through bright field microscopy to determine the corresponding stage of the estrous cycle.
Hormone levels and GnRH and Kisspeptin-10 challenges.
Tail blood was collected from adult male and female metestrus/diestrus littermates between 10 am and 12 pm. Ten min after receiving an i.p. injection of 1 μg/kg GnRH in physiological serum, or 20 min after receiving an i.p. injection of 3 mmoles kisspeptin-10 in sterile saline, a second collection of tail blood was performed. Blood was allowed to clot for 1 h at RT, then centrifuged (RT, 15 min, 2600 g). Serum was collected and stored at −20°C before ELISA analysis of luteinizing hormone (LH) and follicle-stimulating hormone (FSH). Samples were run as singlet on MILIPLEX® (#MPTMAG-49K, Millipore, Baltimore). LH: lower detection limit: 5.6 pg/mL, intra-ACOV 15.2% and inter-ACOV 4.7%. FSH: lower detection limit: 24.9 pg/mL, intra-ACOV 13.7% and inter-ACOV 3.9%.
IHC and Hematoxylin & Eosin (H&E) staining.
IHC in the brain was performed as previously described [55]. Briefly, the primary antibody used was rabbit anti-GnRH (Thermo Scientific #PA1–121, dilution 1/1000, RRID:AB_325077), rabbit anti-GnRH (Novus #NBP2–22444, 1:1000, or Immunostar #20075, dilution 1/2000, RRID: AB_572248), which specifically stain for GnRH [55]. GnRH-positive neurons were counted throughout the brain. VIP staining was performed in the SCN using the rabbit-anti-VIP antibody (Immunostar 20077, 1/1000 RRID:AB_572270). Semi-quantification of the SCN area containing VIP staining was done using Fiji-Image Software (NIH). Image intensity (grey scale) for VIP staining represents the average VIP expressing area of the SCN at Bregma −0.34; −0.60 and −0.82. H&E in adult brain tissue was done on 10 μm coronal tissue sections using H&E (Vector lab). For immunostaining the retinas, enucleated eyes were fixed for 3 h in 4% PFA in PBS. The retinas were extracted, flat mounted on microscope slides, anti-mouse BRN3a primary antibody (Millipore, MAB1595, RRID: AB_94166) and secondary AlexaFluor 555 Anti-mouse (Invitrogen, A32727).
In situ hybridization.
Histological analyses of Vax1KO at P0 were performed according to Bharti, et al. [56]. The Avp in situ hybridization probe has previously been validated [41].
Collection of uterus, ovary, testis, and gonadal histology.
Ovaries and uteri from diestrus females and testes from males were dissected and weighed from animals of 2.8–4 months of age.
Statistical analysis.
Statistical analyses were performed using either Student’s t-test, One-Way ANOVA or Two-Way ANOVA, followed by post hoc analysis by Tukey or Bonferroni as indicated in figure legends, with p < 0.05 to indicate significance using GraphPad Prism 7 and 8.
Results
Vax1 is Required for SCN Development
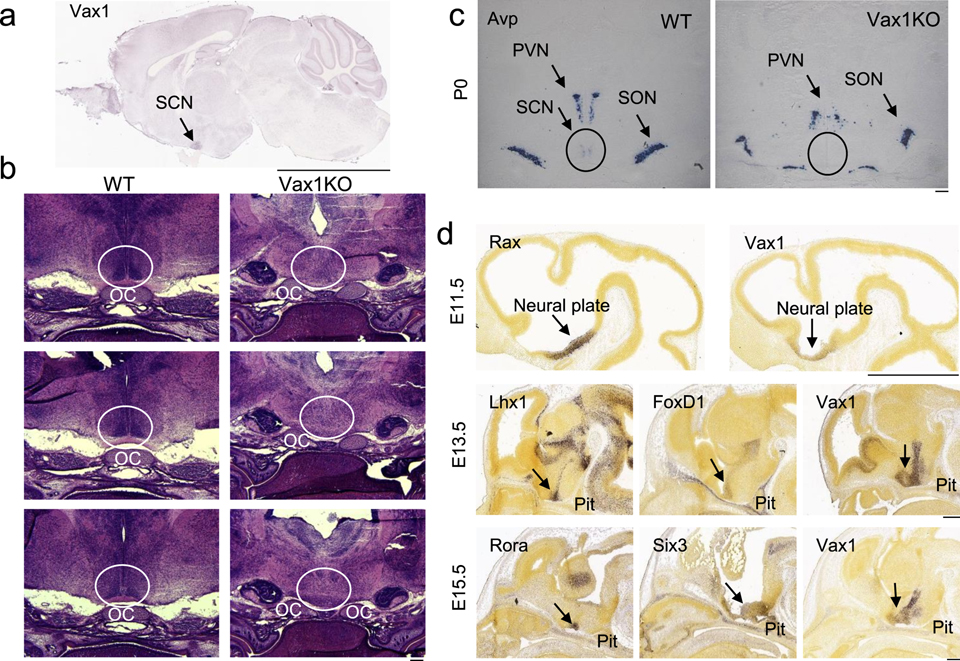
Based on the critical role of Vax1 for ventral forebrain development, we asked if Vax1 expression would be maintained in the brain after birth into adulthood. We consulted the Allen Brain Atlas, and found that in 56-day old males, Vax1 expression was restricted to the SCN (Fig. 1a). To determine if VAX1 was required for SCN development, H&E was performed in wild-type (WT) and Vax1 knock-out (KO) mice at birth (P0). At P0, a time point where the high cell density of the SCN is easily recognizable (Fig. 1b, WT), Vax1KO newborns lack the characteristic SCN morphology (Fig. 1b, Vax1KO), and the SCN marker Avp was undetectable by in situ hybridization in the SCN (Fig. 1c). To determine when Vax1 is expressed in the region of the neural plate giving rise to the terminal hypothalamus, Vax1 expression patterns were compared to known markers of the primordial SCN such as Rax, Lhx1, FoxD1, Rora, and Six3 [57] using the Allen Brain Atlas. Rax is a transcription factor required for the development of the anterior hypothalamus, including the SCN [22, 58]. At E11.5, Vax1 and Rax are expressed within the same area of the neuroepithelium (Fig. 1d,), although Vax1 expression level was relatively low as compared to Rax (Fig. 1d, E11.5). Between E12 and E15, a major proliferation of the neuroepithelium allows the formation of the primordial SCN. Based on this we evaluated expression patterns at E13.5 for Lhx1, FoxD1, and Vax1. We found these SCN markers had similar expression patterns in the neuroepithelium, with high expression of Vax1 in the entire area that gives rise to the SCN (Fig. 1d). The high Vax1 expression is maintained at E15.5, where Vax1 expression patterns in the region of the neuroepithelium giving rise to the terminal hypothalamus overlaps with the SCN markers Six3 and Rora (Fig. 1d).
Fig. 1. Vax1 is required for SCN development.
a Sagittal in situ hybridization images from Allen Brain Atlas showing expression of Vax1 in the adult SCN, scale bar represents 0.5 cm. Allen Mouse Brain Atlas (2004), image 18, Vax1-RP_090303_04_G09-sagittal, http://mouse.brain-map.org/experiment/siv?id=81655570&imageId=81622614&initImage=ish&coordSystem=pixel&x=7192.5&y=3784.5&z=1. b H&E staining on coronal brain sections of Vax1WT (WT) and Vax1KO mice at P0 showing absence of SCN morphology in Vax1KO (circled; x20). c Avp in situ hybridization in coronal brain sections at P0 in WT and Vax1KO embryos (circled; x20). d Sagittal in situ hybridization images from Allen Brain Atlas showing expression of Rax, Vax1, Lhx1, FoxD1, Rora, and Six3 in the developing mouse brain at the indicated embryonic (E) age, scale bar 1 mm. Allen Developing Mouse Brain Atlas (2008), E11.5, Rax_RP090724_05_F3-sagittal, image 7, http://developingmouse.brain-map.org/experiment/siv?id=100085519&imageId=101449838&initImage=ish&x=4120&y=3200&z=3; Vax1-RP_090707_04_E04-saggital, image 6, http://developingmouse.brain-map.org/experiment/siv?id=100085523&imageId=101449914&initImage=ish&x=3512&y=2936&z=3. E13.5, Lhx1-RP080807_03E10-sagittal, image 18, http://developingmouse.brain-map.org/experiment/siv?id=100031987&imageId=100689017&initImage=ish&x=3936&y=5496&z=3; Foxd1-RP_090724_06_H11-sagittal, image 14, http://developingmouse.brain-map.org/experiment/siv?id=100075832&imageId=101262091&initImage=ish&x=4240&y=5392&z=3; Vax1-RP-090303_04_G09-sagittal, image 14, http://developingmouse.brain-map.org/experiment/siv?id=100072475&imageId=101210859&initImage=ish&x=3616&y=5600&z=3. E15.5, Rora-RP_071203_02_H06-sagittal, image 16, http://developingmouse.brain-map.org/experiment/siv?id=100053213&imageId=101049221&initImage=ish&x=6144&y=7552&z=2; Six3-RP_080807_03_E12-sagittal, image 18, http://developingmouse.brain-map.org/experiment/siv?id=100032020&imageId=100689426&initImage=ish&x=3904&y=6488&z=2; Vax1-RP090303_04_G09-sagittal, image 14, http://developingmouse.brain-map.org/experiment/siv?id=100072496&imageId=101211230&initImage=ish&x=5152&y=7248&z=2 Abbreviations. OC: optic chiasm, SCN: suprachiasmatic nucleus, PVN: paraventricular nucleus, SON: supraoptic nucleus, Pit: pituitary.
Vax1 is Required for Normal SCN Function
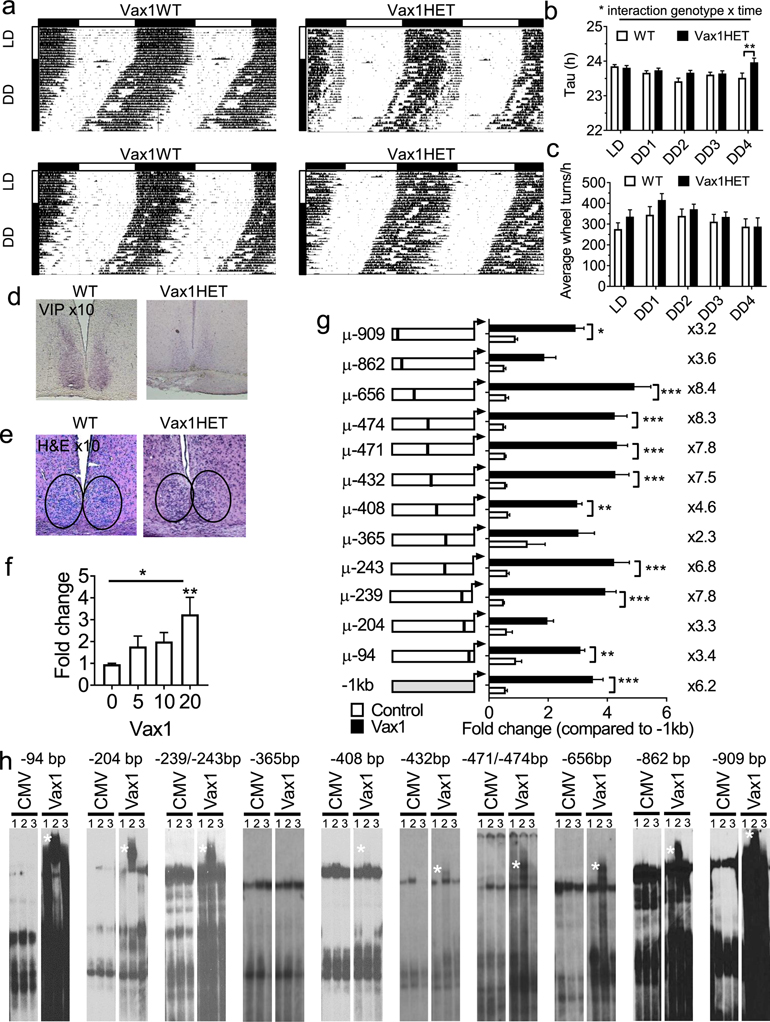
Vax1KO is neonatal lethal [40, 47]. We therefore tested SCN output in female mice by assessing wheel-running activity in Vax1 heterozygotes (HET), a mouse model that we previously found to be subfertile due to Vax1 haploinsufficiency on GnRH neuron development [9, 12]. Both WT and Vax1HET females entrained normally to light on a 12h light:12h dark (LD12:12) light cycle (Fig. 2a, LD). After 4 weeks in constant darkness (DD), but not earlier, Vax1HET females had a slightly, but statistically significant, longer free-running period (τ) than controls (Fig. 2a, b), and had comparable activity levels to controls as evaluated by the average number of wheel-revolutions/h (Fig. 2c). Due to the high expression of Vax1 in the SCN in adulthood (Fig. 1a), we hypothesized that VAX1 in the adult brain potentially regulated VIP expression, a SCN neuropeptide that is expressed in the mature SCN and that is required for normal SCN function and female fertility [24]. Using a semi-quantitative approach, we established that the VIP expression area in the SCN was reduced by ~26% in the Vax1HET (VIP expressing area Vax1WT = 74596.89 ± 5555 pixels, n=3; Vax1HET = 55042 ± 16168 pixels, n=3, Fig. 2d). This reduction in the VIP area was accompanied by a reduction of VIP staining intensity (Fig. 2d, visual evaluation of all the slides of the SCN). This reduction in VIP peptide and expression area did not appear to have a major impact on SCN morphology as assessed by H&E (Fig. 2e).
Fig. 2. Vax1HET females have abnormal SCN output.
a Running-wheel patterns in Vax1WT and Vax1HET females. Double plotted actograms show activity with 10 days in LD12:12 (LD) followed by 28 days in constant darkness (DD). Data are presented as percentile. Horizontal bar above the actograms indicates lights on (white) and lights off (black) during the LD12:12 cycle. b Seven day average free-running periods (Tau) as established by Chi-square (ClockLab), and c average wheel-revolutions per hour for 7 days of data during LD, week 1 in DD (DD1), until week four in DD (DD4). Statistical analysis by Two-way ANOVA followed by a Tukey’s multiple comparison test; *, p<0.05; ** p<0.01, n=8–10. d Representative images of IHC for VIP in the SCN of adult WT and Vax1HET females, n=4, magnification x10. e H&E in coronal hypothalamic brain sections encompassing the SCN from females, magnification x10. f, g Luciferase assays in NIH3T3 cells transiently transfected with the 1 Kb mouse Vip promoter driving luciferase with and without cotransfection with the mouse Vax1 expression vector. f Dose-response of VAX1 effect on Vip-luciferase expression (One-way ANOVA, followed by a Dunnet’s test, **, p<0.01, n-3–5 in triplicate), g Site-directed mutagenesis of ATTA and ATTA-like sites in the 1 Kb mouse Vip-luciferase driven plasmid (Two-way ANOVA, followed by a Tukey multiple comparison test, *, p<0.05; **, p<0.01; ***, p<0.001, n=3–5 in triplicate).h EMSA assay of COS-1 cells transiently transfected with pCMV-Flag (CMV, empty vector) or Vax1-Flag (example gel of n=3). White star indicates super shift. Lane 1: H20, lane 2 anti-Flag-antibody (αFlag), lane 3: IgG.
To determine if the reduction of VIP expression was caused by a direct regulation by VAX1 of the Vip promoter, we transiently transfected NIH3T3 cells with the ~1 Kb Vip mouse promoter driving luciferase expression along with increasing concentrations of a Vax1 expression plasmid. Increasing concentrations of the Vax1 overexpression plasmid, had a dosage effect on Vip-luciferase expression (Fig. 2f). Based on these data, the remaining experiments were performed with 20 ng of Vax1. To determine if the VAX1-induced increase in Vip-luciferase expression was mediated by VAX1 binding to the Vip promoter, we mutated ATTA and ATTA-like sites in the Vip promoter (Table 1). Site-directed mutagenesis of ATTA-like sites at −656 bp, −474 bp, −471 bp, −432 bp, and −239 bp enhanced VAX1 regulated Vip-luciferase induction, whereas mutation of ATTA-like sites at −909 bp, −365 bp, −204 bp, and −94 bp reduced VAX1 regulated Vip-luciferase induction (Fig. 2g). Although VAX1-regulated Vip-luciferase expression was only abolished at site −365, the significant increase (sites at −656 bp, −474bp, −471 bp, −432 bp, and −239 bp), and reduction (sites at −909 bp, −365 bp, −204 bp, and −94 bp) of VAX1-regulated Vip-luciferase expression lead us to test if VAX1 directly binds to any of these sites using electromobility shift assays (EMSAs, for probe sequences see table 1). Nuclear extracts from COS-1 cells transiently transfected with Vax1-flag formed a complex (Fig. 2h) that was supershifted (Fig. 2h, supershift is indicated by a white star) in presence of an α-flag antibody (lane 2). In contrast, no supershift was observed in samples with empty vector (CMV), or site −365 bp (Fig. 2h). The specificity of the supershift was confirmed by the lack of supershift in samples with IgG (Fig. 2h, lane 3).
Ectopic Expression of Gnrhcre in the Developing SCN Confirms Vax1 as Critical in SCN Function
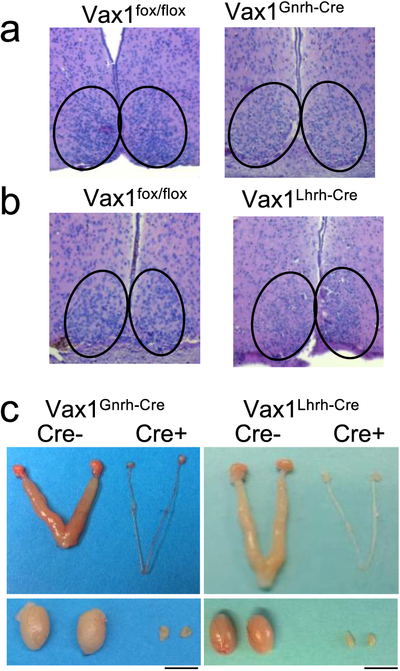
Normal SCN function is required for the LH surge that promotes ovulation and fertility in females. We recently found that a Gnrh-promoter driven Cre-allele (here referred to as Gnrhcre [49]), which we previously used to delete Vax1 within GnRH neurons [46], also targets the SCN [55]. This is important as this Gnrhcre allele would delete Vax1 during SCN embryogenesis and could cause defects in SCN function. This ectopic expression of the Gnrhcre allele was not observed in the Gnrh-promoter driven-Cre allele developed using a bacterial artificial chromosome, here referred to as the Lhrhcre [55]. To rule out that part of the reproductive phenotype in the Vax1flox/flox:Gnrhcre (Vax1Gnrh-cre) females was caused by impaired SCN function, we performed H&E staining of the SCN. We found that the SCN morphology in Vax1Gnrh-cre (Fig. 3a), but not in Vax1Lhrh-cre (Fig. 3b), was comparable to controls (Vax1flox/flox or Gnrhcre) although the cell density appeared to be slightly reduced in Vax1Gnrh-cre.
Fig. 3. Impact of conditional deletion of Vax1 using LHRHcre and GnRHcre on SCN and gonadal morphology.
a, b H&E in coronal hypothalamic brain sections encompassing the SCN from female mice of the indicated genotypes, magnification x10. c Images of male and female Vax1flox/flox, Vax1Gnrh-cre, and Vax1Lhrh-cre gonads and uteri. Scale bar 0.5 cm. n = 3–5 mice.
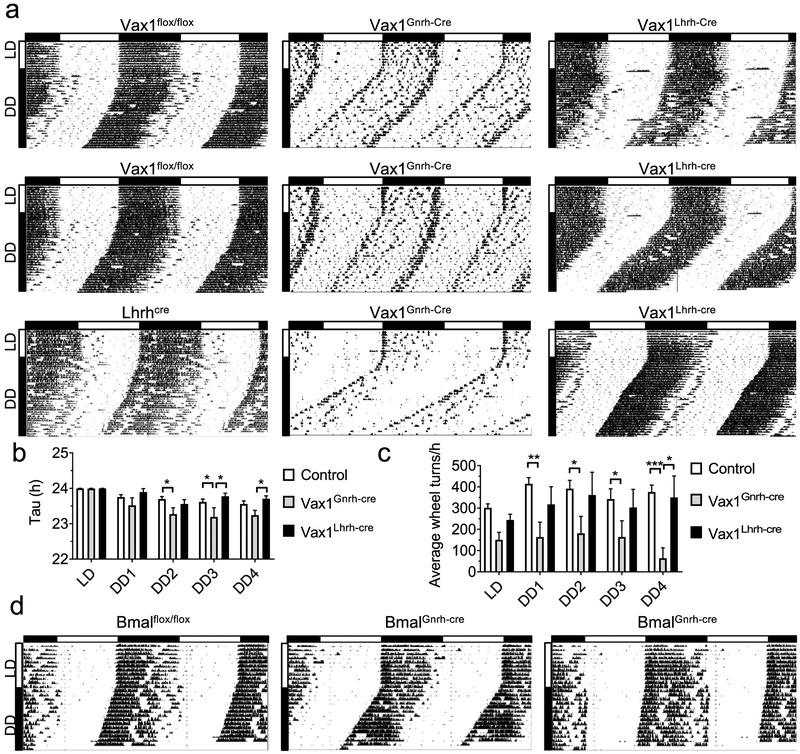
Because gonadal sex steroids can impact wheel-running levels [59, 60] and running levels impact free-running period (Tau, τ) [61], we evaluated wheel-running levels (wheel revolutions and τ) in the hypogonadal Vax1Gnrh-cre and Vax1Lhrh-cre mice (Fig. 3c). Wheel running activity was compared between Vax1Gnrh-cre, Vax1Lhrh-cre, and control mice (Vax1flox/flox, Gnrhcre, and Lhrhcre). Based on our recent characterization of neurons targeted by the Gnrhcre and Lhrhcre alleles, we would not expect the Vax1flox/flox allele to be deleted in the SCN when crossing it with the Lhrhcre mouse (here termed Vax1Lhrh-cre), whereas we would expect the Vax1flox/flox allele to be deleted in parts of the SCN from mid-late embryonic development using the Gnrhcre mouse [48, 55]. To determine if Vax1Gnrh-cre had abnormal SCN function, circadian wheel-running patterns were evaluated in controls (Vax1flox/flox, Lhrhcre; Fig. 4a) and Gnrhcre (an example of Gnrhcre wheel-running is included in Fig. 6a) and compared to Vax1Gnrh-cre and Vax1Lhrh-cre (Fig. 4a). All of the mice studied entrained normally to light in LD12:12 (Fig. 4a, 10 days in LD indicated by vertical white bar). In constant darkness (DD), controls and Vax1Lhrh-cre had similar activity levels and presented with comparable free-running periods (τ < 24h, Fig. 4a–c). In contrast, Vax1Gnrh-cre had shorter τ than controls and Vax1Lhrh-cre (Fig. 4a, b). In addition, overall activity levels were lower in Vax1Gnrh-cre than controls (Fig. 4a, c). Surprisingly, we did not detect lower activity levels in the hypogonadal Vax1Lhrh-cre (Fig. 4a, c), suggesting that the reduced activity in Vax1Gnrh-cre is not solely caused by reduced sex steroid levels. To establish to what degree the impaired wheel running activity in Vax1Gnrh-cre was due to impaired SCN circadian rhythms, we deleted the brain and muscle aryl hydrocarbon receptor nuclear translocator-like protein 1 (Bmal1), a required transcription factor for circadian rhythm generation, using the Gnrhcre (BmalGnrh-cre; [62]). BmalGnrh-cre males entrained normally to LD12:12, and had comparable wheel running activity to controls in constant darkness (Fig. 4d).
Fig. 4. Ectopic expression of Gnrhcre in the SCN disrupts SCN output in Vax1Gnrh-cre mice.
a Running-wheel patterns in controls (Vax1flox/flox and Lhrhcre), Vax1Gnrh-cre, and Vax1Lhrh-cre females (n=8–10), and d in Bmalflox/flox and BmalGnrh-cre males (n=4). Actograms show double plotted data, expressed as percentage, with 10 days in LD12:12 (LD) followed by (a) 28 days, or (d) 14 days in constant darkness (DD). Horizontal bar above the actograms indicates lights on (white) and lights off (black) during the LD12:12 cycle. b) Seven day average free-running periods (Tau) as established by Chi-square (ClockLab) and c average wheel-revolutions per hour for 7 days of data during LD, week one on DD (DD1), until week four in DD (DD4). Statistical analysis by Two-way ANOVA followed by a Tukey’s multiple comparison, * p<0.05; ** p<0.01, *** p<0.001.
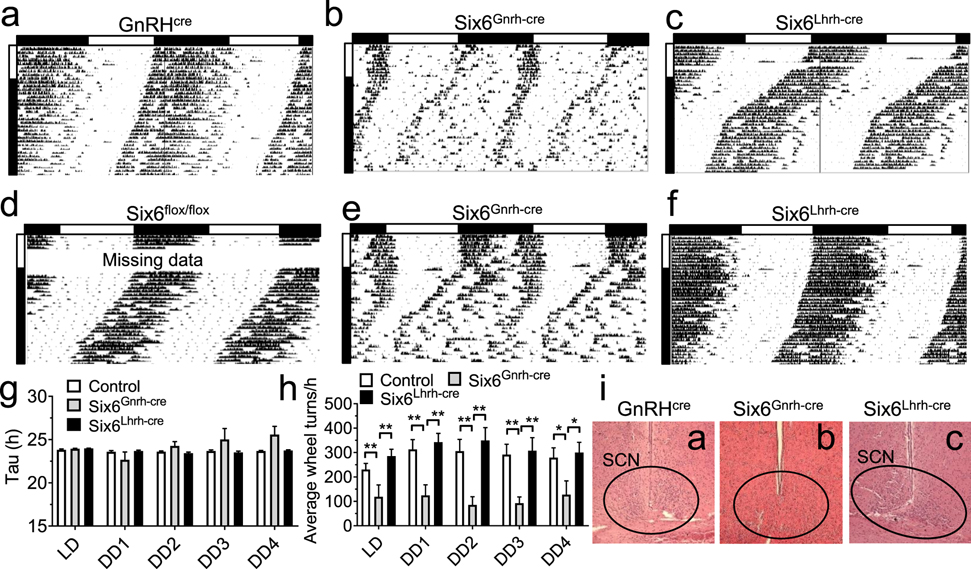
Fig. 6. Six6Gnrh-cre but not Six6Lhrh-cre females have disrupted SCN output.
A–f Running-wheel patterns in GnRHcre, Six6flox/flox, Six6Gnrh-cre, and Six6Lhrh-cre females. Actograms show double plotted data expressed as percentage with 10 days in LD12:12 (LD) followed by 28 days in constant darkness (DD). Horizontal bar above the actograms indicated lights on (white) and lights off (black) during the LD12:12 cycle. g Seven-day average free-running periods (Tau) as established by Chi-square (ClockLab) and h average wheel-revolutions per hour for 7 days of data during LD, and DD week one through 4 (DD1–4). Statistical analysis by Two-way ANOVA followed by a Tukey’s multiple comparison test, *, p<0.05; **, p<0.01; n=5–10. i Illustrative H&E of coronal sections showing presence [i(a), and i(c), and absence of SCN, i(b)]. Magnification x10.
Disrupted SCN Function in Vax1Gnrh-cre Does Not Measurably Exacerbate the Infertility of This Mouse Model
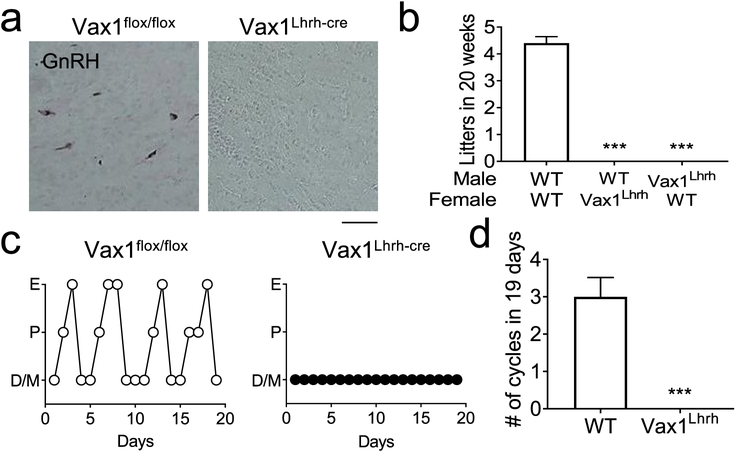
To determine if the impaired SCN function in Vax1Gnrh-cre contributed to the infertility of these mice, we repeated the fertility study in Vax1Lhrh-cre mice which have Vax1 deleted within GnRH neurons, but not in the SCN [55]. First, we confirmed that the hypogonadism of Vax1Lhrh-cre originated at the level of the GnRH neurons by performing IHC for GnRH in adult Vax1Lhrh-cre mice. Evaluating GnRH neuron numbers throughout the adult brain, we identified only a very small number of GnRH expressing neurons in Vax1Lhrh-cre mice (Fig. 5a, Table 2). The almost complete absence of GnRH neurons caused delayed pubertal onset in males and females (Table 2) and a significant reduction in ovary, testis, and uterine size in Vax1Lhrh-cre mice (Fig. 3c, Table 2). Gonadal maturation and function require pulsatile release of LH and follicle-stimulating hormone (FSH) from the pituitary. Indeed, LH and FSH were significantly reduced in both adult males and females (Table 2). To confirm that the low LH and FSH levels were caused by absence of GnRH, we performed hormonal challenges with kisspeptin and GnRH to test GnRH neuron and pituitary gonadotrope function, respectively. The GnRH challenge significantly increased LH release in both controls and Vax1Lhrh-cre confirming normal function of the gonadotropes. In stark contrast, a kisspeptin challenge only elicited LH release in controls (Table 2), confirming the hypogonadism of Vax1Lhrh-cre is triggered by lack of GnRH. As expected, the low LH and FSH levels caused complete infertility in Vax1Lhrh-cre males and females (Fig. 5b), and absence of estrous cyclicity in Vax1Lhrh-cre females (Fig. 5c, d).
Fig. 5. Vax1Lhrh-cre mice are infertile and lack GnRH expression.
a IHC for GnRH on coronal sections in adulthood. Scale bar 100 μm. b A 20-week fertility study evaluating the number of litters born showed complete infertility of Vax1Lhrh-cre males and females. One-Way ANOVA, as compared to WT; *** p<0.001, n=3–5. c Estrus cycling was monitored daily in adult 14–18-week-old females. M, Metestrus; E, estrus; P, proestrus; D, diestrus. d Average cycle length in WT and Vax1Lhrh-cre females. n= 4–6. Statistical analysis by Student’s t-test; ** p<0.001.
Table 2. Fertility parameters in Vax1Lhrh-cre mice.
The total number of GnRH neurons in the brain was counted using IHC for GnRH in adult mice, # every second slide was quantified. Pubertal onset was evaluated by vaginal opening (VO) in females and preputial separation (PPS) in males. Gonadal weights were assessed in 12–20-week-old males and females. Circulating LH and FSH levels were measured in male and diestrus females. Fold change in circulating LH was evaluated in males and diestrus/metestrus females after GnRH and kisspeptin ip injections. Statistical analysis by Student’s t-test. * p < 0.05; ** p < 0.01, *** p < 0.001.
| Vax1flox/flox | Vax1Lhrh-cre | P | |
|---|---|---|---|
| GnRH neurons (adult)# | 208 ± 45, n=4 | 5 ± 3, n=31 | 0.013 (*) |
| Age at VO | 29.56 ± 0.5771, n=16 | 42.57 ± 5.009, n=7 | 0.0008 (***) |
| Age at PPS | 32.88 ± 0.7899, n=16 | 63 ± 13.48, n=5 | 0.0005 (***) |
| Ovary weight (g) | 0.050 ± 0.015, n=7 | 0.006 ± 0.003, n=6 | 0.02 (*) |
| Uterus weight (g) | 0.038 ± 0.012, n=5 | 0.004 ± 0.001, n=6 | 0.0114 (*) |
| Testis weight (g) | 0.224 ± 0.009, n=5 | 0.0341 ± 0.031, n=3 | 0.0003 (***) |
| FSH (ng/ml) male | 12.86 ± 1.46, n=6 | 0.26 ± 0.10, n=8 | 0.001 (***) |
| LH (ng/ml) male | 3.74 ± 1.36, n=6 | 0.36 ± 0.11, n=11 | 0.0038 (**) |
| FSH (ng/ml) female | 1.24 ± 0.24, n=24 | 0.27 ± 0.09, n=22 | 0.0006 (***) |
| LH (ng/ml) female | 0.39 ± 0.04, n=25 | 0.19 ± 0.04, n=26 | 0.0013 (**) |
| Fold change of LH in response to GnRH (male) | 16.99 ± 3.03, n=9 | 8.28 ± 2.50, n=5 | 0.0778 |
| Fold change of LH in response to GnRH (female) | 4.75 ± 1.44, n=5 | 21.21 ± 6.27, n=4 | 0.0242 (*) |
| Fold change of LH in response to kisspeptin (male) | 5.01 ± 0.69, n=12 | 1.84 ± 0.53, n=3 | 0.046 (*) |
| Fold change of LH in response to kisspeptin (female) | 8.29 ± 1.31, n=12 | 0.70 ± 0.10, n=8 | 0.0002 (***) |
Gnrhcre Deletion of Six6flox/flox Disrupts SCN Development and Function and Causes Sporadic Blindness
To determine if the impaired SCN function in Vax1Gnrh-cre was a rare phenotype produced by a modest ectopic expression of the Gnrhcre allele in the developing SCN, or if this ectopic expression should be considered an important contributor to the phenotype observed in animals with flox alleles deleted using the Gnrhcre mouse, we deleted the homeodomain transcription factor Six6 using the Gnrhcre and the Lhrhcre alleles, here referred to as Six6Gnrh-cre and Six6Lhrh-cre, respectively. We chose to delete Six6, as this transcription factor is known to be required for both GnRH neuron and SCN development [45, 43, 44]. To determine if Six6Gnrh-cre and Six6Lhrh-cre had impaired circadian wheel-running patterns in DD, we placed Six6Gnrh-cre and Six6Lhrh-cre females in running wheels in LD12:12 (LD) for 10 days followed by 28 days in constant darkness (DD). All 8 controls, 6/6 Six6Lhrh-cre and 6/10 Six6Gnrh-cre mice entrained normally to light (Fig. 6a–f). The overall activity level of Six6Gnrh-cre females was much lower than controls and Six6Lhrh-cre (Fig. 6h). The ectopic targeting to the SCN of the Gnrhcre allele was obvious in Six6Gnrh-cre mice in DD where Six6Gnrh-cre (Fig. 6b, e), but not Six6Lhrh-cre mice (Fig. 6c, f), had abnormal total wheel revolutions (Fig. 6h). The Six6Gnrh-cre free-running period trended towards longer than controls and Six6Lhrh-cre, although a great variation in Six6Gnrh-cre wheel-running rhythms was observed, where some mice presented with a very short free-running period and others with very long free-running periods ranging from 4.33 h to 36.20 h (mice with τ <20 and >30 h were considered arrhythmic and excluded from the statistical analysis and the histogram, Fig. 6b, e, g, and 7). The abnormal wheel-running patterns in DD in Six6Gnrh-cre correlated with absence of SCN morphology [Fig. 6i, where 6i(a) is from the mouse in 6a, 6i(b) from mouse 6b, and 6i(c) from 6c], such that ectopic expression of Gnrhcre in Six6flox mice was sufficient to prevent developmental formation of normal SCN morphology.
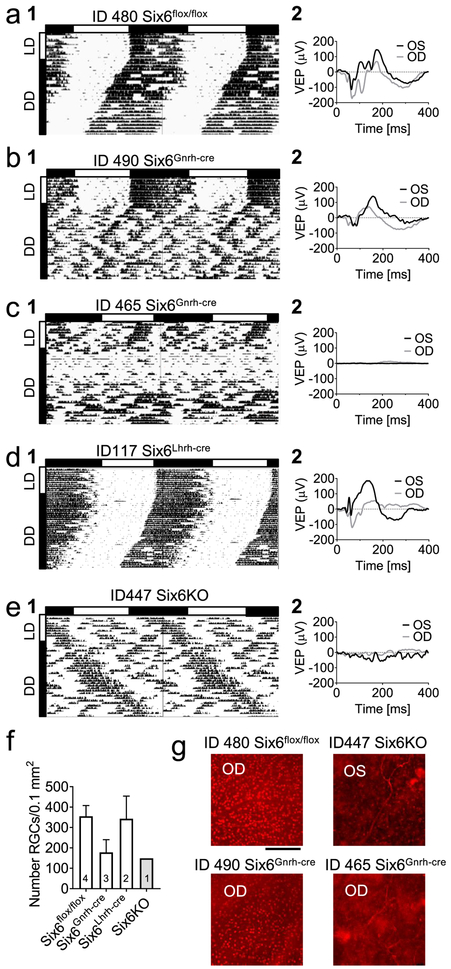
Fig. 7. Ectopic targeting of Gnrhcre causes severe SCN impairment, and abnormal light sensitivity in Six6Gnrh-cre mice.
a–e Examples of (1) wheel-running activity in 10 days LD followed by 28 days in DD (shown as percentile), and (2) vision evoked potentials (VEP) recorded at the termination of the study. f Quantification and g illustrative images of RGCs in the retina as evaluated by IHC. The number of mice per data point is shown in histogram bar (scale bar 100 μM). No statistical analysis was done due to low “n”. Abbreviations: OS: oculus sinister, OD: oculus dexter.
In addition to abnormal wheel running activity in DD of Six6Gnrh-cre females, 4/10 Six6Gnrh-cre females (Fig. 6 and 7) did not entrain to light (Fig. 7c) and presented a wheel-running activity pattern comparable to Six6KO (compare Fig. 7c and 7e). This is in sharp contrast to the 0/17 controls and 5/5 Six6Lhrh-cre females who all entrained normally to light. This suggests that the Gnrhcre allele may be occasionally expressed in the eye or optic nerve rendering the mice insensitive to light. To confirm the impact of the deletion on the general visual system, we performed visual evoked potentials at the end of the running-wheel study followed by staining for retinal ganglion cells (RGC), a population of the retinal neurons whose axons form the optic nerve, which transmits the light information to the brain [63]. We found that the mice that were unable to entrain to light in the running wheel (Fig. 7c #1, e #1) lacked visual evoked potentials (Fig. 7 #2), which was associated with heavily affected number of RGCs (Fig. 7f, g). In contrast, mice that entrained to light had normal visual evoked potentials response and normal RGC cell numbers in the retina (Fig. 7a–c, f, g).
Gnrhcre Deletion of Six6flox/flox Causes Complete Infertility Due to Absence of GnRH Neurons
To determine the impact on GnRH neuron development of Six6Gnrh-cre, we counted GnRH neurons during embryogenesis and in adulthood. Six6Gnrh-cre had a ~90% reduction of GnRH neurons at both E13.5 and E17.5 (Table 3), a reduction that was maintained into adulthood (Table 3). As expected, this severe reduction in the number of GnRH neurons caused delayed pubertal onset in male and female Six6Gnrh-cre (Table 3), hypogonadism (Table 3, Fig. 8a), and complete infertility in adulthood (Fig. 8b, c). As expected, the low circulating LH and FSH (Table 3) levels in Six6Gnrh-cre were associated with an absence of estrous cyclicity (Fig. 8d). To confirm that this infertility arose at the level of the GnRH neuron, we used GnRH and kisspeptin challenges to test for pituitary and GnRH neuron responsiveness, respectively. Unexpectedly, all groups studied had a normal fold-change in LH release in response to GnRH and kisspeptin (Table 3), suggesting the ~10% remaining GnRH neurons in Six6Gnrh-cre were sufficient to induce an increase in LH when GnRH neurons were stimulated in synchrony by kisspeptin.
Table 3. Fertility parameters in Six6Gnrh-cre mice.
The total number of GnRH neurons in the brain was counted using IHC for GnRH at embryonic day 13.5 (E13.5), E17.5 and in adult mice. Pubertal onset was evaluated by vaginal opening (VO) in females and preputial separation (PPS) in males. Gonadal weights were assessed in 12–20-week-old males and females. Circulating LH and FSH levels were measured in male and diestrus females. Fold change in circulating LH was evaluated in males after GnRH and kisspeptin ip injections. Statistical analysis by Student’s t-test. * p < 0.05; ** p < 0.01, *** p < 0.001.
| Six6flox/flox | Six6Gnrh-cre | P | |
|---|---|---|---|
| GnRH neurons (E13.5) | 1234 ± 32 n=3 | 375 ± 38, n=4 | 0.0001 (***) |
| GnRH neurons (E17.5) | 809 ± 44, n=4 | 85 ± 10, n=3 | 0.0001 (***) |
| GnRH neurons (adult) | 487.8 ± 47, n=6 | 340.5 ± 16, n=6 | 0.0001 (***) |
| Age at VO | 29.45 ± 0.34, n=11 | 43.40 ± 2.06, n=5 | 0.0001 (***) |
| Age at PPS | 30.18 ± 0.26, n=11 | 47.40 ± 1.28, n=5 | 0.0001 (***) |
| Ovary weight (g) | 0.014 ± 0.001, n=3 | 0.009 ± 0.001, n=3 | 0.0445 (*) |
| Uterus weight (g) | 0.090 ± 0.015, n=3 | 0.076 ± 0.009, n=3 | 0.454 |
| Testis weight (g) | 0.061 ± 0.004, n=3 | 0.014 ± 0.003, n=3 | 0.0007 (***) |
| FSH (ng/ml) male | 14.01 ± 1.23, n=7 | 4.33 ± 1.63, n=5 | 0.0007 (***) |
| LH (ng/ml) male | 0.15 ± 0.04, n=6 | 0.08 ± 0.04 n=5 | 0.2716 |
| FSH (ng/ml) female | 2.33 ± 1.18, n=4 | 0.87 ± 0.21, n=5 | 0.214 |
| LH (ng/ml) female | 0.04 ± 0.01, n=5 | 0.28 ± 0.11, n=4 | 0.0472 (*) |
| Fold change of LH in response to GnRH (male) | 4.34 ± 1.86, n=4 | 43.94 ± 20.94, n=5 | 0.1401 |
| Fold change of LH in response to kisspeptin (male) | 1.86 ± 0.17, n=4 | 3.70 ± 0.89, n=4 | 0.1154 |
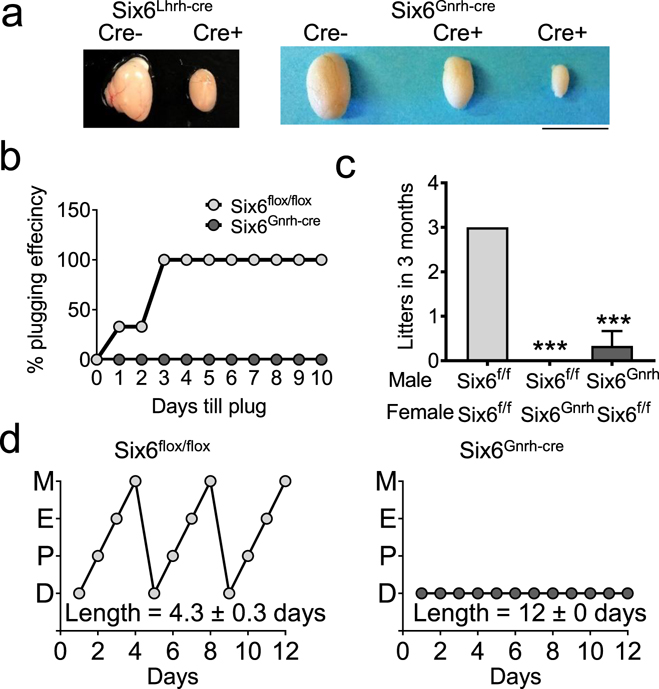
Fig. 8. Six6Gnrh-cre mice are hypogonadal.
a Images of testes from Six6Gnrh-cre and Six6Lhrh-cre mice. Scale bar 0.5 cm. b A 10-day plugging assay of Six6flox/flox and Six6Gnrh-cre males plugging Six6flox/flox females. c Three-month fertility study evaluating the number of litters born, n=3–6. Statistical analysis by One-Way ANOVA, as compared to Six6flox/flox (Six6f/f), * p<0.001. d Estrous cycling was monitored daily for 12 days in adult females. M, Metestrus; E, estrus; P, proestrus; D, diestrus in Six6flox/flox and Six6Gnrh-cre females. Statistical analysis by Student’s t-test; *** p<0.001, n=4–6.
Discussion
Vax1 is a Novel Transcription Factor Required for SCN Development and Function
Despite relatively advanced understanding of the role of the SCN in circadian rhythm generation and synchronization of body tissues to the environment, the transcriptional program directing specification of the SCN remains poorly defined [64]. Only a few transcription factors of the homeodomain family have been identified as required for SCN development. Namely, deletion of transcription factors Six3 [22], Six6 [44], and Lhx1 [22, 17] prior to or during the rapid cell proliferation occurring in the neuroepithelium (~E12-E15) each disrupts the formation of a histotypical SCN in mice [42, 65, 66]. Vax1 may now be added to the short list of transcription factors required for SCN differentiation. Vax1 is expressed in overlapping areas of neuroepithelium expressing Rax, Dlx2, Fzd5, Six6, Six3, and Lhx1 from the interval of neuronal proliferation from E11.5-E15.5. Additionally, Vax1KO hypothalami at P0 lack SCN morphology and localized expression of the SCN marker Avp, despite normal expression of that neuropeptide in other hypothalamic structures (e.g., supraoptic and paraventricular nuclei). Thus, VAX1 likely plays roles both in the proliferation of the neuroepithelium as well as in the specification of neuropeptide phenotypes of SCN neurons.
Vax1 Haploinsufficiency Causes Reduced VIP Expression in the SCN
Vax1 dosage determines the number of GnRH neurons in the brain [46, 9]. Because the extent of Vax1HET subfertility was far greater than might be expected as a consequence of only a 50% reduction in the number of GnRH neurons [9, 43, 67], we questioned whether reproductive effects of Vax1 haploinsufficiency resulted from changes to other neuronal populations, such as those expressing VIP. SCN VIP neurons directly innervate [68–71, 29, 72, 32] and modulate activity [73, 27] of GnRH neurons through VIP receptor 2 (VIPR2). Additionally, knock-out of Vip transcript [24, 74] or VIP neuron ablation [75, 76, 28] disrupts the LH surge, estrous cyclicity and ovulation. Here we found VAX1 to be an activator of Vip-luciferase expression in vitro through direct binding of VAX1 to distinct ATTA-like sites in the proximal mouse Vip promoter. In vivo, Vax1HET mice had approximately a 26% reduction in VIP expression area, in addition to a reduction in VIP neuropeptide in the SCN, indicating that Vax1 dosage sets the level of VIP expression in the adult SCN. The severe Vax1HET subfertility, therefore, could be caused by a combination of reduced GnRH neuron numbers and VIP neuropeptide expression. A single Vax1 allele also appears sufficient to allow normal embryonic SCN neuron proliferation as we observed no impact on SCN morphology in Vax1HET mice, but a total absence of SCN morphology in Vax1KO mice at birth. Functionally, circadian function as measured in LD entrainment and DD free-running assays was essentially intact, but a modest period phenotype was detected in the 4th week of exposure to DD. Though a modest effect, this would represent the first report to our knowledge of a developmental transcription factor that impacts SCN function in its heterozygotic state.
Ectopic Expression of the Gnrhcre Allele Causes Abnormal SCN Function and Sporadic Light-Insensitivity
CRE recombination is an increasingly important neurobiological tool to induce cell-specific deletion of genes in vivo. Despite numerous advantages, this strategy also has limitations such as ectopic CRE expression and germline recombination, creating mosaic and gene knock-out offspring. Two commonly used Gnrh-driven CRE-expressing mice, here termed Gnrhcre [49] and Lhrhcre [48] both target >95% of GnRH neurons. We reported recently, however, that beyond GnRH neurons, the Gnrhcre also targets the SCN, septum, medial thalamus, and other brain regions [55], and both mouse models frequently cause germline recombination [55]. Indeed, as early as E12.5, Gnrhcre targets broad areas of the anterior and ventral forebrain and from E17.5 through adulthood has distinct expression in the SCN [55]. This broader expression pattern is not observed in the Lhrhcre [55].
Although the Gnrhcre targeting of the SCN appeared to be relatively modest [55], it was sufficient nevertheless to reliably suppress activity levels and shorten free-running period in Vax1Gnrh-cre compared to the more restricted Lhrhcre allele. As reduction of wheel-running is known in mice to cause lengthening of free-running period [61], the true effect on circadian period may be substantially larger than was measured with the wheel-running assay. An even more profound contrast between Gnrhcre versus Lhrhcre was apparent for deletions of Six6, a transcription factor previously shown to be required for SCN development [44]. Besides reduced running levels, ectopic expression of the Gnrhcre allele in Six6Gnrh-cre mice was sufficient to prevent entrainment of 40% of animals to LD and decreased rhythm quality in DD.
The altered or impaired SCN function in Vax1Gnrh-cre and Six6Gnrh-cre was associated with reduced to absent SCN morphology suggesting an effect on the developing SCN prior to E15. An inability of some Six6Gnrh-cre mice to entrain to light is potentially consistent with a circadian pacemaker defect and/or a lack of functional light input. The latter is supported by the lack in some animals of visual evoked potentials and an abnormal number of RGCs in the retina, indicating that Gnrhcre has targets outside the brain. Six6 has a previously well-described role in eye and retina development [77, 78]. All mice not able to entrain to light were also lacking the visual evoked potentials. At this point, it is not possible to dissociate the role of Six6 in the SCN from the eye using the Six6flox:Gnrhcre mouse line. Regardless, these results confirm and extend our previous study identifying Gnrhcre targeting of the SCN [55] and establishing functional roles of Six6 [44] and Vax1 (this study) in circadian rhythmicity and entrainment.
Ectopic Expression of Gnrhcre Does Not Contribute to the Infertility of Mice with Vax1 or Six6 Deleted within GnRH Neurons
Our previously published detailed comparison of the adult brain structures targeted by the two Gnrh promoter-driven Cre alleles used in this study revealed extensive expression of CRE in areas known to be important in reproduction such as the olfactory bulb, septum, and parts of the hypothalamus, including the SCN [55]. Although in this study we found that the two Cre alleles yielded different circadian phenotypes when paired with deletions of Six6 and Vax1 as described above, the development and number of GnRH neurons was not different for the two Gnrh-promoter driven Cre alleles [46, 45]. Both caused complete infertility and hypogonadism [46, 45]. In contrast, a more severe fertility phenotype was observed for Otx2 deletion using Gnrhcre versus Lhrhcre [55, 79]. In these studies as well, GnRH neuron development did not differ depending on use of Lhrhcre versus Gnrhrcre to conditionally delete Otx2. We attributed the difference in fertility to a high frequency of germline recombination of these alleles, potentially in combination to Otx2 deletion in the septum, a structure with high Otx2 which is targeted by the Gnrhcre and which is important in sexual behavior [55].
Taken as a whole, these data illustrate a more widespread ectopic expression of the Gnrhcre than we described previously [55] and underscore the importance of considering functional impacts of flox-allele deletion arising from such ectopic expression. As Gnrhcre has been widely used to study the role of genes in GnRH neuron development, function and fertility [80–84], it is important to consider if the reported flox-alleles are expressed in the SCN, retina etc. and if this undesired and uncharacterized gene deletion may have contributed to the observed phenotype. If, on the other hand, the studied flox-allele is not expressed in the broader ventral forebrain affected by Gnrhcre, this allele retains an important advantage over the Lhrhcre insofar as the former targets ~100% of GnRH neurons compared to ~95–100% with Lhrhcre.
Conclusions
We have identified Vax1 as a novel transcription factor expressed in the developing neuroepithelium where it is required for SCN development and SCN function. Importantly, Vax1 haploinsufficiency caused abnormal SCN output and decreased VIP expression. The ectopic deletion of the Gnrhcre allele in the SCN confirmed the critical role of both Vax1 and Six6 in SCN development and function and fertility.
Acknowledgements
This work was supported by National Institutes of Health (NIH) Grants R01 HD072754 and R01 HD082567 (to P.L.M.). It was also supported by NIH/Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) P50 HD012303 as part of the National Centers for Translational Research in Reproduction and Infertility (P.L.M.). P.L.M. was also partially supported by P30 DK063491, P30 CA023100, and P42 ES010337. H.M.H. was partially supported by K99/R00 HD084759 and the United States Department of Agriculture National Institute of Food and Agriculture Hatch project 1018024. E.C.P. was partially supported by NIH R25 GM083275 and F31 HD098652. T.T. was partially supported by the Endocrine Society and R.H. was partially supported by the Howell Foundation and the Frontiers of Innovation Scholars Program, UC San Diego. J.A.B. was partially supported by the Frontiers of Innovation Scholars Program, UC San Diego. Work in the M.R.G. laboratory was supported by Office of Naval Research #N00014-13-1-0285, and work in the D.S.K. laboratory is supported by NIH/National Eye Institute award R01EY027011, RPB Special Scholar Award, Atkinson laboratory funds as well as by RPB Unrestricted Grant to Shiley Eye Institute. K.B. was supported by NIH/ National Institute of Neurological Disorders and Stroke IRP funds. The University of Virginia, Center for Research in Reproduction, Ligand Assay and Analysis Core, is supported by the NIH/NICHD Grant P50 HD028934. We thank Karen J. Tonsfeldt, Erica L. Schoeller, Alexandra Yaw, Sabrina Baretto, Ichiko Saotome, and Jason D. Meadows for assistance. We thank Dr. Catherine Dulac (Harvard University, Cambridge, MA, USA) for the Lhrh-cre mice, and Dr. Andrew Wolfe (Johns Hopkins University) for the Gnrh-cre mice. The Vip-luciferase plasmid was kindly provided by Dr. Satchidananda Panda (Salk Institute, La Jolla, CA, USA).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of interest: The authors declare no competing financial interests
References
- 1.Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, Qi L, Qian J, Blackshaw S (2010) A genomic atlas of mouse hypothalamic development. Nat Neurosci 13 (6):767–775. doi:nn.2545 [pii] 10.1038/nn.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445 (7124):168–176 [DOI] [PubMed] [Google Scholar]
- 3.Chen R, Wu X, Jiang L, Zhang Y (2017) Single-Cell RNA-Seq Reveals Hypothalamic Cell Diversity. Cell Rep 18 (13):3227–3241. doi: 10.1016/j.celrep.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, Alberta JA, Cheng LP, Liu Y, Stenman JM, Valerius MT, Billings N, Kim HA, Greenberg ME, McMahon AP, Rowitch DH, Stiles CD, Ma Q (2004) Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306 (5705):2255–2257. doi: 10.1126/science.1104935 [DOI] [PubMed] [Google Scholar]
- 5.Pasquier L, Dubourg C, Blayau M, Lazaro L, Le Marec B, David V, Odent S (2000) A new mutation in the six-domain of SIX3 gene causes holoprosencephaly. Eur J Hum Genet 8 (10):797–800. doi: 10.1038/sj.ejhg.5200540 [DOI] [PubMed] [Google Scholar]
- 6.Dubourg C, Lazaro L, Pasquier L, Bendavid C, Blayau M, Le Duff F, Durou MR, Odent S, David V (2004) Molecular screening of SHH, ZIC2, SIX3, and TGIF genes in patients with features of holoprosencephaly spectrum: Mutation review and genotype-phenotype correlations. Human mutation 24 (1):43–51. doi: 10.1002/humu.20056 [DOI] [PubMed] [Google Scholar]
- 7.Pasquier L, Dubourg C, Gonzales M, Lazaro L, David V, Odent S, Encha-Razavi F (2005) First occurrence of aprosencephaly/atelencephaly and holoprosencephaly in a family with a SIX3 gene mutation and phenotype/genotype correlation in our series of SIX3 mutations. J Med Genet 42 (1):e4. doi: 10.1136/jmg.2004.023416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larder R, Kimura I, Meadows J, Clark DD, Mayo S, Mellon PL (2013) Gene dosage of Otx2 is important for fertility in male mice. Mol Cell Endocrinol 377 (1–2):16–22. doi: 10.1016/j.mce.2013.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann HM, Tamrazian A, Xie H, Perez-Millan MI, Kauffman AS, Mellon PL (2014) Heterozygous deletion of ventral anterior homeobox (vax1) causes subfertility in mice. Endocrinology 155 (10):4043–4053. doi: 10.1210/en.2014-1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandolfi EC, Hoffmann HM, Schoeller EL, Gorman MR, Mellon PL (2018) Haploinsufficiency of SIX3 abolishes male reproductive behavior through disrupted olfactory development, and impairs female fertility through disrupted GnRH neuron migration. Mol Neurobiol 55 (11):8709–8727. doi: 10.1007/s12035-018-1013-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng X, Acosta S, Lagutin O, Gil H, Oliver G (2016) Six3 dosage mediates the pathogenesis of holoprosencephaly. Development 143 (23):4462–4473. doi: 10.1242/dev.132142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann H, Pandolfi E, Larder R, Mellon P (2018) Haploinsufficiency of Homeodomain Proteins Six3, Vax1, and Otx2, Causes Subfertility in Mice Via Distinct Mechanisms. Neuroendocrinology in press. doi: 10.1159/000494086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis SW, Castinetti F, Carvalho LR, Ellsworth BS, Potok MA, Lyons RH, Brinkmeier ML, Raetzman LT, Carninci P, Mortensen AH, Hayashizaki Y, Arnhold IJ, Mendonca BB, Brue T, Camper SA (2010) Molecular mechanisms of pituitary organogenesis: In search of novel regulatory genes. Molecular and cellular endocrinology 323 (1):4–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesan D, Kurrasch DM (2016) Genetic programs of the developing tuberal hypothalamus and potential mechanisms of their disruption by environmental factors. Mol Cell Endocrinol 438:3–17. doi: 10.1016/j.mce.2016.09.031 [DOI] [PubMed] [Google Scholar]
- 15.Sladek M, Sumova A, Kovacikova Z, Bendova Z, Laurinova K, Illnerova H (2004) Insight into molecular core clock mechanism of embryonic and early postnatal rat suprachiasmatic nucleus. Proc Natl Acad Sci U S A 101 (16):6231–6236. doi: 10.1073/pnas.0401149101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, Zhuang X (2018) Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362 (6416). doi: 10.1126/science.aau5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedont JL, LeGates TA, Slat EA, Byerly MS, Wang H, Hu J, Rupp AC, Qian J, Wong GW, Herzog ED, Hattar S, Blackshaw S (2014) Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep 7 (3):609–622. doi: 10.1016/j.celrep.2014.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Challet E (2010) Interactions between light, mealtime and calorie restriction to control daily timing in mammals. J Comp Physiol B 180 (5):631–644. doi: 10.1007/s00360-010-0451-4 [DOI] [PubMed] [Google Scholar]
- 19.Challet E (2015) Keeping circadian time with hormones. Diabetes Obes Metab 17 Suppl 1:76–83. doi: 10.1111/dom.12516 [DOI] [PubMed] [Google Scholar]
- 20.Alvarez JD, Hansen A, Ord T, Bebas P, Chappell PE, Giebultowicz JM, Williams C, Moss S, Sehgal A (2008) The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. Journal of biological rhythms 23 (1):26–36. doi: 10.1177/0748730407311254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moller-Levet CS, Archer SN, Bucca G, Laing EE, Slak A, Kabiljo R, Lo JC, Santhi N, von Schantz M, Smith CP, Dijk DJ (2013) Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci U S A 110 (12):E1132–1141. doi: 10.1073/pnas.1217154110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.VanDunk C, Hunter LA, Gray PA (2011) Development, maturation, and necessity of transcription factors in the mouse suprachiasmatic nucleus. J Neurosci 31 (17):6457–6467. doi: 10.1523/JNEUROSCI.5385-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahoney MM (2010) Shift work, jet lag, and female reproduction. Int J Endocrinol 2010:813764. doi: 10.1155/2010/813764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh DH, Kuljis DA, Azuma L, Wu Y, Truong D, Wang HB, Colwell CS (2014) Disrupted reproduction, estrous cycle, and circadian rhythms in female mice deficient in vasoactive intestinal peptide. J Biol Rhythms 29 (5):355–369. doi: 10.1177/0748730414549767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hickok JR, Tischkau SA (2010) In vivo circadian rhythms in gonadotropin-releasing hormone neurons. Neuroendocrinology 91 (1):110–120. doi: 10.1159/000243163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosko SS, Moore RY (1979) Neonatal ablation of the suprachiasmatic nucleus. Effects on the development of the pituitary-gonadal axis in the female rat. Neuroendocrinology 29 (5):350–361 [DOI] [PubMed] [Google Scholar]
- 27.Christian CA, Moenter SM (2008) Vasoactive intestinal polypeptide can excite gonadotropin-releasing hormone neurons in a manner dependent on estradiol and gated by time of day. Endocrinology 149 (6):3130–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams WP 3rd, Kriegsfeld LJ (2012) Circadian control of neuroendocrine circuits regulating female reproductive function. Frontiers in endocrinology 3:60. doi: 10.3389/fendo.2012.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vida B, Deli L, Hrabovszky E, Kalamatianos T, Caraty A, Coen CW, Liposits Z, Kallo I (2010) Evidence for suprachiasmatic vasopressin neurones innervating kisspeptin neurones in the rostral periventricular area of the mouse brain: regulation by oestrogen. J Neuroendocrinol 22 (9):1032–1039. doi: 10.1111/j.1365-2826.2010.02045.x [DOI] [PubMed] [Google Scholar]
- 30.Russo KA, La JL, Stephens SB, Poling MC, Padgaonkar NA, Jennings KJ, Piekarski DJ, Kauffman AS, Kriegsfeld LJ (2015) Circadian Control of the Female Reproductive Axis Through Gated Responsiveness of the RFRP-3 System to VIP Signaling. Endocrinology 156 (7):2608–2618. doi: 10.1210/en.2014-1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smarr BL, Gile JJ, de la Iglesia HO (2013) Oestrogen-independent circadian clock gene expression in the anteroventral periventricular nucleus in female rats: possible role as an integrator for circadian and ovarian signals timing the luteinising hormone surge. J Neuroendocrinol 25 (12):1273–1279. doi: 10.1111/jne.12104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams WP 3rd, Jarjisian SG, Mikkelsen JD, Kriegsfeld LJ (2011) Circadian control of kisspeptin and a gated GnRH response mediate the preovulatory luteinizing hormone surge. Endocrinology 152 (2):595–606. doi: 10.1210/en.2010-0943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schafer D, Kane G, Colledge WH, Piet R, Herbison AE (2018) Sex- and sub region-dependent modulation of arcuate kisspeptin neurones by vasopressin and vasoactive intestinal peptide. J Neuroendocrinol 30 (12):e12660. doi: 10.1111/jne.12660 [DOI] [PubMed] [Google Scholar]
- 34.Piet R, Dunckley H, Lee K, Herbison AE (2016) Vasoactive Intestinal Peptide Excites GnRH Neurons in Male and Female Mice. Endocrinology 157 (9):3621–3630. doi: 10.1210/en.2016-1399 [DOI] [PubMed] [Google Scholar]
- 35.Piet R, Fraissenon A, Boehm U, Herbison AE (2015) Estrogen permits vasopressin signaling in preoptic kisspeptin neurons in the female mouse. J Neurosci 35 (17):6881–6892. doi: 10.1523/JNEUROSCI.4587-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann HM, Mellon PL (2018) Regulation of GnRH Gene Expression In: Herbison AE, Plant TM (eds) The GnRH Neuron and its Control. Wiley Blackwell, Hoboken, N.J., pp 95–120 [Google Scholar]
- 37.Bedont JL, Newman EA, Blackshaw S (2015) Patterning, specification, and differentiation in the developing hypothalamus. Wiley Interdiscip Rev Dev Biol 4 (5):445–468. doi: 10.1002/wdev.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chappell PE, White RS, Mellon PL (2003) Circadian gene expression regulates pulsatile gonadotropin-releasing hormone (GnRH) secretory patterns in the hypothalamic GnRH-secreting GT1–7 cell line. J Neurosci 23 (35):11202–11213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forni PE, Wray S (2015) GnRH, anosmia and hypogonadotropic hypogonadism--where are we? Front Neuroendocrinol 36:165–177. doi: 10.1016/j.yfrne.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hallonet M, Hollemann T, Pieler T, Gruss P (1999) Vax1, a novel homeobox-containing gene, directs development of the basal forebrain and visual system. Genes Dev 13 (23):3106–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertuzzi S, Hindges R, Mui SH, O’Leary DD, Lemke G (1999) The homeodomain protein vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes Dev 13 (23):3092–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altman J, Bayer SA (1986) The development of the rat hypothalamus. Adv Anat Embryol Cell Biol 100:1–178 [PubMed] [Google Scholar]
- 43.Larder R, Clark DD, Miller NL, Mellon PL (2011) Hypothalamic dysregulation and infertility in mice lacking the homeodomain protein Six6. J Neurosci 31 (2):426–438. doi:31/2/426 [pii] 10.1523/JNEUROSCI.1688-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark DD, Gorman MR, Hatori M, Meadows JD, Panda S, Mellon PL (2013) Aberrant development of the suprachiasmatic nucleus and circadian rhythms in mice lacking the homeodomain protein six6. J Biol Rhythms 28 (1):15–25. doi: 10.1177/0748730412468084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandolfi EC, Tonsfeldt KJ, Hoffmann HM, Mellon PL (2019) Deletion of the Homeodomain Protein Six6 from GnRH Neurons Decreases GnRH Gene Expression Resulting in Infertility. Endocrinology 160:in press. doi: 10.1210/en.2019-00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffmann HM, Trang C, Gong P, Kimura I, Pandolfi EC, Mellon PL (2016) Deletion of Vax1 from GnRH neurons abolishes GnRH expression and leads to hypogonadism and infertility. J Neurosci 36 (12):3506–3518. doi: 10.1523/JNEUROSCI.2723-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bharti K, Gasper M, Bertuzzi S, Arnheiter H (2011) Lack of the ventral anterior homeodomain transcription factor VAX1 leads to induction of a second pituitary. Development 138 (5):873–878. doi: 10.1242/dev.056465 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon H, Enquist LW, Dulac C (2005) Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell 123 (4):669–682 [DOI] [PubMed] [Google Scholar]
- 49.Wolfe A, Divall S, Singh SP, Nikrodhanond AA, Baria AT, Le WW, Hoffman GE, Radovick S (2008) Temporal and spatial regulation of CRE recombinase expression in gonadotrophin-releasing hormone neurones in the mouse. J Neuroendocrinol 20 (7):909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ridder WH 3rd, Nusinowitz S (2006) The visual evoked potential in the mouse--origins and response characteristics. Vision Res 46 (6–7):902–913. doi: 10.1016/j.visres.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann HM, Gong P, Tamrazian A, Mellon PL (2018) Transcriptional interaction between cFOS and the homeodomain-binding transcription factor VAX1 on the GnRH promoter controls Gnrh1 expression levels in a GnRH neuron maturation specific manner. Mol Cell Endocrinol 461:143–154. doi: 10.1016/j.mce.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hatori M, Gill S, Mure LS, Goulding M, O’Leary DD, Panda S (2014) Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. eLife 3:e03357. doi: 10.7554/eLife.03357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Givens ML, Rave-Harel N, Goonewardena VD, Kurotani R, Berdy SE, Swan CH, Rubenstein JL, Robert B, Mellon PL (2005) Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. The Journal of biological chemistry 280 (19):19156–19165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffmann HM (2018) Determination of reproductive competence by confirming pubertal onset and performing a fertility assay in mice and rats. J Vis Exp (140):e58352. doi: 10.3791/58352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoffmann HM, Larder R, Lee JS, Hu RJ, Trang C, Devries BM, Clark DD, Mellon PL (2019) Differential CRE expression in Lhrh-Cre and Gnrh-Cre alleles and the impact on fertility in Otx2-flox mice. Neuroendocrinology 108:328–342. doi: 10.1159/000497791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bharti K, Liu W, Csermely T, Bertuzzi S, Arnheiter H (2008) Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development 135 (6):1169–1178. doi: 10.1242/dev.014142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bedont JL, Blackshaw S (2015) Constructing the suprachiasmatic nucleus: a watchmaker’s perspective on the central clockworks. Front Syst Neurosci 9:74. doi: 10.3389/fnsys.2015.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plageman TF Jr., Lang RA (2012) Generation of an Rx-tTA: TetOp-Cre knock-in mouse line for doxycycline regulated Cre activity in the Rx expression domain. PLoS One 7 (11):e50426. doi: 10.1371/journal.pone.0050426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blattner MS, Mahoney MM (2014) Estrogen receptor 1 modulates circadian rhythms in adult female mice. Chronobiol Int 31 (5):637–644. doi: 10.3109/07420528.2014.885528 [DOI] [PubMed] [Google Scholar]
- 60.Bailey M, Silver R (2014) Sex differences in circadian timing systems: implications for disease. Front Neuroendocrinol 35 (1):111–139. doi: 10.1016/j.yfrne.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar DM, Kilduff TS, Martin CE, Dement WC (1991) Influence of running wheel activity on free-running sleep/wake and drinking circadian rhythms in mice. Physiol Behav 50 (2):373–378 [DOI] [PubMed] [Google Scholar]
- 62.Tonsfeldt KJ, Schoeller EL, Brusman LE, Cui LJ, Lee J, Mellon PL (2019) The Contribution of the Circadian Gene Bmal1 to Female Fertility and the Generation of the Preovulatory Luteinizing Hormone Surge. J Endocr Soc 3 (4):716–733. doi: 10.1210/js.2018-00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanes JR, Masland RH (2015) The types of retinal ganglion cells: current status and implications for neuronal classification. Annual review of neuroscience 38:221–246. doi: 10.1146/annurev-neuro-071714-034120 [DOI] [PubMed] [Google Scholar]
- 64.Herzog ED, Schwartz WJ (2002) A neural clockwork for encoding circadian time. J Appl Physiol (1985) 92 (1):401–408. doi: 10.1152/japplphysiol.00836.2001 [DOI] [PubMed] [Google Scholar]
- 65.Crossland WJ, Uchwat CJ (1982) Neurogenesis in the chick ventral lateral geniculate and ectomammillary nuclei: relationship of soma size to birthdate. Brain Res 282 (1):33–46 [DOI] [PubMed] [Google Scholar]
- 66.Kabrita CS, Davis FC (2008) Development of the mouse suprachiasmatic nucleus: determination of time of cell origin and spatial arrangements within the nucleus. Brain Res 1195:20–27. doi: 10.1016/j.brainres.2007.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Compston A (1991) Limiting and repairing the damage in multiple sclerosis. J Neurol Neurosurg Psychiatry 54 (11):945–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Beek EM, Horvath TL, Wiegant VM, van den Hurk R, Buijs RM (1997) Evidence for a direct neuronal pathway from the suprachiasmatic nucleus to the gonadotropin-releasing hormone system: combined tracing and light and electron microscopic immunocytochemical studies. The Journal of comparative neurology 384 (4):569–579 [DOI] [PubMed] [Google Scholar]
- 69.Horvath TL, Cela V, van der Beek EM (1998) Gender-specific apposition between vasoactive intestinal peptide-containing axons and gonadotrophin-releasing hormone-producing neurons in the rat. Brain Res 795 (1–2):277–281 [DOI] [PubMed] [Google Scholar]
- 70.Kriegsfeld LJ, Silver R, Gore AC, Crews D (2002) Vasoactive intestinal polypeptide contacts on gonadotropin-releasing hormone neurones increase following puberty in female rats. J Neuroendocrinol 14 (9):685–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ward DR, Dear FM, Ward IA, Anderson SI, Spergel DJ, Smith PA, Ebling FJ (2009) Innervation of gonadotropin-releasing hormone neurons by peptidergic neurons conveying circadian or energy balance information in the mouse. PLoS One 4 (4):e5322. doi: 10.1371/journal.pone.0005322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smarr BL, Morris E, de la Iglesia HO (2012) The dorsomedial suprachiasmatic nucleus times circadian expression of Kiss1 and the luteinizing hormone surge. Endocrinology 153 (6):2839–2850. doi: 10.1210/en.2011-1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith MJ, Jiennes L, Wise PM (2000) Localization of the VIP2 receptor protein on GnRH neurons in the female rat. Endocrinology 141 (11):4317–4320. doi: 10.1210/endo.141.11.7876 [DOI] [PubMed] [Google Scholar]
- 74.Dolatshad H, Campbell EA, O’Hara L, Maywood ES, Hastings MH, Johnson MH (2006) Developmental and reproductive performance in circadian mutant mice. Hum Reprod 21 (1):68–79. doi: 10.1093/humrep/dei313 [DOI] [PubMed] [Google Scholar]
- 75.Legan SJ, Karsch FJ (1975) A daily signal for the LH surge in the rat. Endocrinology 96 (1):57–62 [DOI] [PubMed] [Google Scholar]
- 76.Christian CA, Moenter SM (2010) The neurobiology of preovulatory and estradiol-induced gonadotropin-releasing hormone surges. Endocr Rev 31 (4):544–577. doi: 10.1210/er.2009-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Conte I, Marco-Ferreres R, Beccari L, Cisneros E, Ruiz JM, Tabanera N, Bovolenta P (2010) Proper differentiation of photoreceptors and amacrine cells depends on a regulatory loop between NeuroD and Six6. Development 137 (14):2307–2317. doi: 10.1242/dev.045294 [DOI] [PubMed] [Google Scholar]
- 78.Tetreault N, Champagne MP, Bernier G (2009) The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev Biol 327 (2):541–550. doi: 10.1016/j.ydbio.2008.12.022 [DOI] [PubMed] [Google Scholar]
- 79.Diaczok D, DiVall S, Matsuo I, Wondisford FE, Wolfe AM, Radovick S (2011) Deletion of Otx2 in GnRH neurons results in a mouse model of hypogonadotropic hypogonadism. Mol Endocrinol 25 (5):833–846. doi:me.2010–0271 [pii] 10.1210/me.2010-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kurian JR, Louis S, Keen KL, Wolfe A, Terasawa E, Levine JE (2016) The Methylcytosine Dioxygenase Ten-Eleven Translocase-2 (tet2) Enables Elevated GnRH Gene Expression and Maintenance of Male Reproductive Function. Endocrinology 157 (9):3588–3603. doi: 10.1210/en.2016-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DiVall SA, Herrera D, Sklar B, Wu S, Wondisford F, Radovick S, Wolfe A (2015) Insulin receptor signaling in the GnRH neuron plays a role in the abnormal GnRH pulsatility of obese female mice. PLoS One 10 (3):e0119995. doi: 10.1371/journal.pone.0119995 PONE-D-14–29677 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Novaira HJ, Sonko ML, Hoffman G, Koo Y, Ko C, Wolfe A, Radovick S (2014) Disrupted kisspeptin signaling in GnRH neurons leads to hypogonadotrophic hypogonadism. Mol Endocrinol 28 (2):225–238. doi: 10.1210/me.2013-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu S, Divall S, Hoffman GE, Le WW, Wagner KU, Wolfe A (2011) Jak2 is necessary for neuroendocrine control of female reproduction. J Neurosci 31 (1):184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Divall SA, Williams TR, Carver SE, Koch L, Bruning JC, Kahn CR, Wondisford F, Radovick S, Wolfe A (2010) Divergent roles of growth factors in the GnRH regulation of puberty in mice. J Clin Invest 120 (8):2900–2909. doi: 10.1172/JCI41069 41069 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]