Abstract
Neuroinflammation and accompanying microglial dysfunction are now appreciated to be involved in Alzheimer’s disease (AD) pathogenesis. Critical to the process of neuroinflammation are the type-I interferon (IFN) family of cytokines. Efforts to phenotypically characterize microglia within AD identify distinct populations associated with type-I IFN signalling, yet how this affects underlying microglial function is yet to be fully elucidated. Here we demonstrate that Aβ1–42 exposure increases bioactive levels of type-I IFN produced by primary microglia alongside increased expression of type-I IFN related genes. Primary microglia isolated from brains of APPswePS1ΔE9 mice with ablated type-I IFN signalling show an increased phagocytic ability to uptake FITC-Aβ1–42. Correlative assessment of plaque sizes in aged APPswePS1ΔE9 mice with abrogated type-I IFN signalling show unchanged deposition levels. Microglia from these mice did however show alterations in morphology. This data further highlights the role of type-I IFN signalling within microglia and identifies a role in phagocytosis. As such, targeting both microglial and global type-I IFN signalling presents as a novel therapeutic strategy for AD management.
Subject terms: Cellular neuroscience, Microglia
Introduction
Alzheimer’s disease (AD) is now recognised as the most common form of dementia and is now estimated to be the 5th largest cause of death globally1. AD has been classically characterised by its two hallmark pathologies, neurofibrillary tangles composed of hyper phosphorylated tau, and extracellular senile plaques composed of amyloid beta (Aβ). The immune response to these pathologies, neuroinflammation is now appreciated to be involved in disease progression. Regulated forms of neuroinflammation are viewed as protective, and indeed required for homeostatic function. In contrast, a dysregulated form of this process is present in AD2. This dysregulated form is now recognised as a contributor to AD pathogenesis.
Fundamental to this neuroinflammation are microglia, the resident innate immune cells within the central nervous system (CNS). Critically, microglia mount the initial neuroinflammatory response and further maintain it3. Upon recognition of noxious stimuli, microglia release a number of cytokines including interleukin (IL) 1β, IL6, and tumor necrosis factor alpha (TNFα) to create an inflammatory microenvironment4. This is seen in conjunction with chemokine secretion to recruit additional microglia. Microglia also have a number of roles outside immune-related functions. In particular, microglia are often viewed as macrophages of the CNS due to a shared hierarchal lineage and roles in phagocytosis5.
Due to diverse roles, microglia adopt a number of varied phenotypes with differential corresponding functions throughout the CNS. As such, large efforts have been made to better phenotypically characterize microglia under both normal and AD settings, primarily through transcriptomic-based approaches6,7. In the 5× familial AD mouse model, a single cell ribonucleic acid (RNA) -seq approach was used to investigate all immune cells, from which a unique population termed disease-associated microglia (DAM) was identified8. This population was associated with changes in phagocytosis and lipid metabolism, and primarily localized near Aβ plaques. DAM and DAM-like microglial populations have been further identified within other AD models9. In addition, investigations into microglial morphology have also been conducted. Structure follows function, and it is established that efficient and rapid morphological remodelling is required for microglia to perform varied tasks10. Recent work has also identified other brain residing macrophages that reside between the barriers of the brain11. Under certain conditions, these cells can enter the parenchyma and contribute to the overall pool of myeloid cells and may also contribute to phenotypical differences.
Dysregulation of microglial function is seen throughout AD. Genome wide association studies (GWAS) identify a number of coding variants highly expressed in microglia12. Of the GWAS identified risk factors, cluster of differentiation (CD) 33 and triggering receptor expressed on myeloid cells 2 (TREM2) have been shown to be involved in phagocytosis and response to stimuli when further examined both in vitro and in vivo4,13,14. Microglia harvested from aged dual amyloid precursor protein/presenilin 1 (APPswePS1ΔE9) model of AD also show decreased expression of Aβ binding receptors and Aβ degrading enzymes15.
Similar to microglia, the type-I interferon (IFN) cytokine family are also critical in neuroinflammation. Named for their capacity to “interfere” with viral replication, IFNs are now recognised to have distinct roles outside of infection16. There are two predominant types of type-I IFNs, α with 14 subtypes, and the single subtype β17. The type-I IFNs signal through their cognate receptor, the interferon α/β receptor (IFNAR) which is composed of two subunits, IFNAR1 and IFNAR2. Ablation of the IFNAR1 subunit alone has been demonstrated to abrogate conventional signalling18. Downstream signalling cascades involve induction of a number of signal inducer and transduction complexes and interferon regulatory factors (IRFs). These proteins all mediate different effects. IRF9 is critical for downstream mediation, whilst IRF7 is critical for the induction of further type-I IFNs19,20. These lead to transcription of a wide array of interferon-stimulated genes (ISGs). However, the exact role of type-I IFNs within the CNS remains poorly understood21–23. Increased levels of IFNα and a number of ISGs are seen in individuals in both Aicardi-Goutières syndrome and systemic lupus erythematosus, demonstrating a link between type-I IFNs and CNS disorders24–26. Type-I IFNs are indeed able to regulate levels of IL1β, IL6 and TNFα which are consistently upregulated within AD3,27–29.
Of critical note however, are the observed links between microglia and type-I IFNs30. Blockade of type-I IFN signalling in a murine lupus model alters both the number of CD68 positive reactive microglia and microglial engulfment of synapses26. Furthermore, IRF7 has been shown to be required for early-stage microglial development11. Within AD, a number of transcriptomic studies have shown that type-I IFNs are critical modulators of specific microglial populations8,31,32. A meta-analysis of microglial transcripts from 69 individual disease states, including AD, revealed a co-regulated IFN gene set across all disease states observed7. Whilst these studies have been able to demonstrate an association, the functional implications of microglial type-I IFN signalling within the context of AD are yet to be fully explored.
We posit that it is microglial type-I IFN signalling that contributes to neuroinflammation in AD, and that type-I IFN signalling is involved in phenotypical and functional change in microglia. We make use of murine models lacking IFNAR1 and subsequent type-I IFN signalling alongside APPswePS1ΔE9 model of AD to achieve this. Here we are able to demonstrate for the first time that type-I IFN is involved in phagocytosis and alters the microglial response to Aβ1–42. Lack of type-I IFN signalling also alters microglial morphology in APPswePS1ΔE9 mice. Collectively, these results are the first to elucidate the precise phenotypic and functional role of type-I IFN signalling in microglia and provide a baseline for future bodies of work.
Results
Treatment of microglial cells with type-I IFN inhibits phagocytosis
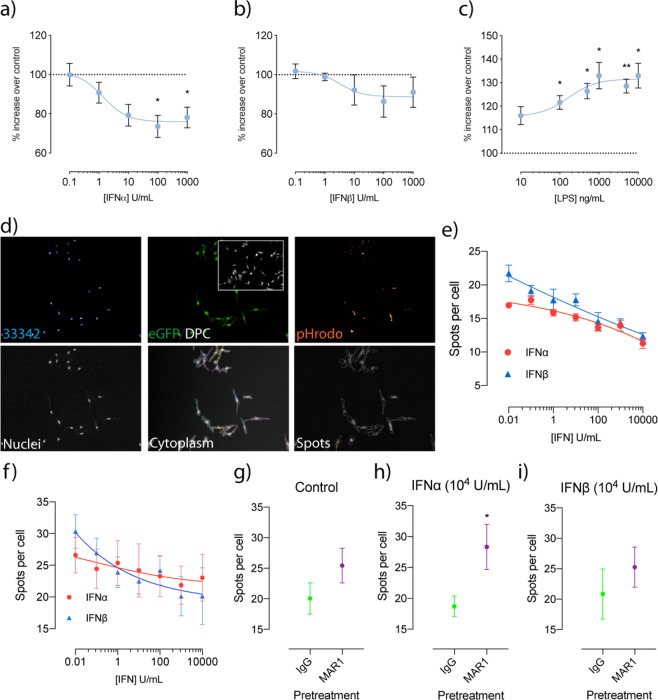
Recently, type-I IFNs have been reported to be involved in the modulation of microglial phenotype. To investigate how type-I IFN stimulation directly affects microglial function, a phagocytosis assay utilizing pHrodo E. coli bioparticles was performed with immortalised BV-2 murine microglial-like cells. Dose response curves revealed that IFNα significantly decreased phagocytosis at concentrations of 100 (untreated: 100 ± 0% vs. 100U: 73.57 ± 5.656%, p = 0.0297, n = 5) and 1000 units (untreated: 100 ± 0% vs. 1000U: 78.07 ± 5.215%, p = 0.0423, n = 5) (Fig. 1a). Regression analysis also showed a negative relationship between phagocytosis and concentration. IFNβ treatments showed a similar trend for decreased phagocytosis with increasing concentration, however this was not statistically significant (Fig. 1b). To compare this to a classical pro-inflammatory response, this assay was repeated with lipopolysaccharide (LPS) (10–10,000 ng/mL) as a stimulus (Fig. 1c). Contrasting to the type-1 IFNs, significant increases in phagocytosis were seen with LPS concentrations greater than 100 ng/mL (untreated: 100 ± 0% vs. 100 ng/mL: 121.6 ± 2.89%, p = 0.0157, 500 ng/mL: 126.4 ± 3.249%, p = 0.0124, 1000 ng/mL: 133 ± 5.99%, p = 0.0305, 5000 ng/mL: 128.5 ± 2.978%, p = 0.0077, 10,000 ng/mL: 133 ± 5.2961%, p = 0.0261). This suggests that type-I IFNs are indeed eliciting a unique functional effect on microglia that differs from a classical pro-inflammatory response. For increased sensitivity and to confirm these findings, we also performed experiments using a high content imaging platform. The workflow is described in Fig. 1d. Regression analysis showed a negative relationship for both IFNα and IFNβ treatments in number of spots per cell, confirming our initial findings. We then repeated these experiments using primary CX3C chemokine receptor 1 (CX3CR1eGFP/+) microglia. Similarly, we are able to show a negative dose-response relationship. To then investigate if this inhibition can be reversed, cells were pre-treated with either IgG controls or an anti-IFNAR1 (MAR1) antibody before treatment with either 104 units IFNα/β. MAR1 was able to increases the number of spots per cell in IFNα treated cells (9.632 ± 4.023, p = 0.0436).
Figure 1.
Treatment with IFN inhibits phagocytosis in both BV-2 and primary CX3CR1eGFP/+ primary microglia, with IFNα inhibition reversible with an anti-IFNAR1 neutralising antibody. Phagocytosis was determined using pHrodo E. coli fluorescent particles and is represented as % increases over untreated samples following varying concentrations of (a) IFNα, (b) IFNβ and (c) LPS (*p < 0.05, **p < 0.01, 1-way ANOVA, Dunnett’s multiple comparisons test, n = 5–6). (d) Workflow of imaging analysis for pHrodo E.coli treatments for both BV-2 and CX3CR1eGFP/+ primary microglia. Analysis of spots per cell following varying concentrations of IFN in (e) BV-2 cells and (f) primary CX3CR1eGFP/+ microglia. Analysis of spots per cell following IgG and MAR1 pre-treatment after which cells were exposed to (g) control (h) 104 U IFNα and (i) 104 U IFNβ (*p < 0.05, Students unpaired t-test, n = 5–6). Data is expressed as mean ± SEM.
Monomeric Aβ1–42 triggers a type-I IFN response in wild type microglia that is ameliorated in IFNAR1−/− microglia
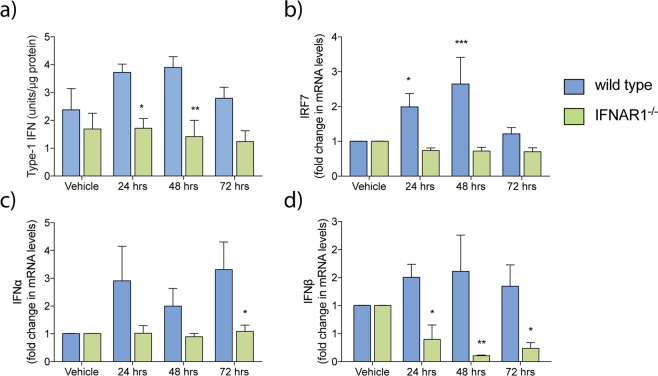
We have previously demonstrated that ablation of type-I IFN signalling in APPswePS1ΔE9 mice slowed cognitive decline and altered global glial phenotype33. Here, we focused specifically on examining the microglial response. To confirm that microglia do indeed mount a type-I IFN response, primary microglia of wild type and IFNAR1−/− genotypes were subjected to monomeric Aβ1–42 treatment in vitro. Media was collected and analysed by a bioactive type-I IFN measurement assay (Fig. 2a). IFNAR1−/− microglia showed decreased levels of bioactive type-I IFN compared to wild type at both 24 (wild type: 3.719 ± 0.299 U/mg total protein vs. IFNAR1−/−: 1.716 ± 0.585 U/mg total protein, p = 0.0427, n = 5–6) and 48 (wild type: 3.889 ± 0.391 U/mg total protein vs. IFNAR1−/−: 1.416 ± 0.585 U/ mg total protein, p = 0.0094, n = 5–6) hours after treatment.
Figure 2.
Aβ1–42 elicits a type-I IFN response in microglia that is ameliorated by removal of IFNAR1. Primary wild type and IFNAR1−/− microglia were treated with 10 μM monomeric Aβ1–42 treatment (24–72 hours). (a) Collected media was analysed via reporter cell assay to measure levels of bioactive type-I IFN. Cells were then analysed via QPCR to measure transcript levels of (b) IRF7, as well as levels of both (c) IFNα and (d) IFNβ. Data is displayed as mean ± SEM (*p < 0.05, **p < 0.01, 2-way ANOVA, Sidak’s multiple comparisons test, wild type vs. IFNAR1−/–, n = 5–6).
We further investigated this type-I IFN response by analysing a number of type-I IFN related genes via quantitative polymerase chain reaction (QPCR), including IFNα, IFNβ, and IRF7 (Fig. 2b–d). Levels of IFNα were decreased in IFNAR1−/− microglia at 72 hours only (wild type: 3.315 ± 0.992 fold vs. IFNAR1−/−: 1.087 ± 0.225 fold, p = 0.0322, n = 4–5). IFNβ was decreased at all time points measured (24 hours - wildtype: 1.503 ± 0.234 fold vs. IFNAR1−/−: 0.397 ± 0.258 fold, p = 0.0489, 48 hours – wild type: 1.609 ± 0.648 vs IFNAR1−/−: 0.105 ± 0.013, p = 0.0042, 72 hours – wild type: 1.345 ± 0.381 vs. IFNAR1−/−: 0.238 ± 0.104, p = 0.0486, n = 5–6). IRF7 was also decreased at 24 (wild type: 2.196 ± 0.217 vs. IFNAR1−/−: 0.723 ± 0.157, p = 0.0033, n = 5–6) and 48 hours (wild type: 2.769 ± 0.709 vs. IFNAR1−/−: 0.773 ± 0.141, p < 0.0001, n = 5–6). Levels of IRF3 remained unchanged between genotypes. This data suggests that the type-I IFN response induced by Aβ is IFNα, IFNβ and IRF7 mediated.
IFNAR1−/− microglia exhibit an altered inflammatory profile following monomeric Aβ1–42 treatment
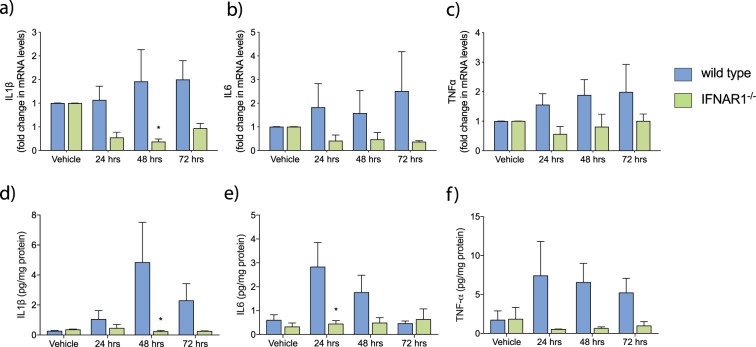
To further investigate the inflammatory response in IFNAR1−/− microglia in response to Aβ treatment, levels of IL6, IL1β and TNFα were determined by QPCR. These cytokines have been shown to be critical in the neuroinflammatory response and are elevated in AD patients3. Transcript levels of IL1β (Fig. 3a) were significantly decreased in IFNAR1−/− microglia at both 48 (wild type: 1.462 ± 0.669 fold vs. IFNAR1−/−: 0.174 ± 0.048 fold, p = 0.0076, n = 6) and 72 hours (wild type: 1.502 ± 0.399 fold vs. IFNAR1−/−: 0.383 ± 0.113 fold, p = 0.0245, n = 6) following Aβ treatment. No significant changes were detected in transcripts for either IL6 (Fig. 3b) or TNFα (Fig. 3c).
Figure 3.
The inflammatory response to Aβ1–42 is attenuated in IFNAR1−/− primary microglia. Transcript levels of (a) IL1β, (b) IL6 and (c) TNFα were analysed by QPCR analysis following 10 μM monomeric Aβ1–42 treatment (24–72 hours). Collected media was also analysed via ELISA for the corresponding cytokines (d) to (f). Data is displayed as mean ± SEM (*p < 0.05, 2-way ANOVA, Sidak’s multiple comparisons test, wild type vs. IFNAR1−/–, n = 5).
To confirm if these changes in messenger RNA (mRNA) expression translated to alterations in protein levels, enzyme-linked immunosorbent assays (ELISA) were performed on media from these same samples. Due to differences in purity between commercially available Aβ1–42, all results are expressed as a ratio compared to total media protein levels as measured by Bradford analysis. Between genotypes, significant differences were observed in levels of IL6 (Fig. 3d) at 24 hours (wild type: 2.83 ± 1.026 pg/mg total protein vs. IFNAR1−/−: 0.44 ± 0.144 pg/mg total protein, p = 0.0164, n = 5–6) and IL1β (Fig. 3e) at 48 hours (wild type: 4.83 ± 2.682 pg/mg total protein vs. IFNAR1−/−: 0.228 ± 0.077 pg/mg total protein). TNFα measurements showed a trend for decreased levels in IFNAR1−/− microglia across all observed timepoints (Fig. 3f). This further confirms the pro-inflammatory response elicited by Aβ1–42 in vitro and identifies type-I IFNs as potential regulators of this response.
APPswePS1ΔE9 × IFNAR1−/− microglia exhibit increased phagocytic capacity towards FITC conjugated Aβ1–42, but not FITC Aβ42–1
To further investigate the role between type-I IFN signalling and microglial function, wild type, IFNAR1−/−, APPswePS1ΔE9 and APPswePS1ΔE9 × IFNAR1−/−genotypes were treated with fluorescein isothiocyanate (FITC) conjugated Aβ1–42 peptide (2 mg/mL) for 1 and 3 hours, after which cells were analysed by flow cytometry. All genotypes showed similar percentage levels of FITC positive cells over 1 and 3 hours (Fig. 4e). However, the mean fluorescent intensity as assessed through geometric mean showed a marked increase in the APPswePS1ΔE9 × IFNAR1−/− microglia compared to all genotypes (1 hour: APPswePS1ΔE9 × IFNAR1−/−: 1532.400 ± 143.406 vs. wild type: 911 ± 159.868, vs. IFNAR1−/−: 870.00 ± 107.699, p < 0.01, APPswePS1ΔE9 × IFNAR1−/− vs. all genotypes, n = 3–6, 3 hours: APPswePS1ΔE9 × IFNAR1−/−: 2870.600 ± 175.241 vs wild type: 1695.250 ± 98.447, vs. IFNAR1−/−: 1801.667 ± 135.451, vs. APPswePS1ΔE9: 1923.000 ± 135.451, p < 0.0001, APPswePS1ΔE9 × IFNAR1−/− vs. all genotypes, n = 3–6) (Fig. 4f). This data demonstrates that on an individual cellular basis, APPswePS1ΔE9 × IFNAR1−/− microglia have an enhanced ability to phagocytose FITC conjugated Aβ1–42. Interestingly, this is unique to these APPswePS1ΔE9 × IFNAR1−/− microglia, with IFNAR1−/− microglia alone showing no differences.
Figure 4.
APPswePS1ΔE9 × IFNAR1−/− exhibit enhanced phagocytosis towards FITC-Aβ1–42, but not the reverse peptide FITC-Aβ42–1. Cells were treated with either FITC conjugated Aβ1–42 or the reverse peptide Aβ42–1 for 1 and 3 hours before being measured by flow cytometry. Representative distributions for each genotype for Aβ1–42 are shown in (a). Measures of percentage of parent and geometric mean are shown for 1–42 in (b). Representative distributions for each genotype for Aβ42–1 are shown in (c). Measures of percentage of parent and geometric mean are shown for 42–1 in (d). Data is displayed as mean ± SEM (****p < 0.0001, **p < 0.01, 2-way ANOVA, Sidak’s multiple comparisons test, wild type vs. IFNAR1−/− vs. APPswePS1ΔE9 vs. APPswePS1ΔE9 × IFNAR1−/−, n = 3–5).
To confirm if this finding was specific to Aβ or an increase in overall phagocytic ability, we then performed the same experiment using the reverse peptide FITC Aβ42–1. Interestingly, APPswePS1ΔE9 × IFNAR1−/− microglia showed no similar increases in uptake, but rather IFNAR1−/− microglia showed significant increases in both percentage of parent (1 hour: IFNAR1−/−: 73.100 ± 5.254 vs. wild type: 21.700 ± 3.889, vs. APPswePS1ΔE9: 22.627 ± 15.448, vs. APPswePS1ΔE9 × IFNAR1−/−: 21.620 ± 3.565, p < 0.0001, IFNAR1−/− vs. all genotypes, n = 3–5, 3 hours: IFNAR1−/−: 80.300 ± 6.585 vs. wild type: 30.933 ± 3.203, vs. APPswePS1ΔE9: 19.957 ± 9.466, vs. APPswePS1ΔE9 × IFNAR1−/−: 38.100 ± 3.102, p < 0.0001, IFNAR1−/− vs. all genotypes, n = 3–5) and geometric mean (1 hour: IFNAR1−/−: 1287.100 ± 47.548 vs. wild type: 664.333 ± 46.423, vs. APPswePS1ΔE9: 676.333 ± 173.680, vs. APPswePS1ΔE9 × IFNAR1−/−: 659.000 ± 48.461, p < 0.0001, IFNAR1−/− vs. all genotypes, n = 3–5, 3 hours: IFNAR1−/−: 1504.000 ± 79.618 vs. wild type: 770.333 ± 38.176, vs. APPswePS1ΔE9: 665.333 ± 105.035, vs. APPswePS1ΔE9 × IFNAR1−/−: 866.000 ± 35.940, p < 0.0001, IFNAR1−/− vs. all genotypes, n = 3–5) at both 1 and 3 hours.
Plaque burden in 9 and 13-month APPswePS1ΔE9 × IFNAR1−/− mice is unchanged
To investigate whether the observed increased Aβ phagocytosis translated in vivo, we performed immunofluorescence analysis of Aβ plaques in both 9 and 13-month APPswePS1ΔE9 and APPswePS1ΔE9 × IFNAR1−/− mice. Sagittal brain sections were stained with an anti-Aβ antibody and imaged before being subjected to an automated analysis utilizing a watershed segmentation approach (Fig. 5b). Both APPswePS1ΔE9 and APPswePS1ΔE9 × IFNAR1−/− mice show significant deposition throughout the brain. However, no significant difference was observed in either 9 or 13-month old mice when quantitation of plaque size, plaques per mm2 or % of plaque burden was performed in either the cortical or hippocampal areas (Fig. 5c–h). Both APPswePS1ΔE9 and APPswePS1ΔE9 × IFNAR1−/− showed similar age-related increases in plaques. These findings suggest that removal of IFNAR1 does not alter plaque burden in either 9 and13-month old APPswePS1ΔE9 mice.
Figure 5.
Aged 9 and 13-month APPswePS1ΔE9 × IFNAR1−/− brains exhibit unaltered Aβ burden compared to APPswePS1ΔE9 mice. Brain sections from 9 and 13-month old APPswePS1ΔE9 and APPswePS1ΔE9 × IFNAR1−/− were stained with anti-Aβ antibodies for automated plaque analysis. Representative hippocampal and cortical sections for each age of APPswePS1ΔE9 and APPswePS1ΔE9 × IFNAR1−/− are shown in (a). Measurements of (b) cortical plaque size, (c) cortical plaque burden and (d) plaques per mm2 are shown. Measurements of hippocampal measures for each are shown in (e) to (g) respectively. Data is displayed as mean ± SEM, n = 3–6). Scale bar = 2 mm.
13-month, but not 9 month, APPswePS1ΔE9 × IFNAR1−/− microglia adopt a stellate morphology
To further elucidate the effect that type-I IFN signalling and ageing has on microglial phenotype, we then performed a morphological analysis on both 9 and 13-month old APPswePS1ΔE9 and APPswePS1ΔE9 × IFNAR1−/− mice (Fig. 6a,b). Here, we examined microglia not in direct contact with Aβ plaques, as the heightened clustering of microglia around plaques in aged mice poses difficulties in ascribing particular branches to individual microglia. Example traces are shown in Fig. 6c,d. At 13 months however, significant increases in the cell radius (APPswePS1ΔE9: 19.14 ± 1.123 μm vs. APPswePS1ΔE9 × IFNAR1−/−: 33.69 ± 6.472 μm, p = 0.0250, n = 6–8) (Fig. 6e) and cell area (APPswePS1ΔE9: 108.6 ± 11.06 μm2 vs. APPswePS1ΔE9 × IFNAR1−/−: 337.3 ± 96.5 μm2, p = 0.0179, n = 6–8) (Fig. 6f) in the APPswePS1ΔE9 × IFNAR1−/− microglia were detected. Combined with an unchanged level in branch length, these findings combined suggest that these microglia adopt a stellate like morphology. Such a morphology suggests an anti-inflammatory phenotype4.
Figure 6.
13-month APPswePS1ΔE9 × IFNAR1−/− microglia adopt a stellate-like morphology. 30 μm thick brain sections of 9 and 13-month old APPswePS1ΔE9 and APPswePS1ΔE9 × IFNAR1−/− mice were stained with both IBA-1 and WO2 antibodies before 15 μm z-stacks were taken from of the hippocampal region. Representative images are shown for 13-month APPswePS1ΔE9 and APPswePS1ΔE9 × IFNAR1−/− in (a) and (b) respectively. A minimum spanning tree algorithm was then employed to measure morphological characteristics with an overview of the skeletonising process in (c) and (d) for both genotypes respectively. Measurements are then shown for (e) total branch length, (f) cell area and (g) cell radius. Data is displayed as mean ± SEM (*p < 0.05, Students t-test, APPswePS1ΔE9 vs. APPswePS1ΔE9 × IFNAR1−/−, n = 3–8). Scale bar = 500 μm.
Discussion
Neuroinflammation is seen as a chronic and detrimental process within AD, and it is suggested that microglia are the critical cell type responsible for this response34,35. This is also seen in congruence with microglial dysfunction. As such the identification of key regulators of this neuroinflammatory process present as novel therapeutic targets36. Type-I IFNs are known to act as “master regulators” of the innate immune response and are able to regulate levels of IL1β, IL6 and TNFα which are consistently upregulated within AD brains3,27–29. Recent work investigating microglial phenotype has identified type-I IFNs as regulators of unique and conserved microglial populations in both ageing and disease8,31,32,37. Here, we further examine the modulation of these phenotypic microglia by the type-I IFNs, critically focussing on investigating their functional roles.
We firstly identified that type-I IFN treatment decreases the ability of microglial phagocytosis in a dose-dependent manner. Through genetic abrogation of type-I IFN signalling by targeting its receptor IFNAR1, we identify that IFNAR1−/− primary microglia exhibit a decreased IL1β, IL6 and TNFα response to monomeric Aβ1–42, and that targeting IFNAR1 within primary microglia isolated from the APPswePS1ΔE9 AD model leads to an enhanced ability to phagocytose FITC Aβ1–42. These aged APPswePS1ΔE9 × IFNAR1−/− mice also show altered microglial morphology in vivo.
We have previously reported that ablation of type-I IFN signalling within the APPswePS1ΔE AD model is neuroprotective33. In this model, reduced type-I IFN signalling was associated with an attenuated whole-brain inflammatory profile and a rescue in cognition as assessed via the Morris water maze. The cell-specific contributions that resulted in this altered response were unknown. This suggested that microglia were indeed the cell type responsible.
Microglial function follows phenotype, and phagocytosis is recognised as a critical function of microglia38. We first employed a phagocytosis assay on microglial cells to investigate how type-I IFN stimulation affects this process. A negative phagocytic dose-response relationship was observed in cells treated with IFNα, with a trend for a decrease in IFNβ also seen. Previous work investigating IFN and phagocytosis has shown differing results. In an autoimmune encephalomyelitis model of multiple sclerosis, IFNβ was shown to increase microglial phagocytosis of myelin39. Furthermore, increased myelin was observed within the brains of IFNAR1−/− knockout mice. Phagocytosis is not limited to a single pathway, and both myelin and Aβ may indeed invoke separate cellular mechanisms that result in engulfment40. There may also be other kinetic measures that explain these differences which are masked in end-point assays. Investigation in macrophages and other mononuclear phagocytes also demonstrate a role for type-I IFNs in promoting phagocytosis41–43. Similarly, these studies also use different particles to measure phagocytosis. Regardless, further work is required to investigate these findings. Our results confirm work that type-I IFNs are involved in microglial function, with data demonstrating that IFNα in particular affects phagocytosis. Interestingly, these decreases contrasted to the increases seen with the pro-inflammatory LPS. To further investigate this finding, we also performed similar experiments using a high content imaging platform for increased sensitivity. We are able to confirm our initial findings with BV-2 cells. Levels of type-I IFNs are known to increase with normal ageing, with our study supporting previous data identifying an impaired phagocytic function of aged microglia31,44. We are able to demonstrate that type-I IFNs elicit differential functional effects on microglia when compared to classical inflammatory stimuli, suggesting that type-I IFNs are able to shift microglia to a unique phenotype. This is in line with recent findings identifying unique microglia populations within the CNS that are enriched with a number of IFN regulated genes45.
Consistent with our previous findings examining Aβ treatments on primary mixed glial cultures, our study confirmed that Aβ1–42 elicits a type-I IFN response in wild type microglia, with this response attenuated in IFNAR1−/− microglia. Increases were seen in levels of bioactive type-I IFN at 24 and 48 hours between genotypes, which is mirrored in the mRNA transcripts of both IFNα and IFNβ. Within IFNAR1−/− microglia transcript levels of IFNα were observed to remain constant, but IFNβ levels actively downregulated. This suggests differing roles for IFNα and IFNβ in AD, and that IFNα is the critical subtype that affects microglia within AD. Type-I IFNs are known to amplify their responses in an autocrine manner and elicit differing responses depending on both type and subtype46. In congruence, levels of IL1β and IL6 are actively decreased within these same IFNAR1−/− microglia. This suggests that expression of these hallmark proinflammatory cytokines may be downstream of type-I IFN signalling. Furthermore, alteration of microglial phenotype through reduction of these hallmark pro-inflammatory cytokines is well established in shifting microglial function47. Levels of IRF7 were upregulated between wild type and IFNAR1−/− genotypes at both 24 and 48 hours respectively. In primary macrophages, it has been reported that IRF7 is able to elicit differential type-I IFN responses, with IRF7 associated with an overall heightened response level48. Furthermore, IRF7 is known to be a major IRF induced downstream of type-I IFN responses and is critical in further induction of the IFN response19.
Phagocytosis is a critical functional process of microglia impaired in AD3. Here we observed that APPswePS1ΔE9 × IFNAR1−/− microglia, but not IFNAR1−/− microglia alone, exhibited greater phagocytic ability towards FITC Aβ1–42. This was compounded by the increases in uptake on a per-cell basis of IFNAR1−/− microglia of the reverse peptide, FITC Aβ42–1. Whether this result is a unique interaction between APPswePS1ΔE9 and IFNAR1 or indeed due to other processes remains unknown, and as such warrants further investigation. One such explanation may be microglial “priming”, a concept explored within neuroinflammation and AD49. Levels of Aβ have been observed in culture from transfected cells that contain human APP, which may be responsible for alterations in microglial phenotype50. Bias for particular Aβ species may also be involved. The nature of FITC conjugation denotes that aggregation dynamics differ between it and unlabelled peptides51. This oligomerisation bias may also explain the observations in vivo, with APPswePS1ΔE9 × IFNAR1−/− mice showing decreases in monomeric levels of Aβ1–42 only33. The observed increases in the IFNAR1−/− microglia may indeed be due to differential phagocytic processes. Critically, the reverse peptide has differential aggregation dynamics when compared to the forward peptide sequence52. Multiple receptors are involved in microglial phagocytosis that differ in alterations in cytoskeletal elements, phagosome maturation and inflammatory responses in response to binding38,40. It is important to note that previous in vitro work investigating immune-related gene knockouts and fluorescent Aβ phagocytosis have not considered the role of their respective AD genotype13,53.
Due to these observed differences in uptake of Aβ1–42, we then examined Aβ plaque burden as a surrogate measure of in vivo microglial phagocytosis. Critically, we expand our analysis from our previous work. We observed no differences between genotypes, which follow our previous findings33. It is well established that plaque burden is not a measure of disease state, as levels do not correlate with cognition. Furthermore, plaque reduction does not alter pathology and cognitively normal individuals containing plaques within their brains54–56.
We further investigate these mice though use of a sophisticated, and critically, verified, algorithm to examine a number of morphological features57. Increased cellular radius and diameter without increases in branch length indicate that APPswePS1ΔE9 × IFNAR1−/− microglia adopt a stellate-like morphology. The exact nature of how this relates to underlying phenotype however remains unknown. Classically, ramified-like morphologies have classically been used to identify “quiescent” or “resting” microglial states and amoeboid-like morphologies for “reactive” states58. Similar morphologies can however exhibit altered phenotypes59. As such, the extent to which this classic paradigm holds true remains unknown. However, quantified morphological analyses add an additional trait to further our understanding of microglia. Further work is required to link various morphological features and microglial functions, both in normal and disease states, as well as across ageing.
Our observations combined with our in vitro data suggest that these microglia possess an altered anti-inflammatory phenotype. The observation of altered morphology at 13 but not 9 months of age in APPswePS1ΔE9 × IFNAR1−/− microglia is notable and calls for further examination. It is established that APPswePS1ΔE9 mice begin to show a decreased cognitive phenotype at 6 months, and as such 13-months is an advanced stage for disease60. This heightened disease state may overcome protective effects due to loss of IFNAR1−/− as we have previously observed, in turn altering microglial phenotype and subsequent morphology. Future work will focus on the direct analysis of microglia isolated from these mice with a transcriptomic based approach utilised, similar to that reported in other related studies focused on microglial phenotypes8,31. This combined with morphological data will allow for deeper insight into how the type-I IFN signalling pathway is involved in regulating microglial function.
Conclusions
These results demonstrate that type-I IFNs are involved in the modulation of both microglial phenotype and function. This work further expands on emerging data demonstrating a link between microglia, type-I IFNs and AD. Type-I IFNs present as a much-needed novel therapeutic target for the management of AD.
Methods
Animals
C57BL/6J wild type mice were sourced from the Animal Resource Centre. IFNAR1−/− mice were initially generated by Hwang, et al.18. APPswePS1ΔE9 transgenic mice were originally sourced from JAX61. APPswePS1ΔE9 transgenic mice lacking IFNAR1−/− were generated by Minter et al.33. All mice were housed in sterile micro-isolator cages and fed ad-libitum. All animal procedures were performed in accordance with the University of Melbourne animal care committee’s regulations and conducted in compliance with the Australian National Health and Medical Research Guidelines. All experiments were approved by the University of Melbourne Animal Ethics Committee (Ethics ID: 1613905).
Genotyping
Genotyping was performed commercially using Transnetyx™.
pHrodo phagocytosis assay
BV-2 microglial cells (a generous donation from Dr. Sherif Boulos, Western Australian Neuroscience Research Institute) were plated within black flat-bottomed 96 well plates (Corning, 3603) at 2.5 × 103 cells per well. Fresh culture media was applied to cells (Dulbeccos modified eagle medium (DMEM)) (10569–044, Gibco), 5% FBS, 1% penicillin/streptomycin) 24 hours before cells were incubated with serum free DMEM containing various concentrations of IFNα, IFNβ and LPS. Cells were then incubated for 1 hour at 37 °C and 5% CO2 before each well was rinsed with 100 μl of DMEM before media was replaced with DMEM containing pHrodo (ThermoFisher, P35361) diluted 1:30. Plates were viewed under a fluorescence microscope to ensure phagocytosis had occurred before being read using a Flexstation II (Molecular Devices) or Operetta High Content Imaging machine (Perkin Elmer).
Mixed cortical and hippocampal glial isolation
Mixed cortical and hippocampus glial cultures were isolated from P0-P1 pups as described previously33. Briefly, cortices and hippocampal structures were isolated, and meninges then surgically removed. This cleaned tissue was then placed into a solution of hanks buffered saline solution (HBSS) containing trypsin ethylenediaminetetraacetic acid (EDTA) (1 × final concentration, 59418 C, Sigma) and deoxyribonuclease (DNAse) (1 mg/mL, D5025, Sigma) for 15 minutes, after which the supernatant was collected. Remaining undigested tissue was then subject to a second round of digestion before the supernatants were combined. Cells were plated at a density of 1.25 × 104 cells/mL in culture medium (DMEM containing 20% foetal bovine serum (FBS), 1% penicillin/streptomycin). Media was replaced at days 2, 7 and 14 after which the glial cells formed a monolayer.
Microglial isolation
Microglia were isolated from mixed glial cultures at 18 days in vitro. Isolation was performed using the Saura, et al.62 mild trypsinization method and fluorescence activated cell sorting (FACS) for CX3CR1eGFP/+ microglia. Conditioned media was collected from each individual plate before undergoing a wash with DMEM. Media was then replaced with a 1:1 mixture of DMEM and 0.25% trypsin EDTA (25200-072, Gibco) and left for 30 minutes. Once detachment of the upper glial layer was confirmed, cells were again washed with DMEM before the collected conditioned media was returned to the individual plates. Cells were given a further 2 days incubation before use in experiments. Microglial purity was approximately 95% as analysed periodically through flow cytometry using a FITC conjugated CD11b antibody (130-113-796, Miltenyi Biotech).
Amyloid-β preparation and treatment
The Aβ1–42 peptide (A-42-T-1, GenicBio) was reconstituted in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) (1 mg/mL, 105228, Sigma) before combined monomerization and lyophilisation in a Heto Maxi Dry Lyo DC40 Dynavac freeze-drying centrifuge (12000 × G, room temperature, 30 minutes). Peptide was then dissolved in ice-cold 5 mM NaOH (in PBS), after which final working concentrations were measured via absorbance spectroscopy at 214 nm on a NanoDrop 3000 spectrophotometer.
FITC-conjugated amyloid-β treatment
Cells were treated with serum free DMEM containing 2 mg/mL of FITC conjugated Aβ1–42 or Aβ42–1 (1% sodium azide, PBS) for 1 and 3 hours respectively. Cells were rinsed before being scraped in PBS and centrifuged (1500 × G, 5 minutes, room temperature) to form a pellet. Resulting pellets were then resuspended in FACS buffer (1% BSA, 100 ng/mL 4′,6-diamidino-2-phenylindole (DAPI), in PBS) and transferred into 5 mL cytometer tubes. Samples were read on a BD Fortessa flow cytometer.
B16 IFN assay
B16-Blue cells (bb-ifnt1, InvivoGen) were used to measure bioactive type-I IFN in collected media as per manufacturers instructions. Briefly, cells were cultured in T75cm2 flasks in Roswell Park Memorial Institute 1640 (21870–092, Gibco) medium containing 5% FBS, 1% penicillin/streptomycin and zeocin (100 μg/ml). Cells were then plated in a 96 well plate at 75,000 cells/well. Collected media was added to these cells for 24 hours alongside IFNα (0–1000 U/mL) to generate a standard curve. QUANTI-Blue was then added in 1:1 v/v ratio for a further 24 hours. Following this, 200 μL of media was transferred into a 96 well plate and absorbance was measured on a Multiskan Ascent plate reader.
ELISA
Murine Il1β (559603, BD Biosciences) IL6 (555240, BD Biosciences), and TNFα (DY410, R&D Systems) were used to detect protein levels in collected media as per manufacturer’s instructions. Briefly, 96-well plates were coated overnight in capture antibodies diluted in assay diluent (1:1000) at 4°. Plates were washed thrice, then blocked in assay diluent. Wells were then filled with either recombinant standards or 100 μL media from collected samples (2 hours, room temperature. Plates were then washed 5 times after which 100 μL working detector (detection antibody + SAv-HRP reagent) was added (1 hour, room temperature). Plates were then washed 7 times and a 1:1 mixture of hydrogen peroxide and 3,3′, 5,5′ tetramethylbenzidine was added (30 minutes, room temperature). The reaction was then stopped using sulphuric acid (160 mM) and absorbance measured at 450 nm in a Multiskan Ascent spectrophotometer (Thermo Scientific)
RNA isolation
All samples underwent a modified RNA isolation protocol using both Trizol® (Invitrogen) and the illustra RNAspin mini kit (25050071, GE Health Sciences). Cultured cells were rinsed once in DMEM before being scraped in 1 mL Trizol® and transferred into a fresh autoclaved RNAse/DNAse free tube. Samples were then homogenized using a 23 g needle and 1 ml syringe. 0.2 mL chloroform was then added and samples shaken vigorously for 15 seconds before incubation for 3 minutes at room temperature. Samples were centrifuged (12000 × G, 15 minutes, 4 °C), after which the clear supernatant was collected and transferred into a new ribonuclease (RNAse)/DNAse free tube. 70% ethanol was then added to the supernatant at a ratio of 1:1, after which the mixture was placed onto the illustra RNAspin collection column. Isolation was then performed as per manufacturer’s instructions. Sample concentration and purity was assessed using a NanoDrop 1000 spectrophotometer. (ThermoScientific). All samples measured underwent on-column DNAse treatment and had 260/280 ratios above 2.
Reverse transcription
1–2 μg of total RNA was reverse transcribed using the High Capacity complementary DNA (cDNA) Reverse Transcription Kit (4368814, Applied Biosciences) as per manufacturer’s instructions. Individual reactions were prepared, and PCR was performed in a thermal cycler (Eppendorf) under the following conditions: 10 minutes at 25 °C, 120 minutes at 37 °C, 5 minutes at 85 °C and 10 minutes at 4 °C. Each sample group also contained an additional reaction without the addition of reverse transcriptase enzyme (-RT). Final cDNA samples were diluted 1:10 for each 1 μg of initial RNA.
QPCR
QPCR was performed within 384 well plates (4309849, Applied Biosystems) using a QuantStudio 6 PCR system (Applied Biosystems) with reactions performed in triplicate. Two detections system were used within this study: Taqman-based detection utilizing 5(6)-carboxyfluorescein (FAM) fluorescence reporting and oligomer-based detection utilizing fluorescent SYBR green DNA binding. For Taqman based detection all primers were purchased commercially (431182, Applied Biosystems), with details of each outlined in Table 1. Reactions were performed in 5 μL volumes and were composed of the following: 2 μL diluted cDNA, 0.25 μL Taqman primer, 0.25 μL RNAse DNAse free H2O and 2.5 μLm universal master mix reagent (SsoFast Probes, Biorad). For SYBR green detection, all oligomers were synthesized commercially (Geneworks, Integrated DNA Technologies) and our outlined in Table 2. Reactions were performed in 5μL volumes and were composed of the following: 2 μL diluted cDNA, 0.25 μL gene-specific forward primer, 0.25 μL gene-specific reverse primer and 2.5 μL fast SYBR green master mix (4385612, Applied Biosystems). The cycle threshold (Ct) values were normalized to a control gene (B2M) and analysed using the ΔΔCt method to generate fold change values.
Table 1.
List of Taqman primers used in this study.
| Gene | Species | Refseq | Amplicon length (bp) | Catalogue no. |
|---|---|---|---|---|
| B2M | Mouse | NM_009735.3 | 77 | Mm00437762_m1 |
| IRF7 | Mouse | NM_001252600.1 | 67 | Mm00516788_m1 |
| NM_001252601.1 | ||||
| NM_016850.3 | ||||
| IFNβ | Mouse | NM_010510.1 | 69 | Mm00439552_s1 |
| Il1β | Mouse | NM_008361.3 | 63 | Mm01336189_m1 |
| IL6 | Mouse | NM_031168.1 | 78 | Mm00446190_m1 |
| TNFα | Mouse | NM_001278601.1 | 81 | Mm00443258_m1 |
| NM_013693.3 |
Table 2.
List of SYBR primers used in this study.
| Gene | Forward primer | Reverse primer(s) |
|---|---|---|
| B2M | GGCTCACACTGAATTCACCCCCAC | ACATGTCTCGATCCCAGTAGACGGT |
| IFNα | SAWCYCTCCTAGACTCMTTCTGCA | TATDTCCTCACAGCCAGCAG |
| TATTTCTTCATAGCCAGCTG |
Immunofluorescence
Mice were anesthetized via intra-peritoneal injection of a combinatorial mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg). Deep anaesthesia was confirmed through absence of paw withdrawal reflex. Mice then underwent cardiac perfusion with PBS to remove blood from the cerebral vasculature, after which 4% paraformaldehyde (PFA) was then delivered to fix the brain. Brains were then excised and placed into a solution of 4% PFA for 24 hours at 4 °C after which they were transferred into 30% sucrose solution for 24 hours at 4 °C for cryoprotection. Brains were then submerged in optimal cutting temperature media (OCT) (Sakura) within 2 cm2 plastic containers and frozen by immersion within an isopropanol dry-ice bath. 30 μm sections were cut sagittally on a cryostat from the initiation of the hippocampal structure to its completion. Sections were then placed into an individual well of a 24 well plate containing PBS for downstream staining. Collected sections were rinsed once in PBS for 5 minutes, after which they were simultaneously permeabilized and blocked by being placed in 200 μL of a solution containing 1.5% Triton-x (Sigma) in 10% goat block (G9023) in PBS (2 hours, room temperature, constant rocking at 20RPM). Sections were then washed thrice in PBS (room temperature, 4 minutes per wash) before incubation with primary antibodies (4 °C, overnight, constant rocking at 20RPM). Details of antibodies and concentrations used are listed in Table 3. Sections were then washed thrice (room temperature, 5 minutes per wash) before incubation with goat fluorescent secondary antibodies occluded from light (1:1000 dilution in PBS, room temperature, A-11012, A-11001, Invitrogen). Sections were washed thrice a final time (room temperature, 5 minutes per wash) before being rinsed in MilliQ H2O. Using a wide orifice pipette, sections were transferred onto slides and allowed to dry completely before coverslips were mounted using DAPI containing mounting media (H-1500, Vectashield). Images were obtained using the Zeiss Axio Observer.Z1 or Zeiss LSM 780 (Carl Zeiss imaging) inverted fluorescence microscopes with manipulation used only to increase contrast. Details of antibodies used are listed below in Table 3:
Table 3.
List of antibodies used in this study.
| Antibody | Origin | Concentration | Vendor | Catalogue no. |
|---|---|---|---|---|
| Anti-IBA1 | Rabbit | 1 in 200 | Wako | 019–19741 |
| Anti-Aβ | Rabbit | 1 in 500 | Cell Signalling | #8243 |
| WO2 | Mouse | 1 in 500 | 63 | N/A |
| Alex Fluor 488 Goat anti-mouse | Goat | 1 in 1000 | Invitrogen | A-11001 |
| Alex Fluor 594 Goat anti-rabbit | Goat | 1 in 1000 | Invitrogen | A-11012 |
Plaque analysis
Entire brain sections were imaged on the Zeiss Axio Observer.Z1 wide-field microscope, after which associated Zeiss ZEN software was used to trace brain regions of interest. Final analysis was then done on ImageJ using the built-in watershed function.
Morphological analysis
30 μm sagittal brain sections were collected approximately 1400 μm from midline. A single field was imaged in the CA1 region of the hippocampus with 15 μm z-stacks at 0.96 μm per slice, after which images then underwent maximum intensity projections before analysis. Images were analysed using a proprietary spanning tree path finding based algorithm with a minimum of 5 microglia per image analysed57. Trace images were then analysed on Matlab v2013b using built-in functions.
Statistical analysis
All statistical analysis was performed using GraphPad Prism (Version 7). For pHrodo assay data, a 1-way analysis of variance (ANOVA) was performed followed by a Dunnett’s multiple comparisons post-hoc test. For bioactive IFN, QPCR, ELISA and FITC-Aβ data, a 2-way ANOVA was performed with a Sidaks multiple comparisons post-hoc test. Amyloid quantification data and morphological analysis utilized a Students t-test. All tests used considered a p < 0.05 statistically significant.
Author contributions
All authors contributed to and approved the manuscript. Z.M. was involved in all aspects of the project and experimental procedures. F.R.W. was involved in morphological data analysis. F.M. was involved in data analysis. Z.M., J.M.T. and P.J.C. were responsible for the overall experimental design.
Data availability
Data from this study is included in this published article. Further information is available from the corresponding author on request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Juliet M. Taylor, Email: juliett@unimelb.edu.au
Peter J. Crack, Email: pcrack@unimelb.edu.au
References
- 1.Nichols E, et al. Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:88–106. doi: 10.1016/s1474-4422(18)30403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xanthos DN, Sandkuhler J. Neurogenic neuroinflammation: inflammatory CNS reactions in response to neuronal activity. Nat. Rev. Neurosci. 2014;15:43–53. doi: 10.1038/nrn3617. [DOI] [PubMed] [Google Scholar]
- 3.Heneka MT, et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/s1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prinz M, Jung S, Priller J. Microglia Biology: One Century of Evolving Concepts. Cell. 2019;179:292–311. doi: 10.1016/j.cell.2019.08.053. [DOI] [PubMed] [Google Scholar]
- 5.Ginhoux F, Lim S, Hoeffel G, Low D, Huber T. Origin and differentiation of microglia. Front. Cell Neurosci. 2013;7:45. doi: 10.3389/fncel.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ajami B, et al. Single-cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nat. Neurosci. 2018;21:541–551. doi: 10.1038/s41593-018-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman BA, et al. Diverse Brain Myeloid Expression Profiles Reveal Distinct Microglial Activation States and Aspects of Alzheimer’s Disease Not Evident in Mouse Models. Cell Rep. 2018;22:832–847. doi: 10.1016/j.celrep.2017.12.066. [DOI] [PubMed] [Google Scholar]
- 8.Keren-Shaul H, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 2017;169(1276–1290):e1217. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Deczkowska A, et al. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell. 2018;173:1073–1081. doi: 10.1016/j.cell.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Walker FR, et al. Dynamic structural remodelling of microglia in health and disease: A review of the models, the signals and the mechanisms. Brain, Behavior, Immun. 2014;37:1–14. doi: 10.1016/j.bbi.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Li Q, Barres BA. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2018;18:225–242. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- 12.Sims R, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griciuc A, et al. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carmona S, et al. The role of TREM2 in Alzheimer’s disease and other neurodegenerative disorders. Lancet Neurol. 2018;17:721–730. doi: 10.1016/s1474-4422(18)30232-1. [DOI] [PubMed] [Google Scholar]
- 15.Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer’s disease mice. J. Neurosci. 2008;28:8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc. R. Soc. London. Ser. B-Biological Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 17.Weerd NA, Nguyen T. The interferons and their receptors—distribution and regulation. Immunology Cell Biol. 2012;90:483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang SY, et al. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc. Natl Acad. Sci. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes. Immun. 2011;12:399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majoros A, et al. Canonical and Non-Canonical Aspects of JAK-STAT Signaling: Lessons from Interferons for Cytokine Responses. Front. Immunol. 2017;8:29. doi: 10.3389/fimmu.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegal FP, et al. The Nature of the Principal Type 1 Interferon-Producing Cells in Human Blood. Sci. 1999;284:1835. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 22.Ng CT, Mendoza, Juan L, Garcia KC, Oldstone MBA. Alpha and Beta Type 1 Interferon Signaling: Passage for Diverse Biologic Outcomes. Cell. 2016;164:349–352. doi: 10.1016/j.cell.2015.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice GI, et al. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirou KA, et al. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50:3958–3967. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 26.Bialas AR, et al. Microglia-dependent synapse loss in type I interferon-mediated lupus. Nat. 2017;546:539–543. doi: 10.1038/nature22821. [DOI] [PubMed] [Google Scholar]
- 27.Kawai T, Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nat. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García-Sastre A, Biron CA. Type 1 Interferons and the Virus-Host Relationship: A Lesson in Détente. Sci. 2006;312:879. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 30.Baruch K, et al. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Sci. 2014;346:89. doi: 10.1126/science.1252945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deczkowska A, et al. Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nat. Commun. 2017;8:0. doi: 10.1038/s41467-017-00769-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hickman SE, et al. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minter MR, et al. Deletion of the type-1 interferon receptor in APPSWE/PS1ΔE9 mice preserves cognitive function and alters glial phenotype. Acta Neuropathologica Commun. 2016;4:72. doi: 10.1186/s40478-016-0341-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heneka MT, Kummer MP, Latz E. Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 2014;14:463–477. doi: 10.1038/nri3705. [DOI] [PubMed] [Google Scholar]
- 35.Sarlus H, Heneka MT. Microglia in Alzheimer’s disease. J. Clin. Invest. 2017;127:3240–3249. doi: 10.1172/JCI90606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cummings JL, Morstorf T, Zhong K. Alzheimer’s disease drug-development pipeline: few candidates, frequent failures. Alzheimer’s Res. Ther. 2014;6:37–37. doi: 10.1186/alzrt269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathys H, et al. Temporal Tracking of Microglia Activation in Neurodegeneration at Single-Cell Resolution. Cell Rep. 2017;21:366–380. doi: 10.1016/j.celrep.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu R, Shen Q, Xu P, Luo JJ, Tang Y. Phagocytosis of Microglia in the Central Nervous System Diseases. Mol. Neurobiol. 2014;49:1422–1434. doi: 10.1007/s12035-013-8620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kocur M, et al. IFNbeta secreted by microglia mediates clearance of myelin debris in CNS autoimmunity. Acta Neuropathol. Commun. 2015;3:20. doi: 10.1186/s40478-015-0192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aderem A, Underhill DM. Mechanisms of Phagocytosis In Macrophages. Annu. Rev. Immunology. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 41.Yoshie O, Mellman IS, Broeze RJ, Garcia-Blanco M, Lengyel P. Interferon action: Effects of mouse α and β interferons on rosette formation, phagocytosis, and surface-antigen expression of cells of the macrophage-type line RAW 309Cr.1. Cell. Immunology. 1982;73:128–140. doi: 10.1016/0008-8749(82)90441-5. [DOI] [PubMed] [Google Scholar]
- 42.Rollag H, Degré M, Sonnenfeld G. Effects of interferon-alpha/beta and interferon-gamma preparations on phagocytosis by mouse peritoneal macrophages. Scand. J. Of. Immunology. 1984;20:149–155. doi: 10.1111/j.1365-3083.1984.tb00988.x. [DOI] [PubMed] [Google Scholar]
- 43.Grunwell JR, et al. TGF-beta1 Suppresses the Type I IFN Response and Induces Mitochondrial Dysfunction in Alveolar Macrophages. J. Immunol. 2018;200:2115–2128. doi: 10.4049/jimmunol.1701325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rawji KS, et al. Immunosenescence of microglia and macrophages: impact on the ageing central nervous system. Brain. 2016;139:653–661. doi: 10.1093/brain/awv395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sala Frigerio C, et al. The Major Risk Factors for Alzheimer’s Disease: Age, Sex, and Genes Modulate the Microglia Response to Abeta Plaques. Cell Rep. 2019;27(1293–1306):e1296. doi: 10.1016/j.celrep.2019.03.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Honda K, Takaoka A, Taniguchi T. Type I Inteferon Gene Induction by the Interferon Regulatory Factor Family of Transcription Factors. Immun. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Cherry JD, Olschowka JA, O’Banion MK. Neuroinflammation and M2 microglia: the good, the bad, and the inflamed. J. Neuroinflammation. 2014;11:98. doi: 10.1186/1742-2094-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solis M, et al. Distinct functions of IRF-3 and IRF-7 in IFN-alpha gene regulation and control of anti-tumor activity in primary macrophages. Biochem. Pharmacol. 2006;72:1469–1476. doi: 10.1016/j.bcp.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Perry VH, Holmes C. Microglial priming in neurodegenerative disease. Nat. Rev. Neurol. 2014;10:217–224. doi: 10.1038/nrneurol.2014.38. [DOI] [PubMed] [Google Scholar]
- 50.Shoji M, et al. Production of the Alzheimer amyloid beta protein by normal proteolytic processing. Sci. 1992;258:126. doi: 10.1126/science.1439760. [DOI] [PubMed] [Google Scholar]
- 51.Jungbauer LM, Yu C, Laxton KJ, LaDu MJ. Preparation of fluorescently-labeled amyloid-beta peptide assemblies: the effect of fluorophore conjugation on structure and function. J. Mol. Recognit. 2009;22:403–413. doi: 10.1002/jmr.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butterfield DA, Kanski J. Methionine residue 35 is critical for the oxidative stress and neurotoxic properties of Alzheimer’s amyloid β-peptide 1–42. Peptides. 2002;23:1299–1309. doi: 10.1016/S0196-9781(02)00066-9. [DOI] [PubMed] [Google Scholar]
- 53.Liu S, et al. TLR2 is a primary receptor for Alzheimer’s amyloid beta peptide to trigger neuroinflammatory activation. J. Immunol. 2012;188:1098–1107. doi: 10.4049/jimmunol.1101121. [DOI] [PubMed] [Google Scholar]
- 54.McLean Catriona A., Cherny Robert A., Fraser Fiona W., Fuller Stephanie J., Smith Margaret J., Konrad Vbeyreuther, Bush Ashley I., Masters Colin L. Soluble pool of A? amyloid as a determinant of severity of neurodegeneration in Alzheimer's disease. Annals of Neurology. 1999;46(6):860–866. doi: 10.1002/1531-8249(199912)46:6<860::AID-ANA8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 55.Fagan AM, et al. Cerebrospinal fluid tau and ptau<sub>181</sub> increase with cortical amyloid deposition in cognitively normal individuals: Implications for future clinical trials of Alzheimer's disease. EMBO Mol. Med. 2009;1:371. doi: 10.1002/emmm.200900048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Dyck CH. Anti-Amyloid-beta Monoclonal Antibodies for Alzheimer’s Disease: Pitfalls and Promise. Biol. Psychiatry. 2018;83:311–319. doi: 10.1016/j.biopsych.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdolhoseini, M., Walker, F. & Johnson, S. In 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). 1208–1211 (2016). [DOI] [PubMed]
- 58.Kettenmann H, Hanisch UK, Noda M, Verkhratsky A. Physiology of microglia. Physiol. Rev. 2011;91:461–553. doi: 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- 59.Wes PD, Sayed FA, Bard F, Gan L. Targeting microglia for the treatment of Alzheimer’s Disease. Glia. 2016;64:1710–1732. doi: 10.1002/glia.22988. [DOI] [PubMed] [Google Scholar]
- 60.Trinchese F, et al. Progressive age-related development of Alzheimer-like pathology in APP/PS1 mice. Ann. Neurol. 2004;55:801–814. doi: 10.1002/ana.20101. [DOI] [PubMed] [Google Scholar]
- 61.Jankowsky JL, et al. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: evidence for augmentation of a 42-specific γ secretase. Hum. Mol. Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 62.Saura J, Tusell JM, Serratosa J. High‐yield isolation of murine microglia by mild trypsinization. Glia. 2003;44:183–189. doi: 10.1002/glia.10274. [DOI] [PubMed] [Google Scholar]
- 63.Wun KS, et al. Crystallization and preliminary X-ray diffraction analysis of the Fab fragment of WO2, an antibody specific for the Abeta peptides associated with Alzheimer’s disease. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 2008;64:438–441. doi: 10.1107/S1744309108011718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data from this study is included in this published article. Further information is available from the corresponding author on request.