Dear Editor,
The geminiviruses are small single-stranded plant DNA viruses belonging to the family Geminiviridae, which cause serious diseases in many economically important crops world-wide. Geminiviruses typically infect the differentiated leaf cells or vascular tissues, and multiply progeny DNA in the nucleus of infected cells. Geminiviruses encoding replication initiator protein (Rep) is essential for viral genome replication, but Rep does not possess DNA polymerase activity and relies heavily on host DNA replication machine for replication (Hanley-Bowdoin et al. 2000). Therefore, exploring the host factors participating in geminivirus replication is of great significance in understanding geminivirus replication mechanism and demystifying the processes of plant DNA replication. However, many drawbacks in using plant host as a model block the research on the effects of host factors on viral DNA replication, for example, plants with some functional gene mutants lead to embryo lethal, the complexities of plant protoplasts transfection and culture inhibit the research progress, and so on. The existence of the above disadvantages urges us to seek a novel approach to identify and characterize the geminivirus-interacting host factors.
Yeast (Saccharomyces cerevisiae) is a eukaryotic model organism with highest percentage of characterized genes, which are relatively conserved among high plants. Indeed, studies with plant viruses, such as brome mosaic virus (BMV) and tomato bushy stunt virus (TBSV) as well as some animal viruses including flock house virus (FHV) and influenza virus have shown that certain viruses can complete almost the replication cycle in yeast cells (Janda and Ahlquist 1993; Price et al. 2000; Panavas et al. 2008; Naito et al. 2007). Raghavan and colleagues firstly reported that mungbean yellow mosaic India virus (MYMIV), a bipartite geminivirus that could replicate in S. cerevisiae, and established the yeast system to study geminivirus replication (Raghavan et al. 2004). Since the yeast essential gene temperature-sensitive (ts) mutant library was constructed (Li et al. 2011), it has been widely used to study virus-host interaction, yeast and other eukaryotic gene functions, DNA replication and so forth (Nawaz-ul-Rehman et al. 2013; Prasanth et al. 2016; Salim et al. 2017).
Geminivirus replication is initiated by the affinity binding of Rep to the iteron-like elements, which makes a nicking in the invariant 9-mer sequence in the predicted loop region (Fontes et al. 1994; Pant et al. 2001). Then several host DNA replication factors are recruited to form a DNA replication machine to fulfill viral DNA replication (Settlage et al. 1996; Singh et al. 2007; Suyal et al. 2013). To identify more host factors that may interact with geminivirus Rep protein and to make a better understanding on host factors that regulate geminivirus replication, we used MYMIV as a model virus in this study to perform the following experiments. First, we constructed a recombinant plasmid dpRS316-MYMIV-1.65A based on the wild vector plasmid pRS316 (Supplementary Materials) and confirmed that the transformed BY4741 yeast cells with the plasmids were able to grow well on SC/-Uracil (Li et al. 2015). But they grow more slowly than yeast cells transformed with pRS316, and the generated colonies seem to be much smaller and erose (Supplementary Fig. S1A). The relative replication efficiency (the colon generation efficiency) of dpRS316-MYMIV-1.65A transformed yeast was about 40% of that of pRS316 transformed yeast (Supplementary Fig. S1B). Cell morphology of BY4741 yeast cells transformed with pRS316, or dpRS316-MYMIV-1.65A was similar to the untransformed cells (Supplementary Fig. S1C). The expression of replication initiator protein (Rep) in yeast cells was confirmed by Western blot assay (Supplementary Fig. S1D).
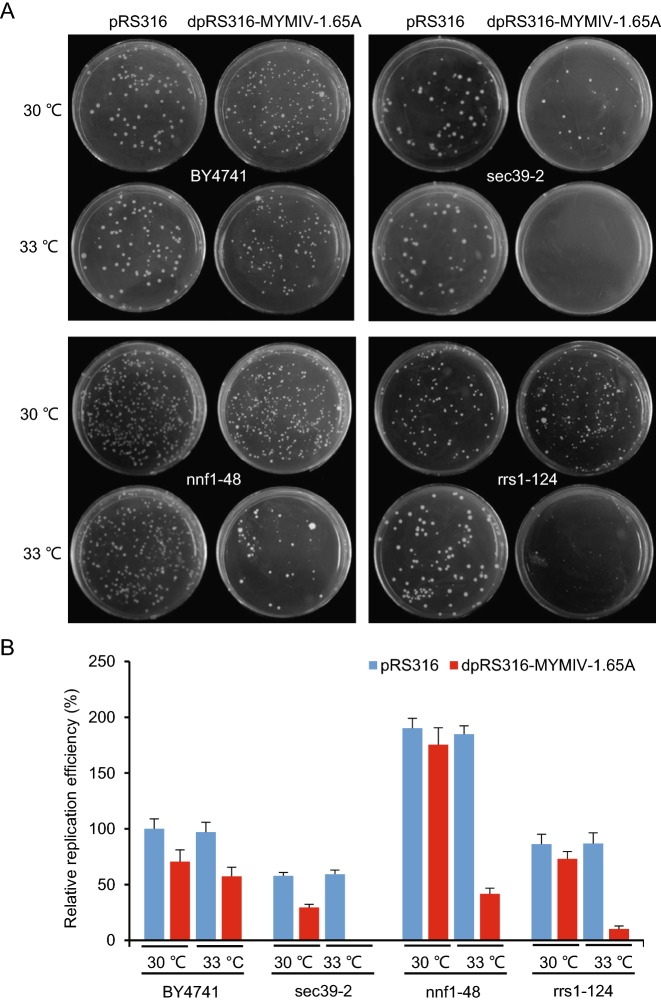
We then screened a novel yeast ts mutant library containing 787 yeast mutants, which contains the individual mutations of about 500 single essential yeast genes (Li et al. 2011). We transformed yeast ts mutants and reference strain BY4741 with wild type plasmid pRS316 or the recombinant plasmid dpRS316-MYMIV-1.65A, followed by culturing at permissive temperature (30 °C in the following example) or semi-permissive temperature (usually 4–5 °C below the nonpermissive temperature, 33 °C in the following example) according to the ts mutants’ nonpermissive temperature (Li et al. 2011) for 3–5 days. The relative replication efficiency was compared by counting the number of colonies formed in BY4741 and ts mutants transformed with pRS316 or dpRS316-MYMIV-1.65A. Four representative examples were shown in Fig. 1A. There was no difference of the relative replication efficiency in pRS316 transformants at permissive temperature and semi-permissive temperature, nor the difference exists in the dpRS316-MYMIV-1.65A transformants when using the reference strain BY4741 (Fig. 1A), suggesting that MYMIV can efficiently replicate in BY4741 reference strain, and the temperature has no effect on its replication. When transformed into yeast ts mutants, the pRS316 transformed cells can grow well at permissive or semi-permissive temperature, and there was no obvious difference of the replication efficiency between them (Fig. 1A). The mutant yeast cells transformed with dpRS316-MYMIV-1.65A can grow well at permissive temperature, but much fewer and smaller colonies formed at semi-permissive temperature due to the increasing temperature (Fig. 1B), indicating that the corresponding yeast gene is partly functional incapacitated and may specifically affect MYMIV replication. Based on this theory that the semi-permissive temperature had a specific effect on the growth of the mutant yeast cells transformed with pRS316-MYMIV-1.65A, we found 141 ts mutants specifically affected MYMIV replication (Table 1). After getting rid of the repeated genes, there were 131 corresponding distinct yeast genes that affect viral replication, and almost all the mutant genes decreased MYMIV replication by more than 50% at semi-permissive temperature when compared with the BY4741 reference strain (Fig. 1B).
Fig. 1.
The effect of representative temperature-sensitive (ts) mutants on MYMIV replication. (A) The plasmids pRS316 and dpRS16-MYMIV-1.65A were transformed into yeast BY4741 reference strain and yeast ts mutants (sec39-2, nnf1-48 and rrs1-124), respectively. The transformants were plated onto SC/-Uracil medium and cultured at permissive temperature (30 °C) and semi-permissive temperature (33 °C). Photographs were taken at 5 days post transformation. (B) Relative replication efficiency of the different transformants. pRS316 in yeast strain BY4741 cultured at permissive temperature (30 °C) was assigned as control and given a value of 100%. The transformations with the plasmids pRS316 and dpRS316-MYMIV-1.65A were performed using the indicated yeast strains with three independent times, and the results were calculated based on the average numbers of yeast colonies (diameter > 1 mm) emerging on the SC/-Uracil media after 5 d of transformation.
Table 1.
Yeast genes affecting MYMIV replication identified by screening the yeast ts mutants.
| Category | Numbera | Numberb | Genesc |
|---|---|---|---|
| Structural protein | 16 | 10 | ACT1(5), ARC35(1), ARP3(3), CCT4(1), PFY1, SCD5, SDA1, SRV2, STU1, STU2 |
| Vesicular transport and protein secretory | 16 | 16 | BET2, BOS1, COG3, EXO70, MYO2, RET3, SEC4, SEC8, SEC12, SEC16, SEC20, SEC26, SEC39, SEC59, SFT1, USO1 |
| DNA replication | 13 | 13 | CDC2(POL3), CDC7, CDC45, CDC47, DBF4, DNA2, HYS2, ORC2, ORC5, POB3, POL1, PSF1, TAH11 |
| Cell division cycle | 17 | 14 | CDC3, CDC21, CDC24(3), CDC28, CDC31, CDC33, CDC43, CDC48, CKS1, DBF2, GPA1, MET30(2), SFH1, SFI1 |
| Pre-mRNA processing | 15 | 15 | CEG1, CET1, PCF11, PRP11, PRP16, PRP18, PRP19, PRP21, PRP24, PRP38, RNA15, RSE1, SAD1, SNU13, SNU114 |
| Transcription initiation | 10 | 9 | RAD3, SSL1, TAF7, TAF11, TAF12(2), TAF13, TFC1, TFC3, TFC6 |
| Chromosome segregation | 6 | 6 | MTW1, NNF1, NSL1, SMC2, SMC3, YCS4 |
| Centromere Protein | 5 | 5 | CEP3, DSN1, SGT1, SKP1, SPC29 |
| RNA modification and degradation | 5 | 5 | ABD1, NOP2, DIS3, MTR3, MTR4 |
| Proteasome | 5 | 5 | PRE3, RPN6, RPN7, RPT2, RPT3 |
| RNA polymerase II mediator complex | 3 | 3 | MED7, MED11, NUT2 |
| Nuclear transport | 3 | 3 | BRL1, RRS1, YRB1 |
| DNA repair | 3 | 3 | NSE1, NSE4, SMC6 |
| Protein kinase | 2 | 2 | PKC1, SGV1 |
| rRNA biogenisis | 2 | 2 | NOP4, SQT1 |
| RNA polymerase | 2 | 2 | RPO31, SRB4 |
| Translation | 1 | 1 | HYP2 |
| Ubiquitin-conjugating enzyme | 1 | 1 | MMS2 |
| Others | 16 | 16 | CMD1, DBP5, FCP1, GAB1, GPI2, GPI13, SMP3, LCB2, MOB2, MSS4, SPT15, TYS1, UBC9, UFD1, URA6, YAH1 |
| Total | 141 | 131 |
aNumber of ts-mutants
bNumber of ts-mutant genes
cNumber in brackets means the number of ts-mutants in this gene
The proteins encoded by those identified yeast mutant genes were classified into 20 distinct kinds based on their known cellular/biochemical functions, among which 13 different proteins (10% of the total number of proteins) are involved in DNA replication, 14 proteins (11%) are participated in cell division cycle regulation, and 15 proteins (11%) are responsible for pre-mRNA processing. Besides, we found that some proteins involved in protein transport and secretion (16 proteins, 12%) may also affect MYMIV replication (Supplementary Fig. S2).
Geminiviruses have limited coding efficiency owing to their small genomes, and the only viral replication essential protein Rep does not possess DNA polymerase activity. Therefore, geminiviruses need to recruit various host factors to fulfill viral genome propagation (Gutierrez 2000; Hanley-Bowdoin et al. 2000). Many challenges exist in the current study of geminivirus replication, which lead to the slow progress made in this research field. Here we used the yeast ts mutant library including essential gene mutants to screen host proteins affecting MYMIV DNA-A replication, and we finally identified 131 yeast genes that probably specifically regulate viral replication. Among these genes, 39 of the 131 candidates were also found to be involved in the replication of TBSV, a positive RNA virus, in a previous study (Nawaz-ul-Rehman et al. 2013), indicating that these genes may affect plant virus replication in a conserved manner. The identified yeast genes are classified into 20 different groups based on their known biological and biochemical roles, with a relative small part of these genes (10%) related to DNA replication, while large amounts of genes are involved in RNA synthesis, protein transportation and secretion, and some other biologic processes, which implies that numerous proteins may take part in geminivirus replication besides the replication related proteins, and the detailed mechanism needs further excavation.
Yeast ts mutant library as a tool to identify the host factors involved in viral replication has lots of advantages. It could partially inhibit protein function by setting semi-permissive temperature. Besides, the known information on mutants in the domain of host factors can provide us the insights about the particular function that regulates viral replication (Li et al. 2011; Nawaz-ul-Rehman et al. 2013). However, there also exist some disadvantages of the ts mutant library, such as a multifunctional protein may affect viral replication as well as some other cell biology processes, which makes it difficult to differentiate and study its particular role on viral replication.
In conclusion, a high throughput screening of yeast genes which affecting geminivirus replication has been established by using yeast ts mutant library, which makes it more efficiently to study geminivirus replication mechanism. More efforts are needed to illuminate the detailed mechanism on how these identified host factors regulate MYMIV replication in yeast and plant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (31390422 and U1136606), the National Key Basic Research and Development Program of China (2012CB114004).
Author Contributions
XZ and YW designed the experiments. FL, XX, ZL and YW performed experiments and wrote the manuscript. YW collected, analyzed the data, and revised the manuscript. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Fangfang Li and Xiongbiao Xu have contributed equally to the article.
References
- Fontes EP, Eagle PA, Sipe PS, Luckow VA, Hanley-Bowdoin L. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J Biol Chem. 1994;269:8459–8465. [PubMed] [Google Scholar]
- Gutierrez C. DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J. 2000;19:792–799. doi: 10.1093/emboj/19.5.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Settlage SB, Orozco BM, Nagar S, Robertson D. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit Rev Biochem Mol Biol. 2000;35:105–140. [PubMed] [Google Scholar]
- Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-D. [DOI] [PubMed] [Google Scholar]
- Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, Vizeacoumar FS, Min R, Vandersluis B, Bellay J, Devit M, Fleming JA, Stephens A, Haase J, Lin ZY, Baryshnikova A, Lu H, Yan Z, Jin K, Barker S, Datti A, Giaever G, Nislow C, Bulawa C, Myers CL, Costanzo M, Gingras AC, Zhang Z, Blomberg A, Bloom K, Andrews B, Boone C. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Xu X, Huang C, Gu Z, Cao L, Hu T, Ding M, Li Z, Zhou X. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol. 2015;208:555–569. doi: 10.1111/nph.13473. [DOI] [PubMed] [Google Scholar]
- Naito T, Kiyasu Y, Sugiyama K, Kimura A, Nakano R, Matsukage A, Nagata K. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proc Natl Acad Sci USA. 2007;104:18235–18240. doi: 10.1073/pnas.0705856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz-ul-Rehman MS, Reddisiva Prasanth K, Baker J, Nagy PD. Yeast screens for host factors in positive-strand RNA virus replication based on a library of temperature-sensitive mutants. Methods. 2013;59:207–216. doi: 10.1016/j.ymeth.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Panavas T, Serviene E, Pogany J, Nagy PD. Genome-wide screens for identification of host factors in viral replication. Methods Mol Biol. 2008;451:615–624. doi: 10.1007/978-1-59745-102-4_41. [DOI] [PubMed] [Google Scholar]
- Pant V, Gupta D, Choudhury NR, Malathi VG, Varma A, Mukherjee SK. Molecular characterization of the Rep protein of the blackgram isolate of Indian mung bean yellow mosaic virus. J Gen Virol. 2001;82:2559–2567. doi: 10.1099/0022-1317-82-10-2559. [DOI] [PubMed] [Google Scholar]
- Prasanth KR, Kovalev N, de Castro Martín IF, Baker J, Nagy PD. Screening a yeast library of temperature-sensitive mutants reveals a role for actin in tombusvirus RNA recombination. Virology. 2016;489:233–242. doi: 10.1016/j.virol.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Price BD, Roeder M, Ahlquist P. DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J Virol. 2000;74:11724–11733. doi: 10.1128/JVI.74.24.11724-11733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan V, Malik PS, Choudhury NR, Mukherjee SK. The DNA-A component of a plant geminivirus (Indian mung bean yellow mosaic virus) replicates in budding yeast cells. J Virol. 2004;78:2405–2413. doi: 10.1128/JVI.78.5.2405-2413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim D, Bradford WD, Freeland A, Cady G, Wang J, Pruitt SC, Gerton JL. DNA replication stress restricts ribosomal DNA copy number. PLoS Genet. 2017;13:e1007006. doi: 10.1371/journal.pgen.1007006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settlage SB, Miller AB, Hanley-Bowdoin L. Interactions between geminivirus replication proteins. J Virol. 1996;70:6790–6795. doi: 10.1128/JVI.70.10.6790-6795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Islam MN, Choudhury NR, Karjee S, Mukherjee SK. The 32 kDa subunit of replication protein A (RPA) participates in the DNA replication of Mung bean yellow mosaic India virus (MYMIV) by interacting with the viral Rep protein. Nucleic Acids Res. 2007;35:755–770. doi: 10.1093/nar/gkl1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyal G, Mukherjee SK, Srivastava PS, Choudhury NR. Arabidopsis thaliana MCM2 plays role(s) in Mungbean yellow mosaic India virus (MYMIV) DNA replication. Arch Virol. 2013;158:981–992. doi: 10.1007/s00705-012-1563-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.