Abstract
Premise
The Climate Reconstruction Analysis using Coexistence Likelihood Estimation (CRACLE) method utilizes a robust set of modeling tools for estimating climate and paleoclimate from vegetation using large repositories of biodiversity data and open access R software.
Methods
Here, we implement a new R package for the estimation of climate from extant and fossil vegetation. The ‘cRacle’ package implements functions for data access, aggregation, and modeling to estimate climate from plant community compositions. ‘cRacle’ is modular and includes many best‐practice features.
Results
Performance tests using modern vegetation survey data from North and South America shows that CRACLE outperforms alternative methods. CRACLE estimates of mean annual temperature are usually within 1°C of the actual values when optimal model parameters are used. Generalized boosted regression (GBR) model correction improves CRACLE estimates by reducing bias.
Discussion
CRACLE provides accurate estimates of climate based on the composition of modern plant communities. Non‐parametric CRACLE modeling coupled with GBR model correction produces the most accurate results to date. The ‘cRacle’ R package streamlines the estimation of climate from plant community data, which will make this modeling more accessible to a wider range of users.
Keywords: climate, CRACLE, ecological niche model, fossil, GBIF, paleoclimate, R
For the past four years, the Climate Reconstruction Analysis using Coexistence Likelihood Estimation (CRACLE) code existed only as a single R script published as a supplement to the original paper (Harbert and Nixon, 2015). Despite this, the method has proven to be a useful tool to the community, yielding informative paleoclimate estimates for several fossil systems. Here, we present the ‘cRacle’ R package, an open‐source R software package designed to streamline the process of CRACLE modeling, data acquisition, the pre‐processing of primary biodiversity data (from the Global Biodiversity Information Facility [GBIF] and other related databases), and CRACLE result visualization. The ‘cRacle’ R package is built from modular functions that perform segments of the modeling protocol, allowing for easy‐to‐read R scripts as well as future modular developments of related methods (e.g., Ballantyne et al., 2010; Greenwood et al., 2017).
The primary application of CRACLE and related methods is in the reconstruction of paleoclimate from fossil floras. The association of plant communities with climate features has long been used in the reconstruction of paleoclimates from fossil floras based on modern climatic distributions (Hickey et al., 1988; Kershaw and Nix, 1988; Kershaw, 1997). All current applications of CRACLE make assumptions about climatic niche stability through time because they rely on the nearest living relative (NLR) approach to infer the elements of fossil taxon niche dimensions by observing the niche space occupied by a closely related extant relative (Mosbrugger and Utescher, 1997; Harris et al., 2014; Utescher et al., 2014). NLR assumptions are most valid when the fossil taxa can be placed as members of extant species and least accurate when fossils are members of extinct lineages. For this reason, NLR and CRACLE will produce the most reliable reconstructions of relatively recent (e.g., Pleistocene and Pliocene) paleoclimates, but may also produce reasonable results for much older fossil assemblages where the plant fossil taxa can be reliably placed into extant groups with well‐sampled modern distributions (e.g., Harris et al., 2014).
The use of fossil floras to estimate climate regularly provides insights that are complementary to other sources of paleoclimatic inference. Here, we will summarize some of the current research using CRACLE and related methods that could benefit from the computational tools presented in ‘cRacle’ and any future standardization of the methods therein. CRACLE modeling was recently applied to the estimation of the paleoclimate in western North America using Late Quaternary (<50,000 years ago) plant macrofossils in packrat (Neotoma spp.) middens (Harbert and Nixon, 2018). The analysis of the packrat midden paleoclimate estimates revealed a history of rapid climate change during and just after the Late Pleistocene deglaciation, followed by Holocene warm and dry periods of 1–2°C above the modern (1970–2000) averages (Harbert and Nixon, 2018). The CRACLE‐derived packrat midden plant macrofossil proxy climate record was similar to other results based on biological and isotopic proxies from the region, but at a temporal coverage (>30,000 years) unavailable from other records (Harbert and Nixon, 2018).
CRACLE was used to analyze community assemblages from excavated permafrost at five Pliocene sites in the Canadian Arctic Archipelago (Fletcher et al., 2017). This work revealed that the Canadian Arctic during the Early Pliocene (~3.6 mya) was up to 22°C warmer than today and supported many cool‐temperate plant taxa. Fletcher et al. (2017) carefully compared results derived from CRACLE estimates using species‐only identifications vs. generic‐level identifications and found that care must be taken when interpreting CRACLE results based on generic‐level identifications to avoid being misled by model imprecision. Further testing of how taxonomic classification impacts CRACLE model output is needed to understand the effect of fossil identification uncertainty on paleoclimate estimates. Paleoclimate estimates for the Paleocene–Eocene Thermal Maximum (~55 mya) and the Eocene Thermal Maximum (53.5 mya) have also been generated using Gaussian probability bioclimatic envelope methods similar to CRACLE (Ballantyne et al., 2010; Greenwood et al., 2017; Hyland et al., 2018; Willard et al., 2019).
CRACLE models have been applied to non‐plant communities as well, but performance in these systems has not been broadly tested. Pliocene Arctic climate estimates revealed notable biases in beetle‐based CRACLE reconstructions relative to plant‐based CRACLE reconstructions and stable isotope methods (Fletcher et al., 2019). These results suggest that plant communities may be more directly influenced by climate than beetle communities and, therefore, that CRACLE will be most reliable when using plant system data.
For comparison with CRACLE, we have also implemented the Thompson's Mutual Climatic Range (MCR) method in the ‘cRacle’ package (Thompson et al., 2012). This method estimates climate based on the climate range intersection rather than probability density functions. We include both the weighted and unweighted MCR methods as representatives of the many available MCR or coexistence approach range‐intersect methods (Mosbrugger and Utescher, 1997; Sinka and Atkinson, 1999; Utescher et al., 2014) for the estimation of climate from vegetation as part of the ‘cRacle’ R package. The ‘cRacle’ package is distributed under an MIT License, and all code is available on GitHub (https://www.github.com/rsh249/cRacle).
METHODS
Full documentation of the ‘cRacle’ R code is available with the package code (https://github.com/rsh249/cRacle). Below is an outline of the major functionality of the ‘cRacle’ package and an explanation of many of the core functions available.
‘cRacle’ package functions
Data acquisition
Downloading primary biodiversity data from the GBIF (https://www.gbif.org/), iNaturalist (https://www.inaturalist.org/), and BISON (https://bison.usgs.gov/) databases is supported in ‘cRacle’ through the functions gbif_get(), get_bison(), and inat(). Each of these functions accepts arguments for a single taxon (genus, species, or family) to query and a maximum number of records to return. Data pre‐processing, including cleaning for statistical climatic outliers and spatial bias reduction and climate data extraction for a set of georeferenced occurrence data, is done through the extraction() function. Data downloading for multiple taxa and pre‐processing functionality can be done in one step using the getextr() function, which is implemented with parallel computing capability to reduce user wait time.
Likelihood modeling
The primary modeling required by CRACLE is the calculation of probability density functions using both parametric (Gaussian normal) and non‐parametric (kernel density) estimates. These calculations can be performed with ‘cRacle’ directly from the output of extraction() using the function(s) dens_obj() and densform() for multiple and single taxa, respectively. Note that dens_obj() is simply a wrapper function for densform() designed to simplify multi‐species likelihood model building. Both parametric and non‐parametric likelihood functions are calculated and stored in the object produced by densform() and dens_obj(), meaning all calculations are done in tandem for a data set so the results for both methods can be examined at the end of the CRACLE process.
The likelihood modeling carried out in the densform() function includes several options that can be manipulated by the user. The kernel density estimation employed by CRACLE can now be implemented with several kernel estimators, including the standard Gaussian, Epanechnikov, cosine, optcosine, biweight, rectangular, and triangular kernels, based on the R ‘stats’ package function density() (R Core Team, 2018). The kernel bandwidth can be optimized using standard criteria via the argument ‘bw’ in the densform() and dens_obj() functions. Likelihoods can be calculated based on a standard or conditional probability via the ‘manip’ argument, in which conditional probabilities take into account the background distribution of each climate parameter using a set of randomly sampled points from within a normalized distance from each primary occurrence location or from a user‐defined set of climate data. Finally, the resulting likelihood distributions can be trimmed to empirical ranges or confidence intervals to avoid unintended extrapolation using the ‘clip’ argument.
CRACLE joint likelihood
After the likelihood functions are estimated, the next key CRACLE step is to calculate first the joint likelihood and then the maximum of the joint likelihood distribution. ‘cRacle’ implements the functions and_fun() for calculating the joint likelihood and or_fun() for creating unions between taxa to merge likelihood features between groups (i.e., between species in a clade). The output of and_fun() is a set of joint likelihood features for all parameters, which can be summarized by the function get_optim(). The output of get_optim() is the optimal climate value for each parameter, providing the user with CRACLE results.
Example pseudocode
The general CRACLE modeling flow uses the following functions in order: getextr(), dens_obj(), and_fun(), get_optim(). This produces climate estimates from a list of taxa by downloading data and extracting climate data, estimating likelihood distributions, calculating the joint likelihood, and finding the optimum of the joint likelihood for each climate variable analyzed.
Visualizing likelihood and joint likelihood functions
‘cRacle’ provides a set of functions for visualizing the likelihood functions as standard probability density distributions. These functions are densplot() for plotting single distribution objects (i.e., for one taxon/climate parameter pair), addplot() for adding to an existing densplot() figure, and multiplot() for plotting distributions for multiple taxa given a single climate parameter. For example, a user may plot the distributions for all taxa in their study for mean annual temperature using multiplot() and the output from dens_obj(), and then add the joint likelihood function using addplot() and the model object output from and_fun().
Issue tracking and versioning
The repository at https://github.com/rsh249/cRacle.git will maintain the current development version of ‘cRacle.’ Future major releases will be submitted to the Comprehensive R Archive Network (CRAN). Users should notify the authors of problems encountered with running ‘cRacle’ code by using the GitHub repository's ‘Issues’ feature (https://github.com/rsh249/cRacle/issues) or by email to the corresponding author of this publication.
Data access
Primary biodiversity data, occurrence coordinates for your target taxa, and climate data are required for CRACLE modeling. We recommend primary biodiversity data downloaded from GBIF and support such data access through ‘cRacle’ functions. Climate data must be in the form of a raster object readable by the R ‘raster’ package (Hijmans, 2018). Climate data from the WorldClim project (Hijmans et al., 2005; Fick and Hijmans, 2017) are recommended, but users may also use raster data downloaded from other sources such as CHELSA (Karger et al., 2017) and ENVIREM (Title and Bemmels, 2018).
Estimation of climate from vegetation
As originally described, CRACLE consists of two methods for likelihood estimation: parametric and non‐parametric probability functions. ‘cRacle’ implements these two approaches in tandem for each step of the likelihood analysis, and has implemented options about whether to calculate the likelihoods from standard probability or conditional probability. For comparison, we have implemented calculations for the MCR method for the estimation of climate from vegetation described by Thompson et al. (2012), although the unweighted MCR described is analogous to the coexistence approach described elsewhere (Mosbrugger and Utescher, 1997).
CRACLE estimates the climate variable likelihood function for a set of taxa (t) as follows. For any given set of climate values ‘x’, the CRACLE parametric (normal) probabilities are estimated as:
whereas the CRACLE non‐parametric (kernel density estimation) probabilities are estimated as:
where K is a kernel function with an area of 1 and ‘h’ is the kernel bandwidth, a smoothing parameter with a value > 0. We recommend using either the “optcosine” or “gaussian” kernels and a bandwidth calculated using Silverman's Rule of Thumb (Silverman, 1986). ‘cRacle’ allows for the calculation of probability density functions conditioned by the probability density of a random background sample for each climate parameter, defined as:
where ‘c’ is the set of background climates given by a sample of points from within the study area. The conditional probability for a taxon (t) is defined as:
For the estimation of climate from vegetation, CRACLE modeling calculates the joint likelihood for multiple taxon probability functions as:
where the maximum value of L(x|t 1:i) corresponds to the value of the climate parameter ‘x’ most likely to lead to the coexistence of that set of taxa (t 1:i).
Note that ‘cRacle’ also implements the calculation of a weighted mean by variance, which can be substituted for the parametric CRACLE functions. A weighted mean is calculated from a set of taxon means (x 1:i) and standard deviations (σ1:i):
Modern validation
To test the performance of CRACLE for paleoclimate modeling, we developed two experimental analyses to test aspects of the CRACLE modeling process. Model performance here is evaluated as the absolute difference between CRACLE modeled values and the WorldClim estimated values for the site location using the 2.5 arcminute WorldClim version 2.0 model data (http://www.worldclim.org/bioclim). The communities being analyzed are based on modern (within the past century) data and, therefore, the CRACLE estimates are compared to the WorldClim modern (1970–2000) climate averages.
For the first experiment, we queried iNaturalist for putatively co‐occurring species of plants across North America to test the performance of parametric and non‐parametric CRACLE as well as weighted and unweighted MCR. For each of these methods, we also tested the effect of varying the scale of spatial thinning in the primary occurrence data on model output to examine the impact of spatial sampling bias on climate predictions. These species lists serve to provide a quick and easy way to generate preliminary lists of species that belong to a wide range of plant communities. Although these lack expert validation, they do represent the range of missing and incorrect community data that might be expected when analyzing fossil plant communities; therefore, the quantification of CRACLE error rates in this experiment should be conservative estimates for real‐world applications.
To obtain preliminary plant community data, we used a grid search to sample the iNaturalist data for North America to identify plant species coexisting in 0.1 × 0.1‐degree (approximately 10 × 10 km) bounded search areas using the ‘rinat’ R package (Barve and Hart, 2017). Each of these search areas returned potential lists of co‐occurring species. These community lists are taken with the caveat that the iNaturalist data are contributed by a range of users, including both expert and amateur naturalists. These lists thus provide only preliminary community composition data sufficient for testing CRACLE, but neither the community composition nor the climate estimates should be considered definitive. Using these lists, we built CRACLE estimates for mean annual temperature using the modern WorldClim 2.0 data set at a resolution of 2.5 arcminutes (~4 × 4 km) (Fick and Hijmans, 2017), using parametric and non‐parametric CRACLE as well as weighted and unweighted MCR methods. Primary biodiversity data for CRACLE model fitting were obtained from GBIF using the ‘cRacle’ get_dist_all() function and the GBIF RESTful JSON‐based API. Model overfitting for occurrence data was tested by spatially thinning the raw GBIF data using the ‘cRacle’ extraction() function for factors of 2, 4, 6, 8, and 10 times the dimensions of the 2.5‐arcminute WorldClim raster cells. The ‘cRacle’ thinning procedure is an imperfect but efficient spatial thinning method consistent with best practices for limiting the effects of spatial sampling bias in the ecological niche modeling (ENM) literature (Aiello‐Lammens et al., 2015). The CRACLE and MCR model outputs for each thinning level were summarized and reported for general guidance. The R code for the iNaturalist grid search test is available at https://github.com/rsh249/cracle_examples/tree/master/inat_grid_search.
In the second experiment, we queried expert‐validated vegetation plot survey data to test the model performance for the 19 bioclimatic variables (http://www.worldclim.org/bioclim) and to test the model bias correction using generalized boosted regression (GBR) models. The Botanical Information and Ecology Network (BIEN 4.0) database was queried for published vegetation plot surveys from North and South America using the ‘rbien’ R package (Maitner et al., 2018). BIEN provides expert‐curated data for georeferenced plant occurrence data, vegetation plot surveys, trait data, phylogenetic trees, and gridded distribution maps. The vegetation plot surveys used for this section come from well‐documented surveys conducted by experts and mobilized through BIEN. Thinning factors were held constant and non‐parametric CRACLE was used for this experiment to simplify the downstream correction modeling. Future work would require similar analyses for other modeling choices if GBR correction is expanded for use across all CRACLE modeling.
Errors in CRACLE are non‐random (Harbert and Nixon, 2015); therefore, non‐linear regression modeling could account for and correct common CRACLE errors. For plot surveys with more than five plant taxa with distribution data available from GBIF, we estimated climate parameters for all 19 bioclimatic variables from the WorldClim 2.0 data (Fick and Hijmans, 2017) using parametric CRACLE methods and spatial thinning to a factor of two times the 2.5‐arcminute climate grid. CRACLE results for 70,391 plot surveys were then partitioned 50:50 into training (n = 35,196) and testing (n = 35,195) sets by random sampling without replacement. GBR models were fit to the training data for each of the 19 bioclimatic parameters using the R ‘gbm’ package (Greenwell et al., 2019). The GBR models were used to adjust the climate estimates in the test. Using independently trained GBR models on the test data set helped to determine whether error patterns in CRACLE could be predicted by GBR and corrected for in independent analyses. The R code for the BIEN experiment and GBR model fitting is available from https://github.com/rsh249/cracle_examples/tree/master/cracle_bien.
RESULTS
Modern validation
iNaturalist
The grid search of iNaturalist in North America enabled climate estimations to be made using the CRACLE (both parametric and non‐parametric) and MCR (both unweighted and weighted) approaches for 285 sites at spatial thinning factors of 0, 2, 4, 6, 8, and 10 times the 2.5‐arcminute climate raster. A total of 1,204,925 unique records were accessed from GBIF for this analysis, for a total of 1541 species. The sampled localities ranged in mean annual temperature from 0.4–26.1°C. The top‐performing (lowest error rates) estimates were the CRACLE non‐parametric (‘kde’ in Fig. 1) and the MCR weighted (‘mcr.w’ in Fig. 1) methods. The mean errors for CRACLE were ~1°C, while the median errors were generally less than the means. Spatial thinning did not change the mean or median error rates for any of the tested methods (Fig. 1B, C).
Figure 1.
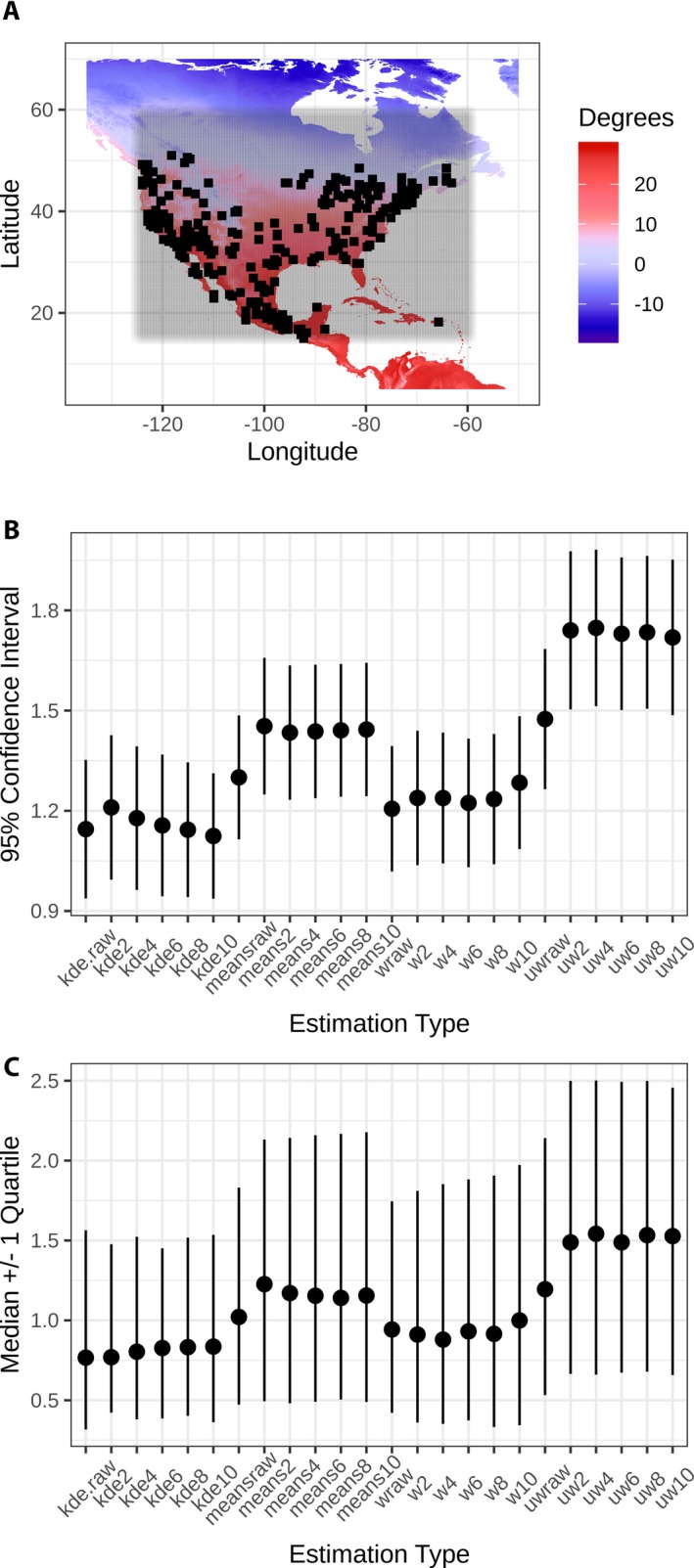
iNaturalist grid search climate estimation results. (A) Geographic search area (shading) with successful climate estimation locations marked (black squares), (B) mean anomaly rates, and (C) median anomaly rates for CRACLE (kde = non‐parametric, means = parametric) and MCR (uw = unweighted, w = weighted) results for factor levels of 0 (“raw”), 2, 4, 6, 8, and 10 times the 2.5‐arcminute climate raster.
BIEN
Non‐parametric CRACLE estimates of all 19 bioclimatic variables from the WorldClim 2.0 data set (Fick and Hijmans, 2017) were generated for the 70,391 vegetation plot surveys accessed through the BIEN 4.0 database using a total of 2,560,261 georeferenced specimen records from GBIF. These CRACLE results produced generally accurate estimates of climate across a broad geographic range with many distinct climatic regions (Fig. 2).
Figure 2.
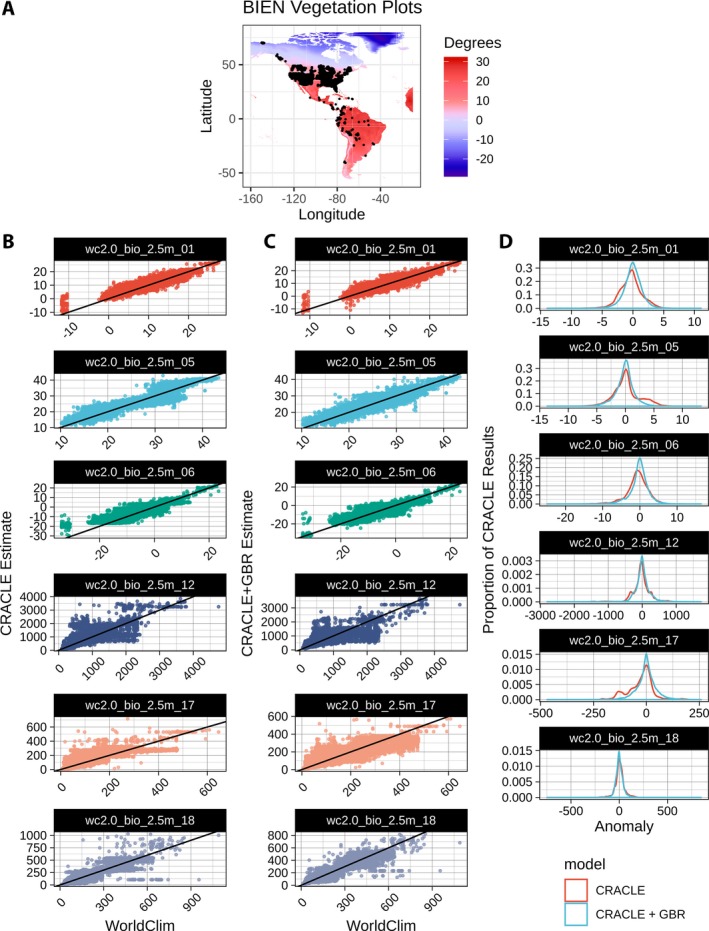
CRACLE estimates and generalized boosted regression (GBR) error correction demonstration for BIEN vegetation plot surveys and representative climate parameters. BIEN vegetation plot surveys from 70,391 unique localities (A) were analyzed with CRACLE (B) to estimate the 19 bioclimatic variables (showing BIO1 [mean annual temperature], 5 [maximum temperature of warmest month], 6 [minimum temperature of coldest month], 12 [annual precipitation], 17 [precipitation of driest quarter], and 18 [precipitation of coldest quarter] here as top‐performing examples) from the WorldClim 2.0 data set (Fick and Hijmans, 2017). GBR error correction modeling was trained and tested (C) on independent subsets of plot data. The GBR correction yielded overall reduced error rates and less biased estimates in many cases (D).
The mean absolute anomaly rates (i.e., the amount by which CRACLE differs from the WorldClim 2.0 model data) for all temperature parameters were between 1°C and 2°C with Pearson's correlation coefficients greater than 0.9 in the best scenarios (Table 1), but up to 5°C and with correlation coefficients <0.7 for estimates of mean temperature during the wettest and driest quarters of the year (BIO8 and BIO9). The mean absolute anomaly rate for mean annual precipitation (BIO12) was 179 mm with a correlation of 0.79 (Table 1).
Table 1.
Test data set CRACLE performance statistics for 19 bioclimatic parameters estimated for BIEN vegetation plot data (n = 35,195 plots).
| Climate_Parameter | Mean anomaly | Mean absolute anomaly | Median absolute anomaly | RMSE | NRMSE | Pearson's r | Spearman's r | Units |
|---|---|---|---|---|---|---|---|---|
| wc2.0_bio_2.5m_01 | 0.34 | 1.40 | 1.09 | 1.86 | 0.05 | 0.95 | 0.96 | °C |
| wc2.0_bio_2.5m_02 | −0.31 | 1.01 | 0.74 | 1.39 | 0.07 | 0.82 | 0.79 | °C |
| wc2.0_bio_2.5m_03 | −0.25 | 2.51 | 1.80 | 3.44 | 0.04 | 0.87 | 0.87 | °C |
| wc2.0_bio_2.5m_04 | −23.93 | 64.64 | 42.35 | 92.68 | 0.06 | 0.84 | 0.85 | °C |
| wc2.0_bio_2.5m_05 | −0.19 | 1.71 | 1.19 | 2.32 | 0.07 | 0.92 | 0.95 | °C |
| wc2.0_bio_2.5m_06 | 0.64 | 2.13 | 1.57 | 2.93 | 0.05 | 0.91 | 0.92 | °C |
| wc2.0_bio_2.5m_07 | −0.94 | 2.42 | 1.97 | 3.19 | 0.07 | 0.79 | 0.83 | °C |
| wc2.0_bio_2.5m_08 | 1.00 | 5.60 | 3.27 | 7.81 | 0.18 | 0.69 | 0.63 | °C |
| wc2.0_bio_2.5m_09 | −1.00 | 5.64 | 2.43 | 8.80 | 0.16 | 0.60 | 0.63 | °C |
| wc2.0_bio_2.5m_10 | 0.03 | 1.30 | 0.97 | 1.74 | 0.06 | 0.94 | 0.96 | °C |
| wc2.0_bio_2.5m_11 | 0.31 | 1.89 | 1.36 | 2.66 | 0.05 | 0.92 | 0.93 | °C |
| wc2.0_bio_2.5m_12 | 26.20 | 179.17 | 111.69 | 275.09 | 0.06 | 0.79 | 0.83 | mm/year |
| wc2.0_bio_2.5m_13 | 7.32 | 25.04 | 11.31 | 47.92 | 0.07 | 0.70 | 0.75 | mm/month |
| wc2.0_bio_2.5m_14 | 9.89 | 15.49 | 9.72 | 21.79 | 0.10 | 0.77 | 0.76 | mm/month |
| wc2.0_bio_2.5m_15 | −7.11 | 11.66 | 8.30 | 16.12 | 0.15 | 0.69 | 0.71 | — |
| wc2.0_bio_2.5m_16 | 21.84 | 68.96 | 31.48 | 135.63 | 0.08 | 0.69 | 0.76 | mm/3*month |
| wc2.0_bio_2.5m_17 | 26.80 | 45.61 | 27.49 | 64.92 | 0.08 | 0.80 | 0.76 | mm/3*month |
| wc2.0_bio_2.5m_18 | −6.20 | 34.76 | 23.16 | 52.69 | 0.05 | 0.90 | 0.91 | mm/3*month |
| wc2.0_bio_2.5m_19 | 34.97 | 99.35 | 73.22 | 142.96 | 0.10 | 0.57 | 0.69 | mm/3*month |
NRMSE = normalized root mean standard error; RMSE = root mean standard error.
GBR models were used to correct the CRACLE estimates using independent random samples without replacement of 50% of the data for the training and testing data sets. GBR correction improved CRACLE performance in the test set by 20–50% (Tables 1, 2; Fig. 2). The CRACLE+GBM‐corrected anomalies were smaller and more symmetrically distributed around ‘0’ than the non‐corrected CRACLE results (Fig. 2D), and CRACLE+GBM estimates were more strongly correlated with the WorldClim data than the raw CRACLE results (Tables 1, 2).
Table 2.
Generalized boosted regression–corrected test data set CRACLE performance statistics for 19 bioclimatic parameters estimated for BIEN vegetation plot data (n = 35,195 plots).
| Climate_Parameter | Mean anomaly | Mean absolute anomaly | Median absolute anomaly | RMSE | NRMSE | Pearson's r | Spearman's r | Units |
|---|---|---|---|---|---|---|---|---|
| wc2.0_bio_2.5m_01 | −0.06 | 1.11 | 0.85 | 1.51 | 0.04 | 0.96 | 0.96 | °C |
| wc2.0_bio_2.5m_02 | 0.00 | 0.84 | 0.60 | 1.18 | 0.06 | 0.86 | 0.81 | °C |
| wc2.0_bio_2.5m_03 | −0.01 | 2.08 | 1.48 | 2.89 | 0.04 | 0.91 | 0.89 | °C |
| wc2.0_bio_2.5m_04 | 1.03 | 51.65 | 36.94 | 72.73 | 0.05 | 0.90 | 0.87 | °C*100 |
| wc2.0_bio_2.5m_05 | −0.11 | 1.18 | 0.83 | 1.64 | 0.05 | 0.96 | 0.95 | °C |
| wc2.0_bio_2.5m_06 | −0.16 | 1.75 | 1.18 | 2.49 | 0.04 | 0.93 | 0.93 | °C |
| wc2.0_bio_2.5m_07 | −0.03 | 1.85 | 1.33 | 2.57 | 0.06 | 0.85 | 0.86 | °C |
| wc2.0_bio_2.5m_08 | −0.38 | 4.43 | 3.00 | 6.17 | 0.15 | 0.74 | 0.70 | °C |
| wc2.0_bio_2.5m_09 | 0.41 | 4.86 | 2.38 | 7.66 | 0.14 | 0.71 | 0.69 | °C |
| wc2.0_bio_2.5m_10 | −0.06 | 0.97 | 0.73 | 1.32 | 0.05 | 0.97 | 0.96 | °C |
| wc2.0_bio_2.5m_11 | −0.15 | 1.61 | 1.14 | 2.32 | 0.04 | 0.94 | 0.94 | °C |
| wc2.0_bio_2.5m_12 | 12.84 | 147.83 | 95.59 | 225.75 | 0.05 | 0.86 | 0.86 | mm/year |
| wc2.0_bio_2.5m_13 | 1.52 | 19.52 | 10.52 | 35.44 | 0.05 | 0.78 | 0.82 | mm/month |
| wc2.0_bio_2.5m_14 | 0.04 | 10.83 | 7.50 | 15.50 | 0.08 | 0.85 | 0.86 | mm/month |
| wc2.0_bio_2.5m_15 | 2.11 | 7.56 | 4.77 | 11.17 | 0.11 | 0.75 | 0.73 | — |
| wc2.0_bio_2.5m_16 | 3.25 | 51.70 | 28.72 | 97.37 | 0.06 | 0.79 | 0.83 | mm/3*month |
| wc2.0_bio_2.5m_17 | −0.80 | 32.85 | 22.45 | 46.69 | 0.07 | 0.87 | 0.87 | mm/3*month |
| wc2.0_bio_2.5m_18 | 2.48 | 28.98 | 21.01 | 42.83 | 0.04 | 0.93 | 0.92 | mm/3*month |
| wc2.0_bio_2.5m_19 | 11.46 | 70.76 | 46.68 | 114.25 | 0.08 | 0.71 | 0.76 | mm/3*month |
NRMSE = normalized root mean standard error; RMSE = root mean standard error.
DISCUSSION
The modern validation analyses conducted here should provide a good baseline for expected CRACLE performance going forward. More thorough testing in future studies is certainly welcome, but most modeling choices should be made by the user to reflect any unique properties of their study system. The results we provide here indicate that the best estimates generated using the ‘cRacle’ software are the CRACLE non‐parametric estimates with spatially thinned data. These yield mean errors of approximately 0.5°C less than the MCR method implemented by ‘cRacle’ (Fig. 1), a method analogous to the widely used coexistence approach (Mosbrugger and Utescher, 1997). Furthermore, the CRACLE results presented here compare favorably, although indirectly, to recent applications of the weighted average partial least squares (WA‐PLS) model, commonly used for the analysis of fossil pollen samples, which uses modern analog communities to build proxy models (Montade et al., 2019). A direct comparison of the CRACLE and WA‐PLS methods is necessary in future work.
Through our analysis of BIEN vegetation plot surveys, we show that CRACLE can produce accurate climate estimates for a variety of both temperature and precipitation parameters (Fig. 2), although some parameters are better predicted than others (Table 1). Notably, BIO8 (mean temperature of the warmest quarter), BIO9 (mean temperature of the coldest quarter), BIO13 (precipitation of the wettest month), and BIO15 (precipitation seasonality – coefficient variable) are relatively poorly predicted by CRACLE, possibly due to less direct impact of those variables on plant distributions. In contrast, estimates of BIO1 (mean annual temperature), BIO5 (maximum temperature), BIO6 (minimum temperature), and BIO18 (precipitation of the driest quarter) yield the highest correlation with known values (Table 1). We also show that CRACLE model correction via GBR can lower error rates from the CRACLE baseline (Fig. 2, Table 2). The GBR model correction based on the model testing presented here is implemented in ‘cRacle’ get_optim() as an option for users to correct non‐parametric CRACLE estimates for the WorldClim 2.0 bioclimatic variables. Further testing and fitting of GBR models to test data sets can expand on the options for model correction to include other climate parameters and model choices in the ‘cRacle’ modeling suite.
Resources and tutorials
We are actively developing resources and tutorials in support of the ‘cRacle’ R package. Demonstration code and short projects will be maintained on a continuing basis at https://github.com/rsh249/cracle_examples. We aim to provide a series of web tutorials for various CRACLE modeling tasks to guide beginner users through the process. Tutorials and issue tracking are distributed through the main ‘cRacle’ repository: https://github.com/rsh249/cRacle.git.
Conclusions
‘cRacle’ is a new and actively maintained resource for climate estimation from biological community compositions. These estimates are particularly relevant to the study of fossil systems, where often the best indication of past climate is the community of plant and animal fossils. We show that users of ‘cRacle’ should expect accurate estimates (e.g., within 1°C for mean annual temperature) when applying best practices.
AUTHOR CONTRIBUTIONS
A.A.B. conceived of and implemented the iNaturalist grid search and CRACLE analysis and provided valuable testing, benchmarking, and feedback data on the ‘cRacle’ R package. R.S.H. designed the BIEN vegetation plots CRACLE study and implemented the generalized boosted regression model correction analysis. R.S.H. designed and wrote the ‘cRacle’ R package code.
ACKNOWLEDGMENTS
The authors would like to thank Stonehill College for their support of undergraduate research that made this study possible. We also thank the contributions of our reviewers and the Applications in Plant Sciences editorial board for comments and suggestions that significantly improved this manuscript.
Harbert, R. S. , and Baryiames A. A.. 2020. cRacle: R tools for estimating climate from vegetation. Applications in Plant Sciences 8(2): e11322.
DATA AVAILABILITY
Primary biodiversity data used in this study was accessed through the Global Biodiversity Information Facility (GBIF) API (http://api.gbif.org/v1/), iNaturalist (http://www.inaturalist.org), and the Botanical Information and Ecology Network (BIEN; http://bien.nceas.ucsb.edu/bien/). Code to repeat data downloads with current versions of these databases is available at https://github.com/rsh249/cracle_examples/blob/master/cracle_bien/cracle.R and https://github.com/rsh249/cracle_examples/blob/master/inat_grid_search/get_coord_loop.R. Development of data access functions in ‘cRacle’ is currently implementing support for new GBIF API support for assigning DOI numbers for downloads with ‘rgbif’. The ‘cRacle’ package is distributed under an MIT License, and all code and documentation are available on GitHub (https://www.github.com/rsh249/cRacle); the current development version will be maintained at https://github.com/rsh249/cRacle.git.
LITERATURE CITED
- Aiello‐Lammens, M. E. , Boria R. A., Radosavljevic A., Vilela B., and Anderson R. P.. 2015. spThin: An R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 38: 541–545. [Google Scholar]
- Ballantyne, A. P. , Greenwood D. R., Sinninghe Damsté J. S., Csank A. Z., Eberle J. J., and Rybczynski N.. 2010. Significantly warmer Arctic surface temperatures during the Pliocene indicated by multiple independent proxies. Geology 38(7): 603–606. [Google Scholar]
- Barve, V. , and Hart E.. 2017. ‘rinat’: Access iNaturalist data through APIs. R package version 0.1.5. Website https://cran.r-project.org/src/contrib/Archive/rinat/ [accessed 23 January 2020].
- Fick, S. E. , and Hijmans R. J.. 2017. WorldClim 2: New 1‐km spatial resolution climate surfaces for global land areas. International Journal of Climatology 37: 4302–4315. [Google Scholar]
- Fletcher, T. , Feng R., Telka A. M., Matthews J. V., and Ballantyne A.. 2017. Floral dissimilarity and the influence of climate in the Pliocene High Arctic: Biotic and abiotic influences on five sites on the Canadian Arctic Archipelago. Frontiers in Ecology and Evolution 5: 19. [Google Scholar]
- Fletcher, T. L. , Csank A. Z., and Ballantyne A. P.. 2019. Identifying bias in cold season temperature reconstructions by beetle mutual climatic range methods in the Pliocene Canadian High Arctic. Palaeogeography, Palaeoclimatology, Palaeoecology 514: 672–676. [Google Scholar]
- Greenwell, B. , Boehmke B., Cunningham J., and GBM Developers . 2019. ‘gbm’: Generalized boosted regression models. R package version 2.1.5. Website https://cran.r-project.org/package=gbm [accessed 12 January 2020].
- Greenwood, D. R. , Keefe R. L., Reichgelt T., and Webb J. A.. 2017. Eocene paleobotanical altimetry of Victoria's Eastern Uplands. Australian Journal of Earth Sciences 64(5): 625–637. [Google Scholar]
- Harbert, R. S. , and Nixon K. C.. 2015. Climate reconstruction analysis using coexistence likelihood estimation (CRACLE): A method for the estimation of climate using vegetation. American Journal of Botany 102: 1277–1289. [DOI] [PubMed] [Google Scholar]
- Harbert, R. S. , and Nixon K. C.. 2018. Quantitative Late Quaternary climate reconstruction from plant macrofossil communities in western North America. Open Quaternary 4: 8. [Google Scholar]
- Harris, A. J. , Papeş M., Yun‐Dong G. A. O., and Watson L.. 2014. Estimating paleoenvironments using ecological niche models of nearest living relatives: A case study of Eocene Aesculus L. Journal of Systematics and Evolution 52: 16–34. [Google Scholar]
- Hickey, L. J. , Johnson K. R., and Dawson M. R.. 1988. The stratigraphy, sedimentology, and fossils of the Haughton Formation: A post‐impact crater‐fill, Devon Island, N.W.T., Canada. Meteoritics 23: 221–231. [Google Scholar]
- Hijmans, R. J. 2018. raster: Geographic data analysis and modeling. R package version 2.8‐4. Website https://CRAN.R-project.org/package=raster [accessed 12 January 2020].
- Hijmans, R. J. , Cameron S. E., Parra J. L., Jones P. G., and Jarvis A.. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978. [Google Scholar]
- Hyland, E. G. , Huntington K. W., Sheldon N. D., and Reichgelt T.. 2018. Temperature seasonality in the North American continental interior during the early Eocene climatic optimum. Climate of the Past Discussions 14: 1391–1404. [Google Scholar]
- Karger, D. N. , Conrad O., Böhner J., Kawohl T., Kreft H., Soria‐Auza R. W., Zimmermann N. E., et al. 2017. Climatologies at high resolution for the earth's land surface areas. Scientific Data 4: 170122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw, A. P. 1997. A bioclimatic analysis of Early to Middle Miocene brown coal floras, Latrobe Valley, south‐eastern Australia. Australian Journal of Botany 45: 373. [Google Scholar]
- Kershaw, A. P. , and Nix H. A.. 1988. Quantitative palaeoclimatic estimates from pollen data using bioclimatic profiles of extant taxa. Journal of Biogeography 15: 589–602. [Google Scholar]
- Maitner, B. S. , Boyle B., Casler N., Condit R., Donoghue J., Durán S. M., Guaderrama D., et al. 2018. The bien r package: A tool to access the Botanical Information and Ecology Network (BIEN) database. Methods in Ecology and Evolution 9: 373–379. [Google Scholar]
- Montade, V. , Peyron O., Favier C., Francois J. P., and Haberle S. G.. 2019. A pollen‐climate calibration from western Patagonia for palaeoclimatic reconstructions. Journal of Quaternary Science 34: 76–86. [Google Scholar]
- Mosbrugger, V. , and Utescher T.. 1997. The coexistence approach: A method for quantitative reconstructions of Tertiary terrestrial palaeoclimate data using plant fossils. Palaeogeography, Palaeoclimatology, Palaeoecology 134: 61–86. [Google Scholar]
- R Core Team . 2018. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: Website https://www.R-project.org/ [accessed 12 January 2020]. [Google Scholar]
- Silverman, B. W. 1986. Density estimation for statistics and data analysis. Chapman & Hall/CRC, London, United Kingdom. [Google Scholar]
- Sinka, K. J. , and Atkinson T. C.. 1999. A mutual climatic range method for reconstructing palaeoclimate from plant remains. Journal of the Geological Society 156(2): 381–396. [Google Scholar]
- Thompson, R. S. , Anderson K. H., Pelltier R. T., Strickland L. E., Bartlein P. J., and Shafer S. L.. 2012. Quantitative estimation of climatic parameters from vegetation data in North America by the mutual climatic range technique. Quaternary Science Reviews 51: 18–39. [Google Scholar]
- Title, P. O. , and Bemmels J. B.. 2018. ENVIREM: An expanded set of bioclimatic and topographic variables increases flexibility and improves performance of ecological niche modeling. Ecography 41: 291–307. [Google Scholar]
- Utescher, T. , Bruch A. A., Erdei B., François L., Ivanov D., Jacques F. M. B., Kern A. K., et al. 2014. The Coexistence Approach—theoretical background and practical considerations of using plant fossils for climate quantification. Palaeogeography, Palaeoclimatology, Palaeoecology 410: 58–73. [Google Scholar]
- Willard, D. A. , Donders T. H., Reichgelt T., Greenwood D. R., Sangiorgi F., Peterse F., Nierop K. G. J., et al. 2019. Arctic vegetation, temperature, and hydrology during Early Eocene transient global warming events. Global and Planetary Change 178: 139–152. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Primary biodiversity data used in this study was accessed through the Global Biodiversity Information Facility (GBIF) API (http://api.gbif.org/v1/), iNaturalist (http://www.inaturalist.org), and the Botanical Information and Ecology Network (BIEN; http://bien.nceas.ucsb.edu/bien/). Code to repeat data downloads with current versions of these databases is available at https://github.com/rsh249/cracle_examples/blob/master/cracle_bien/cracle.R and https://github.com/rsh249/cracle_examples/blob/master/inat_grid_search/get_coord_loop.R. Development of data access functions in ‘cRacle’ is currently implementing support for new GBIF API support for assigning DOI numbers for downloads with ‘rgbif’. The ‘cRacle’ package is distributed under an MIT License, and all code and documentation are available on GitHub (https://www.github.com/rsh249/cRacle); the current development version will be maintained at https://github.com/rsh249/cRacle.git.


