Significance
Oncogenic signaling by small GTPases enhances malignant phenotypes in both breast and lung cancer. The SmgGDS splice variant isoforms SmgGDS-607 and SmgGDS-558 activate oncogenic small GTPases by promoting their prenylation. We report that a pathologic ratio of SmgGDS splice variants is expressed in cancer cells and is associated with decreased survival of breast cancer patients. We developed a splice-switching oligonucleotide (SSO) to target this oncogenic SmgGDS isoform ratio. This SSO suppresses prenylation of small GTPases, induces apoptotic cell death of breast and lung cancer cells, and diminishes tumorigenesis in an aggressive mouse model of breast cancer. These results define a strategy to suppress the prenylation of small GTPases and diminish malignancy by modulating SmgGDS splicing.
Keywords: SmgGDS, RAP1GDS1, small GTPases, prenylation, alternative splicing
Abstract
The chaperone protein SmgGDS promotes cell-cycle progression and tumorigenesis in human breast and nonsmall cell lung cancer. Splice variants of SmgGDS, named SmgGDS-607 and SmgGDS-558, facilitate the activation of oncogenic members of the Ras and Rho families of small GTPases through membrane trafficking via regulation of the prenylation pathway. SmgGDS-607 interacts with newly synthesized preprenylated small GTPases, while SmgGDS-558 interacts with prenylated small GTPases. We determined that cancer cells have a high ratio of SmgGDS-607:SmgGDS-558 (607:558 ratio), and this elevated ratio is associated with reduced survival of breast cancer patients. These discoveries suggest that targeting SmgGDS splicing to lower the 607:558 ratio may be an effective strategy to inhibit the malignant phenotype generated by small GTPases. Here we report the development of a splice-switching oligonucleotide, named SSO Ex5, that lowers the 607:558 ratio by altering exon 5 inclusion in SmgGDS pre-mRNA (messenger RNA). Our results indicate that SSO Ex5 suppresses the prenylation of multiple small GTPases in the Ras, Rho, and Rab families and inhibits ERK activity, resulting in endoplasmic reticulum (ER) stress, the unfolded protein response, and ultimately apoptotic cell death in breast and lung cancer cell lines. Furthermore, intraperitoneal (i.p.) delivery of SSO Ex5 in MMTV-PyMT mice redirects SmgGDS splicing in the mammary gland and slows tumorigenesis in this aggressive model of breast cancer. Taken together, our results suggest that the high 607:558 ratio is required for optimal small GTPase prenylation, and validate this innovative approach of targeting SmgGDS splicing to diminish malignancy in breast and lung cancer.
SmgGDS (gene name RAP1GDS1) is a chaperone protein that promotes malignancy by binding and promoting the prenylation of multiple small GTPases that contain a C-terminal polybasic region (PBR), such as Rac1, RhoA, KRas, and Rap1 (1–8). These small GTPases are commonly mutated or overexpressed in different types of cancer, which accelerates malignant transformation and tumor development (9–11). The oncogenic potential of SmgGDS is indicated by its elevated expression in breast (3), nonsmall cell lung (12), and prostate (13) cancer, and by the demonstration that SmgGDS depletion diminishes breast and lung tumorigenesis in mouse models (3, 14).
We previously identified 2 SmgGDS isoforms, SmgGDS-607 and SmgGDS-558, that are generated through alternative RNA splicing (1). These isoforms function together to regulate the entrance of PBR-containing small GTPases into the prenylation pathway and their subsequent membrane trafficking (1, 2, 15). SmgGDS-607 binds and delivers preprenylated small GTPases to prenyltransferases (PTases), while SmgGDS-558 binds small GTPases only after they have been prenylated. The ability of these SmgGDS isoforms to promote the prenylation and oncogenic activities of small GTPases makes SmgGDS a novel therapeutic candidate in cancer, but strategies to limit SmgGDS activity have not been developed. There have been many therapeutic attempts to directly inhibit the prenylation of oncogenic small GTPases using farnesyltransferase (FTase) inhibitors and geranylgeranyltransferase (GGTase) inhibitors, but these approaches have proven to be less clinically effective than anticipated. The failure of these drugs to inhibit malignancy therapeutically is largely due to the ability of certain GTPases such as KRas to accept a geranylgeranyl isoprenoid group upon treatment with FTase inhibitors, allowing for full functionality to be sustained (16). Since SmgGDS acts as a global regulator of the prenylation pathway for both farnesylated and geranylgeranylated small GTPases (1, 2), targeting the splice variants of SmgGDS may provide a new way to effectively inhibit the malignancy of cancers that are sensitive to the disruption of small GTPase prenylation.
We report here that cancer cell lines and tumor tissues have an oncogenic splicing program that generates more SmgGDS-607 than SmgGDS-558, resulting in an abnormally high ratio of SmgGDS-607:SmgGDS-558 (607:558 ratio). Previous studies to diminish SmgGDS-607 or SmgGDS-558 expression in cancer cells have had mixed results. The RNA interference (RNAi)-mediated depletion of SmgGDS-607 inhibits cell proliferation in the NCI-H1703 lung cancer cell line (14), but not in other lung and breast cancer cell lines (3, 14), most likely because SmgGDS-607 expression could not be reduced enough in these lines to suppress its functions. In contrast, the RNAi-mediated depletion of SmgGDS-558 significantly slows proliferation and inhibits tumorigenesis of breast and lung cancer cells (3, 14), indicating that cancer cells require a threshold level of SmgGDS-558 to maintain the malignant state. In this study, we used a different approach to suppress the oncogenic functions of SmgGDS, by targeting the splicing program that generates the high 607:558 ratio. The targeting of aberrant alternative splicing for cancer-associated genes is an attractive therapeutic approach, as oncogenic splice isoforms are highly prevalent in cancer and provide growth and survival advantages that drive tumorigenesis (17–19).
We have developed a splice-switching oligonucleotide (SSO) as a therapeutic strategy to redirect SmgGDS splicing and reduce the high oncogenic 607:558 ratio of cancer cells. We demonstrate that this SSO, called SSO Ex5, decreases the 607:558 ratio and significantly suppresses global protein prenylation. SSO Ex5 induces apoptotic cell death in breast and lung cancer cell lines, and diminishes mammary tumorigenesis in transgenic MMTV-PyMT mice. Collectively, these findings suggest that cancer cells require a high 607:558 ratio for optimal prenylation of oncogenic small GTPases. The effectiveness of SSO Ex5 in modulating SmgGDS splicing to diminish malignant phenotypes and inhibit tumor growth provides a strong rationale for targeting the 607:558 ratio in the treatment of breast and lung cancer.
Results
An Elevated 607:558 Splice Variant Ratio Is Expressed in Cancer Cells and Tumor Tissue and Correlates with Decreased Survival of Patients with Breast Cancer.
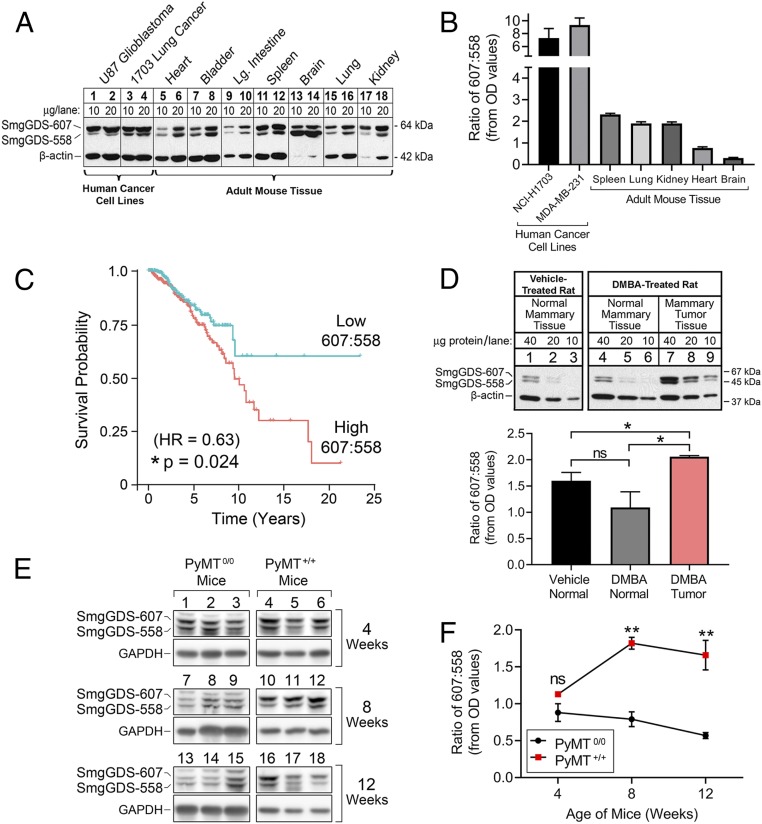
SmgGDS total expression is elevated in cancer (3, 12, 13) and correlates with reduced survival of breast cancer patients (3). We investigated the expression of the individual SmgGDS splice variants by immunoblot in proliferative versus nonproliferative tissue types. The 607:558 ratio is higher in tissues composed of proliferating cells, such as the spleen (Fig. 1A, lanes 11 and 12, and Fig. 1B), but much lower in tissues consisting mainly of nonproliferating and terminally differentiated cells, such as the heart and brain (Fig. 1A, lanes 5, 6, 13, and 14, and Fig. 1B). Most notably, the 607:558 ratio is highest in human cancer cells (Fig. 1A, lanes 1 to 4, and Fig. 1B).
Fig. 1.
Elevated 607:558 ratio is associated with malignancy. (A) Lysates prepared from human cancer cell lines (lanes 1 to 4) and adult mouse tissues (lanes 5 to 18) were run on SDS/PAGE at the indicated amount of protein per lane and immunoblotted for SmgGDS and β-actin. (B) Densitometry of immunoblots was performed to quantify the 607:558 ratio in human cancer cell lines (n = 6) and adult mouse tissues (n = 3). Results are presented as mean ± SEM. OD, optical density. (C) TCGA SpliceSeq data from 710 female patients with breast cancer were used to produce a Kaplan–Meier plot of survival probability based on expression levels of SmgGDS-558 mRNA that generate either a high or low 607:558 mRNA ratio. Data were analyzed by Cox regression to assess statistical significance (*P < 0.05). (D) Rats were treated with vehicle or DMBA and killed after tumors formed in the DMBA-treated rats (n = 4 rats per treatment group). Lysates prepared from either normal mammary tissue (lanes 1 to 6) or mammary tumor tissue (lanes 7 to 9) were run on SDS/PAGE at the indicated amount of protein per lane and immunoblotted for SmgGDS and β-actin. Representative immunoblots from 2 rats are shown (Top). Densitometry of the immunoblots from all rats was performed to quantify the 607:558 ratio in each tissue type (Bottom). Results are presented as mean ± SEM, and statistical significance was determined by unpaired, 2-tailed Student t test (ns, not significant; *P < 0.05). (E) Normal mammary and mammary tumor tissues were collected from MMTV-PyMT0/0 and MMTV-PyMT+/+ mice at 4, 8, and 12 wk of age. Lysates were generated and immunoblotted for SmgGDS and GAPDH (n = 3 mice per genotype and age cohort). (F) Densitometry of the immunoblots from E was performed to calculate the 607:558 ratio. Results are presented as mean ± SEM, and statistical significance was determined by unpaired, 2-tailed Student t test at each age time point (ns, not significant; **P < 0.01).
To assess the translational importance of the 607:558 ratio in cancer outcome, we compared patient survival with SmgGDS isoform transcript levels from tumor RNA-seq (sequencing) data of 710 female breast cancer cases (The Cancer Genome Atlas [TCGA] SpliceSeq database, BRCA cohort). This analysis revealed a significant correlation between the expression of the shorter splice variant isoform SmgGDS-558 and increased patient survival (HR [hazard ratio] = 0.63, P = 0.024) (Fig. 1C). In comparison, the survival probability of patients with breast tumors that have lower expression levels of SmgGDS-558, and thus a higher 607:558 ratio, is significantly reduced (Fig. 1C). These data suggest that the 607:558 ratio is translationally relevant to breast cancer patient survival, with a lower 607:558 ratio in tumor tissue being associated with a more favorable outcome.
To determine if the 607:558 ratio is altered in animal models of cancer, we utilized 2 different model systems. The carcinogen DMBA (7,12-dimethyl-benz[a]-anthracene) has been well-characterized for generating mammary tumors in rats to model human breast cancer (20–22). We treated rats with a single dose of either vehicle control or DMBA and killed the rats after mammary tumors had developed in the DMBA-treated animals. Interestingly, not only does the total expression level of SmgGDS increase in the mammary tumors of DMBA-treated rats, but the 607:558 ratio is also significantly elevated (Fig. 1D). We also assessed the 607:558 ratio using MMTV-PyMT mice, in which aggressive mammary tumors develop as early as 5 wk of age with 100% incidence by 6 mo (23). Tumor progression occurs in this model in 3 distinct stages: hyperplasia from 4 to 6 wk; adenoma/mammary intraepithelial neoplasia (MIN) from 6 to 8 wk; and early/late carcinoma by 8 to 12 wk (24, 25). We compared cancer-imminent MMTV-PyMT+/+ female mice with healthy age-matched MMTV-PyMT0/0 female mice and assessed the 607:558 ratio in their mammary glands or tumor tissues at 4, 8, and 12 wk of age. The 607:558 ratio is significantly elevated in the mammary tissue of MMTV-PyMT+/+ mice at 8 and 12 wk of age compared with healthy MMTV-PyMT0/0 mice (Fig. 1 E and F). Taken together, these findings indicate that a high 607:558 ratio may act as a driver of malignant transformation and tumor progression in multiple animal models of cancer.
A Splice-Switching Oligonucleotide Targeted to Exon 5 of SmgGDS pre-mRNA Reduces the 607:558 Ratio in Human Cancer Cells.
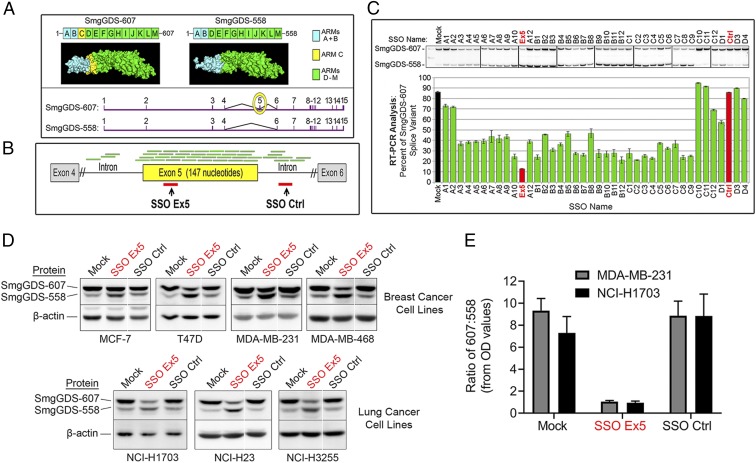
To test the oncogenic potential of a high 607:558 ratio, we developed SSOs that target the exon 5 region of SmgGDS pre-mRNA (messenger RNA) and reduce the 607:558 ratio. SmgGDS pre-mRNA consists of 15 exons, and the 607:558 ratio is dependent on alternative splicing of exon 5, which encodes armadillo (ARM) domain C that is included only in SmgGDS-607 (1, 2) (Fig. 2A). Crystallography studies recently confirmed these structural differences in ARM domains between the 2 isoforms (26, 27).
Fig. 2.
SmgGDS-607:SmgGDS-558 ratio is reduced in cancer cells by splice-switching oligonucleotides. (A) PyMOL homology models of SmgGDS-607 and SmgGDS-558 protein structures (2) (Top) and mRNA exon sequences (Bottom) are shown. SmgGDS-607 contains 13 ARM domains, labeled A to M, and differs from SmgGDS-558 only by the presence of ARM C (yellow shading). SmgGDS pre-mRNA is composed of 15 exons, labeled 1 to 15, and exon 5 (yellow circle) encodes ARM C. (B) Green lines depict 40 oligonucleotides aligning with SmgGDS pre-mRNA that were screened for regulation of exon 5 splicing. Red lines depict regions targeted by SSO Ex5 and SSO Ctrl. (C) Semiquantitative 32P RT-PCR PAGE analysis of SmgGDS-607 and SmgGDS-558 mRNA levels was conducted after MDA-MB-231 cells were treated for 72 h with transfection reagent alone (mock) or 1 of the oligonucleotides depicted in B. Results are presented as mean ± SEM (n = 3). (D) The indicated breast and lung cancer cell lines were transfected with mock, SSO Ex5 (50 nM), or SSO Ctrl (50 nM) for 72 h and lysates were immunoblotted for SmgGDS and β-actin. Representative immunoblots are shown. For some immunoblot images, white lines between lanes are used to indicate protein lanes on the same gel that were nonadjacent. (E) Densitometry was performed for MDA-MB-231 and NCI-H1703 immunoblots to quantify the 607:558 ratio after treatment with mock, SSO Ex5, or SSO Ctrl for 72 h. Results are presented as mean ± SEM (n = 6 for each cell line).
We screened 40 separate SSOs surrounding and encompassing exon 5 in MDA-MB-231 breast cancer cells. Each antisense SSO base pairs with a specific target region of SmgGDS pre-mRNA to potentially occlude the binding of undefined spliceosome factors and redirect the splicing pattern of SmgGDS. We identified SSO Ex5 that targets exon 5 and reduces the 607:558 RNA ratio most significantly (Fig. 2 B and C and SI Appendix, Fig. S1). Additionally, an SSO targeted to the downstream intronic region, called SSO Ctrl, acts as a negative control, as it does not alter the 607:558 RNA ratio compared with cells treated with transfection reagent alone (mock) (Fig. 2 B and C and SI Appendix, Fig. S1). We then assessed whether the observed SmgGDS RNA isoform alterations are reflected in protein production by treating 4 different human breast cancer cell lines and 3 different human nonsmall cell lung cancer (NSCLC) cell lines (SI Appendix, Table S1) with mock, SSO Ex5, or SSO Ctrl. SSO Ex5 efficiently reduces the 607:558 protein ratio in all cell lines, while SSO Ctrl has a minimal effect on the 607:558 protein ratio (Fig. 2 D and E). The total level of SmgGDS protein was not significantly altered by either SSO Ex5 or SSO Ctrl (SI Appendix, Fig. S2).
SSO Ex5 Diminishes the Proliferation of Breast and Lung Cancer Cell Lines and Induces Apoptotic Cell Death.
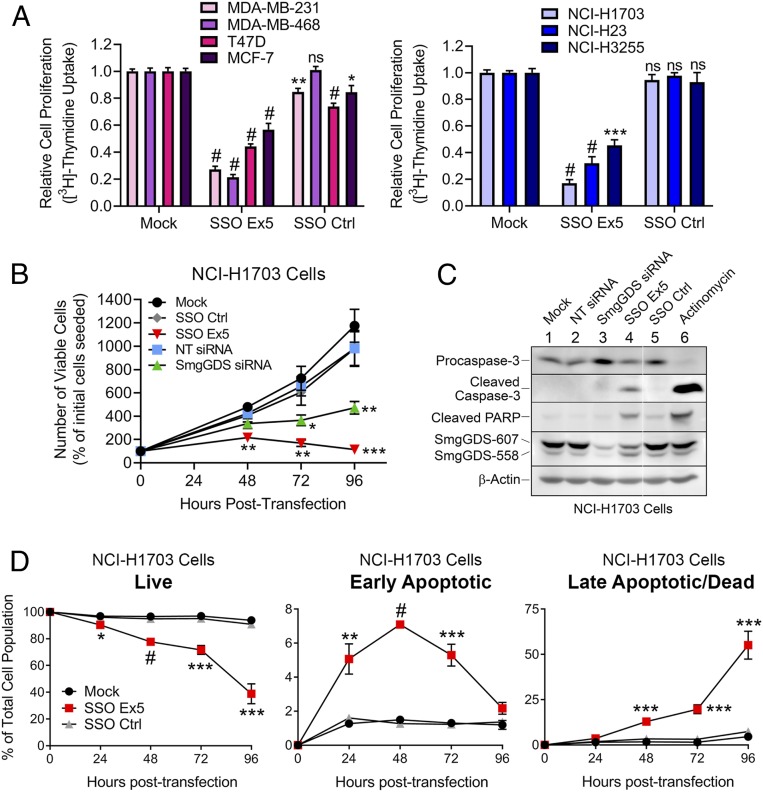
To investigate changes in cell proliferation induced by SSO Ex5, we tested the effects of SSO Ex5 on uptake of [3H]thymidine, which is an indicator of cell-cycle progression (3, 12, 14). We found that [3H]thymidine uptake is significantly inhibited by SSO Ex5 in 7 different cancer cell lines (Fig. 3A). Furthermore, treatment with SSO Ex5 diminishes the number of viable cells in cultures of NCI-H1703 lung cancer cells (Fig. 3B) and MDA-MB-231 breast cancer cells (SI Appendix, Fig. S3). Interestingly, cell proliferation is inhibited more by SSO Ex5 than by SmgGDS small interfering RNA (siRNA) (Fig. 3B and SI Appendix, Fig. S3), which depletes both SmgGDS isoforms and was previously found to diminish proliferation by inducing a G1-phase cell-cycle arrest (3, 14, 28). This result demonstrates a higher sensitivity of cancer cells to the reduction of the 607:558 ratio compared with the depletion of SmgGDS total levels by siRNA.
Fig. 3.
SSO Ex5 diminishes the proliferation of breast and lung cancer cell lines and induces apoptotic cell death. (A) The indicated breast cancer (Left) and lung cancer (Right) cell lines were transfected with mock, SSO Ex5, or SSO Ctrl for 72 h and incubated with [3H]thymidine for 3 h, followed by scintillation counting to measure relative proliferation. Values are normalized to [3H]thymidine uptake in mock-treated cells and are the mean ± SEM (n ≥ 3 for each cell line), conducted in quadruplicate and assessed by 1-way ANOVA followed by Dunnett’s multiple comparisons post hoc test relative to mock (ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001). (B) NCI-H1703 cells were transfected with SmgGDS siRNA (25 nM) or SSOs (50 nM) and viable cells were counted at 48, 72, and 96 h. Values shown represent the number of viable cells as a percentage of the cells that were initially seeded before treatment, and are the mean ± SEM (n = 3). Statistical significance was determined by 1-way ANOVA followed by Dunnett’s multiple comparisons post hoc test relative to mock at each time point (*P < 0.05, **P < 0.01, ***P < 0.001). (C) NCI-H1703 cells were transfected with SmgGDS siRNA (25 nM) or SSOs (50 nM) for 96 h and immunoblotting was performed to examine the expression of apoptotic protein markers. Treatment of cells with actinomycin (250 nM, 16 h) was included as a positive control for induction of apoptotic factors. Mean normalized densitometry values for cleaved caspase-3 and cleaved PARP are shown in SI Appendix, Fig. S4A (n = 3). The white line between lanes is used to indicate protein lanes on the same gel that were nonadjacent. (D) Flow cytometry was used to assess NCI-H1703 apoptotic cell death induced by SSO Ex5 treatment at 24, 48, 72, and 96 h. Cells were stained with annexin V-FITC and the vital dye PI prior to flow cytometry. Values are the mean ± SEM (n = 3), and statistical significance was determined by 1-way ANOVA followed by Dunnett’s multiple comparisons post hoc test relative to mock at each time point (*P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001).
We next investigated whether the reduced proliferation and viability of SSO Ex5-treated cells is caused by apoptotic cell death. We found that SSO Ex5 significantly elevates the expression of the apoptotic marker cleaved caspase-3 and its downstream target, cleaved PARP, in NCI-H1703 cells (Fig. 3C and SI Appendix, Fig. S4A) and in MDA-MB-231 cells (SI Appendix, Fig. S4 B and C). For both cell lines, SmgGDS siRNA treatment did not significantly increase cleaved caspase-3 or cleaved PARP levels (Fig. 3C and SI Appendix, Fig. S4). This result indicates that unlike SSO Ex5, SmgGDS depletion using siRNA does not induce apoptosis, consistent with our previous report (14). We further evaluated apoptosis induced by SSO Ex5 by using fluorescein isothiocyanate (FITC)-labeled annexin V and propidium iodide (PI) staining, followed by flow cytometry. SSO Ex5 significantly induces apoptosis from 24 to 96 h in NCI-H1703 cells (Fig. 3D). MDA-MB-231 cells exhibit a more subtle apoptotic response, but similar trends for live and dead cell populations are observed (SI Appendix, Fig. S5).
Apoptosis induced by SSO Ex5 may be caused by the combined effects of reduced SmgGDS-607 expression and increased SmgGDS-558 expression. We previously reported that the RNAi-mediated depletion of SmgGDS-607 diminishes the malignant phenotype of NCI-H1703 cells (14), supporting the conclusion that loss of SmgGDS-607 expression contributes to the antimalignant effect of SSO Ex5. To test the effects of increased SmgGDS-558 levels on apoptosis, we ectopically expressed SmgGDS-558 to reduce the 607:558 ratio. Intriguingly, we found that increasing SmgGDS-558 levels alone induces apoptosis in both NCI-H1703 and MDA-MB-231 cells (SI Appendix, Fig. S6). Collectively, these findings indicate that both the loss of SmgGDS-607 and the gain of SmgGDS-558 likely suppress the malignant phenotype when cells are treated with SSO Ex5.
SSO Ex5 Inhibits the Prenylation and Downstream Signaling of Multiple Small GTPases.
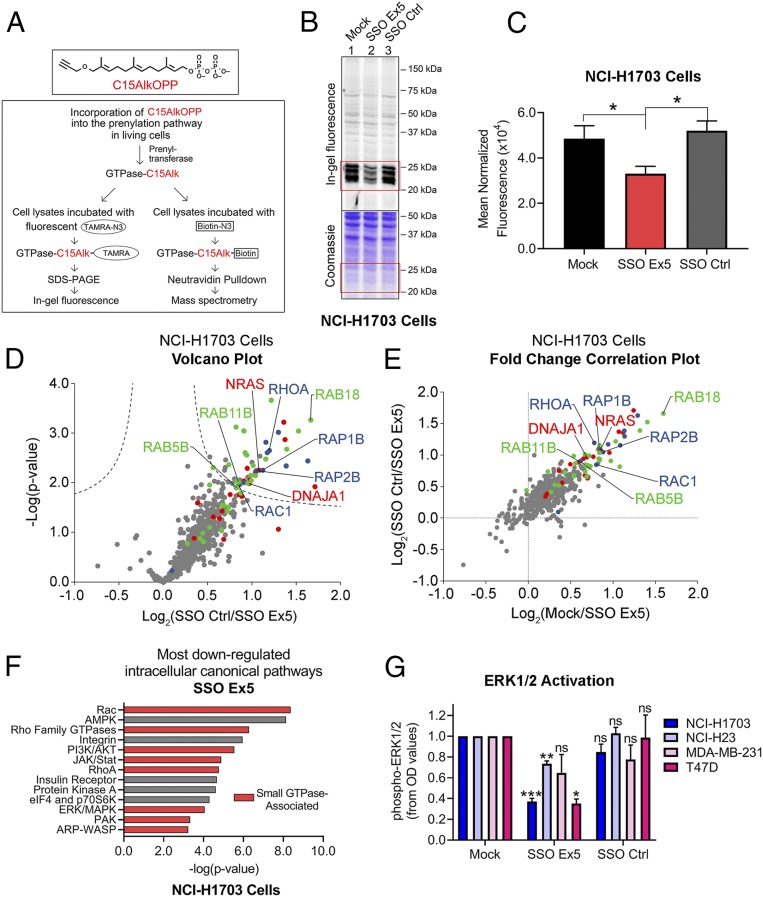
To assess whether reducing the 607:558 ratio disrupts prenylation, we tested the incorporation of a synthetic isoprenoid probe, C15AlkOPP (29), after SSO treatment and conducted analysis by in-gel fluorescence and mass spectrometry (Fig. 4A). In-gel fluorescence analysis indicated that SSO Ex5 diminished probe incorporation in both NCI-H1703 cells (Fig. 4 B and C) and MDA-MB-231 cells (SI Appendix, Fig. S7).
Fig. 4.
SSO Ex5 suppresses small GTPase prenylation and signaling. (A) The diagram depicts the experimental approaches used to assess SSO Ex5 effects on prenylation using the synthetic isoprenoid probe C15AlkOPP. (B) In-gel fluorescence analysis of C15AlkOPP probe incorporation (10 μM, 12 h) was conducted after NCI-H1703 cells were treated with mock, SSO Ex5, or SSO Ctrl for 72 h. (C) Coomassie-normalized fluorescence values of the 20- to 30-kDa region (highlighted in red boxes in B) for NCI-H1703 cells are shown as mean ± SEM (n = 3). Statistical significance was determined by paired, 2-tailed Student t test relative to SSO Ex5 (*P < 0.05). (D) A volcano plot was generated from quantitative proteomic analysis of C15AlkOPP-treated SSO Ctrl and SSO Ex5 NCI-H1703 samples (FDR = 0.01, s0 = 0.1). Proteins with statistically significant suppression of C15AlkOPP isoprenoid incorporation upon SSO Ex5 treatment are primarily composed of the 3 different classes of prenylation substrates: farnesylation (red), geranylgeranylation type I (blue), and Rab geranylgeranylation type II (green). Examples of significantly suppressed proteins from each substrate class are labeled, and gray dots represent nonprenylation substrates. (E) A comparison of fold changes log2(SSO Ctrl/SSO Ex5) vs. log2(mock/SSO Ex5) demonstrates similar enrichment profiles for both treatment comparisons, and indicates suppression of C15AlkOPP incorporation with SSO Ex5 compared with both mock and SSO Ctrl. (F) IPA of RNA-seq microarray data from NCI-H1703 cells treated with SSO Ex5 for 72 h identified the most down-regulated intracellular canonical signaling pathways. Red bars indicate small GTPase-associated signaling networks. (G) Densitometric analysis was conducted for activated phospho-ERK1/2 from immunoblots for the indicated cancer cell lines following treatment with mock, SSO Ex5, or SSO Ctrl for 72 h. Values were calculated by normalizing phospho-ERK1/2 and total ERK1/2 OD values to β-actin, and dividing the normalized phospho-ERK1/2 value by the normalized total ERK1/2 value. Values were then normalized to phospho-ERK1/2 in mock-treated cells, and results are presented as mean ± SEM (n ≥ 3). Statistical significance was determined by 1-way ANOVA followed by Dunnett’s multiple comparisons post hoc test relative to mock (ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001). Representative immunoblots for each cell line are shown in SI Appendix, Fig. S10.
Prenylation analysis using pull-down enrichment and mass spectrometry proteomics identified multiple small GTPases and other prenylated proteins that have reduced probe incorporation in cells treated with SSO Ex5 (Fig. 4 D and E and Datasets S1 and S2). Prenylated proteins were found to dominate the portion of enriched proteins, as about 75% of proteins with significantly suppressed labeling are known prenylation substrates. Surprisingly, the population of proteins that exhibit significantly decreased prenylation with SSO Ex5 treatment consists largely of substrates that have never been reported to interact with SmgGDS or associate with SmgGDS signaling, and include proteins that are substrates for all 3 of the primary PTase enzymes: FTase, geranylgeranyltransferase type I (GGTase-I), and Rab geranylgeranyltransferase type II (GGTase-II) (Fig. 4 D and E).
To validate these findings using another approach, we tested whether SSO Ex5 alters the prenylation of Rac1, which is a well-documented binding partner of SmgGDS (1, 30). Using a mevastatin block-and-release assay (1, 4, 31), we found that SSO Ex5 moderately decreases the accumulation of Rac1 in the detergent phase after mevastatin is removed and isoprenoid production resumes (SI Appendix, Fig. S8). This result is consistent with SSO Ex5 slowing the conversion of Rac1 from the preprenylated form (which fractionates in the aqueous phase) to the prenylated form (which fractionates in the detergent phase). Together, these results indicate that reducing the 607:558 ratio with SSO Ex5 inhibits the prenylation of small GTPases known to interact with SmgGDS isoforms, and also diminishes the global prenylation of other protein substrates.
To further investigate the effect that SSO Ex5 has on pathways involving small GTPases and oncogenic signaling, we conducted RNA-seq analysis. Ingenuity Pathway Analysis (IPA) revealed that the top intracellular and second-messenger pathways that are down-regulated by SSO Ex5 consist mainly of canonical small GTPase signaling networks that include Rac, RhoA, PI3K/AKT, and ERK/MAPK (Fig. 4F, SI Appendix, Fig. S9, and Datasets S3 and S4). Moreover, phospho-ERK1/2 activation levels are significantly diminished at steady state following SSO Ex5 treatment in multiple cancer cell lines (Fig. 4G and SI Appendix, Fig. S10), suggesting an involvement of decreased Ras and Rac signaling in the antimalignant effects produced by reducing the 607:558 ratio.
SSO Ex5 Causes Endoplasmic Reticulum Stress and Activates the Unfolded Protein Response.
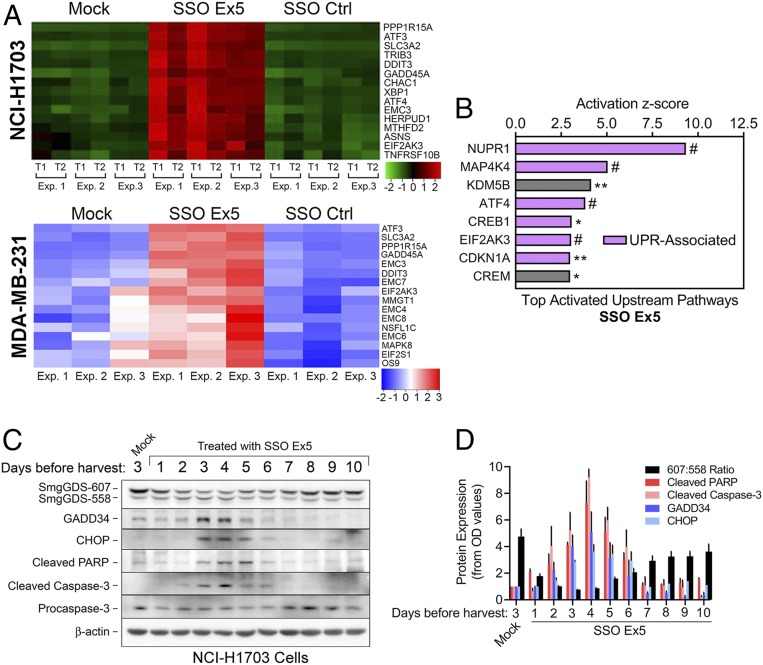
In addition to the identified small GTPase pathways that are down-regulated by SSO Ex5, RNA-seq analysis also revealed multiple endoplasmic reticulum (ER) stress- and unfolded protein response (UPR)-associated factors that are among the top up-regulated genes in NCI-H1703 and MDA-MB-231 cells following SSO Ex5 treatment (Fig. 5A and Datasets S3 and S4). These factors include key canonical ER stress and UPR genes such as ATF4, DDIT3 (CHOP), PPP1R15A (GADD34), GADD45A (GADD45), XBP1, EIF2AK3 (PERK), and ATF3. Furthermore, IPA of NCI-H1703 cells treated with SSO Ex5 indicated that the top activated upstream pathways consist of gene networks that have been directly or indirectly linked to ER stress and the UPR [NUPR1 (32–34), z score = 9.34, P = 2.01 × 10−11; MAP4K4 (35, 36), z score = 5.04, P = 3.54 × 10−10; ATF4 (37, 38), z score = 3.82, P = 4.56 × 10−10; CREB1 (39), z score = 3.06, P = 0.0174; EIF2AK3 (37, 38), z score = 3.04, P = 2.54 × 10−7; CDKN1A (40), z score = 2.99, P = 0.00145] (Fig. 5B).
Fig. 5.
SSO Ex5 activates the unfolded protein response. (A) Heatmaps of RNA-seq microarray data display changes in expression of genes involved with ER stress and the UPR that were most significantly altered in NCI-H1703 (Top) and MDA-MB-231 (Bottom) cells treated with mock, SSO Ex5, or SSO Ctrl for 72 h. For the NCI-H1703 heatmap, data were generated in 2 technical replicates (T1 and T2) from 3 independent experiments (Exp. 1, 2, and 3). For the MDA-MB-231 heatmap, data were generated from 3 independent experiments (Exp. 1, 2, and 3). (B) IPA of RNA-seq microarray data from NCI-H1703 cells treated with SSO Ex5 for 72 h identified significant activation of the indicated upstream signaling pathways. Purple bars indicate pathways that are directly associated with the canonical UPR, or pathways that have been reported to be indirectly associated with ER stress and the UPR. Pathways are ordered by their corresponding z score, and labeled for IPA gene overlap significance (*P < 0.05, **P < 0.01, #P < 0.0001). (C) NCI-H1703 cells were treated once with mock or SSO Ex5, and lysates were collected at the indicated days after treatment for immunoblot analysis of apoptotic and UPR factors. (D) Densitometric analysis of immunoblots from C was performed by normalizing the OD values of cleaved PARP, cleaved caspase-3, GADD34, and CHOP to β-actin. Values were then normalized to mock-treated cells and plotted, along with the 607:558 ratio. Results are presented as mean ± SEM (n = 3).
To validate the RNA-seq analysis, we conducted immunoblotting to examine the effects of SSO Ex5 on protein expression of GADD34 and CHOP. We compared the expression of these UPR proteins with the apoptotic markers cleaved caspase-3 and cleaved PARP, since a prolonged or overactive UPR can lead to apoptosis and cell death, primarily through caspase activation (37, 38, 41–44). These UPR and apoptotic proteins are expressed simultaneously as the 607:558 ratio is lowered by SSO Ex5 over a 10-d period (Fig. 5 C and D and SI Appendix, Fig. S11). All 4 factors peak in their relative expression at 4 d posttransfection of SSO Ex5 (Fig. 5D), which is when we observe the largest percentage of NCI-H1703 cells in late apoptosis (Fig. 3D). These data provide strong evidence for implicating the UPR in the apoptotic mechanism induced by splice-switching SmgGDS.
SSO Ex5 Reduces Mammary Tumorigenesis in MMTV-PyMT Mice.
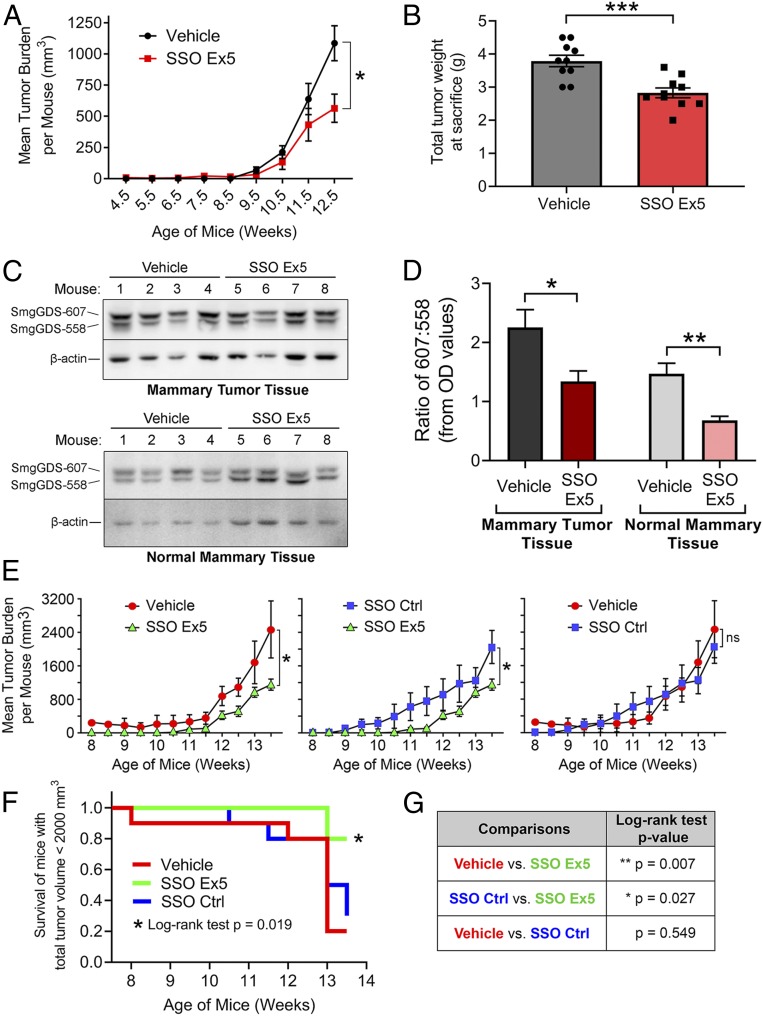
Since the 607:558 ratio increases as malignancy progresses in MMTV-PyMT+/+ mice (Fig. 1 E and F), a high 607:558 ratio may be required for tumor development. Alternatively, the high 607:558 ratio may not play a driving role in tumorigenesis, but merely results from splicing abnormalities that arise during transformation. To examine these possibilities, we tested the effects of SSO Ex5 on tumorigenesis in MMTV-PyMT+/+ mice. SSO Ex5 and SSO Ctrl are ideal for these studies, since they are chemically optimized for in vivo delivery due to a modified phosphorothioate backbone and a 2′-O-methoxyethyl sugar modification that improve stability, increase potency, and decrease toxicity (45). To determine if SmgGDS splicing is a crucial early event in the development of malignancy, we treated female MMTV-PyMT+/+ mice with phosphate-buffered saline (PBS) vehicle or SSO Ex5 (n = 10 mice per group) by intraperitoneal (i.p.) injection beginning at 3.5 wk of age, before neoplastic transformation typically occurs (24, 25). SSO Ex5 diminishes tumor volume by 12.5 wk of age (Fig. 6A), reduces the mean ex vivo tumor weight burden by nearly 1 g (Fig. 6B), and significantly lowers the mean tumor burden normalized to body weight (SI Appendix, Fig. S12). We also observed a significant reduction in the 607:558 ratio in tumor tissue from SSO Ex5-treated mice (Fig. 6 C and D). Interestingly, SSO Ex5 lowers the 607:558 ratio of mammary tumor tissue to similar levels observed in the normal mammary tissue of vehicle-treated mice (Fig. 6 C and D), suggesting that SSO Ex5 normalizes the 607:558 ratio.
Fig. 6.
SSO Ex5 significantly reduces mammary tumorigenesis in MMTV-PyMT mice. (A) Female MMTV-PyMT mice were injected i.p. 2 times per wk with PBS vehicle or SSO Ex5 beginning at 3.5 wk of age (n = 10 mice per treatment group). Mean tumor volume was calculated at each time point using calipers to measure the volume of all tumors arising in each mouse. Results are presented as mean ± SEM, and statistical significance was determined by unpaired, 2-tailed Student t test at each time point (*P < 0.05). (B) The graph depicts the mean ex vivo total tumor weight of each treatment group from A, immediately following killing. Results are presented as mean ± SEM (n = 10 mice per treatment group), and statistical significance was determined by unpaired, 2-tailed Student t test (***P < 0.001). (C) Mammary tumor tissue (Top) and normal mammary tissue (Bottom) were harvested from the same 4 mice in each treatment group, and lysates were immunoblotted for SmgGDS and β-actin. (D) Densitometry of the immunoblots from C was performed to quantify the 607:558 ratio in each tissue type and between treatment groups. Results are presented as mean ± SEM (n = 4), and statistical significance was determined by unpaired, 2-tailed Student t test (*P < 0.05, **P < 0.01). (E) Female MMTV-PyMT mice were injected i.p. 2 times per wk with PBS vehicle, SSO Ex5, or SSO Ctrl beginning at 6.5 wk of age (n = 10 mice per treatment group). Mean tumor volume was calculated at each time point using calipers to measure the volume of all tumors arising in each mouse. Results are presented as mean ± SEM, and statistical significance was determined by unpaired, 2-tailed Student t test at each time point (ns, not significant; *P < 0.05). (F) A Kaplan–Meier plot was generated to show the time to exceed a total tumor volume threshold of 2,000 mm3 for each treatment group from E. A log-rank test was used to make an overall comparison between all 3 treatment groups (*P < 0.05). (G) Separate log-rank tests were conducted for pairwise treatment group comparisons of survival shown in F (*P < 0.05, **P < 0.01).
To assess if we could inhibit tumorigenesis at a later stage, we conducted an independent follow-up study where treatment was initiated at 6.5 wk of age, after adenomas have formed and MIN is developing (24, 25). This experiment included the addition of mice treated with SSO Ctrl to ensure that there are no significant differences between vehicle and SSO Ctrl due to general effects of SSOs. SSO Ex5 significantly diminished the mean tumor volume burden by 13.5 wk of age compared with both control treatment groups, while SSO Ctrl had no effect on tumor burden compared with vehicle-treated mice (Fig. 6E). Additionally, survival analysis of tumor volume burden indicated a significant delay in reaching a total tumor volume threshold of 2,000 mm3 for SSO Ex5-treated mice compared with vehicle- and SSO Ctrl-treated mice (Fig. 6 F and G). As expected, we observed a significant reduction in the 607:558 ratio in whole mammary tumor lysates treated with SSO Ex5 compared with both controls (SI Appendix, Fig. S13). For both in vivo studies, SSO-treated MMTV-PyMT+/+ mice displayed no obvious physiological side effects or behavioral abnormalities, and no differences in weight gain were detected between all treatment groups over the experimental timeline (SI Appendix, Fig. S14). Collectively, these studies demonstrate the effectiveness of using SSO Ex5 to inhibit tumor development, and highlight the importance of a high 607:558 ratio in driving malignant transformation.
Discussion
Our findings identify an oncogenic SmgGDS splicing program and present insights into how the high ratio of SmgGDS isoforms is crucial for cancer cell biology. This study validates the critical role of a high 607:558 ratio in supporting malignant phenotypes and tumor development, and describes a unique approach for targeting the splicing of SmgGDS using SSOs.
The translational relevance of the SmgGDS splice variant ratio in cancer is indicated by our discovery that an elevated 607:558 ratio is a poor prognostic indicator for breast cancer patients. The in vivo rodent studies also validate the importance of the 607:558 ratio in malignancy, since the 607:558 ratio increases as mammary tumors develop in these animal models. MMTV-PyMT mice provide a reliable model of breast cancer progression, as they share transcriptional prognostic biomarkers and morphological features with the human disease (24, 25). Our finding that SSO Ex5 reduces the 607:558 ratio and diminishes mammary tumorigenesis in this aggressive cancer model suggests that the high 607:558 ratio is a molecular driver for tumorigenesis rather than merely a consequence of the transformed state. This high 607:558 ratio may drive malignancy by providing enough SmgGDS-607 to facilitate the prenylation of the many small GTPases that are overexpressed in cancer cells (9–11). Since normal cells may not be as dependent on SmgGDS-607 expression, the loss of SmgGDS-607 induced by SSO Ex5 may be less deleterious for normal cells than for cancer cells. This proposal is consistent with the absence of any noticeable side effects in mice treated with SSOs, supporting the therapeutic potential of SSO Ex5.
In addition to suppressing malignancy by diminishing SmgGDS-607 expression, SSO Ex5 also likely suppresses malignancy by increasing SmgGDS-558 levels. This possibility is supported by our finding that lowering the 607:558 ratio by overexpressing SmgGDS-558 induces apoptosis. SmgGDS-558 can promote the cytosolic accumulation of prenylated small GTPases (46–48), which occurs by SmgGDS-558 shielding the prenyl group of the GTPase from the aqueous environment of the cytosol (27). Other chaperones of prenylated GTPases, including PDEδ (49, 50), PRA1 (49, 51), VPS35 (52), and RhoGDI (53), similarly solubilize prenylated GTPases by shielding the prenyl group of the GTPase. An excessive amount of SmgGDS-558 in SSO Ex5-treated cells may enhance the solubilization of prenylated GTPases, diminishing signaling by the GTPases at the membrane and resulting in apoptosis. Excessive SmgGDS-558 might also successfully compete with other chaperones for prenylated GTPases, disrupting the proper trafficking of the small GTPases and promoting apoptosis. Taken together, both the reduction in SmgGDS-607 and the increase in SmgGDS-558 likely contribute to the apoptotic effects of SSO Ex5.
Proteomics analysis of cells treated with SSO Ex5 allowed us to define prenylation changes of known substrates from all 3 of the prenylation classes. SmgGDS-607 and SmgGDS-558 are best characterized for their binding to PBR-containing small GTPases that are substrates for GGTase-I, including RhoA, Rac1, and Rap1 (1–4). The tight regulatory role of SmgGDS isoforms for this class of small GTPases was validated by our finding that nearly all detected GGTase-I substrates containing a PBR exhibit suppressed isoprenoid incorporation in cells treated with SSO Ex5. Unexpectedly, however, we discovered that SSO Ex5 also suppresses the prenylation of GTPases that lack a PBR, as well as other proteins that are not known to interact directly with SmgGDS, such as DNAJA1. This finding might be explained by the proposed model (54) that the delivery of an unprenylated substrate to the PTase promotes the release of a prenylated substrate from the PTase. Based on this model, the delivery of an unprenylated GTPase by SmgGDS-607 to the PTase may promote the release of prenylated substrates from PTases. When SmgGDS-607 levels are low, such as in cells treated with SSO Ex5, prenylated substrates may remain associated for longer times with the PTase, potentially slowing the rate of prenylation of a broad range of substrates.
Intriguingly, García-Torres and Fierke (8) recently proposed that SmgGDS-607 accelerates the farnesylation and release of the non–PBR-containing small GTPase HRas by forming a ternary complex with HRas and the FTase. This proposal has similarities to the model discussed above, in which SSO Ex5 inhibits prenylation because there is no longer sufficient SmgGDS-607 to enhance the release of non–PBR-containing GTPases (or other CAAX-containing proteins) from the FTase. This event may then cause FTase enzymes to become saturated with substrate, creating a global inhibitory effect for other farnesylated proteins.
In contrast to HRas prenylation, García-Torres and Fierke (8) concluded that KRas4B prenylation is only minimally regulated by SmgGDS-607. This conclusion is consistent with our finding that SSO Ex5 did not significantly inhibit prenylation of KRas. However, SmgGDS was recently shown to impact KRas4B prenylation by forming a trimeric complex with KRas and a truncated form of the small GTPase RabL3, which was found to be associated with hereditary pancreatic cancer (7). Further examining the role of SmgGDS splice variants in the regulation of KRas prenylation and trafficking remains a crucial focus for fully understanding the biological relevance of SmgGDS in KRas-driven cancers.
It was also surprising to find that SSO Ex5 broadly suppresses the prenylation of Rab-family small GTPases that are substrates of the GGTase-II enzyme, since SmgGDS is not known to interact with or regulate Rab proteins. The only reported mechanism for regulating Rab protein prenylation involves the Rab escort protein, which helps newly synthesized Rab small GTPases receive their dual prenylation from GGTase-II (55, 56). The global changes in Rab prenylation might be due to unknown feedback mechanisms that are triggered by the many downstream signaling alterations that occur when SSO Ex5 inhibits prenylation of multiple GGTase-I and FTase substrates. Alternatively, our discoveries may have uncovered a novel aspect of SmgGDS intracellular functions, implicating the 607:558 ratio in the direct regulation and control of prenylation for the GGTase-II class of substrates.
We found that reducing the 607:558 ratio with SSO Ex5 produces a significant induction of ER stress-related genes and activation of the UPR. This result is consistent with previous reports indicating that the global suppression of protein prenylation by statins induces ER stress and the UPR (57–59). SSO Ex5 may induce this stress response by inhibiting the prenylation of Rab proteins, since Rab proteins regulate intracellular vesicle trafficking to and from the ER and other organelles (60), and disruption of this process is linked to UPR activation due to excessive accumulation of proteins in the ER (61).
Reduced ERK activity caused by SSO Ex5 may also promote ER stress and the UPR. The small GTPases RhoA, Rac1, and NRas are reported to protect cells from ER stress and the UPR by activating ERK (62–67), and activation of ERK promotes cell survival following ER stress (68). Since SSO Ex5 suppresses the prenylation of these small GTPases, the resulting decrease in their downstream signaling may diminish ERK activity and induce ER stress and the UPR. Consistent with this proposal, inhibition of Rho signaling induces expression of ATF3, ATF4, and DDIT3 (63), which are UPR genes that are elevated with SSO Ex5 treatment. Together, these findings support the idea that decreased ERK activity due to the suppression of small GTPase prenylation and subsequent loss of signaling promotes apoptosis through prolonged ER stress and the UPR.
Our study describes the development of a SmgGDS-targeted approach for inhibiting the prenylation of oncogenic small GTPases. The crystal structure of SmgGDS was recently determined in complex with a small GTPase (26, 27), making it more feasible to develop small-molecule inhibitors that target the binding surface between SmgGDS isoforms and small GTPase substrates to disrupt their prenylation and activation. Our unique strategy of inhibiting prenylation by redirecting the splicing of SmgGDS to reduce the 607:558 ratio highlights the effectiveness of SSOs in diminishing malignant phenotypes, and demonstrates how targeting SmgGDS to regulate small GTPase prenylation is a valid anticancer approach that should be further pursued.
Materials and Methods
All experimental procedures, including cell-culture conditions, transfection protocols, lysate preparation and immunoblotting, cell proliferation and apoptosis assays, TCGA analysis, prenylation analysis, RNA-seq analysis, animal experiments, and statistical analyses are presented in SI Appendix, Materials and Methods.
Data Availability Statement.
All additional data and information are included in SI Appendix as Materials and Methods, figures, additional figure legends, tables, datasets, and references.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute (NCI) R01 CA188871 (to C.L.W.), NCI R01 CA193343 (to M.J.F.), National Institute of General Medical Sciences R01 GM084152 (to M.D.D.), a Medical College of Wisconsin Cancer Center Multi-Principal Investigator Pilot Grant (to M.T.M. and M.J.F.), Kathy Duffey Fogarty Award for Breast Cancer Research KDF-13 (to M.T.M.), Wisconsin Breast Cancer Showhouse BST-15 (to M.T.M.), METAvivor Foundation (M.J.F.), and Mary Kay Foundation Award Grant 024.16 (to M.J.F.). Additional support was provided by the Joan K. Van Deuren Professor in Breast Cancer Research Award (to C.L.W.), Kathleen M. Duffey Fogarty Eminent Scholar in Breast Cancer Research Award (to C.L.W.), and Nancy Laning Sobczak, PhD, Breast Cancer Research Award (to C.L.W.).
Footnotes
Competing interest statement: F.R. is an employee of Ionis Pharmaceuticals. F.R., M.T.M., and C.L.W. have patents pending with the US Patent and Trademark Office for the SSOs and the targeting approach.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914153117/-/DCSupplemental.
References
- 1.Berg T. J., et al. , Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J. Biol. Chem. 285, 35255–35266 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuld N. J., et al. , The chaperone protein SmgGDS interacts with small GTPases entering the prenylation pathway by recognizing the last amino acid in the CAAX motif. J. Biol. Chem. 289, 6862–6876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauser A. D., et al. , The SmgGDS splice variant SmgGDS-558 is a key promoter of tumor growth and RhoA signaling in breast cancer. Mol. Cancer Res. 12, 130–142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ntantie E., et al. , An adenosine-mediated signaling pathway suppresses prenylation of the GTPase Rap1B and promotes cell scattering. Sci. Signal. 6, ra39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizuno T., et al. , A stimulatory GDP/GTP exchange protein for smg p21 is active on the post-translationally processed form of c-Ki-ras p21 and rhoA p21. Proc. Natl. Acad. Sci. U.S.A. 88, 6442–6446 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto T., et al. , Purification and characterization from bovine brain cytosol of proteins that regulate the GDP/GTP exchange reaction of smg p21s, ras p21-like GTP-binding proteins. J. Biol. Chem. 265, 16626–16634 (1990). [PubMed] [Google Scholar]
- 7.Nissim S., et al. , Mutations in RABL3 alter KRAS prenylation and are associated with hereditary pancreatic cancer. Nat. Genet. 51, 1308–1314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García-Torres D., Fierke C. A., The chaperone SmgGDS-607 has a dual role, both activating and inhibiting farnesylation of small GTPases. J. Biol. Chem. 294, 11793–11804 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konstantinopoulos P. A., Karamouzis M. V., Papavassiliou A. G., Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat. Rev. Drug Discov. 6, 541–555 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Alan J. K., Lundquist E. A., Mutationally activated Rho GTPases in cancer. Small GTPases 4, 159–163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong G. S., et al. , Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat. Med. 24, 968–977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tew G. W., et al. , SmgGDS regulates cell proliferation, migration, and NF-kappaB transcriptional activity in non-small cell lung carcinoma. J. Biol. Chem. 283, 963–976 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Zhi H., et al. , SmgGDS is up-regulated in prostate carcinoma and promotes tumour phenotypes in prostate cancer cells. J. Pathol. 217, 389–397 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Schuld N. J., et al. , SmgGDS-558 regulates the cell cycle in pancreatic, non-small cell lung, and breast cancers. Cell Cycle 13, 941–952 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azoulay-Alfaguter I., Strazza M., Mor A., Chaperone-mediated specificity in Ras and Rap signaling. Crit. Rev. Biochem. Mol. Biol. 50, 194–202 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Berndt N., Hamilton A. D., Sebti S. M., Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer 11, 775–791 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Marabti E., Younis I., The cancer spliceome: Reprograming of alternative splicing in cancer. Front. Mol. Biosci. 5, 80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oltean S., Bates D. O., Hallmarks of alternative splicing in cancer. Oncogene 33, 5311–5318 (2014). [DOI] [PubMed] [Google Scholar]
- 19.David C. J., Manley J. L., Alternative pre-mRNA splicing regulation in cancer: Pathways and programs unhinged. Genes Dev. 24, 2343–2364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson H. J., Singh M., Rat models of premalignant breast disease. J. Mammary Gland Biol. Neoplasia 5, 409–420 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Jordan V. C., Laboratory models of breast cancer to aid the elucidation of antiestrogen action. J. Lab. Clin. Med. 109, 267–277 (1987). [PubMed] [Google Scholar]
- 22.Abba M. C., et al. , DMBA induced mouse mammary tumors display high incidence of activating Pik3caH1047 and loss of function Pten mutations. Oncotarget 7, 64289–64299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fantozzi A., Christofori G., Mouse models of breast cancer metastasis. Breast Cancer Res. 8, 212 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai Y., et al. , Transcriptomic dynamics of breast cancer progression in the MMTV-PyMT mouse model. BMC Genomics 18, 185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin E. Y., et al. , Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163, 2113–2126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu H., et al. , Structure-based analysis of the guanine nucleotide exchange factor SmgGDS reveals armadillo-repeat motifs and key regions for activity and GTPase binding. J. Biol. Chem. 292, 13441–13448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu H., Toma-Fukai S., Kontani K., Katada T., Shimizu T., GEF mechanism revealed by the structure of SmgGDS-558 and farnesylated RhoA complex and its implication for a chaperone mechanism. Proc. Natl. Acad. Sci. U.S.A. 115, 9563–9568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonyo P., et al. , SmgGDS is a transient nucleolar protein that protects cells from nucleolar stress and promotes the cell cycle by regulating DREAM complex gene expression. Oncogene 36, 6873–6883 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suazo K. F., et al. , Metabolic labeling of prenylated proteins using alkyne‐modified isoprenoid analogues. Curr. Protoc. Chem. Biol. 10, e46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanning C. C., Ruiz-Velasco R., Williams C. L., Novel mechanism of the co-regulation of nuclear transport of SmgGDS and Rac1. J. Biol. Chem. 278, 12495–12506 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Wilson J. M., Prokop J. W., Lorimer E., Ntantie E., Williams C. L., Differences in the phosphorylation-dependent regulation of prenylation of Rap1A and Rap1B. J. Mol. Biol. 428, 4929–4945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chowdhury U. R., Samant R. S., Fodstad O., Shevde L. A., Emerging role of nuclear protein 1 (NUPR1) in cancer biology. Cancer Metastasis Rev. 28, 225–232 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Jin H. O., et al. , Nuclear protein 1 induced by ATF4 in response to various stressors acts as a positive regulator on the transcriptional activation of ATF4. IUBMB Life 61, 1153–1158 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Ozkaya A. B., Ak H., Aydin H. H., High concentration calcitriol induces endoplasmic reticulum stress related gene profile in breast cancer cells. Biochem. Cell Biol. 95, 289–294 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Chen S., et al. , Sertraline induces endoplasmic reticulum stress in hepatic cells. Toxicology 322, 78–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu C., Watts M. E., Rubin L. L., MAP4K4 activation mediates motor neuron degeneration in amyotrophic lateral sclerosis. Cell Rep. 26, 1143–1156.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Diehl J. A., Fuchs S. Y., Koumenis C., The cell biology of the unfolded protein response. Gastroenterology 141, 38–41, 41.e1–41.e2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hetz C., Papa F. R., The unfolded protein response and cell fate control. Mol. Cell 69, 169–181 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Kikuchi D., Tanimoto K., Nakayama K., CREB is activated by ER stress and modulates the unfolded protein response by regulating the expression of IRE1α and PERK. Biochem. Biophys. Res. Commun. 469, 243–250 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Inoue Y., et al. , The CDK inhibitor p21 is a novel target gene of ATF4 and contributes to cell survival under ER stress. FEBS Lett. 591, 3682–3691 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Tameire F., Verginadis I. I., Koumenis C., Cell intrinsic and extrinsic activators of the unfolded protein response in cancer: Mechanisms and targets for therapy. Semin. Cancer Biol. 33, 3–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang M., Kaufman R. J., The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer 14, 581–597 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Manié S. N., Lebeau J., Chevet E., Cellular mechanisms of endoplasmic reticulum stress signaling in health and disease. 3. Orchestrating the unfolded protein response in oncogenesis: An update. Am. J. Physiol. Cell Physiol. 307, C901–C907 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Fribley A., Zhang K., Kaufman R. J., Regulation of apoptosis by the unfolded protein response. Methods Mol. Biol. 559, 191–204 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Havens M. A., Hastings M. L., Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 44, 6549–6563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawamura S., Kaibuchi K., Hiroyoshi M., Hata Y., Takai Y., Stoichiometric interaction of smg p21 with its GDP/GTP exchange protein and its novel action to regulate the translocation of smg p21 between membrane and cytoplasm. Biochem. Biophys. Res. Commun. 174, 1095–1102 (1991). [DOI] [PubMed] [Google Scholar]
- 47.Kawamura M., Kaibuchi K., Kishi K., Takai Y., Translocation of Ki-ras p21 between membrane and cytoplasm by smg GDS. Biochem. Biophys. Res. Commun. 190, 832–841 (1993). [DOI] [PubMed] [Google Scholar]
- 48.Nakanishi H., Kaibuchi K., Orita S., Ueno N., Takai Y., Different functions of Smg GDP dissociation stimulator and mammalian counterpart of yeast Cdc25. J. Biol. Chem. 269, 15085–15091 (1994). [PubMed] [Google Scholar]
- 49.Bhagatji P., Leventis R., Rich R., Lin C. J., Silvius J. R., Multiple cellular proteins modulate the dynamics of K-ras association with the plasma membrane. Biophys. J. 99, 3327–3335 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dharmaiah S., et al. , Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc. Natl. Acad. Sci. U.S.A. 113, E6766–E6775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Figueroa C., Taylor J., Vojtek A. B., Prenylated Rab acceptor protein is a receptor for prenylated small GTPases. J. Biol. Chem. 276, 28219–28225 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Zhou M., et al. , VPS35 binds farnesylated N-Ras in the cytosol to regulate N-Ras trafficking. J. Cell Biol. 214, 445–458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Mata R., Boulter E., Burridge K., The ‘invisible hand’: Regulation of RHO GTPases by RHOGDIs. Nat. Rev. Mol. Cell Biol. 12, 493–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Long S. B., Casey P. J., Beese L. S., Reaction path of protein farnesyltransferase at atomic resolution. Nature 419, 645–650 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Müller M. P., Goody R. S., Molecular control of Rab activity by GEFs, GAPs and GDI. Small GTPases 9, 5–21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shinde S. R., Maddika S., Post translational modifications of Rab GTPases. Small GTPases 9, 49–56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mörck C., et al. , Statins inhibit protein lipidation and induce the unfolded protein response in the non-sterol producing nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 106, 18285–18290 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J. C., Wu M. L., Huang K. C., Lin W. W., HMG-CoA reductase inhibitors activate the unfolded protein response and induce cytoprotective GRP78 expression. Cardiovasc. Res. 80, 138–150 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Mollazadeh H., et al. , The effect of statin therapy on endoplasmic reticulum stress. Pharmacol. Res. 137, 150–158 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Thurnher M., Nussbaumer O., Gruenbacher G., Novel aspects of mevalonate pathway inhibitors as antitumor agents. Clin. Cancer Res. 18, 3524–3531 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Tsvetanova N. G., The secretory pathway in control of endoplasmic reticulum homeostasis. Small GTPases 4, 28–33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bouchecareilh M., Marza E., Caruso M. E., Chevet E., Small GTPase signaling and the unfolded protein response. Methods Enzymol. 491, 343–360 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Evelyn C. R., et al. , Small-molecule inhibition of Rho/MKL/SRF transcription in prostate cancer cells: Modulation of cell cycle, ER stress, and metastasis gene networks. Microarrays (Basel) 5, e13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yaari-Stark S., et al. , Ras inhibits endoplasmic reticulum stress in human cancer cells with amplified Myc. Int. J. Cancer 126, 2268–2281 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Bright M. D., Clarke P. A., Workman P., Davies F. E., Oncogenic RAC1 and NRAS drive resistance to endoplasmic reticulum stress through MEK/ERK signalling. Cell. Signal. 44, 127–137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caruso M. E., et al. , GTPase-mediated regulation of the unfolded protein response in Caenorhabditis elegans is dependent on the AAA+ ATPase CDC-48. Mol. Cell. Biol. 28, 4261–4274 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blum R., et al. , Gene expression signature of human cancer cell lines treated with the Ras inhibitor salirasib (S-farnesylthiosalicylic acid). Cancer Res. 67, 3320–3328 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Darling N. J., Balmanno K., Cook S. J., ERK1/2 signalling protects against apoptosis following endoplasmic reticulum stress but cannot provide long-term protection against BAX/BAK-independent cell death. PLoS One 12, e0184907 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All additional data and information are included in SI Appendix as Materials and Methods, figures, additional figure legends, tables, datasets, and references.