Significance
Since their discovery more than 50 years ago, dinucleoside tetraphosphates have been found in all realms of life, at levels correlated with a variety of intriguing physiological changes. However, their molecular mechanism of action has largely remained an enigma. Here we present evidence that in bacteria they are incorporated as an alternative chain-initiating nucleotide during transcription by RNA polymerase, generating RNAs that bear a novel nucleoside tetraphosphate cap at the 5′ end. Their assimilation into nascent transcripts is remarkably efficient, up to nine times the efficiency of nucleoside triphosphates. These and other findings suggest that dinucleoside tetraphosphates influence bacterial physiology not by signaling via a hypothetical receptor, but rather by modifying cellular RNAs and impacting their function.
Keywords: diadenosine tetraphosphate, Ap4A, Ap3A, non-canonical initiating nucleotide, ApaH
Abstract
Stresses that increase the cellular concentration of dinucleoside tetraphosphates (Np4Ns) have recently been shown to impact RNA degradation by inducing nucleoside tetraphosphate (Np4) capping of bacterial transcripts. However, neither the mechanism by which such caps are acquired nor the function of Np4Ns in bacteria is known. Here we report that promoter sequence changes upstream of the site of transcription initiation similarly affect both the efficiency with which Escherichia coli RNA polymerase incorporates dinucleoside polyphosphates at the 5′ end of nascent transcripts in vitro and the percentage of transcripts that are Np4-capped in E. coli, clear evidence for Np4 cap acquisition by Np4N incorporation during transcription initiation in bacterial cells. E. coli RNA polymerase initiates transcription more efficiently with Np4As than with ATP, particularly when the coding strand nucleotide that immediately precedes the initiation site is a purine. Together, these findings indicate that Np4Ns function in bacteria as precursors to Np4 caps and that RNA polymerase has evolved a predilection for synthesizing capped RNA whenever such precursors are abundant.
The biological function of dinucleoside 5′, 5′′′-P1, P4-tetraphosphates (Np4Ns) has largely remained a mystery since they were first discovered more than 50 years ago (1, 2). Present in both eukaryotes and prokaryotes (3, 4), they have long been thought to function as second messengers despite a paucity of supporting evidence. Indeed, a pathway for signaling by Np4Ns has been clearly delineated in only one cell type (5).
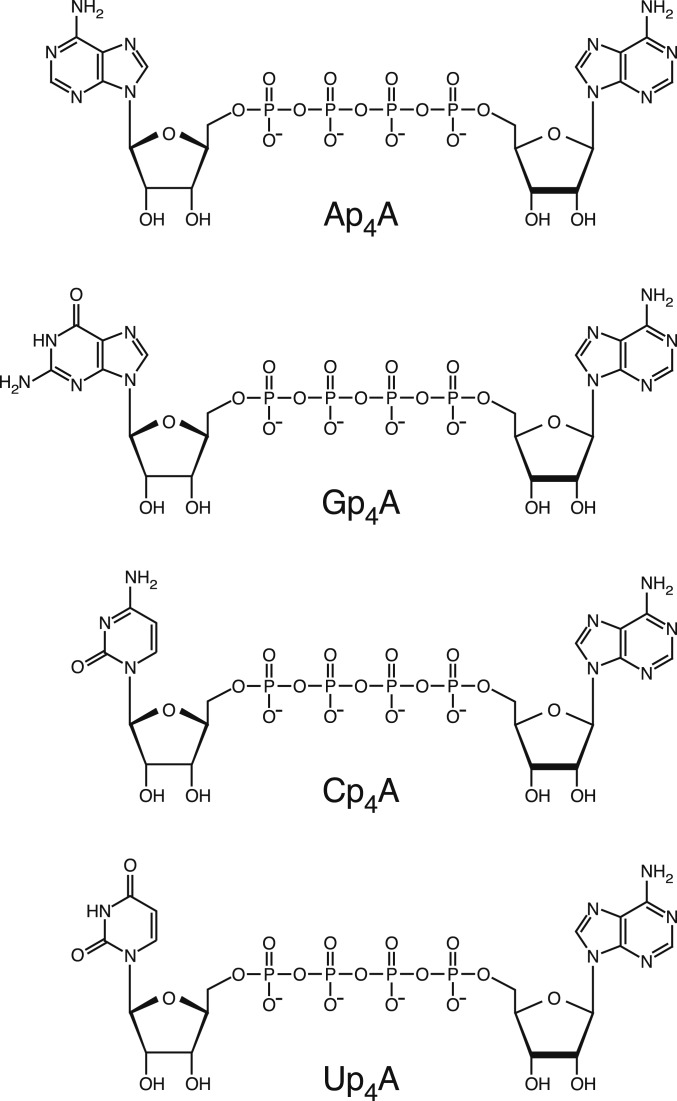
The predominant dinucleoside tetraphosphates in bacteria are Ap4A, Gp4A, Cp4A, and Up4A (Fig. 1) (6). Generated as byproducts of tRNA aminoacylation when the aminoacyl-adenylate intermediate reacts with a nucleoside triphosphate (2, 3), these dinucleoside tetraphosphates are particularly abundant in bacterial cells experiencing stress, especially disulfide stress induced by treatment with cysteine cross-linkers, such as cadmium or diamide (6–8). For this reason, Np4As have been deemed alarmones, that is, second messengers that signal stress (9). They are also present at elevated concentrations in mutant strains lacking ApaH, the principal proteobacterial Np4A hydrolase (10–13), whose absence results in a variety of intriguing phenotypes that have been attributed to signaling by Np4As. These mutant phenotypes include diminished motility, increased sensitivity to heat shock and oxidative stress, greater vulnerability to aminoglycosides, reduced antibiotic tolerance, diminished sporulation, impaired regulation of biofilm formation, disrupted coupling of DNA replication and cell division, and a decreased ability to invade mammalian cells (11, 14–20). However, no receptor that mediates the influence of Np4As has ever been identified in bacteria (13).
Fig. 1.
Major dinucleoside tetraphosphates in E. coli. The structures of the dinucleoside tetraphosphates Ap4A, Gp4A, Cp4A, and Up4A are shown. Ap5A, Ap3A, and Gp3A are identical to their dinucleoside tetraphosphate counterparts except for the number of bridging phosphates.
The assumption that Np4As signal via a receptor has been challenged by the recent discovery that, under conditions that elevate cellular Np4A concentrations, diverse Escherichia coli RNAs acquire a cognate nucleoside tetraphosphate (Np4) cap, in which the cap nucleoside may be A, G, C, or U, among others (21). In wild-type cells subjected to disulfide stress or in unstressed ∆apaH mutants, such caps appear to be present at a high level on most primary transcripts whose 5′-terminal transcribed nucleotide is A, C, or U and at a lower level on transcripts that begin with G. By contrast, few if any Np4-capped transcripts are present in unstressed wild-type cells. These caps are removed in E. coli by either of two enzymes, ApaH or the RNA pyrophosphohydrolase RppH, thereby triggering rapid RNA degradation (21). Of the two, ApaH appears to play a greater role in decapping, and its inactivation by inducers of disulfide stress helps to explain the elevated concentration and prolonged lifetime of capped RNAs in cells subjected to this stress. The importance of ApaH for cap removal and subsequent RNA decay raises the possibility that the various phenotypes observed for mutants lacking this enzyme might result not from receptor-mediated signaling by Np4As, but rather from altered RNA lifetimes and consequent changes in gene expression.
The mechanism by which Np4 caps are acquired by bacterial transcripts is unknown. In principle, they could be acquired at the moment of RNA synthesis if RNA polymerase initiates transcription with a dinucleoside tetraphosphate in place of a nucleoside triphosphate. They could also be added posttranscriptionally, either by aminoacyl-tRNA synthetases if the aminoacyl-adenylate intermediate reacts with the triphosphorylated 5′ end of cellular transcripts, or by some other enzyme. The feasibility of each of these capping mechanisms has been demonstrated in vitro with purified E. coli RNA polymerase and E. coli lysyl-tRNA synthetase (21, 22), but their relevance to Np4 cap acquisition in vivo has never been investigated.
We have now tested whether Np4 caps are acquired during transcription in E. coli. Our data show that the promoter sequence upstream of the site of transcription initiation influences both Np4A incorporation into nascent transcripts in vitro and levels of Np4 capping in vivo. In fact, E. coli RNA polymerase prefers Np4As over ATP as the chain-initiating nucleotide. These findings indicate that RNA polymerase makes a major contribution to Np4 capping in E. coli and that Np4As function as precursors to Np4 caps in bacterial cells.
Results
Effect of the Promoter Sequence on Cap Acquisition in E. coli.
Np4-capped transcripts are abundant in E. coli cells that are experiencing disulfide stress or that lack the principal Np4-decapping enzyme ApaH. By contrast, few Np3- or NAD-capped RNAs and almost no Np5-capped transcripts are present under those conditions (21, 23).
The origin of Np4 caps in bacterial cells is unknown. Theoretically, they might result from the incorporation of Np4As at the 5′ end of nascent transcripts during initiation of transcription by RNA polymerase. Alternatively, such caps might be added posttranscriptionally by aminoacyl-tRNA synthetases or other enzymes. To test the former possibility, we investigated whether mutating the promoter sequence upstream of the site of transcription initiation affects the percentage of transcripts that are Np4-capped in E. coli. We reasoned that sequence changes there might differentially influence the ease with which RNA polymerase can initiate transcription with a dinucleoside tetraphosphate vs. a nucleoside triphosphate by altering the structure of the open promoter complex to which the Np4A binds, but that such changes should not affect the ability of other enzymes to cap or decap the RNA 5′ end posttranscriptionally.
As a model system for these studies, we chose the yeiP transcript, which encodes a paralog of translation elongation factor EF-P and is among the most highly capped RNAs in E. coli under inducing conditions (76% capped in cadmium-treated cells and 36% capped in ∆apaH cells) (21). To determine whether the promoter affects levels of Np4 capping in vivo, we altered the DNA sequence at positions −3, −2, and −1 upstream of the transcription initiation site (position +1) of a plasmid-borne yeiP gene, introduced the resulting plasmid variants into an E. coli strain from which both the apaH gene and the chromosomal yeiP gene had been deleted, and extracted RNA during unstressed exponential growth. Capped and uncapped cellular transcripts were separated from one another by site-specific yeiP cleavage with a 10-23 deoxyribozyme (24), followed by electrophoresis through a polyacrylamide gel matrix modified with boronate side chains, which selectively retard the migration of capped RNAs by transiently forming a covalent adduct with the vicinal diol of the cap nucleoside (25). In this manner, it was possible to resolve bands representing the capped and uncapped yeiP transcripts by Northern blot analysis and to accurately determine the percentage of these transcripts that were Np4-capped (21, 26).
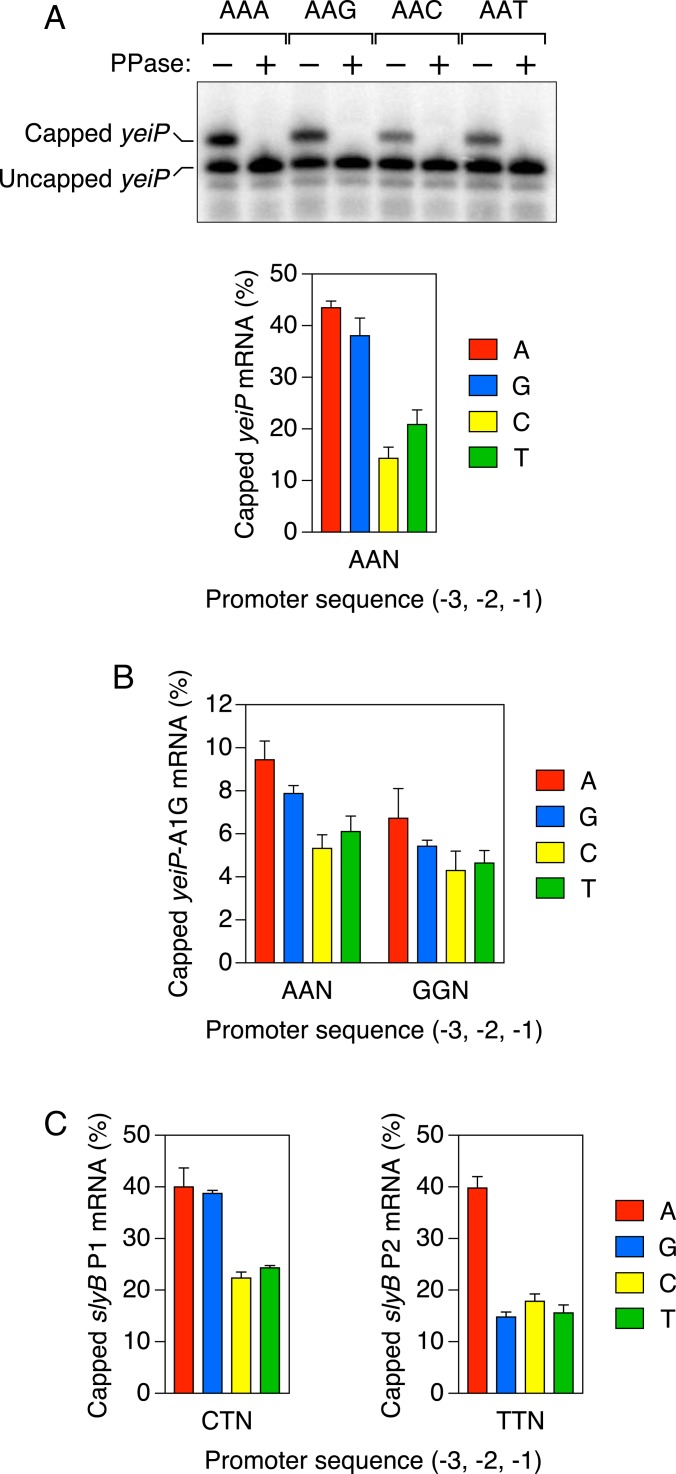
Altering this region of the promoter had a marked effect on the percentage of cellular yeiP transcripts that were Np4-capped in E. coli. Replacing the sequence at positions −3, −2, and −1 (GCG on the coding strand of the wild-type gene) with AAN, where N = A, G, C, or T, resulted in yeiP transcripts that were anywhere from 14% to 44% capped in cells lacking ApaH, depending on the identity of the base pair at position −1 (Fig. 2A). Levels of capping were two to three times greater when the coding strand nucleotide there was a purine (AAA or AAG) rather than a pyrimidine (AAC or AAT). None of these substitutions altered the site of transcription initiation, as determined by RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) analysis of the capped 5′ termini (SI Appendix, Fig. S1), or led to detectable levels of NAD-capped yeiP mRNA, which would have been recognizable by its distinct electrophoretic mobility (21). As expected, the low-mobility band representing Np4-capped yeiP mRNA disappeared when the RNA samples were decapped in vitro before electrophoresis (Fig. 2A). The substantial impact of the promoter nucleotide at position −1 on levels of Np4 capping indicates that such caps are acquired in large part during transcription initiation in E. coli, presumably by a mechanism involving the incorporation of Np4As at the 5′ end of nascent transcripts.
Fig. 2.
Promoter-dependent capping of mRNA in E. coli. (A) Percentage of yeiP transcripts that are Np4-capped in unstressed ∆apaH cells. (Top) Northern blot. Total RNA was extracted from unstressed ∆apaH cells that lacked the chromosomal yeiP gene and instead contained a plasmid-borne yeiP gene in which the sequence of the coding strand at positions −3, −2, and −1 of the promoter had been changed to AAN, where N = A, G, C, or T. The RNA was cleaved site-specifically 69 nt from the yeiP 5′ end with a 10-23 deoxyribozyme (24) so as to generate a 5′-terminal fragment bearing a 2′,3′ cyclic phosphate rather than a vicinal diol at its 3′ end, and samples containing similar amounts of uncapped yeiP RNA were then subjected to electrophoresis on a boronate gel, blotted, and probed with radiolabeled yeiP DNA (26). Equivalent RNA samples that had been decapped in vitro with RppH (PPase) were analyzed in parallel. (Bottom) Graph. For each promoter, the percentage of yeiP transcripts that were capped was calculated from the mean of four or five biological replicates. Error bars represent 1 SD. P ≤ 0.001 for each AAPu vs. AAPy comparison, where Pu = A or G and Py = C or T. (B) Percentage of yeiP-A1G transcripts that are Np4-capped in unstressed ∆apaH cells. Each value corresponds to the mean of five biological replicates for yeiP-A1G mRNA synthesized by transcription from promoter variants in which the sequence of the coding strand at positions −3, −2, and −1 had been changed to AAN or GGN, where N = A, G, C, or T. Error bars represent 1 SD. P ≤ 0.001 for each AAPu vs. AAPy comparison, where Pu = A or G and Py = C or T; P ≤ 0.025 for each GGPu vs. GGPy comparison; P ≤ 0.05 for each cognate AAN vs. GGN comparison. (C) Percentage of slyB P1 and slyB P2 transcripts that are Np4-capped in unstressed ∆apaH cells. Each value represents the mean of five biological replicates for slyB P1 mRNA (Left) or slyB P2 mRNA (Right) synthesized by transcription from promoter variants in which the sequence of the coding strand at position −1 had been changed to A, G, C, or T. Error bars represent 1 SD. The value for slyB P1 bearing an A at position −1 represents a lower limit on the actual percentage of capped transcripts, as this mutation adventitiously created a third slyB promoter whose capped transcript comigrated with the uncapped form of slyB P1 mRNA. P ≤ 0.001 for each CTPu vs. CTPy comparison and each TTA vs. TTPy comparison, where Pu = A or G and Py = C or T.
Because the yeiP transcript begins with A at the 5′ terminus, E. coli RNA polymerase could potentially synthesize this mRNA in vivo with a mixture of Np4 caps by initiating transcription with either Ap4A, Gp4A, Cp4A, or Up4A instead of ATP. To constrain the cap to a single identity (Ap4), we introduced an A-to-G substitution at the site of yeiP transcription initiation (yeiP-A1G) so as to force a two-way competition between Ap4G and GTP as the initiating nucleotide. (Note that Ap4G and Gp4A are synonymous due to molecular symmetry.) As reported previously (21), this substitution greatly diminished the steady-state percentage of these transcripts that were capped (Fig. 2B). Nevertheless, the effect of the promoter nucleotide at position –1 persisted in this context, with cap levels for yeiP-A1G mRNA ranging from 6.1% to 9.5%, depending on the base pair there. Once again, a purine at that position of the coding strand resulted in a higher percentage of capped mRNA than a pyrimidine there.
We also varied the identity of that nucleotide after changing the adjacent DNA sequence at positions −3 and −2 (GGN instead of AAN). In each case, the percentage of capped yeiP-A1G transcripts was modestly reduced (by 0.2- to 0.3- fold), but the differential effect of the nucleotide at position −1 remained, albeit somewhat diminished in magnitude (Fig. 2B).
To further test the generality of these findings, we examined the influence of the nucleotide at position –1 on the capping of two other E. coli transcripts, slyB P1 and slyB P2, which begin with A and encode the same outer membrane lipoprotein (27) but are synthesized from distinct promoters spaced 25 base pairs apart. Those promoters differ in sequence from one another and from the yeiP promoters that we analyzed (SI Appendix, Table S1), including at positions −3 and −2 (CT or TT, respectively, instead of AA or GG). Nevertheless, the differential effect of a purine or pyrimidine at position −1 of the two slyB promoters in cells lacking ApaH resembled that observed for yeiP and yeiP-A1G, except for the relatively low percentage of slyB P2 transcripts that bore a cap when the coding strand nucleotide at that position was G (Fig. 2C).
Taken together, these findings suggest that Np4 capping of E. coli transcripts occurs in large measure through Np4A incorporation during transcription initiation, and that the efficiency of this process is governed, at least in part, by the untranscribed region encompassing positions −3, −2, and −1 of the promoter. This mechanism of cap acquisition appears to be effective on transcripts with diverse promoter sequences and 5′ termini.
Capped Nucleotide Incorporation by E. coli RNA Polymerase In Vitro.
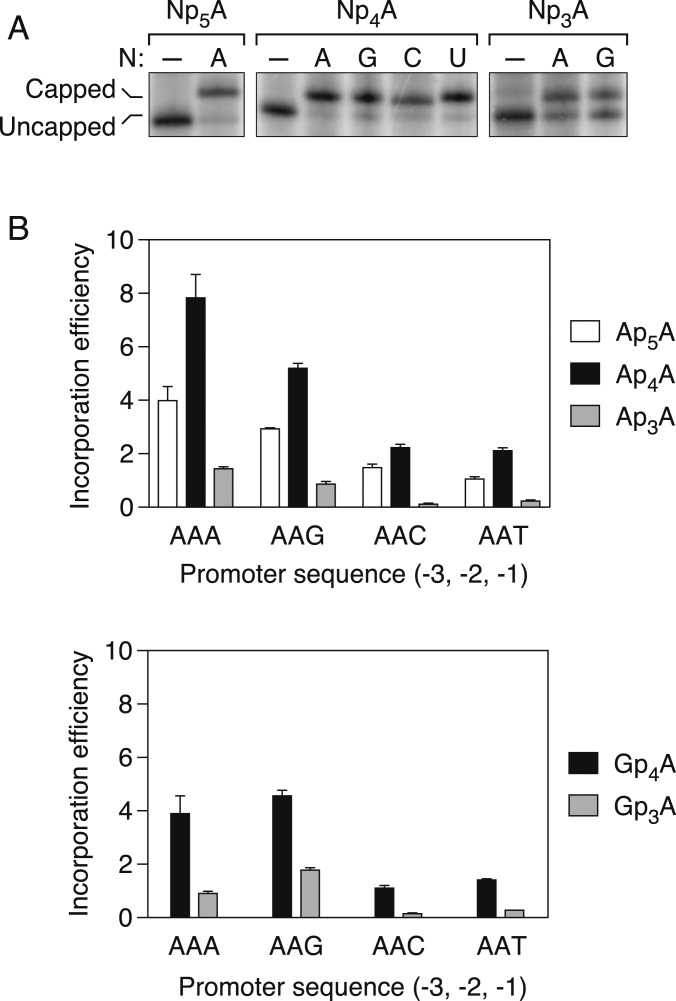
The evidence that the base pair at position −1 of the DNA template influences the efficiency of Np4A assimilation at the 5′ end of nascent transcripts in E. coli raised the possibility that the template nucleotide there might allow RNA polymerase to discriminate among distinct dinucleoside tetraphosphates during transcription initiation. To investigate this possibility, we determined the relative incorporation efficiency of several dinucleoside polyphosphates (NpnA, where N = A, G, C, or U and n = 3, 4, or 5) during transcription initiation by purified E. coli RNA polymerase σ70 holoenzyme on a set of yeiP-like DNA templates that each bore an A at position +1 of the coding strand and differed in sequence only at the preceding promoter nucleotide (AAN at positions −3, −2, and −1 of the coding strand, where N = A, G, C, or T). In each case, a specific dinucleoside polyphosphate was mixed with ATP at a defined molar ratio, and the two were allowed to compete with one another as the initiating nucleotide during the synthesis of a 122-nt transcript. The ratio of capped to uncapped transcripts was then determined by boronate gel electrophoresis.
At an initial NpnA:ATP ratio of 1:1, it was immediately apparent that the various NpnAs are assimilated with disparate efficiencies at the 5′ end of nascent transcripts and that, unexpectedly, most are incorporated more readily than ATP (Fig. 3A). Therefore, the NpnA:ATP ratio was decreased to 0.5:1 for Ap5A and the four Np4As and increased to 2:1 for the two Np3As so that the ratio of capped to uncapped transcription products could be measured more accurately. The incorporation efficiency of each NpnA on each DNA template was then calculated by normalizing the product ratio (capped transcripts/uncapped transcripts) to the substrate ratio (NpnA/ATP).
Fig. 3.
Effect of the number of bridging phosphates on the efficiency of dinucleoside polyphosphate incorporation by E. coli RNA polymerase. (A) Incorporation of dinucleoside polyphosphates during transcription initiation in vitro. RNA was synthesized by purified E. coli RNA polymerase holoenzyme in the presence of an equimolar mixture of each dinucleoside polyphosphate (NpnA) and ATP. The radiolabeled products were then cleaved site-specifically with a 10-23 deoxyribozyme (24) and analyzed by boronate gel electrophoresis and autoradiography (26). The promoter sequence at positions −3, −2, and −1 of the coding strand was AAA, followed by A at position +1 (the first transcribed nucleotide). As a negative control, transcription was also performed in the absence of any NpnA (–). (B) Efficiency of ApnA (Top) or GpnA (Bottom) incorporation during transcription initiation in vitro. RNA was synthesized by purified E. coli RNA polymerase holoenzyme in the presence of a mixture of each dinucleoside polyphosphate and ATP. The promoter sequence at positions −3, −2, and −1 of the coding strand was AAN, where N = A, G, C, or T. In each case, the product ratio (capped transcripts/uncapped transcripts) was normalized to the substrate ratio (NpnA/ATP: 0.50 for Ap5A, Ap4A, and Gp4A and 2.00 for Ap3A and Gp3A). Each value corresponds to the mean of three technical replicates. Error bars represent 1 SD. P ≤ 0.003 for each ApnA comparison and each GpnA comparison involving the same promoter but a different number of phosphates.
The 5′-terminal incorporation efficiency of Ap4A relative to ATP was remarkable: as high as 8 on the AAA template and no less than 2 on any template (Fig. 3B). Irrespective of the DNA template, Ap4A was assimilated up to twice as efficiently as Ap5A and 5 to 16 times more efficiently than Ap3A. Similarly, Gp4A was assimilated three to seven times more efficiently than Gp3A (Fig. 3B). Thus, four bridging phosphates (n = 4) appear to be optimal for NpnA incorporation, regardless of the promoter sequence.
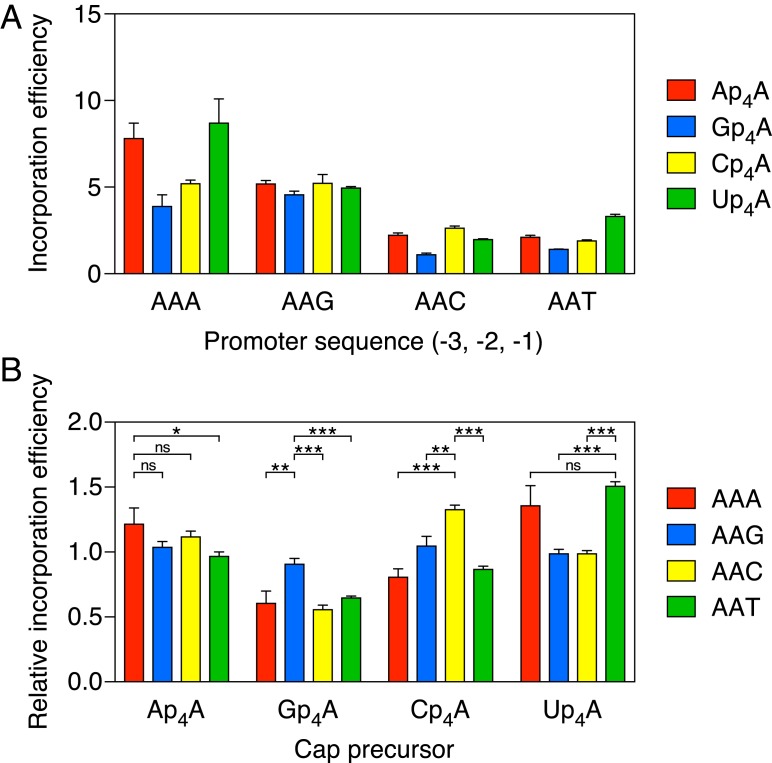
Even among cap precursors with the same number of bridging phosphates, incorporation efficiencies differed substantially, depending on the identity of both the cap nucleoside and the nucleotide at position −1 of the DNA template. Thus, the efficiency of assimilating each of the four dinucleoside tetraphosphates Ap4A, Gp4A, Cp4A, and Up4A varied considerably on the four AAN promoter templates (Fig. 4A). In each case, Np4A incorporation was most efficient (by as much as four-fold) when the nucleotide at position −1 of the coding strand was a purine (AAA or AAG), a finding in accordance with the influence of this promoter nucleotide on yeiP capping in vivo. Similar results were obtained at an Np4A:ATP ratio of 0.25:1 (SI Appendix, Fig. S2).
Fig. 4.
Comparative efficiency of Ap4A, Gp4A, Cp4A, and Up4A incorporation by E. coli RNA polymerase. (A) Absolute efficiency of Np4A incorporation. RNA was synthesized in vitro by E. coli RNA polymerase holoenzyme in the presence of a mixture of each dinucleoside tetraphosphate and ATP. The promoter sequence at positions −3, −2, and −1 of the coding strand was AAN, where N = A, G, C, or T. In each case, the product ratio (capped transcripts/uncapped transcripts) was determined by boronate gel electrophoresis and normalized to the substrate ratio (Np4A/ATP = 0.50). Each value corresponds to the mean of three technical replicates. Error bars represent 1 SD. P ≤ 0.003 for each AAPu vs. AAPy comparison involving the same Np4A, where Pu = A or G and Py = C or T. (B) Relative efficiency of Np4A incorporation. For each promoter in A, the efficiencies of Ap4A, Gp4A, Cp4A, and Up4A incorporation were normalized to the mean efficiency of all four with the same promoter. The relative efficiencies calculated in this manner can range from 0 to 4, with a relative efficiency of 1 being average for that promoter. Each value was calculated by using data from three technical replicates for each of the four Np4As and a particular promoter. Error bars correspond to a confidence level of 68.3%, equivalent to 1 SD. ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05. ns, not significant (P > 0.05).
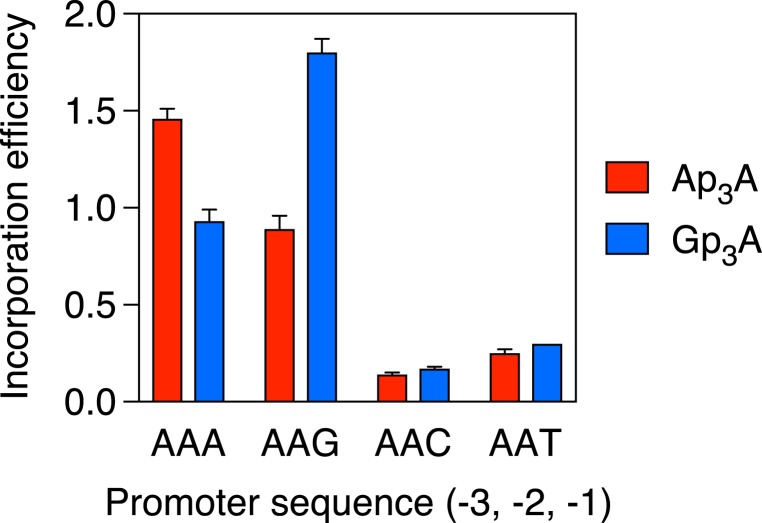
To meaningfully compare the effect of the nucleotide at position –1 on the ability of each Np4A to outcompete its three counterparts during transcription initiation, it was necessary to correct for differences in the overall Np4A assimilation efficiency intrinsic to each promoter sequence. This was accomplished by replotting the data after normalizing the efficiencies of Ap4A, Gp4A, Cp4A, and Up4A incorporation at each individual promoter to the mean incorporation efficiency of all four Np4As at that promoter (Fig. 4B). These calculations revealed that, relative to the average incorporation efficiency of all four Np4As on each AAN template, Gp4A had its greatest competitive advantage when N = G, Cp4A was most competitive when N = C, and Up4A was most competitive when N = T or A (Fig. 4B). (By this measure of competitive advantage, the preference of Ap4A for A at position −1 was not statistically significant.) Although these differences were small (no more than 1.4-fold) for the four Np4As, they were more clearly evident when the same AAN promoter templates were used to compare the assimilation of dinucleoside triphosphates, as Ap3A was significantly favored when N = A, Gp3A was significantly favored when N = G, and neither was incorporated well when N was a pyrimidine (Fig. 5).
Fig. 5.
Comparative efficiency of Ap3A and Gp3A incorporation by E. coli RNA polymerase. RNA was synthesized in vitro by E. coli RNA polymerase holoenzyme in the presence of a mixture of each dinucleoside triphosphate and ATP. The promoter sequence at positions −3, −2, and −1 of the coding strand was AAN, where N = A, G, C, or T. In each case, the product ratio (capped transcripts/uncapped transcripts) was determined by boronate gel electrophoresis and normalized to the substrate ratio (Np3A/ATP = 2.00). Each value corresponds to the mean of three technical replicates. Error bars represent 1 SD. P ≤ 0.001 for each Ap3A vs. Gp3A comparison involving the same AAPu promoter and for each AAA vs. AAG and AAPu vs. AAPy comparison involving the same Np3A, where Pu = A or G and Py = C or T.
Taken together, these findings show that E. coli RNA polymerase can assimilate any Np4A or Np3A at the 5′ end of a nascent A-initiated transcript, and that it does so with a particularly high efficiency when the nucleotide at position −1 of the DNA coding strand is a purine. Only 26% of E. coli mRNAs are synthesized from promoters that have a purine there, a property shared by a similar percentage of the many transcripts whose cellular concentration is significantly affected by cadmium-induced disulfide stress (28, 29). Moreover, most dinucleoside polyphosphates are best able to compete with their average counterpart when the incipient cap has the potential to form a Watson–Crick base pair with the nucleotide at position −1 of the template strand, although this preference is substantial only for Np3As.
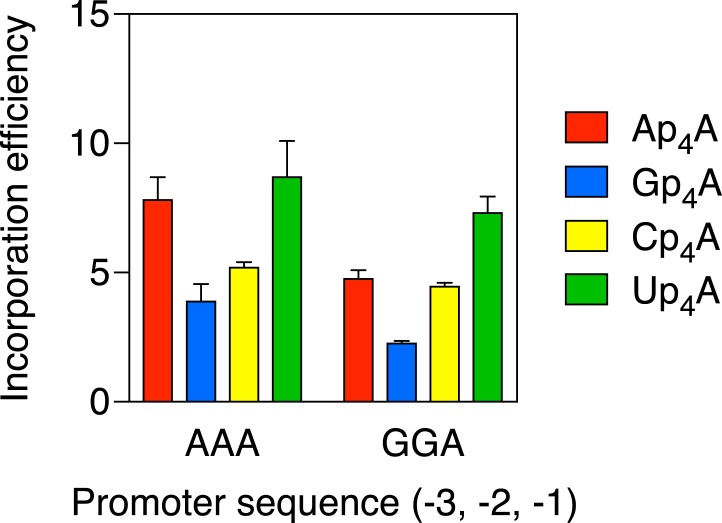
The incorporation efficiencies of Ap4A, Gp4A, Cp4A, and Up4A were also compared on a DNA template (GGA) that differed from the AAA template at positions −3 and −2. This promoter modification diminished the assimilation of each of the Np4As in vitro, especially Ap4A and Gp4A (Fig. 6), an outcome reminiscent of the effect of the same sequence change on capping in vivo (Fig. 2B). These findings confirm that the promoter sequence at positions −3 and −2 modestly affects the ease with which Np4As are incorporated during transcription initiation, particularly when the incipient cap nucleobase is a purine.
Fig. 6.
Effect of the promoter sequence at positions −3 and −2 on the efficiency of Ap4A, Gp4A, Cp4A, and Up4A incorporation by E. coli RNA polymerase. RNA was synthesized in vitro by E. coli RNA polymerase holoenzyme in the presence of a mixture of each dinucleoside tetraphosphate and ATP. The promoter sequence at positions −3, −2, and −1 of the coding strand was AAA or GGA. In each case, the product ratio (capped transcripts/uncapped transcripts) was determined by boronate gel electrophoresis and normalized to the substrate ratio (Np4A/ATP = 0.50). Each value corresponds to the mean of three technical replicates. Error bars represent 1 SD. P ≤ 0.01 for each AAA vs. GGA comparison involving the same Np4A, except Up4A (P = 0.18).
Discussion
An understanding of the cellular functions of Np4As has remained elusive for decades, especially in bacteria. Recently, Np4-capped transcripts were reported to become abundant in E. coli under conditions that cause Np4A levels to rise (21). This correlation could mean either that Np4As serve as cap precursors which at elevated concentrations become incorporated into RNA during transcription initiation by RNA polymerase, or that conditions conducive to Np4A synthesis also enable capping of preexisting triphosphorylated transcripts (e.g., by potentiating their reaction with the aminoacyl-adenylate intermediate of aminoacyl-tRNA synthetases). Previous experiments showing that E. coli RNA polymerase and lysyl-tRNA synthetase can be used to synthesize Np4-capped RNA in vitro (21, 22) demonstrated the feasibility of each of these possible capping mechanisms but did not reveal which, if either, is key for cap acquisition in vivo.
Our evidence that cellular levels of Np4-capped RNA are influenced by the promoter sequence upstream of the site of transcription initiation, and that similar sequence effects are observed during in vitro transcription with purified E. coli RNA polymerase, indicates that a substantial percentage (perhaps the overwhelming majority) of Np4 caps in E. coli are acquired during transcription initiation by a mechanism involving Np4A assimilation at the 5′ end of nascent transcripts by RNA polymerase. As a corollary, we conclude that Np4As function in bacterial cells as precursors to RNA caps. To our knowledge, this is the first biological function that has been empirically substantiated for this enigmatic family of molecules in bacteria, as there is still no evidence that their function in such organisms involves signal transduction via a hypothetical receptor, as was initially assumed (13).
Ap4A, Gp4A, Cp4A, and Up4A are all abundant in E. coli under conditions of disulfide stress (6, 8) or in the absence of ApaH, the principal Np4A hydrolase (10–13, 30). In wild-type cells treated with cadmium or in mutant cells lacking ApaH, the concentration of each Np4A increases by two to three orders of magnitude, approaching that of the corresponding NTP (6, 13). Therefore, Np4A incorporation during transcription initiation by RNA polymerase should result in diverse Np4 cap nucleosides at the 5′ end of A-initiated transcripts and Ap4 caps at the 5′ end of G-, C-, and U-initiated transcripts due to the molecular symmetry of Np4As (Np4A = Ap4N). Consistent with these expectations, Np4 caps have been observed in E. coli on transcripts that begin with A, G, C, or U, and Ap4, Gp4, Cp4, and (probably) Up4 caps have all been detected on E. coli RNA (21).
The finding that RNA polymerase plays a major role in Np4 cap acquisition in E. coli does not preclude a supplementary role for aminoacyl-tRNA synthetases or other cellular enzymes in capping preexisting transcripts by nucleotidyl transfer to the 5′ end. However, the contribution of any such posttranscriptional mechanism is likely to be relatively small in view of the quantitative similarity of the effects of promoter sequence changes in vitro and in vivo. Consonant with this interpretation is the low ratio of Np3-capped to Np4-capped yeiP mRNAs in E. coli (21) despite the prevalence of diphosphorylated over triphosphorylated yeiP 5′ ends (31) as potential substrates for posttranscriptional capping. Furthermore, because aminoacyl-tRNA synthetases would be capable of adding only adenylate caps to RNA, they alone could not explain the diversity of cap nucleotides on E. coli transcripts. Posttranscriptional capping by the nucleotidyltransferases RtcA and RtcB is precluded by the failure of inactivating mutations in both enzymes to affect levels of Np4 capping in E. coli (SI Appendix, Fig. S3).
The efficiency with which E. coli RNA polymerase initiates transcription with Np4As is extraordinary, as much as nine times the efficiency with which it initiates transcription with ATP. The strong preference of this enzyme for Np4As implies a favorable interaction of the incipient cap nucleotide with the active site of RNA polymerase and/or with the DNA template, although it is not yet clear which step in transcription initiation is facilitated. By contrast, the efficiency of Np3A incorporation into nascent transcripts, while still impressive, is similar to or less than that of ATP.
Irrespective of the cap nucleobase, an A or G at position −1 of the coding strand generally results in more efficient Np4A assimilation than a C or T there, both in vitro and in vivo. Thus, a pyrimidine at this position of the template strand creates a better environment for Np4A incorporation than does a purine, regardless of the potential for Watson–Crick base pairing with the incipient cap nucleotide. Nevertheless, compared to the average incorporation efficiency of all four dinucleoside tetraphosphates with each promoter, Np4As are usually most competitive, if sometimes only marginally so, when the incipient cap nucleobase (N) has the potential to form a Watson–Crick base pair with the nucleotide at position −1 of the template strand. Similar results were obtained for Np3A assimilation, except that the impediment to cap incorporation caused by a purine at position −1 of the template strand was greater and the competitive advantage conferred by the potential for Watson–Crick base pairing by the incipient cap nucleobase (A or G) with the template nucleotide there (T or C, respectively) was more pronounced.
These findings suggest two productive modes of dinucleoside polyphosphate binding in the active site of the RNA polymerase holoenzyme, one that is aided by a pyrimidine at position −1 of the template strand (perhaps to avoid steric hindrance or to exploit differential stacking energies) and that does not involve Watson–Crick base pairing by the incipient cap nucleobase with that template nucleotide and another in which the incipient cap forms a Watson–Crick base pair with the template nucleotide there at the same time that the other nucleobase of the dinucleoside polyphosphate base pairs with the template nucleotide at position +1. The former mode of binding may be very favorable for Np4As and therefore predominate for them, helping to explain the high efficiency with which they are assimilated, irrespective of cap complementarity. However, that binding mode may be less favorable for Np3As, thereby accounting for their lower incorporation efficiency and greater reliance on Watson–Crick base pairing. The possibility of simultaneous base pairing by both NpnA nucleosides to adjacent nucleotides on the DNA template despite the 5′-5′ dinucleoside linkage and multiple bridging phosphates is consistent with evidence that both Ap3A and Ap4A free in solution adopt a conformation in which the two nucleobases stack on one another (32).
The promoter sequence at position(s) −3 and/or −2 also influences the efficiency of Np4A incorporation by RNA polymerase (AA > GG), both in vitro and in vivo, but unlike position −1, those positions have a comparatively minor effect on discrimination among Ap4A, Gp4A, Cp4A, and Up4A. Consequently, it seems unlikely that these two upstream nucleotides of the DNA template directly contact the incipient cap nucleobase during transcription initiation. The possible effect of the promoter sequence even further upstream of the transcription initiation site was not investigated.
Previous studies have shown that E. coli RNA polymerase can also incorporate NAD at the 5′ end of nascent transcripts (33–35), although unlike Np4As, NAD can be assimilated only into RNAs whose first transcribed nucleotide is A (34, 36). Like Np4As, the reactivity of NAD relative to ATP is stimulated up to fourfold by the presence of a purine at position −1 of the coding strand and is influenced to a lesser degree by the promoter sequence at positions −3 and −2 (36), evidence that these effects are not specific to a particular type of cap. However, even when an optimal DNA sequence surrounds the transcription initiation site, NAD is assimilated just one-seventh as well as ATP, helping to explain the low abundance of NAD caps in vivo. Thus, E. coli RNA polymerase incorporates Np4As up to 60 times more efficiently than it incorporates NAD. The lower reactivity of NAD may result from the presence of only two bridging phosphates between its component nucleosides. Consistent with this interpretation, E. coli RNA polymerase incorporates ADP just one-fifth as well as ATP at the 5′ terminus of nascent transcripts (31).
The ability of ApaH not only to hydrolyze the dinucleoside tetraphosphate precursors of Np4 caps but also to decap Np4-capped transcripts once they have been made (10, 21) should amplify the influence of this enzyme on the concentration of Np4-capped RNA in E. coli by enabling it to both reduce the synthesis and accelerate the degradation of capped transcripts. Together with the efficiency of Np4A incorporation by RNA polymerase, this dual role helps to explain why Np4-capped RNA is so abundant in ∆apaH cells despite being undetectable in unstressed wild-type cells (21), as it should enable a 300-fold increase in the cellular concentration of Ap4A (13) to be accompanied by a much larger increase in the steady-state level of Np4-capped RNA. Conversely, the lack of detectable Np4-capped RNA in ∆rppH cells (21) reinforces the view that the role of RppH as an Np4A hydrolase and decapping enzyme is much smaller than that of ApaH in the absence of stress.
The extraordinary efficiency of Np4A assimilation during transcription initiation indicates that RNA polymerase has evolved a predilection for synthesizing Np4-capped RNA whenever the cellular concentration of such precursors rivals that of NTPs. This property ensures the production of an abundance of Np4-capped transcripts under conditions that cause cellular Np4A levels to rise substantially, such as when ApaH is inactivated by inducers of disulfide stress. The ancient origin of aminoacyl-tRNA synthetases able to generate Np4As (37) and the remarkable ease with which RNA polymerase incorporates these dinucleotides at the 5′ end of nascent transcripts suggest that Np4 capping by RNA polymerase may well have predated the evolution of mechanisms for the posttranscriptional addition of m7Gppp caps in eukaryotic cells.
Materials and Methods
Analysis of Capped RNA in E. coli.
Capping of the 5′ end of yeiP or yeiP-A1G mRNA was examined in a derivative of the E. coli K-12 strain BW25113 (38) that bore in-frame deletions of the apaH and yeiP coding regions (21) and had been transformed with a derivative of the medium-copy plasmid pYeiP1 (39) or pYeiP1-A1G (21) in which the three coding strand nucleotides immediately upstream of the yeiP transcription initiation site had been changed to AAN or GGN, where N is A, G, C, or T. Capping of the 5′ end of slyB P1 or slyB P2 mRNA was analyzed in a BW25113 derivative that had in-frame deletions of the apaH and slyB coding regions and contained a derivative of the low-copy plasmid pPM30 (40) bearing either the wild-type slyB gene (pSlyB1) or a slyB mutant in which the coding strand nucleotide immediately upstream of the slyB P1 or slyB P2 transcription initiation site had been changed to A, G, C, or T. Total cellular RNA was extracted from liquid cultures of these strains following growth to midlog phase at 37 °C in MOPS-glucose medium (31). Capped and uncapped yeiP, yeiP-A1G, slyB P1, and slyB P2 mRNAs were then detected by boronate gel electrophoresis and Northern blot analysis after site-specific RNA cleavage 69 to 128 nt from the 5′ end with a 10-23 deoxyribozyme (SI Appendix, Table S2), as described previously (21). The percentage of the transcript that was capped was calculated from the ratio of band intensities (capped vs. uncapped), and mean values and SDs were determined from four or five biological replicates (SI Appendix, Tables S3 to S5). To identify the first transcribed nucleotide of capped yeiP mRNA synthesized from each plasmid-borne promoter variant in E. coli, total RNA was treated with calf intestine alkaline phosphatase to removed exposed phosphates and then analyzed by RLM-RACE (21).
Cap Incorporation during In Vitro Transcription by E. coli RNA Polymerase.
The 231-bp DNA template for in vitro transcription by E. coli RNA polymerase comprised a σ70 promoter and transcribed region derived from the E. coli yeiP gene (CTTAAGGTTAACCACCGGATTGATGAGCGCGCACAGTACCACCTTTTTTGCACCAGCAAAAGTGCGAATACCACTTGACAGAAAGGCCCGTCGCGAGTACTTTGTCRRNATATTTTTAATATTTTATATTATTTCGACTACAGGAATTTTTCGATGCCAAGAGCGAACGAAATTAAAAAAGGTATGGTACTGAATTACAACGGCTCGTATCCATATCGAAGAACGCAAAAA, where RR is AA or GG and N is any nucleotide). The 122-bp transcribed region of the template (underscored) encoded the first 86 nt of yeiP mRNA (with a G→A substitution at position +9, a C→T substitution at position +11, and an ATATTATTT insertion at positions +17 to +25) fused to a 27-nt deoxyribozyme-binding site. To facilitate transcription, the −35 region of the yeiP promoter was changed from TTGCCC to TTGACA (21).
Purified E. coli RNA polymerase σ70 holoenzyme (1 pmol, kindly provided by Venu Kamarthapu) (41) was combined with the gel-purified DNA template (1 pmol) and incubated for 5 min at 37 °C in a solution (10 μL) containing Tris⋅HCl pH 8.0 (40 mM), NaCl (50 mM), and MgCl2 (10 mM). A solution (1 μL) containing ATP (10 mM), UTP (2.5 mM), and NpnA (0, 2.5, 5, 10, or 20 mM) was added, and nascent transcripts were allowed to form for 5 min at 37 °C. To radiolabel the RNA at position +26, [α-32P] CTP (4 μCi) was added, and the reaction mixture was left at room temperature for 5 min. Run-off transcription of the DNA template was then completed by adding a solution (2 μL) containing CTP, GTP, and UTP (333 μM each) and allowing RNA synthesis to continue for 5 min at room temperature. The reaction was quenched with EDTA (2 μL of a 120 mM solution), and the enzyme was heat-inactivated by incubation at 70 °C for 20 min (21). The 3′ vicinal diol of the transcript was removed by site-specific cleavage 106 nt from the 5′ end with the 10-23 deoxyribozyme DZyeiP573 (4 μL of a 100 μM solution; SI Appendix, Table S2) in the presence of MgCl2 (2 μL of a 220 mM solution) for 2 h at 37 °C. The cleavage reaction was quenched with a solution (25 μL) containing EDTA pH 8.0 (20 mM), formamide (95% vol/vol), and tracking dyes, and the radioactive products were subjected to boronate gel electrophoresis and detected with a Typhoon Trio imager (GE Healthcare).
The percentage of the transcript that was capped was calculated from the ratio of band intensities (capped vs. uncapped), and mean values and SDs were determined from three technical replicates. Efficiencies of NpnA incorporation were calculated by normalizing the product ratio (capped transcripts/uncapped transcripts) to the substrate ratio (NpnA/ATP) (SI Appendix, Tables S6–S8). In addition, relative efficiencies of Np4A incorporation for each promoter were calculated by normalizing the incorporation efficiency of each Np4A for that promoter to the mean incorporation efficiency of all four Np4As for the same promoter.
Data Availability.
All data are included in the figures and supporting information.
Supplementary Material
Acknowledgments
We thank Rose Levenson-Palmer for testing the rtcBA double mutant and Venu Kamarthapu and Evgeny Nudler for providing purified E. coli RNA polymerase. This research was supported by NIH Fellowship T32AI007180 (to D.J.L.), a fellowship from the Vilcek Endowment (to D.J.L.), and NIH Research Grants R01 GM035769 and R01 GM112940 (to J.G.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914229117/-/DCSupplemental.
References
- 1.Finamore F. J., Warner A. H., The occurrence of P1, P4-diguanosine 5′-tetraphosphate in brine shrimp eggs. J. Biol. Chem. 238, 344–348 (1963). [PubMed] [Google Scholar]
- 2.Zamecnik P. C., Stephenson M. L., Janeway C. M., Randerath K., Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 24, 91–97 (1966). [DOI] [PubMed] [Google Scholar]
- 3.Plateau P., Blanquet S., Dinucleoside oligophosphates in micro-organisms. Adv. Microb. Physiol. 36, 81–109 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Kisselev L. L., Justesen J., Wolfson A. D., Frolova L. Y., Diadenosine oligophosphates (Ap(n)A), a novel class of signalling molecules? FEBS Lett. 427, 157–163 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Lee Y. N., Nechushtan H., Figov N., Razin E., The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity 20, 145–151 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Coste H., Brevet A., Plateau P., Blanquet S., Non-adenylylated bis(5′-nucleosidyl) tetraphosphates occur in Saccharomyces cerevisiae and in Escherichia coli and accumulate upon temperature shift or exposure to cadmium. J. Biol. Chem. 262, 12096–12103 (1987). [PubMed] [Google Scholar]
- 7.Lee P. C., Bochner B. R., Ames B. N., AppppA, heat-shock stress, and cell oxidation. Proc. Natl. Acad. Sci. U.S.A. 80, 7496–7500 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochner B. R., Lee P. C., Wilson S. W., Cutler C. W., Ames B. N., AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 37, 225–232 (1984). [DOI] [PubMed] [Google Scholar]
- 9.Varshavsky A., Diadenosine 5′, 5"′-P1, P4-tetraphosphate: A pleiotropically acting alarmone? Cell 34, 711–712 (1983). [DOI] [PubMed] [Google Scholar]
- 10.Plateau P., Fromant M., Brevet A., Gesquière A., Blanquet S., Catabolism of bis(5′-nucleosidyl) oligophosphates in Escherichia coli: Metal requirements and substrate specificity of homogeneous diadenosine-5′,5′''-P1,P4-tetraphosphate pyrophosphohydrolase. Biochemistry 24, 914–922 (1985). [DOI] [PubMed] [Google Scholar]
- 11.Farr S. B., Arnosti D. N., Chamberlin M. J., Ames B. N., An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 86, 5010–5014 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lévĕque F., Blanchin-Roland S., Fayat G., Plateau P., Blanquet S., Design and characterization of Escherichia coli mutants devoid of Ap4N-hydrolase activity. J. Mol. Biol. 212, 319–329 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Despotović D., et al. , Diadenosine tetraphosphate (Ap4A)–An E. coli alarmone or a damage metabolite? FEBS J. 284, 2194–2215 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Johnstone D. B., Farr S. B., AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 10, 3897–3904 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimura A., et al. , Diadenosine 5′,5′′′-P1,P4-tetraphosphate (Ap4A) controls the timing of cell division in Escherichia coli. Genes Cells 2, 401–413 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Ismail T. M., Hart C. A., McLennan A. G., Regulation of dinucleoside polyphosphate pools by the YgdP and ApaH hydrolases is essential for the ability of Salmonella enterica serovar typhimurium to invade cultured mammalian cells. J. Biol. Chem. 278, 32602–32607 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Hansen S., Lewis K., Vulić M., Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob. Agents Chemother. 52, 2718–2726 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monds R. D., et al. , Di-adenosine tetraphosphate (Ap4A) metabolism impacts biofilm formation by Pseudomonas fluorescens via modulation of c-di-GMP-dependent pathways. J. Bacteriol. 192, 3011–3023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura Y., Tanaka C., Sasaki K., Sasaki M., High concentrations of intracellular Ap4A and/or Ap5A in developing Myxococcus xanthus cells inhibit sporulation. Microbiology 163, 86–93 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Ji X., et al. , Alarmone Ap4A is elevated by aminoglycoside antibiotics and enhances their bactericidal activity. Proc. Natl. Acad. Sci. U.S.A. 116, 9578–9585 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luciano D. J., Levenson-Palmer R., Belasco J. G., Stresses that raise Np4A levels induce protective nucleoside tetraphosphate capping of bacterial RNA. Mol. Cell 75, 957–966 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hudeček O., et al. , Dinucleoside polyphosphates act as 5′-RNA caps in Escherichia coli. bioRxiv:10.1101/563817 (1 March 2019).
- 23.Cahová H., Winz M. L., Höfer K., Nübel G., Jäschke A., NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519, 374–377 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Santoro S. W., Joyce G. F., A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U.S.A. 94, 4262–4266 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Igloi G. L., Kössel H., Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Application of polyacrylamide containing a covalently bound boronic acid. Nucleic Acids Res. 13, 6881–6898 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luciano D. J., Belasco J. G., Analysis of RNA 5′ ends: Phosphate enumeration and cap characterization. Methods 155, 3–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddalo G., et al. , Systematic analysis of native membrane protein complexes in Escherichia coli. J. Proteome Res. 10, 1848–1859 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Thomason M. K., et al. , Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 197, 18–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helbig K., Grosse C., Nies D. H., Cadmium toxicity in glutathione mutants of Escherichia coli. J. Bacteriol. 190, 5439–5454 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cartwright J. L., Britton P., Minnick M. F., McLennan A. G., The IalA invasion gene of Bartonella bacilliformis encodes a (de)nucleoside polyphosphate hydrolase of the MutT motif family and has homologs in other invasive bacteria. Biochem. Biophys. Res. Commun. 256, 474–479 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Luciano D. J., Vasilyev N., Richards J., Serganov A., Belasco J. G., A novel RNA phosphorylation state enables 5′ end-dependent degradation in Escherichia coli. Mol. Cell 67, 44–54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott J. F., Zamecnik P. C., Some optical properties of diadenosine-5′-phosphates. Proc. Natl. Acad. Sci. U.S.A. 64, 1308–1314 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malygin A. G., Shemyakin M. F., Adenosine, NAD and FAD can initiate template-dependent RNA synthesis catalyzed by Escherichia coli RNA polymerase. FEBS Lett. 102, 51–54 (1979). [DOI] [PubMed] [Google Scholar]
- 34.Bird J. G., et al. , The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535, 444–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Julius C., Yuzenkova Y., Bacterial RNA polymerase caps RNA with various cofactors and cell wall precursors. Nucleic Acids Res. 45, 8282–8290 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vvedenskaya I. O., et al. , CapZyme-seq comprehensively defines promoter-sequence determinants for RNA 5′ capping with NAD. Mol. Cell 70, 553–564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rauhut R., Gabius H. J., Engelhardt R., Cramer F., Archaebacterial phenylalanyl-tRNA synthetase. Accuracy of the phenylalanyl-tRNA synthetase from the archaebacterium Methanosarcina barkeri, Zn(II)-dependent synthesis of diadenosine 5′,5′''-P1,P4-tetraphosphate, and immunological relationship of phenylalanyl-tRNA synthetases from different urkingdoms. J. Biol. Chem. 260, 182–187 (1985). [PubMed] [Google Scholar]
- 38.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards J., Luciano D. J., Belasco J. G., Influence of translation on RppH-dependent mRNA degradation in Escherichia coli. Mol. Microbiol. 86, 1063–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meacock P. A., Cohen S. N., Partitioning of bacterial plasmids during cell division: A cis-acting locus that accomplishes stable plasmid inheritance. Cell 20, 529–542 (1980). [DOI] [PubMed] [Google Scholar]
- 41.Nudler E., Gusarov I., Bar-Nahum G., Methods of walking with the RNA polymerase. Methods Enzymol. 371, 160–169 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the figures and supporting information.