Significance
The regions surrounding living marine phytoplankton cells harbor communities of heterotrophic bacteria that play roles in carbon and energy flux in the microbial ocean and have global-scale carbon cycle implications. Yet, the drivers underlying bacterial community assembly remain unclear. In synthetic systems designed to mimic the chemistry and turnover time of natural phycospheres, bacterial community assembly could be predicted as a simple sum of assemblages supported by each individual metabolite. For host phytoplankton cells in the ocean, this implies control over bacterial associates through excreted metabolites, a condition that could favor the evolution of marine microbial interactions and influence heterotrophic carbon processing in the surface ocean.
Keywords: phytoplankton–bacteria interactions, community assembly, phycospheres
Abstract
In the nutrient-rich region surrounding marine phytoplankton cells, heterotrophic bacterioplankton transform a major fraction of recently fixed carbon through the uptake and catabolism of phytoplankton metabolites. We sought to understand the rules by which marine bacterial communities assemble in these nutrient-enhanced phycospheres, specifically addressing the role of host resources in driving community coalescence. Synthetic systems with varying combinations of known exometabolites of marine phytoplankton were inoculated with seawater bacterial assemblages, and communities were transferred daily to mimic the average duration of natural phycospheres. We found that bacterial community assembly was predictable from linear combinations of the taxa maintained on each individual metabolite in the mixture, weighted for the growth each supported. Deviations from this simple additive resource model were observed but also attributed to resource-based factors via enhanced bacterial growth when host metabolites were available concurrently. The ability of photosynthetic hosts to shape bacterial associates through excreted metabolites represents a mechanism by which microbiomes with beneficial effects on host growth could be recruited. In the surface ocean, resource-based assembly of host-associated communities may underpin the evolution and maintenance of microbial interactions and determine the fate of a substantial portion of Earth’s primary production.
The ecological interactions that occur between ocean phytoplankton and bacteria are among the most important quantitative links in global carbon and nutrient cycles. Marine phytoplankton are responsible for half of Earth’s photosynthesis, and heterotrophic marine bacteria process 40 to 50% of this fixed carbon (1–3). Much of the bacterial consumption of recent photosynthate occurs through uptake of dissolved organic carbon released by host phytoplankton cells into surrounding seawater by mechanisms such as leakage and exudation from living cells, as well as from mortality via senescence and predation (4).
In the diffusive boundary layer immediately surrounding phytoplankton cells, termed the phycosphere, the opportunity for transfer of substrates to bacteria is enhanced. Compared to bulk seawater where concentrations of labile metabolites are in the low nanomolar to picomolar range, phycosphere concentrations can reach into the hundreds of nanomolar and remain elevated above bulk seawater concentrations for up to hundreds of microns away (5). The metabolites released by phytoplankton span broad chemical classes, including carboxylic acids, amino acids, carbohydrates, C1 compounds, and organic sulfur compounds (4, 6–8). Yet, the specific mix of metabolites present in a given phycosphere is variable and influenced by phytoplankton taxonomy (8, 9) and physiology (10, 11).
The composition of bacterial communities that consume phytoplankton metabolites impacts the rates and efficiencies of marine organic matter transformation (12–15), with the latter a key factor in ocean–atmosphere CO2 balance. Further, bacterial community composition has cascading effects on food web yield governed by the susceptibility of bacterial taxa to protist grazing and viral infection (16, 17) and also impacts host biology (18, 19). The ecological mechanisms that influence the assembly of phycosphere microbiomes are not well understood, however, in part because of the micrometer scale at which bacterial communities congregate. It remains unclear whether simple rules exist that could predict the composition of these communities.
Phycospheres are short-lived in the ocean, constrained by the 1- to 2-d average life span of phytoplankton cells (20, 21). The phycosphere bacterial communities must therefore form and disperse rapidly within a highly dynamic metabolite landscape (14). We hypothesized a simple rule for assembly in metabolically diverse phycospheres in which communities congregate as the sum of discrete metabolite guilds (22). Each guild is hypothesized to support one to many bacterial species that exploit a metabolite resource either directly or indirectly via intermediate products, and these single-resource guilds form building blocks for the mixed-resource communities. To the extent that communities assemble in this additive fashion, composition is controlled by the host phytoplankton through the metabolites they release. Deviations from predictions of this strict resource-based model would indicate the influence of other drivers of community composition, particularly species–species interactions among the congregating heterotrophic bacteria.
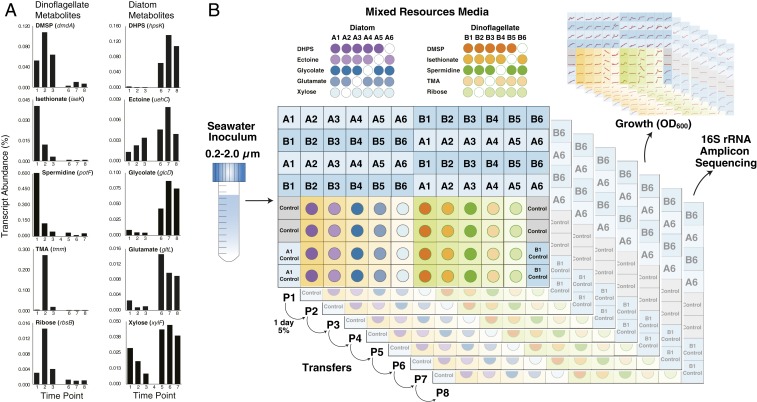
We tested this resource-based model using laboratory systems that mimic phycosphere metabolite composition and turnover time. The synthetic phycospheres contained from one to five compounds, organized into two suites representative of either diatom or dinoflagellate exometabolite mixtures (6, 8, 19). For each phytoplankton type, resource conditions with varying proportions of the five metabolites were inoculated with a natural assemblage of bacterial and archaeal cells concentrated from coastal seawater. Communities were transferred into fresh media once per day for 8 d, and their composition was assessed after the final growth cycle. A weighted-sum (WS) model was used to test for resource-controlled community assembly by summing the taxonomic compositions of the single-metabolite guilds after weighting each by the growth it supports.
Results
Synthetic phycospheres were established in a 96-deep-well-plate format using metabolites that are synthesized by phytoplankton and support growth of associated heterotrophic bacteria (8, 19). One suite of five metabolites represented molecules with higher release rates by the diatom Thalassiosira pseudonana compared to the dinoflagellate Alexandrium tamarense; these were xylose, glutamate, glycolate, ectoine, and dihydroxypropanesulfonate (DHPS) (8). The second suite represented molecules with higher release rates by A. tamarense compared to T. psuedonana; these were ribose, spermidine, trimethylamine (TMA), isethionate, and dimethylsulfoniopropionate (DMSP) (8) (Fig. 1A). The synthetic diatom phycospheres were composed of a single resource (five treatments), a mixture of all five resources (one treatment: A1), or mixtures of four resources (five treatments: A2, A3, A4, A5, and A6); in total, 11 different diatom resource treatments were replicated four times (Fig. 1B). The synthetic dinoflagellate phycospheres similarly contained single, a mixture of five (B1), or mixtures of four (B2, B3, B4, B5, and B6) resources (Fig. 1B). The total carbon concentration in each medium was 7.5 mM, established during pilot tests to maximize biomass for downstream sequencing while maintaining aerobic growth in the stirred wells. In natural phycospheres, metabolite concentrations are estimated to reach 240 nM carbon (5), with bacterial biomass scaled down proportionately compared to our synthetic phycospheres. Each metabolite set contained molecules of roughly similar compound classes: organic nitrogen compounds (glutamate and ectoine in the diatom media; spermidine and TMA in the dinoflagellate media), a sugar monomer (xylose; ribose), an organic sulfur compound (DMSP; isethionate and DHPS), and an osmolyte (ectoine; DMSP).
Fig. 1.
Experimental design of the synthetic phycospheres. (A) Ten metabolites characteristic of diatom or dinoflagellate phycospheres were identified from differential gene expression by a reporter bacterium, R. pomeroyi DSS-3, in a coculture dominated by a dinoflagellate (A. tamarense) for the first 12 d (time points 1, 2, and 3) and by a diatom (T. pseudonana) for the final 15 d (time points 6, 7, and 8). Data from ref. 8. (B) The 96-well plates contained 22 media types containing combinations of diatom metabolites or dinoflagellate metabolites, with each medium replicated four times. Following inoculation with a natural bacterial community from coastal seawater, communities were transferred daily and analyzed by 16S rRNA sequencing after eight growth-dilution cycles (P8). The composition of mixed-resource media A1 through A6 and B1 through B6 is shown in the upper matrix, where an empty dot signifies the absence of a metabolite.
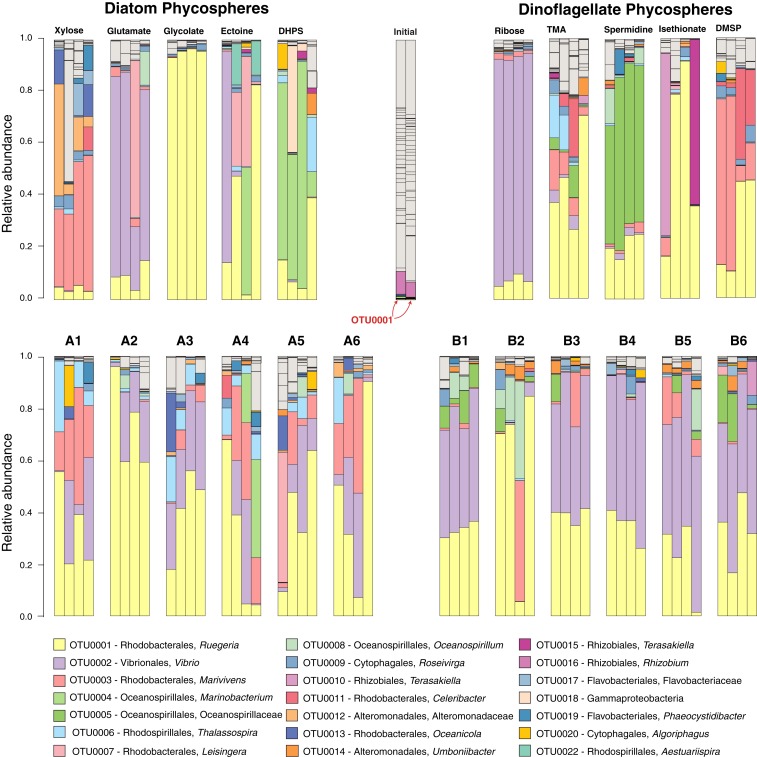
The synthetic phycospheres were inoculated with a microbial community concentrated from coastal seawater (0.2- to 2.0-μm size fraction). To initiate the study, ∼6 × 104 bacterial and archaeal cells were added to each well. Phycospheres were incubated for eight sequential 1-d periods, with a 5% inoculum at each transfer (4.3 doublings per growth-dilution cycle; Fig. 1B). The composition of the phycosphere communities after eight growth-dilution cycles (P8) was analyzed by 16S ribosomal RNA (rRNA) sequencing (23), with operational taxonomic units (OTUs) defined based on 97% sequence identity (Fig. 2). The number of OTUs contributing at least 0.1% of the community averaged 21 ± 4 in the P8 diatom phycospheres after normalizing to sequencing depth and 25 ± 5 in the P8 dinoflagellate phycospheres (Fig. 2), indicating that the synthetic phycosphere systems retained considerable diversity even after eight growth-dilution cycles. The seawater inoculum contained 137 (± 7) OTUs contributing at least 0.1% of the community. Archaea and Cyanobacteria together accounted for ∼12% of the inoculum but were less than 0.1% of the P8 communities.
Fig. 2.
Composition of bacterial communities in the P8 growth cycle of diatom phycospheres (Left), dinoflagellate phycospheres (Right), and the inoculum (Center) based on 97% identity OTUs of 16S rRNA amplicons. Gray coloring indicates OTUs that made up <0.1% of the pooled final bacterial communities. OTU0001 made up 0.4% of the inoculum community.
Single-Resource Communities.
From the diverse coastal inoculum a suite of heterotrophic bacterioplankton lineages emerged that are typical of those associated with marine phytoplankton (18, 24, 25), dominated by lineages in the Alphaproteobacteria, Gammaproteobacteria, and Flavobacteriia. Of the 50 most abundant 16S rRNA sequences from the synthetic phycospheres, 48% had >95% identity to a sequence obtained previously from bacterial associates of marine diatoms or dinoflagellates (SI Appendix, Table S1).
Bacterial community assembly was generally coherent among the replicates of single-guild phycospheres (Fig. 2). In pairwise comparisons of the five diatom single-metabolite treatments, 7 of 10 pairs were statistically different (Adonis test, P < 0.05); the three exceptions were the DHPS, ectoine, and glutamate guilds, which were not statistically distinct from each other because of within-treatment variability. For example, in the ectoine communities a unique bacterial OTU (related to Aestuariispria) was present in only two of four replicates (Fig. 2). This OTU was not represented in the ∼100,000 16S rRNA amplicons sequenced from the seawater inoculum, suggesting bottlenecks in inoculation of low-abundance OTUs. Amplicon sequencing of the complete time series of ectoine communities (growth-dilution cycles P2 through P7) showed that the differences among replicates were already evident in the early transfers and stable communities were established by P8 (SI Appendix, Fig. S1). In pairwise comparisons of the five dinoflagellate single-metabolite treatments, all 10 community pairs were different from one another (Adonis test, P < 0.05). Chemical analysis of spent medium from P8 indicated that most single-metabolite communities fully consumed the substrate between transfers; the exceptions were the diatom metabolite DHPS (26% remained unused) and the dinoflagellate metabolites TMA and isethionate (40% and 20%) (SI Appendix, Fig. S2). Overall, we found that the bacterial communities that formed on different single resources were compositionally distinguishable.
Multiple-Resource Communities.
Communities assembling on mixtures of metabolites were less distinct from one another than single-resource guilds, as expected from the overlap of substrates across mixtures. In pairwise comparisons among the six diatom mixed-metabolite treatments, only 1 out of 15 pairs was statistically distinguishable; among the dinoflagellate mixed-metabolite treatments, only 6 out of 15 were distinguishable (Adonis test, P < 0.05). Although we anticipated that the multiple substrates available in the metabolite mixtures would support higher community richness than single-substrate treatments, as expected from theory (26, 27) and experiments (28), this was not the case (24 ± 5 OTUs for mixed versus 25 ± 6 for single), nor were there differences in community diversity (Shannon index 1.25 ± 0.38 for mixed versus 1.10 ± 0.51 for single) (SI Appendix, Fig. S3). The OTUs with membership in single-resource guilds but missing in mixed-resource communities were biased toward Gammaproteobacteria (Neptunomonas, Marinomonas, or unclassified) and Flavobacteriia (Flavobacteriales) (SI Appendix, Table S2). Chemical analysis of spent medium from P8 indicated that mixed-metabolite communities fully consumed the substrates with the exception that TMA remained in 9 of the 20 replicates that contained it; these were from media types B1, B2, B4, B5, and B6 and averaged 21% of the initial TMA concentration (SI Appendix, Fig. S2).
Test of a Resource-Based Model for Community Assembly.
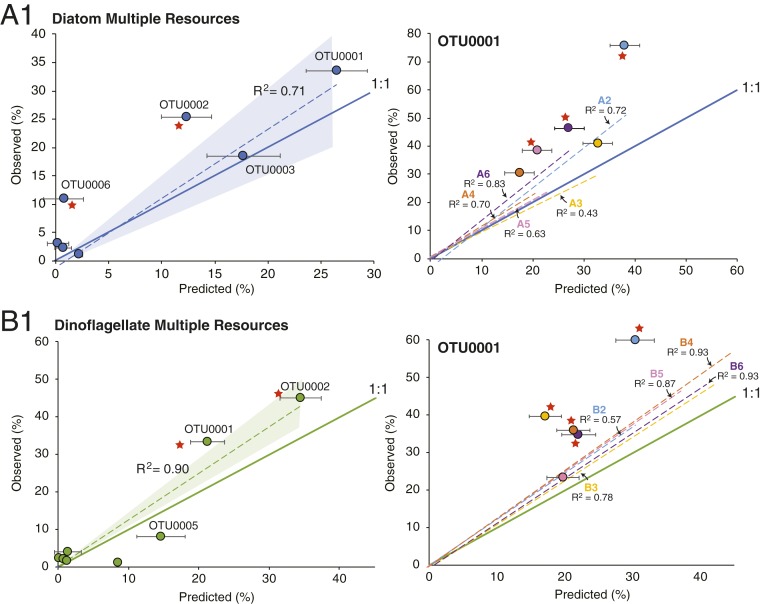
We predicted the composition of mixed-resource communities according to a WS model. To encompass the variability within treatments, one replicate from each single-resource guild was randomly selected to include in the predicted community. OTU abundances were normalized to the growth supported by the selected replicate (measured as optical density at 600 nm, OD600) and to the concentration of the resource in the mixed-metabolite medium. A distribution of the predicted relative abundance of OTUs was obtained by generating 1,000 such predicted communities (SI Appendix, Fig. S4).
Comparisons of observed versus predicted OTU composition showed that resource availability is an effective predictor of community assembly (Fig. 3 and SI Appendix, Fig. S5), with the dominant taxa in multiple-resource communities successfully predicted based on weighted averages of taxa in single-resource guilds. The linear regression coefficients (R2) of observed versus predicted OTU abundance was statistically significant (P ≤ 0.001) for all mixed-resource communities. There was no difference in the strength of the relationship for communities assembling on five (A1 and B1) versus four (A2 through A6; B2 through B6) resources.
Fig. 3.
Observed versus predicted OTU percent abundance in mixed-resource phycosphere communities. Dotted line and shading, linear regression and 95% CI of observed versus predicted based on a WS null model; solid line, 1:1 relationship between observed and predicted. (Left) Data from media A1 and B1 (five metabolite mixtures); only OTUs accounting for ≥ 1% of the community are shown for simplicity. (Right) Data from media A2 through A6 and B2 through B6 (four metabolite mixtures); only OTU0001 is plotted for simplicity (see SI Appendix, Fig. S5 for complete plots). Red stars indicate significant overrepresentation of OTUs in the mixed-resource communities. Error bars indicate ±1 SE.
The WS model predicts a 1:1 relationship between observed and predicted OTU abundance, and therefore a slope of 1.0 is expected if single-resource guilds fully explain the mixed-resource communities after eight growth-dilution cycles. For 7 out of the 12 mixed-resource communities, the 95% confidence interval of regression slopes of observed versus predicted abundances included 1.0, with slopes ranging from 1.4 to 0.9. All but two regression slopes were >1.0, indicating a trend toward higher observed abundances for some OTUs than predicted by the WS model (Fig. 3 and SI Appendix, Fig. S5).
To address the systematic positive deviation from a 1:1 slope, individual OTUs were classified as significant over- or underperformers in the observed communities if the observed mean ± 1 SE fell above or below the 95% confidence intervals of the predicted distribution (SI Appendix, Fig. S4). OTU0001 was overrepresented in 8 of the 12 mixed-resource communities (Fig. 3) and OTU0002 was overrepresented in 5 (SI Appendix, Fig. S6). Overall, slope deviations were driven largely by overperformance of more abundant taxa (SI Appendix, Fig. S5). Thus, while host resources provided good predictability of bacterial community assembly in the synthetic systems, other factors weakened the strict predictions of the WS model. In these simple synthetic systems, these factors could be ecological species interactions occurring among the heterotrophic bacteria, such as competition or mutualism, or additional aspects of resource supply, such as complexity or quality.
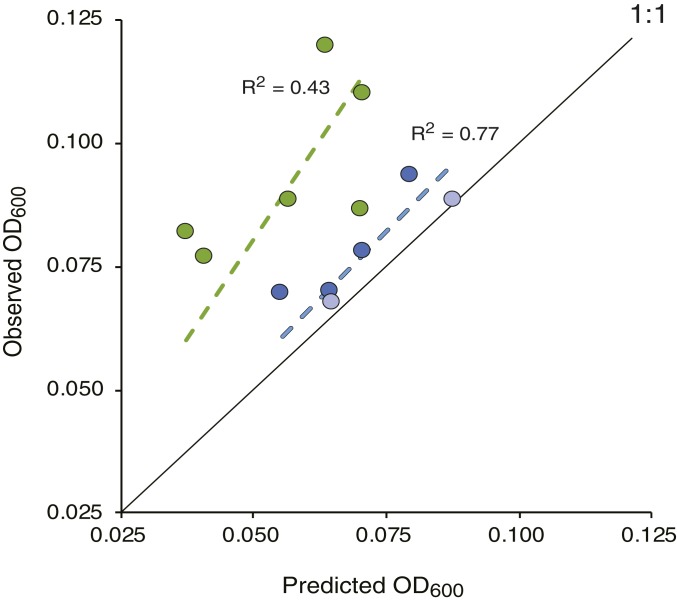
To differentiate between heterotrophic bacterial interactions versus other resource-based factors in deviations from the WS model, the phycosphere system was reestablished with only one bacterial species as the inoculum. This eliminated the possibility of species interactions and allowed us to ask whether systematic OTU overperformance would nonetheless be observed. An isolate with 100% average nucleotide identity to a metagenome-assembled genome (MAG) of OTU0001, the most abundant and frequently overrepresented OTU in P8 communities (Fig. 3), was used as the single bacterium inoculum. Strain Ruegeria pomeroyi DSS-3 was isolated from seawater collected at the same site as this study’s inoculum (29) and has gene content identical to OTU0001 except for 44 genes (out of 4,371 total; SI Appendix, Fig. S7 and Table S3) (30). As for the seawater-inoculated phycosphere system, R. pomeroyi DSS-3 was introduced into the 22 resource conditions, growth was tracked over eight growth-dilution cycles, and final OD600 was predicted from the WS of OD600 in the single resources. The bacterium achieved significantly higher growth in 10 of the 12 mixed-resource conditions compared to WS predictions from single resources, indicating overperformance of this species in multiple-resource conditions in the absence of other heterotrophic bacterial species (Fig. 4).
Fig. 4.
Observed versus predicted OD600 of R. pomeroyi DSS-3 after eight growth-dilution cycles in mixed-resource phycospheres. Blue symbols indicate diatom media (A1 through A6) and green symbols indicate dinoflagellate media (B1 through B6). Dotted lines, linear regression; solid line, 1:1 relationship between observed and predicted. Error bars (± 1 SE) all fall within the symbols. Light blue symbols mark the metabolite mixtures (A5 and A6) for which observed OD600 fell within the 95% CI of the predicted OD600.
Discussion
The release of metabolites from living phytoplankton cells was first described in the 1960s (7, 31, 32) and recognition of the chemical complexity of the phycosphere environment has steadily grown. Phytoplankton metabolites vary based on taxonomy (7–10, 33), physiological state such as nutrient limitation and stress (10, 34), and environmental features such as temperature (34, 35). This variable chemistry offers diverse niches for heterotrophic bacterioplankton but makes predicting the composition of colonizing communities challenging. Nevertheless, we find that simple linear combinations of species abundances on single resources accurately predict assemblies of mixed-resource communities. Similarly, the composition of microbial communities assembling from natural soil, plant, and seawater inocula is highly predictable from resource availability (36, 37). The success of a resource-based model in predicting phycosphere communities does not preclude important effects of interspecies interactions on community composition (38). Rather, it suggests that, if important, they operate largely within single-resource guilds rather than across them. Indeed the diversity of OTUs making up single-resource guilds (SI Appendix, Fig. S3) suggests ample opportunities for within-guild interactions in the form of competition or mutualisms (39–41). An additional level of microbial interactions occurs in natural phycospheres between bacteria and living phytoplankton that is not represented in this synthetic system; such interactions have been shown previously to encompass both mutualistic and antagonistic relationships (39, 42, 43).
The consistent skew of some bacterial taxa away from the 1:1 linear relationship predicted by the WS model could potentially arise from between-guild species interactions. However, our direct test with single-species phycospheres recapitulated the same skew (Fig. 4). Metabolic modeling approaches have suggested that adding resources promotes higher levels of species interactions among microbes (40, 44), but the short half-lives of phycospheres (20), both synthetic and natural, may provide fewer opportunities for between-guild interactions to develop. The observed overperformance of heterotrophic bacteria is consistent with previous evidence of fitness advantages when resources are available together rather than individually (45). Soil microbial ecologists have proposed a priming effect whereby energy from a more labile substrate enhances the ability of heterotrophic microbes to synthesize catabolic enzymes for a less-labile substrate (46). Microbiologists use the term “cometabolism” for a similar phenomenon although typically specify that the less-labile substrate does not support biomass (47), which is not the case in our study (SI Appendix, Fig. S8). Alternatively, species overperformance could arise when high cell numbers achieved on one resource allow procurement of a larger fraction of a second resource that itself only supports slow growth. Finally, overperformance could result when resources share degradation pathways or are regulated through common mechanisms. All three scenarios require energy savings from the simultaneous availability of multiple resources. The first two also require that the less-labile substrate is slowly or incompletely used when provided alone; the three partially consumed metabolites in the single-resource guilds (DHPS, TMA, and isethionate; SI Appendix, Fig. S2) are candidates for such substrates. In a separate analysis of R. pomeroyi growth on single metabolites, the time to maximum specific growth rate (μ) for DHPS, TMA, and isethionate was longer than for the other seven substrates (averaging 58 h versus 25 h) and the maximum biomass achieved was lower (averaging 0.07 OD600 versus 0.12 OD600) (SI Appendix, Fig. S8). These data support the scenarios of enhanced utilization of less-labile resources when catabolized with more-labile ones. In an analysis of resource use by three other abundant strains isolated from the phycosphere communities (OTU0002, OTU0003, and OTU0006), all could grow at the expense of more than one of the provided resources (SI Appendix, Fig. S8).
Finding that the dominant OTU in the synthetic phycospheres was the same bacterial species as isolated 20 y prior from the same inoculum site was unanticipated. We first ruled out contamination based on minor but consistent differences in genome content between the OTU0001 MAG and R. pomeroyi DSS-3 (SI Appendix, Fig. S7 and Table S3). In addition, OTU0001 was present in the seawater inoculum at levels sufficient to add ∼240 16S rRNA genes per well (0.4% of initial 16S rRNA amplicons; Fig. 2). Previous studies of R. pomeroyi DSS-3 in phytoplankton cocultures (8, 19) guided the selection of the phycosphere metabolites used in this study (Fig. 1A), and the nearly identical OTU0001 MAG also has the genetic capability to transport and catabolize all 10 metabolites (SI Appendix, Table S4). The importance of phytoplankton resources in the assembly of bacterial associates is thus reinforced by the specific enrichment of a metabolically optimized bacterium from a diverse seawater inoculum.
Some mechanisms of phytoplankton metabolite release, such as direct excretion, photosynthetic overflow, photorespiration, and redox balancing (4), are controlled by the host; others, including leakage and predation (48), are not host-controlled. Regardless of the release mechanism involved, resource-based bacterial assembly offers a means by which phytoplankton could influence the taxonomic diversity of associated bacteria, and this could explain the consistency of bacterial communities colonizing natural marine phycospheres (24). Phytoplankton have been shown to accrue a number of benefits from heterotrophic bacteria, for example access to vitamins for which many are auxotrophic (49) or to scarce trace elements (50) or oxidative stress enzymes (39). The ability to control the composition of associated bacteria through metabolite release could amplify these benefits, as has been shown for multicellular photosynthetic hosts such as vascular plants that enrich for beneficial rhizosphere communities via root exudates (51). For bacteria, labile compounds occurring in predictable combinations could enhance resource discovery (14) and drive acquisition and coregulation of catabolic pathways that are needed simultaneously. R. pomeroyi/OTU0001 and its marine relatives in the Rhodobacterales are frequently found in association with phytoplankton cells (24, 52, 53) (SI Appendix, Table S1), and their large and well-regulated genomes are proposed to have expanded in content coincident with the diversification of eukaryotic phytoplankton (54). Release of phytoplankton metabolites has been experimentally linked to bacterial-derived benefits in a few cases, such as the polysaccharide fucoidan triggering increased bacterial vitamin B12 production (55) and diatom-derived tryptophan initiating bacterial synthesis of the growth hormone indole-3-acetic acid (56). While our results set the stage for such mutualisms, they could nonetheless be explained without invoking selective mechanisms. For example, bacteria may take advantage of spatially and temporally clumped resources without any underlying coevolutionary relationship with phytoplankton producers.
The simple logic of community assembly observed here suggests that changes in the composition of phytoplankton communities in a future ocean (57) will cascade to heterotrophic bacterial communities. Potential impacts on biogeochemical processes include changes in regeneration of macronutrients (2), formation of climate-active organic molecules (58), and mineralization of a major fraction of Earth’s net primary production (3).
Methods
The bacterial composition of synthetic phycospheres was analyzed by 16S rRNA amplicon sequencing, performed on the Illumina MiSeq platform. Read analysis was carried out with the Mothur (v.1.39.5) pipeline following https://www.mothur.org/wiki/MiSeq_SOP, d.d. 2018-2-8. To predict bacterial community assembly on mixed resources, a WS model was applied to the 50 most abundant taxa (>99.2% of reads). Relative abundance of each taxon on single metabolites normalized by OD600 (SI Appendix, Fig. S9) and metabolite concentration in the medium were used to calculate relative abundances according to the equation
where fp is the predicted relative abundance of a taxon, fi is the observed relative abundance of that taxon in treatment i, and ODi is the measured optical density. This step was bootstrapped 1,000 times to generate distributions of predicted relative abundances. Metabolites in spent media samples from the P8 phycospheres were quantified by 1H NMR spectroscopy using a NEO III (Bruker) with a 1.7-mm cryoprobe. Data were acquired by a one-dimensional 1H experiment. A unique peak region for each compound was defined. Full details of sampling and analysis are given in SI Appendix, Supplementary Methods.
Data Availability.
DNA sequences are available in the NCBI Sequence Read Archive (project PRJNA553557), under accession nos. SRR9668153–SRR9668338 for 16S rRNA amplicons and SRR9668573 and SRR9668574 for MAGs.
Supplementary Material
Acknowledgments
This work was supported by Simons Foundation grants 542391 to M.A.M. and 542385 to J.G. within the Principles of Microbial Ecosystems Collaborative. We thank J. Schreier and C. Smith for assistance, K. G. Ross for statistical advice, and the University of Georgia Complex Carbohydrate Research Center and Georgia Genomics and Bioinformatics Core for instrumentation and services. This is contribution 1083 of the University of Georgia Marine Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: DNA sequences are available in the NCBI Sequence Read Archive (project no. PRJNA553557) under accession numbers SRR9668153–SRR9668338 for 16S ribosomal RNA amplicons and SRR9668573 and SRR9668574 for metagenome-assembled genomes.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917265117/-/DCSupplemental.
References
- 1.Moran M. A., et al. , Deciphering ocean carbon in a changing world. Proc. Natl. Acad. Sci. U.S.A. 113, 3143–3151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azam F., et al. , The ecological role of water-column microbes. Mar. Ecol. Prog. Ser. 10, 257–263 (1983). [Google Scholar]
- 3.Cole J. J., Findlay S., Pace M. L., Bacterial production in fresh and saltwater ecosystems: A cross-system overview. Mar. Ecol. Prog. Ser. 43, 1–10 (1988). [Google Scholar]
- 4.Thornton D. C., Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 49, 20–46 (2014). [Google Scholar]
- 5.Seymour J. R., Amin S. A., Raina J.-B., Stocker R., Zooming in on the phycosphere: The ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2, 17065 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Durham B. P., et al. , Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. U.S.A. 112, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellebust J., Excretion of organic compounds by cultured and natural populations of marine phytoplankton. Estuaries 10, 192–206 (1967). [Google Scholar]
- 8.Landa M., Burns A. S., Roth S. J., Moran M. A., Bacterial transcriptome remodeling during sequential co-culture with a marine dinoflagellate and diatom. ISME J. 11, 2677–2690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker J. W., et al. , Closely related phytoplankton species produce similar suites of dissolved organic matter. Front. Microbiol. 5, 111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen A. E., et al. , Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. U.S.A. 105, 10438–10443 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barofsky A., Vidoudez C., Pohnert G., Metabolic profiling reveals growth stage variability in diatom exudates. Limnol. Oceanogr. Methods 7, 382–390 (2009). [Google Scholar]
- 12.Del Giorgio P. A., Cole J. J., Bacterial growth efficiency in natural aquatic systems. Annu. Rev. Ecol. Syst. 29, 503–541 (1998). [Google Scholar]
- 13.Eiler A., Langenheder S., Bertilsson S., Tranvik L. J., Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl. Environ. Microbiol. 69, 3701–3709 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stocker R., Seymour J. R., Ecology and physics of bacterial chemotaxis in the ocean. Microbiol. Mol. Biol. Rev. 76, 792–812 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X., Polz M. F., Alm E. J., Interactions in self-assembled microbial communities saturate with diversity. ISME J. 13, 1602–1617 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suttle C. A., Marine viruses–Major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Weitz J. S., Wilhelm S. W., Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol. Rep. 4, 17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amin S. A., Parker M. S., Armbrust E. V., Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durham B. P., et al. , Recognition cascade and metabolite transfer in a marine bacteria-phytoplankton model system. Environ. Microbiol. 19, 3500–3513 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Behrenfeld M. J., Falkowski P. G., Photosynthetic rates derived from satellite‐based chlorophyll concentration. Limnol. Oceanogr. 42, 1–20 (1997). [Google Scholar]
- 21.Whitman W. B., Coleman D. C., Wiebe W. J., Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Root R. B., The niche exploitation pattern of the blue‐gray gnatcatcher. Ecol. Monogr. 37, 317–350 (1967). [Google Scholar]
- 23.Moran M. A., Fu H., Mock phycosphere microbial community 16S rRNA reads. NCBI SRA. https://www.ncbi.nlm.nih.gov/sra/PRJNA553557. Deposited 9 July 2019.
- 24.Buchan A., LeCleir G. R., Gulvik C. A., González J. M., Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Goecke F., Thiel V., Wiese J., Labes A., Imhoff J. F., Algae as an important environment for bacteria–phylogenetic relationships among new bacterial species isolated from algae. Phycologia 52, 14–24 (2013). [Google Scholar]
- 26.Schoener T. W., Resource partitioning in ecological communities. Science 185, 27–39 (1974). [DOI] [PubMed] [Google Scholar]
- 27.Tilman D., Resource Competition and Community Structure (Princeton University Press, 1982). [PubMed] [Google Scholar]
- 28.Muscarella M. E., Boot C. M., Broeckling C. D., Lennon J. T., Resource heterogeneity structures aquatic bacterial communities. ISME J. 13, 2183–2195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.González J. M., et al. , Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int. J. Syst. Evol. Microbiol. 53, 1261–1269 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Rivers A. R., Smith C. B., Moran M. A., An updated genome annotation for the model marine bacterium Ruegeria pomeroyi DSS-3. Stand. Genomic Sci. 9, 11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fogg G. E., Nalewajko C., Watt W. D., Extracellular products of phytoplankton photosynthesis. Proc. R. Soc. Lond. B Biol. Sci. 162, 517–534 (1965). [DOI] [PubMed] [Google Scholar]
- 32.Thomas J. P., Release of dissolved organic matter from natural populations of marine phytoplankton. Mar. Biol. 11, 311–323 (1971). [Google Scholar]
- 33.Durham B. P., et al. , Sulfonate-based networks between eukaryotic phytoplankton and heterotrophic bacteria in the surface ocean. Nat. Microbiol. 4, 1706–1715 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Brembu T., Mühlroth A., Alipanah L., Bones A. M., The effects of phosphorus limitation on carbon metabolism in diatoms. Phil. Trans. Royal Soc. B: Biol. Sci. 372, 20160406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toseland A., et al. , The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Clim. Chang. 3, 979–984 (2013). [Google Scholar]
- 36.Enke T. N., et al. , Modular assembly of polysaccharide-degrading marine microbial communities. Current Biol. 29, 1528–1535.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Goldford J. E., et al. , Emergent simplicity in microbial community assembly. Science 361, 469–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman J., Higgins L. M., Gore J., Community structure follows simple assembly rules in microbial microcosms. Nat. Ecol. Evol. 1, 0109 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Morris J. J., Johnson Z. I., Szul M. J., Keller M., Zinser E. R., Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS One 6, e16805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pacheco A. R., Moel M., Segrè D., Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems. Nat. Commun. 10, 103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zengler K., Zaramela L. S., The social network of microorganisms–how auxotrophies shape complex communities. Nat. Rev. Microbiol. 16, 383–390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barak-Gavish N., et al. , Bacterial virulence against an oceanic bloom-forming phytoplankter is mediated by algal DMSP. Sci. Adv. 4, eaau5716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seyedsayamdost M. R., Case R. J., Kolter R., Clardy J., The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3, 331–335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freilich S., et al. , Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2, 589 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Gulvik C. A., Buchan A., Simultaneous catabolism of plant-derived aromatic compounds results in enhanced growth for members of the Roseobacter lineage. Appl. Environ. Microbiol. 79, 3716–3723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guenet B., Danger M., Abbadie L., Lacroix G., Priming effect: Bridging the gap between terrestrial and aquatic ecology. Ecology 91, 2850–2861 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Dalton H., Stirling D. I., Co-metabolism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 297, 481–496 (1982). [DOI] [PubMed] [Google Scholar]
- 48.Flynn K. J., Clark D. R., Xue Y., Modeling the release of dissolved organic matter by phytoplankton 1. J. Phycol. 44, 1171–1187 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Sañudo-Wilhelmy S. A., Gómez-Consarnau L., Suffridge C., Webb E. A., The role of B vitamins in marine biogeochemistry. Annu. Rev. Mar. Sci. 6, 339–367 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Amin S. A., et al. , Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc. Natl. Acad. Sci. U.S.A. 106, 17071–17076 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhalnina K., et al. , Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480 (2018). [DOI] [PubMed] [Google Scholar]
- 52.González J. M., et al. , Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66, 4237–4246 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teeling H., et al. , Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Luo H., Moran M. A., Evolutionary ecology of the marine Roseobacter clade. Microbiol. Mol. Biol. Rev. 78, 573–587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G., Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Amin S. A., et al. , Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Barton A. D., Irwin A. J., Finkel Z. V., Stock C. A., Anthropogenic climate change drives shift and shuffle in North Atlantic phytoplankton communities. Proc. Natl. Acad. Sci. U.S.A. 113, 2964–2969 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simó R., Production of atmospheric sulfur by oceanic plankton: Biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16, 287–294 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences are available in the NCBI Sequence Read Archive (project PRJNA553557), under accession nos. SRR9668153–SRR9668338 for 16S rRNA amplicons and SRR9668573 and SRR9668574 for MAGs.