We thank Tran et al. (1) for their appreciation of our open innovation platform (https://opnme.com/) and interest in our manuscript (2) in which we demonstrate that the small-molecule KRAS inhibitor BI-2852 reduces pERK (EC50 = 6 µM) and inhibits proliferation (EC50 = 7 µM) in NCI-H358 cells. Tran et al. (1) propose that BI-2852 exerts its cellular effects on the MAPK pathway at least in part through stabilization of a nonfunctional KRAS dimer. No cellular data for KRAS dimer induction are provided and the hypothesis is primarily based on KRAS dimer formation observed with size exclusion chromatography using concentrations 1,000 times higher (3 mM) than those in which the cellular effects of BI-2852 are observed. Hence, inhibiting the protein–protein interactions between KRAS and its GEFs, GAPs, and downstream effectors is a more plausible explanation than induction of inactive KRAS dimers for the cellular mode of action for BI-2852 (2).
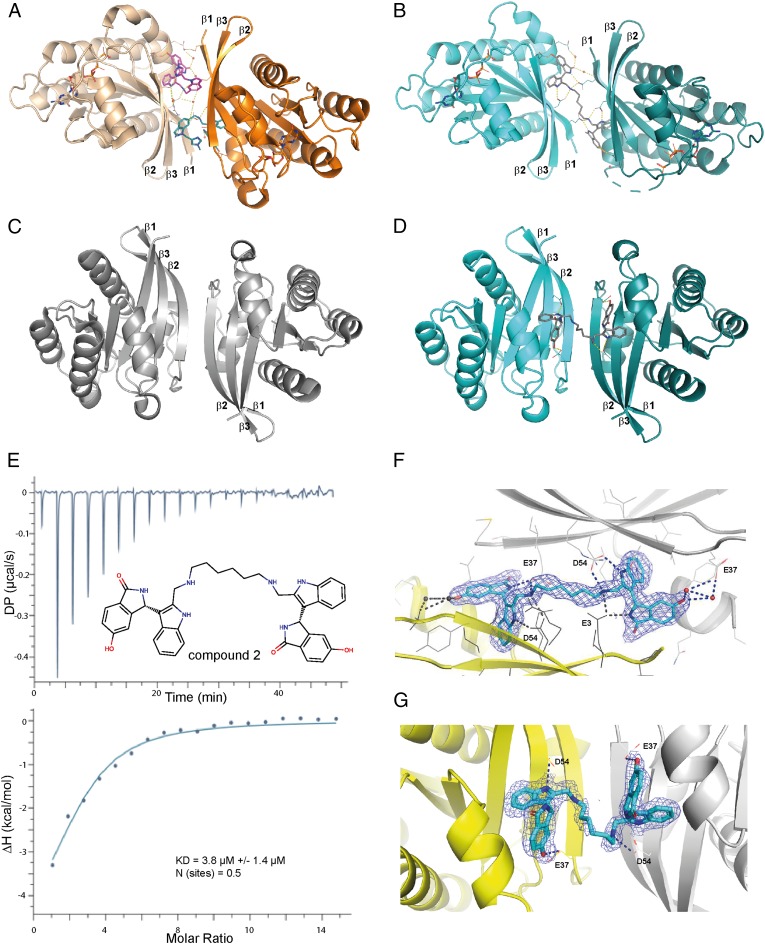
However, we concur that stabilization of nonfunctional KRAS dimers as a potential therapeutic approach to treat KRAS-driven cancers is intriguing. Accordingly, we have synthesized a range of dimeric switch I/II pocket binders in analogy to linkerology (3) in the PROTAC field. We discovered compound 2, which dimerizes KRAS with a KD of 3.8 µM according to isothermal calorimetry measurements (Fig. 1E). Using our high-throughput crystal-soaking system for active KRASG12D (4) (Fig. 1C), we obtained a dimeric KRAS structure with 2 (Fig. 1D) at a resolution of 1.9 Å (Fig. 1G), which represents the inactivating effector lobe dimer form 1 identified by Jang et al. (5) using molecular dynamics calculations. Cocrystallization of 2 with GCP-KRASG12D leads to the same KRAS dimer as observed for BI-2852 (Fig. 1 A and B), which is the second effector lobe form proposed by Jang et al. (5) at a resolution of 1.57 Å (Fig. 1F). In order to investigate the potential of small-molecule stabilization of nonfunctional KRAS dimers for the treatment of KRAS-driven cancers, more permeable and potent compounds will be needed to enable cellular studies.
Fig. 1.
KRAS dimers and biophysical validation. (A) KRAS G12D modeled dimer with BI-2852 (Protein Data Bank [PDB] ID code 6GJ8). The dimer interface was generated with symmetry operations from the published structure, showing a β-sandwich interface. (B) Cocrystal structure of KRAS G12D with compound 2 showing a similar β-sandwich interface as with BI-2852. (C) Published dimeric KRAS-G12D apo system (PDB ID code 6QUU) with an extended β-sheet interface. (D) Structure of compound 2 soaked in published KRAS-G12D soaking system. The compound needs to adapt to the extended β-sheet interface. (E) Isothermal titration calorimetry experiments with compound 2 showing binding to KRAS-G12D. (F) 2FoFc density around cocrystallized compound 2 at 1.57-Å resolution showing unambiguous binding. The interacting amino acids of both KRAS monomers are labeled. (G) 2FoFc density around compound 2 soaked in KRAS G12D at 1.9-Å resolution showing unambiguous binding of the ligand. The interacting amino acids of both KRAS monomers are labeled, respectively.
Footnotes
Competing interest statement: D.K., A.G., M.G., A.M., L.J.M., A.Z., M.M., D.C., S.F., T. Gerstberger, T. Gmaschitz, P.G., D.H., W.H., J.H., J.K.-O., P.K., S.K., M.K., R.K., L.L., F.M., S.M.-M., C.P., J.R., C.S., Y.S., K.S., R.S., A.S., B.S., G.S., B.W., M.Z., M.P., and D.B.M. were employees of Boehringer Ingelheim at the time of the work.
References
- 1.Tran T. H., et al. , The small molecule BI-2852 induces a nonfunctional dimer of KRAS. Proc. Natl. Acad. Sci. U.S.A. 117, 3363–3364 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kessler D., et al. , Drugging an undruggable pocket on KRAS. Proc. Natl. Acad. Sci. U.S.A. 116, 15823–15829 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bondeson D. P., Crews C. M., Targeted protein degradation by small molecules. Annu. Rev. Pharmacol. Toxicol. 57, 107–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergner A., et al. , KRAS binders hidden in nature. Chemistry 25, 12037–12041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang H., Muratcioglu S., Gursoy A., Keskin O., Nussinov R., Membrane-associated Ras dimers are isoform-specific: K-Ras dimers differ from H-Ras dimers. Biochem. J. 473, 1719–1732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]