Significance
Elongation factor Tu (EF-Tu) facilitates rapid and accurate selection of aminoacyl-tRNA (aa-tRNA) by the bacterial ribosome during protein synthesis. We show that EF-Tu dissociates from the ribosome as aa-tRNA navigates the accommodation corridor en route to peptide bond formation. We find that EF-Tu’s release from the ribosome during aa-tRNA selection can be reversible. We also demonstrate that new ternary complex formation, accompanied by futile cycles of GTP hydrolysis, can occur on aa-tRNA bound within the ribosome. These findings inform on the decoding mechanism, the contributions of EF-Tu to the fidelity of translation, and the potential consequences of reduced rates of peptide bond formation on cellular physiology.
Keywords: protein synthesis, single-molecule FRET, elongation factor Tu, aminoacyl-tRNA, translation
Abstract
The substrate for ribosomes actively engaged in protein synthesis is a ternary complex of elongation factor Tu (EF-Tu), aminoacyl-tRNA (aa-tRNA), and GTP. EF-Tu plays a critical role in mRNA decoding by increasing the rate and fidelity of aa-tRNA selection at each mRNA codon. Here, using three-color single-molecule fluorescence resonance energy transfer imaging and molecular dynamics simulations, we examine the timing and role of conformational events that mediate the release of aa-tRNA from EF-Tu and EF-Tu from the ribosome after GTP hydrolysis. Our investigations reveal that conformational changes in EF-Tu coordinate the rate-limiting passage of aa-tRNA through the accommodation corridor en route to the peptidyl transferase center of the large ribosomal subunit. Experiments using distinct inhibitors of the accommodation process further show that aa-tRNA must at least partially transit the accommodation corridor for EF-Tu⋅GDP to release. aa-tRNAs failing to undergo peptide bond formation at the end of accommodation corridor passage after EF-Tu release can be reengaged by EF-Tu⋅GTP from solution, coupled to GTP hydrolysis. These observations suggest that additional rounds of ternary complex formation can occur on the ribosome during proofreading, particularly when peptide bond formation is slow, which may serve to increase both the rate and fidelity of protein synthesis at the expense of GTP hydrolysis.
The bacterial ribosome is a two-subunit RNA–protein assembly that catalyzes the translation of mRNA into protein using aminoacyl-tRNA (aa-tRNA) substrates (Fig. 1 A and B). During the elongation phase of translation, aa-tRNAs are delivered to the aminoacyl (A) site of the ribosome as a ternary complex with an evolutionary conserved, three-domain, G protein, elongation factor-Tu (EF-Tu), and guanosine-5′-triphosphate (GTP) (EF-Tu⋅GTP⋅aa-tRNA) (1–7). By guiding the entry of aa-tRNA into the ribosome, EF-Tu increases both the rate and fidelity of translation (8–14).
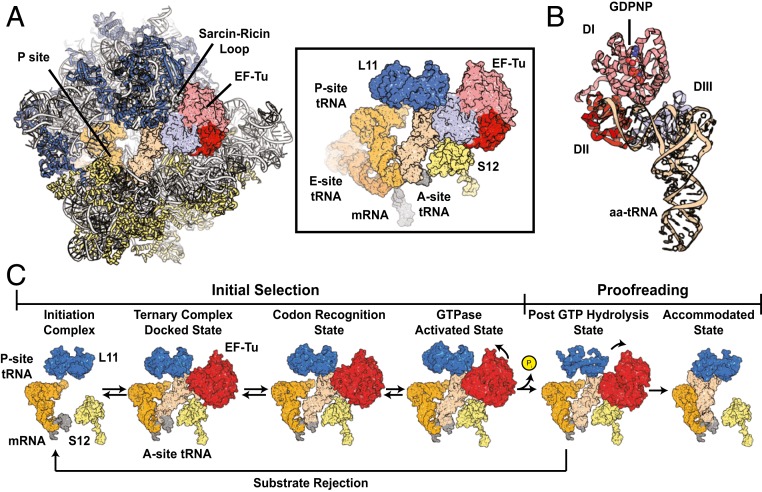
Fig. 1.
Structural models of EF-Tu and the bacterial tRNA selection mechanism. (A) The GTPase activated ribosomal complex with EF-Tu bound and aa-tRNA in the A/T state. Ribosomal RNA is shown in gray; proteins on the large subunit are blue; proteins on the small subunit are highlighted in yellow. Contacts between L11 (blue), S12 (yellow), aa-tRNA (tan), and EF-Tu (pink/red) are shown (Right). (B) Structural model of EF-Tu in ternary complex with GDPNP and aa-tRNA. All three structural domains (DI to DIII) of EF-Tu are indicated. (C) Structural models of the canonical tRNA selection mechanism showing the relative positions of the P-site tRNA (orange), mRNA (gray), L11 (blue), S12 (yellow), aa-tRNA (white), and EF-Tu (red) during initial selection and proofreading. Structural models were obtained from PDB ID codes 5UYL, 5UYM, 5UYK, 4V5G, and 1OB2.
Accurate selection of cognate (correct) aa-tRNA substrates at each mRNA codon arises from a two-step kinetic proofreading mechanism separated by GTP hydrolysis (Fig. 1C) (15–19). In the first step, initial selection, the ribosome can reject incorrect substrates prior to irreversible GTP hydrolysis. In the second step, proofreading, the ribosome is again able to reject aa-tRNA, prior to irreversible peptide bond formation. Ternary complexes bearing near- or noncognate aa-tRNA substrates are preferentially rejected from the ribosome during initial selection, such that they are rapidly released back to the cellular pool prior to energy expenditure (19–25). However, near- and noncognate aa-tRNAs are also selectively rejected during proofreading after GTP hydrolysis (17, 20, 26). The molecular basis of both initial selection and proofreading, which serve to establish the genetic code, have been the subject of intense investigation spanning several decades.
During initial selection, ternary complexes bearing cognate, near-cognate, or noncognate aa-tRNAs compete for binding to the A site, located at the ribosome’s leading edge. Proper base pairing between the mRNA codon and the anticodon of cognate aa-tRNA within the small (30S) subunit decoding region triggers a “domain closure” process, in which the 30S shoulder domain closes to facilitate engagement of the mRNA codon–tRNA anticodon pair (27–29). This process allosterically stimulates EF-Tu to hydrolyze GTP by facilitating the docking of its GTP binding domain (G domain) with the Sarcin-Ricin loop (SRL) at the base of the GTPase activating center (GAC) of the large (50S) subunit (19, 24, 30–36). Proofreading, a rate-determining feature of the aa-tRNA selection mechanism (17, 19, 26), entails conformational rearrangements within and between EF-Tu’s subdomains, subsequent to GTP hydrolysis, that allow aa-tRNA entry into the peptidyl transferase center (PTC) within the large subunit (17, 30, 37–39).
Contrasting models have been offered regarding the reaction coordinate of EF-Tu on the ribosome relative to that of aa-tRNA during proofreading. Structural data suggest a strictly ordered mechanism that involves the obligate dissociation of EF-Tu from the ribosome prior to aa-tRNA accommodation into the A site (30). In this view, GTP hydrolysis triggers conformational rearrangements within EF-Tu that disrupt the factor’s interactions with both the acceptor arm of tRNA and the ribosome (31, 38, 40, 41), thereby precipitating its rapid dissociation. This model specifies that aa-tRNA entry into the PTC is unassisted by EF-Tu. In contrast, ensemble fluorescence and two-color single-molecule fluorescence-resonance energy transfer (smFRET) investigations indicate that aa-tRNA accommodation occurs faster than EF-Tu⋅GDP dissociation (17, 42, 43). Such findings suggest that EF-Tu may remain bound to the ribosome during proofreading and dissociate after peptide bond formation has occurred.
Here, using two- and three-color smFRET imaging, we examine the aa-tRNA selection mechanism from four distinct structural perspectives under presteady-state conditions to determine the order and timing of EF-Tu dissociation from aa-tRNA and the ribosome. Our results demonstrate that EF-Tu dissociates during the proofreading stage of aa-tRNA selection, after aa-tRNA initiates its accommodation into the A site, and prior to peptide bond formation. We do not observe EF-Tu residing on the ribosome after aa-tRNA selection is complete.
Strikingly, EF-Tu bound to GTP was infrequently observed to repetitively reengage cognate aa-tRNA within the ribosome. These findings are consistent with a small subpopulation of cognate aa-tRNAs undergoing multiple rounds of proofreading in the absence of peptide bond formation. Consistent with this hypothesis, pharmacological inhibition of peptide bond formation markedly enhanced the frequency of such events. These findings suggest that ternary complex formation may facilitate proofreading and the accommodation of aa-tRNA substrates that are slow to form a peptide bond via additional energy expenditure. EF-Tu’s capacity to rebind the ribosome may also provide a mechanism for cognate and near-cognate aa-tRNAs to recycle back to the cytosol as ternary complex when rejection occurs after GTP hydrolysis. Futile cycles of ternary complex formation and proofreading may have the potential to lead to spurious consumption of GTP in a manner that may contribute to growth inhibition and cellular stress responses.
Results
Three-Color Imaging of tRNA Selection.
The reaction coordinate of aa-tRNA selection has been extensively characterized using two-color smFRET approaches by measuring the distance between an acceptor-labeled aa-tRNA substrate and donor-labeled reference points within the ribosome (24, 44–46). These studies have revealed that the initial selection and proofreading steps of aa-tRNA selection are defined by rapid and reversible rearrangements of aa-tRNA between distinct configurations within the A site that exhibit distinct FRET values. The transitions between each FRET value, or state, principally reflect pivot-like motions of aa-tRNA axial to the mRNA codon–tRNA anticodon interaction at the decoding site (19, 44, 45, 47).
Here, we extend our investigations of aa-tRNA selection using a three-color smFRET approach (described in detail below) in which the far-red acceptor fluorophore, LD750, is attached to EF-Tu in order to track EF-Tu’s presence and position with respect to aa-tRNA and the ribosome. These investigations include four distinct structural perspectives of the aa-tRNA selection process (Figs. 2 A and D and 3A and SI Appendix, Figs. S1 and S3D), consisting of a donor-labeled (Cy3 or Cy3B) peptidyl-tRNA or ribosome, an acceptor-labeled (LD650) aa-tRNA within the ternary complex, and far-red acceptor-labeled (LD750) EF-Tu.
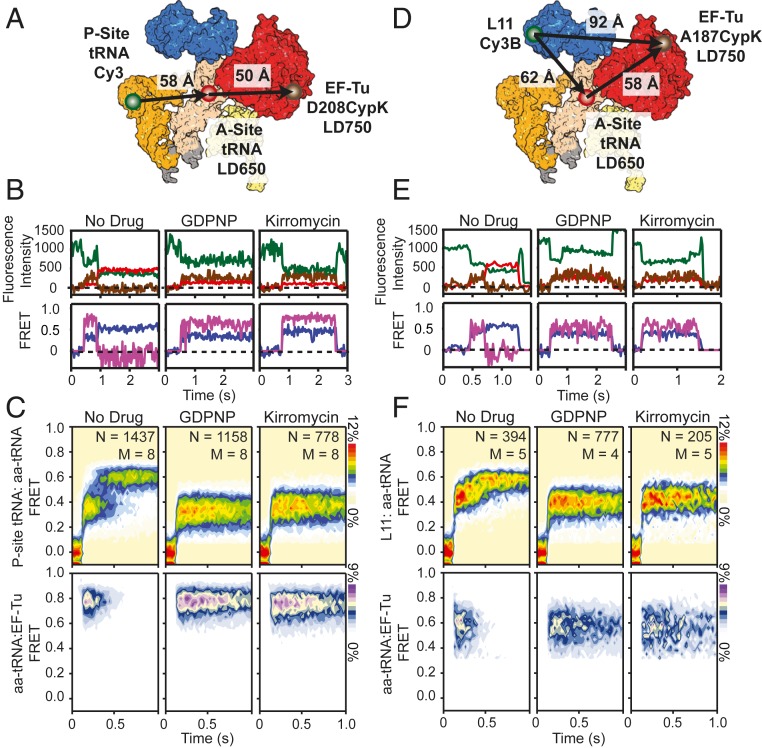
Fig. 2.
Three-color imaging of presteady-state tRNA selection. (A) Structural representation depicting the approximate locations of P-site tRNA (orange), aa-tRNA (white), and EF-Tu (red) during tRNA selection. The locations of Cy3 (green circle), LD650 (red circle), and LD750 (brown circle) are shown. (B, Upper) Fluorescence intensity trajectories of Cy3 (dark green), LD650 (red), and LD750 (brown) and corresponding (Lower) single-molecule trajectories of P-site tRNA(Cy3):aa-tRNA(LD650) FRET (purple) and aa-tRNA(LD650):EF-Tu(LD750) FRET (fuchsia) imaged at 25-ms time resolution during stopped-flow delivery of a dual-labeled ternary complex to P-site–labeled ribosomes at 15 mM Mg2+. (C) Population histograms for P-site tRNA(Cy3):aa-tRNA(LD650) FRET (Upper) and aa-tRNA(LD650):EF-Tu(LD750) FRET (Lower) during presteady-state tRNA selection without drug (Left), with GDPNP (Center), or with kirromycin (Right). (D) Structural representation depicting the approximate locations of L11 (blue), A-site tRNA (white), and EF-Tu (red) in complexes used to study the kinetics of EF-Tu and aa-tRNA from the perspective of the large subunit during tRNA selection. The locations of Cy3 (green circle), LD650 (red circle), and LD750 (brown circle) are shown. (E, Upper) Fluorescence intensity trajectories of Cy3B (dark green), LD650 (red), and LD750 (brown) and corresponding (Lower) single-molecule trajectories of L11(Cy3B):aa-tRNA(LD650) FRET (purple) and aa-tRNA(LD650):EF-Tu(LD750) FRET (fuchsia) imaged at 25-ms time resolution during stopped-flow delivery of a dual-labeled ternary complex to ribosomes with donor-labeled L11 at 15 mM Mg2+. (F) Population histograms for L11(Cy3B):aa-tRNA(LD650) FRET (Upper) and aa-tRNA(LD650):EF-Tu(LD750) FRET (Lower) during presteady-state tRNA selection without drug (Left), with GDPNP (Center), or with kirromycin (Right).
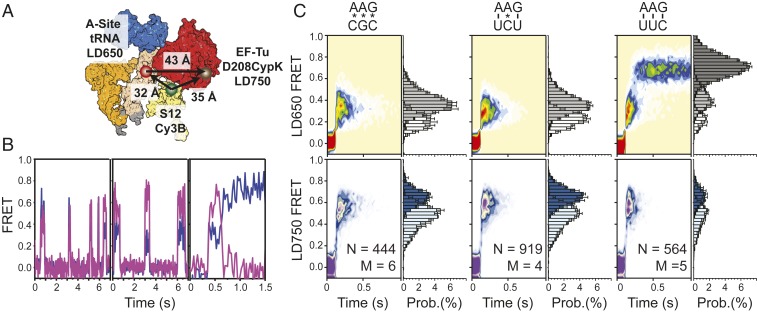
Fig. 3.
Delivery of tRNA to near-cognate and noncognate ribosomal complexes. (A) Structural representations depicting the approximate locations of P-site tRNA (orange), A-site tRNA (white), S12 (light yellow), and EF-Tu (red) during tRNA selection. The locations of Cy3 (green circle), LD650 (red circle), and LD750 (brown circle) are indicated. (B) Representative LD650 FRET (purple) and LD750 FRET (fuchsia) trajectories observed at 25-ms time resolution during noncognate tRNA selection (Left), near-cognate selection (Center), and cognate selection (Right) in S12-labeled complexes at 15 mM Mg2+; individual fluorescence traces for each dye channel are not shown for clarity of presentation. (C) Population LD650 FRET histograms (Upper) and corresponding LD750 histograms (Lower) observed in noncognate (Left), near-cognate (Center), and cognate (Right) tRNA selection.
In the first three structural perspectives, we employed a linear FRET cascade in which FRET is initiated via excitation of either Cy3-labeled Peptidyl (P)-site tRNA or Cy3B-labeled ribosomal protein L11 (uL11) within surface-immobilized ribosome particles (Methods). Upon ternary complex binding to the ribosome, excited donor fluorophores can transfer energy through the LD650 fluorophore on aa-tRNA, which can then transfer to the LD750 fluorophore on EF-Tu (Fig. 2 A and D and SI Appendix, Figs. S1 and S3D). Energy transfer directly between the donor fluorophore on the ribosome and LD750-labeled EF-Tu is negligible given the distance between the dyes (>90 Å) and the Förster radius of the donor-LD750 FRET pair (∼40 Å) (48, 49). In this cascading geometry, two ratiometric fluorescent signals are obtained, which can be interpreted as parallel measurements of FRET efficiency (FRET), and thus distance, between the donor-labeled ribosome and acceptor-labeled aa-tRNA, and between acceptor-labeled aa-tRNA and far-red acceptor-labeled EF-Tu.
In the fourth structural perspective, ribosomal complexes were labeled with a Cy3B on S12 (uS12) (Fig. 3A) to give rise to a triangular FRET geometry, where the donor on S12 and the acceptor on aa-tRNA can both directly excite LD750 on EF-Tu (50). Here, the lifetime of EF-Tu on the ribosome is revealed by the net energy transfer through LD750 (LD750 FRET) and the progression of aa-tRNA is revealed by the net energy transfer through LD650 (LD650 FRET). In this geometry, FRET values can only be qualitatively interpreted in the context of structural knowledge of the tRNA selection mechanism.
Presteady-State Three-Color Single-Molecule Measurements of aa-tRNA Selection.
To delineate the order and timing of events during cognate aa-tRNA selection, we prepared ribosome initiation complexes (70SICs) bearing donor (Cy3)-labeled tRNAfMet in the P site and displaying a UUC phenylalanine codon in the A site (Methods). Purified 70SICs were surface-immobilized within passivated microfluidic chambers and imaged by exciting the donor fluorophore with a single-frequency 532-nm diode laser using a custom-built, prism-based, total internal reflection fluorescence microscope (45). Fluorescence was collected using a 1.27 NA water-immersion objective. Cy3, LD650, and LD750 fluorescence was imaged onto three temporally synchronized sCMOS cameras following optical treatments for emission frequency and spatial separation (Methods).
Measurements of aa-tRNA selection were initiated by stopped-flow delivery of a ternary complex containing LD650-labeled Phe-tRNAPhe, GTP (1 mM), and LD750-labeled EF-Tu, where the fluorophore was positioned at either residue 208 within domain II (D208CypK), residue 187 within domain I (A187CypK), or residue 304 (K304CypK) within domain III. Experiments were performed at 25-ms time resolution with 5 or 15 mM Mg2+. Elevated levels of Mg2+ (15 mM) slows, but does not otherwise alter the aa-tRNA selection mechanism (19). Our use of a relatively low imaging time resolution (25 ms) was necessary to enable photon collection efficiencies for the LD750 fluorophore sufficient to capture salient features of the multistep tRNA selection mechanism. This constraint did, however, lead to reduced detection of short-lived intermediates during aa-tRNA selection, such as the codon recognition (CR) state. Correspondingly, the sequence of FRET states transited during the selection process was largely extracted from individual single-molecule fluorescence and FRET trajectories.
EF-Tu Dissociation Is Coordinated with aa-tRNA Accommodation.
We first examined the aa-tRNA selection mechanism using fluorescently labeled peptidyl- and aa-tRNAs together with EF-Tu site-specifically labeled within domain II. In line with previous investigations (19, 24, 45, 46), productive aa-tRNA delivery events were characterized by rapid transit of aa-tRNA through distinct positions within the A site exhibiting low- (∼0.2), intermediate- (∼0.32), and high- (∼0.62) FRET between P-site tRNA and A-site tRNA (P-site tRNA:aa-tRNA FRET) (Fig. 2 B and C, Left). Here, the low-FRET state reflects initial, transient binding of the codon–anticodon pair, the CR state, prior to EF-Tu docking at the GAC (19, 30). Intermediate FRET reflects GTPase-activated (GA′) states, in which the GAC is first transiently and reversibly sampled, followed by productive docking, wherein aa-tRNA adopts a bent configuration and EF-Tu engages the SRL, facilitating GTP hydrolysis (GA) and ending initial selection (30, 33, 34, 39, 51, 52). High-FRET states reflect tRNA positions in which aa-tRNA transiently samples the universally conserved accommodation corridor within the 50S A site (AC′), as well as the fully accommodated (AC) configuration where peptide bond formation can occur (12, 47, 53). Nonproductive aa-tRNA selection events were observed to be transient in nature, abruptly terminating FRET from predominately low-FRET states.
At 15 mM Mg2+, we observed the average time for aa-tRNA to transit between the CR and AC states to be ∼370 ms (kAC = 2.7 ± 0.06 s−1) (Fig. 2C and SI Appendix, Supplementary Methods, Fig. S3A, and Table S1) (19). The mean transit time decreased to ∼200 ms (kAC = 5.1 ± 0.5 s−1) at 5 mM Mg2+ (Fig. 4C and SI Appendix, Fig. S3A and Table S1). These transit times are longer than previously reported for both magnesium concentrations (19). We attribute such changes to the lower time resolution of the present investigations and the stipulation that the traces selected for analyses must display clear evidence of LD750 fluorescence (Methods).
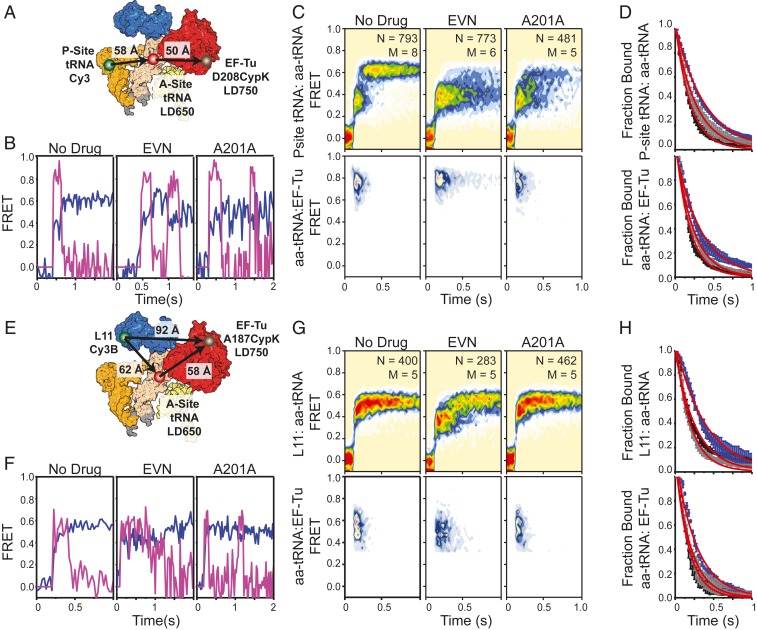
Fig. 4.
Inhibition of tRNA accommodation with antibiotics. (A) Structural diagram depicting the P-site donor-labeled FRET perspective used to determine the effects of A201A and EVN on the residence time of EF-Tu on the ribosome. smFRET trajectories (B) and population P-site(Cy3):aa-tRNA(LD650) FRET histograms (C) showing the evolution of tRNA and EF-Tu during presteady-state imaging at 5 mM Mg2+ without drug (Left), stalled with EVN (Center), or inhibited with A201A (Right) as observed at 25-ms time resolution. (D) Cumulative survivor plots for uninhibited (black), A201-inhibited (gray), and EVN-inhibited (blue) complexes. (E) Structural diagram of the L11 donor-labeled FRET perspective. smFRET trajectories (F) and population L11(Cy3B):aa-tRNA(LD650) FRET histograms (G) showing the evolution of tRNA and EF-Tu in during presteady-state imaging without drug (Left), stalled with EVN (Center), or inhibited with A201A (Right). (H) Cumulative survivor plots for uninhibited (black), A201-inhibited (gray), and EVN-inhibited (blue) complexes.
Ternary complex binding events were generally accompanied by a short-lived burst of LD750 fluorescence (Fig. 2 B and C, Left), indicative of FRET between aa-tRNA and EF-Tu (tRNA:EF-Tu FRET ∼0.75). Due to the finite time resolution of the present investigations, we were unable to detect transient (<25 ms) aa-tRNA selection events occurring within a single imaging frame.
Visual inspection of individual aa-tRNA selection events revealed that FRET between aa-tRNA and EF-Tu dissipated immediately upon formation of the AC state. Analysis of these data revealed that the residence time of EF-Tu on the ribosome closely paralleled the average transit time of aa-tRNA from the CR to AC states at both 15 mM (∼320 ms, kTu = 3.1 ± 0.1 s−1) and 5 mM Mg2+ (∼180 ms, kTu = 5.5 ± 0.3 s−1) (SI Appendix, Fig. S3A and Table S1). The timing of aa-tRNA accommodation and EF-Tu dissociation from the ribosome remained strongly correlated when experiments were performed at increased time resolution (10 ms) (SI Appendix, Fig. S3C). Similar results were also obtained when the LD750 fluorophore was moved to domain III of EF-Tu (K304CypK), 30 Å closer to the A-site tRNA (SI Appendix, Fig. S3 D and E). These findings suggest that aa-tRNA’s passage of the accommodation corridor is either closely timed with EF-Tu’s dissociation from aa-tRNA and the ribosome or rearrangements in EF-Tu that move the site of labeling within domain II to a position distal to the donor fluorophore on aa-tRNA (e.g., outside the FRET range; ∼70 Å).
To ensure that these findings were independent of the labeling positions, we performed analogous experiments using a distinct labeling strategy. Here, we delivered LD650-labeled Phe-tRNAPhe in complex with a LD750-labeled EF-Tu construct site-specifically tagged on domain I (A187Cypk) to ribosomal complexes containing Cy3B-labeled L11 (Methods and Fig. 2D). From this structural perspective, changes in FRET between aa-tRNA and L11 (L11:aa-tRNA FRET) principally reflect repositioning of the tRNA body within the accommodation corridor relative to the large subunit. Changes in FRET between aa-tRNA and EF-Tu reflect rearrangements between aa-tRNA and domain I of EF-Tu.
From the L11:aa-tRNA perspective (Fig. 2D), stopped-flow delivery of LD650-labeled Phe-tRNAPhe again gave rise to three discernable FRET values corresponding to CR (∼0.22), GA (∼0.42), and AC (∼0.55) states. Due to the finite time resolution, only the GA (∼0.42) and AC (∼0.55) states were evident at the population level (Fig. 2 E and F). Here, the transition to the AC state was accompanied by a nearly instantaneous loss of FRET between aa-tRNA and EF-Tu (Fig. 2 D–F and SI Appendix, Fig. S3B). The similarity of these findings regarding the timing of EF-Tu’s separation from aa-tRNA and dissociation from the ribosome collectively indicate that the observations made are largely independent of the sites of fluorophore labeling within the ribosome and EF-Tu. They also suggest that proofreading involves the separation of both domains I and II from aa-tRNA during its transition from the GA (A/T) to the AC (A/A) states, which we observed to occur on similar time scales at the time resolution of our experiments. Faster imaging studies will be needed to potentially resolve the order and timing of events within the 25-ms time window.
Taking advantage of both structural perspectives, we specifically examined the GA (A/T) state of aa-tRNA selection by performing analogous experiments in the presence of saturating concentrations of either the nonhydrolyzable GTP analog GDPNP (1 mM) or the antibiotic kirromycin (2 µM). GDPNP blocks the selection mechanism at the penultimate step of initial selection, while kirromycin inhibits domain rearrangements in EF-Tu, subsequent to GTP hydrolysis, at the onset of proofreading immediately after inorganic phosphate (Pi) release (54). We first delivered LD650-labeled Phe-tRNAPhe and LD750-labeled EF-Tu (D208Cypk) to ribosomal complexes with donor-labeled P-site tRNA. In accordance with two-color smFRET studies (19, 44) and structures of decoding complexes (30, 33, 34), both GDPNP and kirromycin stalled the aa-tRNA selection process in the GA state and extended the residence time of EF-Tu by ∼18-fold to ∼3.3 s (kTu = 0.3 ± 0.03 s−1). These findings are consistent with EF-Tu’s separation from aa-tRNA and the ribosome requiring both GTP hydrolysis and conformational changes in EF-Tu subsequent to Pi release (31, 38, 55) (Fig. 2 B and C, Center and Left and SI Appendix, Fig. S4A).
Notably, the mean FRET efficiency between aa-tRNA and EF-Tu was slightly lower when stalled by kirromycin (∼0.74) compared to GDPNP (∼0.77) (Fig. 2 C, Lower, and SI Appendix, Fig. S4 B, Lower). The width of the FRET distribution was also increased in kirromycin-stalled complexes (SI Appendix, Fig. S4B). The changes in aa-tRNA:EF-Tu FRET occurred with a simultaneous change in FRET efficiency between P-site tRNA and the delivered A-site tRNA from ∼0.31 with GDPNP to ∼0.35 with kirromycin. The effects of GDPNP and kirromycin on the selection mechanism were also apparent when tRNA selection was imaged from the Cy3B-labeled L11 structural perspective (Fig. 2 E and F, Center and Right, and SI Appendix, Fig. S4 C and D). These observations are consistent with domain separation in EF-Tu being required for aa-tRNA to accommodate into the PTC and for EF-Tu⋅GDP to release from the ribosome (19, 31, 55).
EF-Tu and aa-tRNA Dissociate in Parallel from Noncognate and Near-Cognate Complexes.
To assess the residence time of EF-Tu on the ribosome and the timing of EF-Tu dissociation in the context of near and noncognate aa-tRNA decoding, we programmed 70SICs with either non- (CGC) or near- (UCU) cognate mRNA codons and stopped-flow–delivered a ternary complex consisting of LD750-labeled EF-Tu (D208CypK) and LD650-labeled Phe-tRNAPhe to surface-immobilized ribosome complexes labeled with a Cy3B donor fluorophore on the C terminus of ribosomal protein S12 (Methods and Fig. 3A). This system has a triangular geometry in which both Cy3B-labeled S12 and LD650-labeled aa-tRNA excite the LD750 fluorophore on EF-Tu.
Ternary complex binding to ribosomal complexes programmed with noncognate and near-cognate mRNA codons exhibited clear indications of energy transfer through S12 to aa-tRNA, as well as from S12 and aa-tRNA to EF-Tu (Fig. 3B). These findings are consistent with both the predicted FRET geometries and with structures of ternary complexes captured in intermediate states of the aa-tRNA selection process (SI Appendix, Fig. S5A) (33, 34). In both conditions, individual FRET trajectories (Fig. 3B) and population FRET histograms (Fig. 3C) revealed that the appearance and disappearance of both FRET signals were synchronized, indicating that near- and noncognate tRNAs are predominantly rejected by the ribosome as intact ternary complexes during initial selection (19).
Notably, the FRET values exhibited by near- and noncognate complexes were nonuniform. Noncognate ternary complex-binding interactions displayed a greater proportion of events in which FRET via both LD650 and LD750 fluorophores (LD650 and LD750 FRET) were of intermediate values; near-cognate events displayed a greater proportion of events in which LD750 FRET was significantly higher than LD650 FRET (Fig. 3 B, Left and Center). Such tendencies were also captured at the population level (Fig. 3 C, Left and Center). These observations suggest that near-cognate ternary complexes progress further in the aa-tRNA selection reaction coordinate than noncognate, consistent with EF-Tu having a greater probability of docking with the GAC and subsequently hydrolyzing GTP with near- than with noncognate tRNA (19, 23). In line with kinetic models of aa-tRNA selection (19, 56), both the lifetime of the GA (A/T) state as well as the overall residence time of EF-Tu on the ribosome were codon-dependent (SI Appendix, Fig. S5B). These findings are consistent with domain II of EF-Tu shifting toward ribosomal protein S12 and EF-Tu docking at the GAC during 30S domain closure (SI Appendix, Fig. S5A) (30, 33, 34, 57).
EF-Tu Changes Conformation within Ternary Complex Prior to Its Dissociation.
Delivery of the same dual-labeled ternary complex (LD650-labeled Phe-tRNAPhe and LD750-labeled EF-Tu) to S12-labeled ribosomes programmed with a cognate (UUU) mRNA codon exhibited step-wise progressions in LD650 and LD750 FRET prior to the loss of LD750 fluorescence (Fig. 3 B, Right). Inspection of individual traces revealed, albeit rarely, that this progression begins with lower LD750 FRET, which subsequently transitions through high-LD750 FRET, followed by a return to lower-LD750 FRET states prior to the loss of FRET that accompanies EF-Tu dissociation. In these cases, LD650 FRET initiated with an intermediate-FRET value, from which both lower-FRET and higher-FRET states were transited. These data are consistent with aa-tRNA reversibly sampling short-lived (approximately <25 ms) configurations within the A site after GTP hydrolysis, that occur prior to EF-Tu dissociation and complete accommodation into the A site (19, 44).
Given these findings, and the notion that EF-Tu dissociation is somehow coordinated with the formation of the AC state, we hypothesized that EF-Tu release from the ribosome could be coupled to the accommodation of aa-tRNA into the A site. To test this model, we set out to perform three-color aa-tRNA selection studies in the presence of peptidyl transferase inhibitors, which are known to alter the accommodation mechanism (46, 53). We first stopped-flow–delivered a ternary complex comprised of LD650-labeled Phe-tRNAPhe and LD750-labeled EF-Tu (D208CypK) to a P-site tRNA donor-labeled ribosomal complex (Fig. 4A) in the presence of the peptide antibiotic evernimicin (EVN). EVN binds Helices 89 and 91 adjacent to the GAC to sterically inhibit aa-tRNA elbow movement through the accommodation corridor (53), while having no detectable impact on the GTPase activities of translational GTPases (58, 59). Positioning the donor fluorophore in the P site in this manner enabled measurement of aa-tRNA elbow conformation during navigation of the accommodation corridor.
In agreement with previous work, saturating EVN (20 µM) delayed aa-tRNA entry within a GA-like FRET state when compared to untreated complexes (Fig. 4 B and C, Left and Center). Importantly, the delayed accommodation of aa-tRNA into the A site (kAC = 2.6 ± 0.01 s−1 vs. kAC = 5.1 ± 0.5 s−1) was accompanied by a modestly prolonged EF-Tu residence time on the ribosome of ∼320 ms versus ∼180 ms in the absence of the drug (kTu = 3.1 ± 0.4 s−1 vs. kTu = 5.5 ± 0.28 s−1) (Fig. 4 C and D). Visual inspection of individual fluorescence and FRET traces demonstrated that when EF-Tu dissociates from the ribosome it does so following short-lived structural transitions in which the aa-tRNA:P-site tRNA FRET signal reversibly samples high-FRET, AC-like states (Fig. 4 B, Center). As EVN binds distally to the ternary complex binding site, we reasoned that EVN may allosterically regulate the conformational changes in EF-Tu required for its release from aa-tRNA and the ribosome by preventing entry of the aa-tRNA elbow into the accommodation corridor prior to peptide bond formation (58).
To gain further clarity in this regard, we performed analogous experiments in the presence of the antibiotic A201A, which inhibits entry of the 3′CCA end of aa-tRNA into the PTC but not entry of the aa-tRNA elbow into the accommodation corridor (46). In the presence of A201A (20 µM), we observed no measurable impact on the waiting time of aa-tRNA in the GA state or the residence time of EF-Tu on the ribosome (Fig. 4 C and D). We confirmed these results by performing analogous experiments from a distinct structural perspective involving the stopped-flow delivery of LD750-labeled EF-Tu (A187CypK) to L11 donor-labeled complexes (Fig. 4 E–H). From this perspective, we observed step-like decreases in aa-tRNA:EF-Tu FRET in the presence of EVN (Fig. 4 F, Center), indicative of partial separation of the aa-tRNA body from domain I of EF-Tu. These data suggest that EF-Tu dissociation after GTP hydrolysis is linked to entry of the aa-tRNA elbow into the accommodation corridor, such that when its entry is partially blocked by an inhibitor such as EVN, EF-Tu⋅GDP is more likely to remain on the ribosome. In this view, we speculate that a sufficient extent of aa-tRNA elbow separation from EF-Tu is required for the structural rearrangements in EF-Tu required for its release from aa-tRNA and the ribosome.
Notably, in the presence of either A201A or EVN, we observed that EF-Tu was able to reengage the ribosome, demarked by a return of LD750 fluorescence and backward excursions of aa-tRNA to a GA-like state (Fig. 4 B and F). In each instance, EF-Tu rebinding occurred while the A site was filled. Hence, the return of LD750 fluorescence was not the result of a new round of aa-tRNA selection. These observations suggest that the slowing of peptide bond formation in the presence of chemically distinct inhibitors that bind different regions of the accommodation corridor can result in backward excursions of aa-tRNA that are large enough in amplitude for EF-Tu to reengage the the 3′-CCA end.
Strikingly, reinspection of individual cognate aa-tRNA selection smFRET traces revealed that similar rebinding events were also evidenced in the absence of drugs, albeit rarely (∼1% of all traces). Similar levels of EF-Tu rebinding were observed for all three structural perspectives examined (SI Appendix, Fig. S6), indicating that the occurrence of such events is independent of the fluorophore-labeling strategy employed. These findings are consistent with a small population of cognate aa-tRNAs being unable to efficiently terminate proofreading with peptide bond formation, which normally prevents the aa-tRNA excursions or interactions that enable EF-Tu to reengage. Here, the absence of peptide bond formation may result if aa-tRNA is delivered to a small subpopulation of ribosomes bearing deacylated tRNA within the P site or if a small fraction of ribosomes undergoing accommodation are unproductive. Contemporary models of aa-tRNA selection indeed posit that cognate selection events terminate without peptide bond formation at low frequency (56). We conclude from these data that EF-Tu can repetitively engage aa-tRNA bound within the A site of the ribosome both in the absence and presence of specific inhibitors that reduce accommodation efficiency. In this context, we note that EF-Tu rebinding may either reflect the binding of a new EF-Tu⋅GTP complex or reengagement of EF-Tu⋅GDP that has not fully dissociated.
Direct Evidence of Ternary Complex Formation on the Ribosome.
To examine whether the observed EF-Tu rebinding events represent the formation of a new ternary complex on the ribosome with EF-Tu from solution and additional rounds of GTP hydrolysis, we performed two-color smFRET imaging experiments in which we used unlabeled EF-Tu to deliver donor (Cy3B)-labeled Phe-tRNAPhe to surface-immobilized ribosome complexes in the presence of hygromycin A (20 µM), an antibiotic with a similar mode of action as A201A but with higher binding affinity for the PTC (46). Following accommodation of the donor-labeled aa-tRNA into the A site, the flow-cell was flushed with buffer containing hygromycin A (20 µM), and then LD650-labeled EF-Tu (100 nM D208CypK) was delivered in the presence of either GTP or GDPNP (1 mM). In so doing, we observed clear evidence of FRET between donor-labeled aa-tRNA and acceptor-labeled EF-Tu, indicative of ternary complex formation events on the ribosome (Fig. 5A).
Fig. 5.
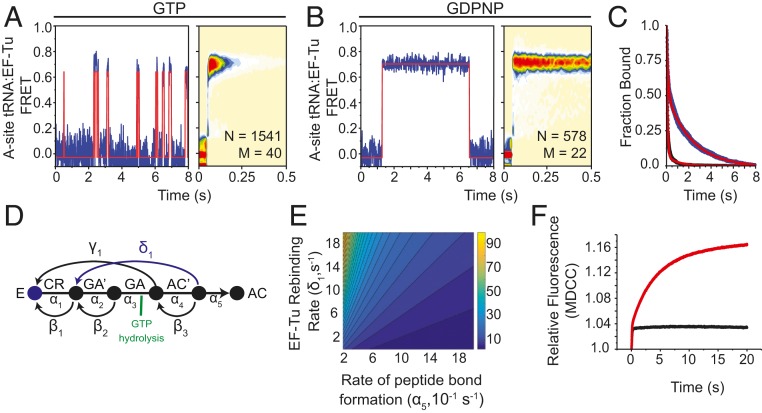
EF-Tu rebinds stalled ribosomes. Representative smFRET trajectories (Left) and tRNA(Cy3B):EF-Tu(LD650) FRET histograms (Right) observed at 10-ms time resolution when (A) EF-Tu⋅GTP or (B) EF-Tu⋅GDPNP is stopped-flow–delivered with 5 mM Mg2+ to A-site–filled ribosomal complexes stalled by 20 µM hygromycin A. smFRET traces were idealized using a hidden Markov model with two states (orange line). (C) Survival plot showing the ensemble lifetime of interactions between stalled ribosomal complexes and either EF-Tu⋅GTP (black line) or EF-Tu⋅GDPNP (blue line). Each condition was fit to a two-term exponential decay model (red line). (D) Markov model of tRNA selection composed of forward (α) and backward (β) elemental rate constants with the proposed EF-Tu rebinding pathway (blue). (E) Contour plot illustrating the relationship between the rates of peptide bond formation, EF-Tu rebinding, and the number of GTP molecules hydrolyzed (z axis). (F) Pi release in tRNA selection without drug (black trace) and with 20 µM hygromycin A (red trace).
In the presence of GTP, the average dwell-time of EF-Tu during rebinding events was ∼160 ms (Fig. 5 A and C). Binding events of this kind were not observed when analogous experiments were performed in the absence of hygromycin A, consistent with new ternary complex formation events being specifically associated with circumstances in which peptide bond formation has yet to occur. In line with the observed FRET events corresponding to bona fide ternary complex formation events on the ribosome that culminate in GTP hydrolysis, the lifetime of the observed FRET states increased by 10-fold (∼1.2-s duration) when acceptor-labeled EF-Tu was instead delivered with GDPNP (Fig. 5 B and C). These findings indicate that aa-tRNA molecules that fail to undergo peptide bond formation after the initial release of EF-Tu are able to reform ternary complex while aa-tRNA remains bound within the 30S subunit.
Hygromycin A Increases GTP Turnover during tRNA Selection.
To better understand the energetic costs associated with reforming ternary complex on the ribosome, we next asked how the rates of EF-Tu rebinding and peptide bond formation influence the magnitude of GTP hydrolysis during tRNA selection. Using previously derived elemental rate constants (19), we constructed a Markov model that mathematically represents the transitions of a single aa-tRNA substrate between individual states during the selection process (Fig. 5D). This model assumes that aa-tRNA selection is complete once the substrate forms a peptide bond (estimated by the rate of achieving the AC state). Based on our findings that EF-Tu⋅GTP can engage stalled aa-tRNA in the A site, accompanied by rapid GTP hydrolysis, we modeled the EF-Tu rebinding pathway as occurring prior to peptide bond formation (AC′ state), causing the system to return to the CR state, where the selection mechanism can resume. Transitions from the GA′ state to the GA state were also modeled as hydrolyzing one GTP molecule. Under these assumptions, the competition between the rates of EF-Tu rebinding (δ1) and peptide bond formation (α5) determines the number of GTP molecules hydrolyzed prior to peptide bond formation (Fig. 5E). At physiological rates of peptide bond formation (approximately >50 s−1) (19, 26), aa-tRNA has a high probability of progressing immediately to peptide bond formation after only one molecule of GTP has been hydrolyzed. However, at lower rates of peptide bond formation (approximately <10 s−1), EF-Tu rebinding dramatically increases the number of GTP molecules hydrolyzed per peptide bond.
Based on these theoretical considerations, and the evidence that multiple rebinding events can be seen in single-molecule investigations, we predicted that inhibition of the tRNA selection mechanism with hygromycin A would lead to elevated levels of GTP hydrolysis. To test this hypothesis, we performed ensemble stopped-flow investigations of aa-tRNA selection in the presence of a fluorescently labeled phosphate binding protein (PBP), which exhibits a large increase in fluorescence intensity upon binding Pi (60). Consistent with our model predictions, the inhibition of aa-tRNA selection with hygromycin A led to a marked increase in relative fluorescence compared to uninhibited complexes, corresponding to at least a fourfold increase in the number of GTP hydrolysis events (Fig. 5F).
EVN Inhibits tRNA Release from EF-Tu.
To gain further insights into the structural events that cause EVN to delay EF-Tu’s release from the ribosome during aa-tRNA selection, we performed all-atom, structure-based molecular dynamics (MD) simulations of the intact ribosome bound to a ternary complex with and without EVN (Fig. 6). These studies were performed to gain a deeper understanding of our smFRET observation that suggest that aa-tRNA fluctuations away from EF-Tu during proofreading initiate EF-Tu release from the ribosome and that EVN binding to the ribosome slows the rate of EF-Tu release (Fig. 4 C, D, G, and H). Previous MD investigations of EF-Tu conformational changes have suggested that aa-tRNA dissociation from EF-Tu entails domain separation and rotation within EF-Tu as well as increased flexibility of switch I (40, 41). Here, we focused our studies on EVN’s potential hindrances of aa-tRNA’s movement through the aa-tRNA accommodation corridor, providing preliminary insights into the nature of the conformational changes between EF-Tu and aa-tRNA during proofreading. Although structure-based methods lack some of the detail present in explicit solvent MD, they are relatively rapid and offer significantly greater conformational sampling. Structure-based methods have also proven excellent at resolving geometric constraints during conformational change across a diverse range of biological systems, including macromolecular systems such as the ribosome (47, 61, 62).
Fig. 6.
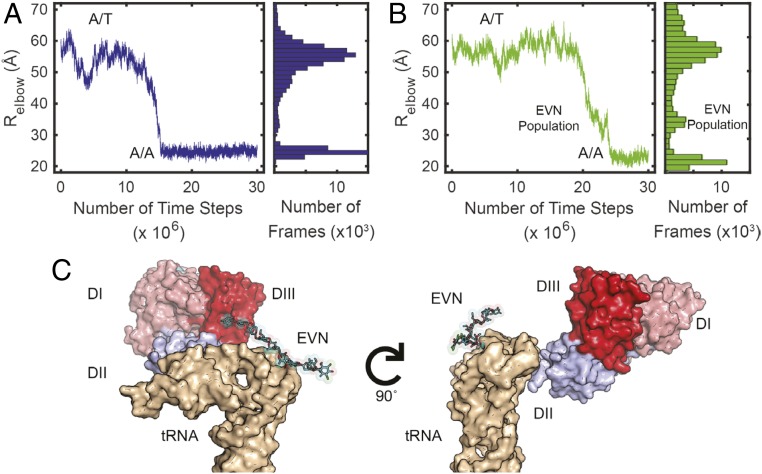
MD simulations demonstrate that aa-tRNA accommodation is sterically blocked by EVN preventing EF-Tu disengagement. aa-tRNA accommodation was measured by the reaction coordinate Relbow (distance between O3′ of U60 and U8 of the A-site and P-site tRNA, respectively), accommodation was considered complete when Relbow of ∼25 Å. (A) Single trace of Relbow during the first 30 million time steps (Left) and a population histogram of 60 simulations (Right) in the absence of antibiotics. (B) Single trace of Relbow during the first 30 million time steps (Left) and a population histogram of 60 simulations (Right) in the presence of EVN. An additional population in the presence of EVN is observed at an Relbow of ∼30 to 38 Å. (C) Structural representation of the additional population observed in the presence of EVN revealing the steric block that EVN imposes on aa-tRNA accommodation (Left) and the interactions that are maintained between domain II of EF-Tu and the aa-tRNA (Right).
Using 120 individual simulations in the presence or absence of EVN, we reduced the dimensionality of aa-tRNA accommodation to a reaction coordinate that describes the distance between the aa-tRNA and P-site tRNA elbows (Relbow, the distance between O3′ of A-site U60 and P-site U8). Relbow has been shown to be an ideal reaction coordinate for investigations of the aa-tRNA selection mechanism as it minimizes the number of transition paths, distinguishes between the start (A/T) and end (A/A) points, is diffusive, and captures the transition-state ensemble during accommodation (63). In these simulations, the ternary complex was bound to the A site of the ribosome in a GDP-bound state (i.e., immediately after GTP hydrolysis and Pi release). Simulations were initiated with EF-Tu in a closed GTP-like conformation (PDB ID code 5UYK), which was subsequently allowed to spontaneously transition to an open, GDP-bound conformation. From this simulated EF-Tu⋅GDP conformation, in which the ternary complex remains in an A/T-like state, aa-tRNA was allowed to spontaneously transition to its fully accommodated position (PDB ID codes 5UYM, 4V66, 1EFC) (33, 64).
In the absence of EVN, aa-tRNA moved from the A/T state (Relbow ∼55 Å) to the A/A state (Relbow ∼25 Å) in a relatively fluid motion, which was only briefly delayed by the steric barrier imposed by H89 (47) (Fig. 6A). The observed delay was evident at an Relbow distance of ∼35 Å (Fig. 6A). In the presence of EVN, aa-tRNA was observed to briefly stall during accommodation at roughly the same barrier (Fig. 6B), consistent with EVN sterically hindering aa-tRNA passage through the accommodation corridor (Figs. 4B and 6B) (58). This finding agrees with our smFRET data, which show that aa-tRNA accommodation in the presence of EVN exhibits a spread of FRET values ranging from the A/T state (∼0.35 FRET) to higher FRET states, approaching, but not reaching the fully accommodated A/A state (∼0.62 FRET) (Fig. 4C). Our MD simulations also reveal that the observed inhibition arises from direct interactions of EVN with the aa-tRNA elbow as it attempts to navigate the accommodation corridor (Fig. 6C). In this stalled conformation, the acceptor stem of aa-tRNA remains engaged with domain II of EF-Tu (Fig. 6C). Given that these particular structure-based simulations do not include electrostatics or specific contacts between EF-Tu and aa-tRNA, it is difficult to ascertain what properties maintain this engagement. More detailed simulations will have to be performed to delineate the precise features of the EF-Tu dissociation mechanism and proofreading reaction coordinate. Nonetheless, the simulations we have performed indicate that EVN sterically hinders aa-tRNA passage through the accommodation corridor by attenuating the amplitude of spontaneous aa-tRNA excursions away from EF-Tu. Based on our experimental finding that EVN stalls the release of EF-Tu from the ribosome during the proofreading mechanism we infer that aa-tRNA fully disengages from EF-Tu during proofreading as aa-tRNA nears, or reaches, the H91 restriction within the accommodation corridor.
Discussion
The mechanism of aa-tRNA selection on the ribosome is central to information transfer between mRNA and protein, a critical target for clinically used antibiotics and a recognized bottleneck in engineering efforts to reprogram the ribosome to synthesize nonnatural polymers (65–67). While recent efforts have provided compelling insights into the initial selection mechanism (56), the rate-limiting features of the proofreading mechanism have yet to be fully resolved.
By implementing a three-color smFRET imaging approach, we have simultaneously measured the kinetics of aa-tRNA accommodation and EF-Tu dissociation during individual, productive aa-tRNA selection events. In so doing, we reveal that EF-Tu dissociates during the proofreading stage of the selection mechanism as aa-tRNA attempts to navigate the accommodation corridor, prior to peptide bond formation. We verified this conclusion by imaging the aa-tRNA selection process from multiple structural perspectives, in which the fluorophores used were attached to distinct components of the system. We also performed a battery of investigations in the presence of aa-tRNA selection inhibitors as well as MD simulations to gain a deeper understanding of the structural rearrangements in aa-tRNA and EF-Tu that accompany the proofreading mechanism. Although the precise nature, timing, and role of conformational events in EF-Tu required for aa-tRNA accommodation and EF-Tu release from the ribosome will require improved imaging time resolution and signal-to-noise ratio in three-color single-molecule measurements, as well as additional MD simulations and mutagenesis studies, the present investigations nonetheless offer important insights into the molecular basis of the proofreading mechanism during aa-tRNA selection.
Our findings suggest that EF-Tu release from the ribosome is triggered by thermally driven excursions of the aa-RNA elbow away from EF-Tu⋅GDP toward the accommodation corridor. These conformational changes likely occur after Pi has dissociated from EF-Tu as they include structural rearrangements in both the catalytic histidine 84 residue as well as the switch I region of EF-Tu, which coordinates the terminal, γ-phosphate residue of GTP (32, 37, 38, 68). Alternatively, the structural changes in EF-Tu that accompany aa-tRNA excursions toward the accommodation corridor could instead contribute to GTP hydrolysis and Pi release by facilitating the productive docking of EF-Tu⋅GTP with the SRL and GAC as the penultimate step in the initial selection process.
EF-Tu’s presence on the ribosome after GTP hydrolysis is expected to increase fidelity during early aspects of the proofreading mechanism (14, 19, 44). For example, fidelity could be increased if the activation barriers required for aa-tRNA to initiate its entry into the accommodation corridor are influenced by EF-Tu’s presence. Here, enhanced selectivity requires that barrier crossing is either less probable or more easily reversed for near- and noncognate aa-tRNAs, consistent with small-subunit domain closure processes contributing to the proofreading fidelity mechanism. Given extant literature, a more stable domain closure process could facilitate aa-tRNA movements away from EF-Tu by enforcing, or increasing, the bending and distortions observed in aa-tRNA when EF-Tu is docked at the GAC (31). Domain closure events that accompany cognate aa-tRNA recognition may also be more efficient at initiating restructuring events in EF-Tu that allow aa-tRNA passage of the accommodation corridor through direct contacts between domain II of EF-Tu and helix 5 (h5) within the 16S rRNA of the small-subunit shoulder domain (38).
High-resolution crystal structures of the ribosome bound to the ternary complex and stalled before GTP hydrolysis by use of nonhydrolyzable GTP analogs or immediately after GTP hydrolysis by the antibiotic kirromycin reveal that Pi release leads to switch I region disorder (38). In our MD simulations, we observe that fluctuations of the aa-tRNA acceptor arm away from EF-Tu are required for it to fully disengage domain II of EF-Tu. We therefore anticipate that these interactions must be broken for EF-Tu to release from the ribosome.
After aa-tRNA moves a sufficient distance from EF-Tu, up to and including partial or complete EF-Tu dissociation, the ribosome must retest the nature of the mRNA codon–tRNA anticodon pair to proofread (14–16). This consideration, together with extant structural data (28–30, 33, 34), implies that proofreading requires reversal of the domain closure process that follows CR during initial selection. While the link between aa-tRNA’s passage of the accommodation corridor, EF-Tu’s presence on the ribosome during proofreading, and the precise kinetic and structural features of the proofreading mechanism require further investigation, the findings presented suggest that aa-tRNA passage through the accommodation corridor, which entails steric interactions between the aa-tRNA elbow and H91 (53, 58), may contribute to reversal of the domain-closure process (domain opening) and thus directly or indirectly to EF-Tu release. Allosteric links of this kind could occur via signal transmission through the aa-tRNA elbow domain and anticodon stem loop or via local rearrangements in the 50S subunit that alter the positioning of neighboring helices 89 and 92, which are in direct contact with EF-Tu’s G domain. Reversible domain opening/closing processes within the 30S subunit and EF-Tu may also contribute to fidelity by facilitating the accommodation of cognate aa-tRNA into the PTC and the rejection of non- and near-cognate tRNAs. Notably, the point of steric contact between aa-tRNA and the ribosome is immediately proximal to the EVN binding site. This positioning may allow EVN to alter the passage of this steric barrier, and thus the process of proofreading after GTP hydrolysis, thereby altering the dissociation of EF-Tu from aa-tRNA and the ribosome.
This structural and kinetic framework helps explain the unexpected observation that EF-Tu can rebind aa-tRNAs that already reside within the A site of the ribosome during the proofreading stage of the selection mechanism. EF-Tu rebinding likely requires aa-tRNA to resample conformations and positions akin to those at the start of the initial selection process. The findings presented suggest that ternary complex formation on the ribosome rarely occurs during cognate aa-tRNA selection in the absence of impediments that decrease accommodation efficiency. In this context, it is interesting to consider circumstances in which the accommodation efficiency and peptide bond formation are known to be slowed, such as at poorly translated mRNA codons, including prolyl-tRNA decoding and nonnatural amino acid incorporation (69), as well as conditions that may be naturally encountered in which peptide bond formation may be slowed (e.g., lower temperature; elevated Mg2+) (70). In such contexts, our findings suggest that ternary complex formation events on the ribosome may occur at relatively high frequency, where additional energy expenditure has the potential to improve the rate and fidelity of translation (SI Appendix, Fig. S7) (71). Previous investigations have reported that synthesis of poly-phenylalanine peptides, which are thought to have difficulty entering the nascent peptide exit tunnel (72), requires multiple rounds of GTP hydrolysis per peptide bond formed (11, 73–75). EF-Tu rebinding may also give unincorporated aa-tRNAs the opportunity to dissociate from the ribosome in an intact ternary complex, reducing the likelihood of spontaneous deacylation, which may slow translation by nonenzymatic binding to the E site (76, 77) or impact other aspects of cellular physiology (78).
In this context, we note that slowed rates of peptide bond formation after EF-Tu release from the ribosome are predicted to result in an increase in GTP hydrolysis (Fig. 5E). Indeed, we observe experimentally that the inhibition of cognate tRNA selection with specific inhibitors of peptide bond formation increases GTP turnover (Fig. 5F). Our findings suggest that a significant portion of this increased turnover results from futile cycles of ternary complex formation on the ribosome and GTP hydrolysis (Fig. 5A). In principle, the consumption of GTP energy stores resulting from such rebinding events could significantly impact cellular homeostasis if a large number of ribosomes were stalled in such a manner and if sufficient levels of free EF-Tu⋅GTP or EF-Tu⋅GTP/Ts were present in the cell. Future studies will be needed to determine whether, and to what extent, futile cycles of ternary complex formation on the ribosome contribute to the translation mechanism, particularly for substrates that are naturally slow to form peptide bonds. It will also be important to gain a deeper understanding of the cellular consequences of pharmacological inhibition of peptide bond formation and the extent to which futile cycles of ternary complex formation trigger cellular responses to stress.
Methods
smFRET Data Acquisition and Analysis.
All smFRET data were acquired at 25 °C using a custom-built prism-based total internal reflection fluorescence microscope. Donor-labeled ribosomes were programmed on 5′-biotinylated mRNA transcripts and surface-immobilized (76). The 20-nM ternary complex consisting of LD750-labeled EF-Tu, LD650-labeled Phe-tRNAphe, and GTP was rapidly stopped-flow–delivered to surface-immobilized ribosomes in Tris Polymix Buffer [50 mM Tris-OAc pH = 7.5, 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 0.1 mM EDTA, 5 mM putrescine, 1 mM spermidine, and 1.5 mM BME] and a mixture of triplet state quenchers (79) (1 mM Trolox, 1 mM nitrobenzyl alcohol, and 1 mM cyclooctatetraene). All experiments were performed with an oxygen scavenging system (80). Unless otherwise stated, each experiment was performed with 5 mM Mg(OAc)2. During stopped-flow delivery of the ternary complex, the donor fluorophore (Cy3 or Cy3B) on the ribosome was directly excited with a 532 nM solid-state laser (Laser Quantum). Three-color data were acquired at 25-ms time-resolution unless otherwise stated.
FRET trajectories were extracted from each experimental movie using SPARTAN, a custom-built software package implemented in MATLAB (45). The fraction of all photons emitted by LD650 (i.e., LD650 FRET) and LD750 (i.e., LD750 FRET) in triangular FRET geometries was calculated as ILD650/(Icy3 + ILD650 + ILD750) and ILD750/(Icy3 + ILD650 + ILD750), respectively. In the cascading FRET geometry, where the donor cannot directly excite LD750, LD650 FRET and LD750 FRET were calculated as (ILD650 + ILD750)/(Icy3 + ILD650 + ILD750) and (ILD750)/(ILD650 + ILD750), respectively. All traces were manually inspected to confirm the presence of fluorescence emission from each of the three fluorophores. Further experimental methods are described in SI Appendix.
Data Availability.
All experimental data, materials and software are available upon request.
Supplementary Material
Acknowledgments
The authors thank all members of the S.C.B. laboratory for their helpful input and comments during the course of this project, and Daniel S. Terry and Emily Rundlet, in particular, for their suggestions on the written manuscript; Dr. Daniel N. Wilson for the generous gifts of evernimicin, hygromycin A, and A201A; and Drs. James B. Munro, Michael R. Wasserman, and Chad M. Kurylo for the preparation of specific ribosome complexes. This work was supported by NIH National Institute of General Medical Sciences Grants R01-GM079238-13 and R01-GM098859-07 (to S.C.B.) and R01-GM072686 (to K.Y.S.); Natural Sciences and Engineering Research Council Discovery Grant RGPIN-2016-05199 (to H.-J.W.); and Alberta Innovates Strategic Chairs Program SC60-T2 (H.-J.W.).
Footnotes
Competing interest statement: S.C.B. and R.B.A. hold equity interests in Lumidyne Technologies.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904469117/-/DCSupplemental.
References
- 1.Thomposon R. C., Dix D. B., Accuracy of protein biosynthesis. A kinetic study of the reaction of poly(U)-programmed ribosomes with a leucyl-tRNA2-elongation factor Tu-GTP complex. J. Biol. Chem. 257, 6677–6682 (1982). [PubMed] [Google Scholar]
- 2.Burnett B. J., et al. , Elongation factor Ts directly facilitates the formation and disassembly of the Escherichia coli elongation factor Tu·GTP·aminoacyl-tRNA ternary complex. J. Biol. Chem. 288, 13917–13928 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnett B. J., et al. , Direct evidence of an elongation factor-Tu/Ts·GTP·Aminoacyl-tRNA quaternary complex. J. Biol. Chem. 289, 23917–23927 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucas-Lenard J., Haenni A. L., Requirement of granosine 5′-triphosphate for ribosomal binding of aminoacyl-SRNA. Proc. Natl. Acad. Sci. U.S.A. 59, 554–560 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravel J. M., Shorey R. L., Shive W., The composition of the active intermediate in the transfer of aminoacyl-RNA to ribosomes. Biochem. Biophys. Res. Commun. 32, 9–14 (1968). [DOI] [PubMed] [Google Scholar]
- 6.Skoultchi A., Ono Y., Waterson J., Lengyel P., Peptide chain elongation. Cold Spring Harb. Symp. Quant. Biol. 34, 437–454 (1969). [DOI] [PubMed] [Google Scholar]
- 7.Lucas-Lenard J., Protein biosynthesis. Annu. Rev. Biochem. 40, 409–448 (1971). [DOI] [PubMed] [Google Scholar]
- 8.Gavrilova L. P., Perminova I. N., Spirin A. S., Elongation factor Tu can reduce translation errors in poly(U)-directed cell-free systems. J. Mol. Biol. 149, 69–78 (1981). [DOI] [PubMed] [Google Scholar]
- 9.Gavrilova L. P., Kostiashkina O. E., Koteliansky V. E., Rutkevitch N. M., Spirin A. S., Factor-free (“non-enzymic”) and factor-dependent systems of translation of polyuridylic acid by Escherichia coli ribosomes. J. Mol. Biol. 101, 537–552 (1976). [DOI] [PubMed] [Google Scholar]
- 10.Thompson R. C., Dix D. B., Karim A. M., The reaction of ribosomes with elongation factor Tu.GTP complexes. Aminoacyl-tRNA-independent reactions in the elongation cycle determine the accuracy of protein synthesis. J. Biol. Chem. 261, 4868–4874 (1986). [PubMed] [Google Scholar]
- 11.Weijland A., Parmeggiani A., Toward a model for the interaction between elongation factor Tu and the ribosome. Science 259, 1311–1314 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Noel J. K., Whitford P. C., How EF-Tu can contribute to efficient proofreading of aa-tRNA by the ribosome. Nat. Commun. 7, 13314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tapio S., Kurland C. G., Mutant EF-Tu increases missense error in vitro. Mol. Gen. Genet. 205, 186–188 (1986). [DOI] [PubMed] [Google Scholar]
- 14.Ieong K.-W., Uzun Ü., Selmer M., Ehrenberg M., Two proofreading steps amplify the accuracy of genetic code translation. Proc. Natl. Acad. Sci. U.S.A. 113, 13744–13749 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopfield J. J., Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. U.S.A. 71, 4135–4139 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ninio J., Kinetic amplification of enzyme discrimination. Biochimie 57, 587–595 (1975). [DOI] [PubMed] [Google Scholar]
- 17.Pape T., Wintermeyer W., Rodnina M. V., Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 17, 7490–7497 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurland C. G., The role of guanine nucleotides in protein biosynthesis. Biophys. J. 22, 373–392 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geggier P., et al. , Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J. Mol. Biol. 399, 576–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson R. C., Stone P. J., Proofreading of the codon-anticodon interaction on ribosomes. Proc. Natl. Acad. Sci. U.S.A. 74, 198–202 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogle J. M., Ramakrishnan V., Structural insights into translational fidelity. Annu. Rev. Biochem. 74, 129–177 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Zaher H. S., Green R., Fidelity at the molecular level: Lessons from protein synthesis. Cell 136, 746–762 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodnina M. V., Gromadski K. B., Kothe U., Wieden H.-J., Recognition and selection of tRNA in translation. FEBS Lett. 579, 938–942 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Blanchard S. C., Gonzalez R. L., Kim H. D., Chu S., Puglisi J. D., tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 11, 1008–1014 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Zhang J., Ieong K.-W., Mellenius H., Ehrenberg M., Proofreading neutralizes potential error hotspots in genetic code translation by transfer RNAs. RNA 22, 896–904 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson M., Bouakaz E., Lovmar M., Ehrenberg M., The kinetics of ribosomal peptidyl transfer revisited. Mol. Cell 30, 589–598 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Ogle J. M., et al. , Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Ogle J. M., Murphy F. V., Tarry M. J., Ramakrishnan V., Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G., A new understanding of the decoding principle on the ribosome. Nature 484, 256–259 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Voorhees R. M., Ramakrishnan V., Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 82, 203–236 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Voorhees R. M., Schmeing T. M., Kelley A. C., Ramakrishnan V., The mechanism for activation of GTP hydrolysis on the ribosome. Science 330, 835–838 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daviter T., Wieden H.-J., Rodnina M. V., Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J. Mol. Biol. 332, 689–699 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Loveland A. B., Demo G., Grigorieff N., Korostelev A. A., Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fislage M., et al. , Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A. Nucleic Acids Res. 46, 5861–5874 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gregory S. T., Carr J. F., Dahlberg A. E., A signal relay between ribosomal protein S12 and elongation factor EF-Tu during decoding of mRNA. RNA 15, 208–214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuette J.-C., et al. , GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 28, 755–765 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kothe U., Rodnina M. V., Delayed release of inorganic phosphate from elongation factor Tu following GTP hydrolysis on the ribosome. Biochemistry 45, 12767–12774 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Schmeing T. M., et al. , The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valle M., et al. , Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 10, 899–906 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Lai J., Ghaemi Z., Luthey-Schulten Z., The conformational change in elongation factor Tu involves separation of its domains. Biochemistry 56, 5972–5979 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Yang H., Perrier J., Whitford P. C., Disorder guides domain rearrangement in elongation factor Tu. Proteins 86, 1037–1046 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Kavaliauskas D., et al. , Structural dynamics of translation elongation factor Tu during aa-tRNA delivery to the ribosome. Nucleic Acids Res. 46, 8651–8661 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu W., et al. , EF-Tu dynamics during pre-translocation complex formation: EF-Tu·GDP exits the ribosome via two different pathways. Nucleic Acids Res. 43, 9519–9528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanchard S. C., Kim H. D., Gonzalez R. L. Jr, Puglisi J. D., Chu S., tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. U.S.A. 101, 12893–12898 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Juette M. F., et al. , Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polikanov Y. S., et al. , Distinct tRNA accommodation intermediates observed on the ribosome with the antibiotics hygromycin A and A201A. Mol. Cell 58, 832–844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitford P. C., et al. , Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA 16, 1196–1204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J., et al. , Single-molecule four-color FRET. Angew. Chem. Int. Ed. Engl. 49, 9922–9925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S., Lee J., Hohng S., Single-molecule three-color FRET with both negligible spectral overlap and long observation time. PLoS One 5, e12270 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hohng S., Joo C., Ha T., Single-molecule three-color FRET. Biophys. J. 87, 1328–1337 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maracci C., Peske F., Dannies E., Pohl C., Rodnina M. V., Ribosome-induced tuning of GTP hydrolysis by a translational GTPase. Proc. Natl. Acad. Sci. U.S.A. 111, 14418–14423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Villa E., et al. , Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 106, 1063–1068 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanbonmatsu K. Y., Joseph S., Tung C.-S., Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. U.S.A. 102, 15854–15859 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogeley L., Palm G. J., Mesters J. R., Hilgenfeld R., Conformational change of elongation factor Tu (EF-Tu) induced by antibiotic binding. Crystal structure of the complex between EF-Tu.GDP and aurodox. J. Biol. Chem. 276, 17149–17155 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Rodnina M. V., Fricke R., Kuhn L., Wintermeyer W., Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J. 14, 2613–2619 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pavlov M. Y., Ehrenberg M., Substrate-induced formation of ribosomal decoding center for accurate and rapid genetic code translation. Annu. Rev. Biophys. 47, 525–548 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Frank J., et al. , The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett. 579, 959–962 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Arenz S., et al. , Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome. Proc. Natl. Acad. Sci. U.S.A. 113, 7527–7532 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krupkin M., et al. , Avilamycin and evernimicin induce structural changes in rProteins uL16 and CTC that enhance the inhibition of A-site tRNA binding. Proc. Natl. Acad. Sci. U.S.A. 113, E6796–E6805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brune M., Hunter J. L., Corrie J. E., Webb M. R., Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 33, 8262–8271 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Eddy N. R., Onuchic J. N., Rotation-activated and cooperative zipping characterize class I viral fusion protein dynamics. Biophys. J. 114, 1878–1888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Q., et al. , Molecular origin of the weak susceptibility of kinesin velocity to loads and its relation to the collective behavior of kinesins. Proc. Natl. Acad. Sci. U.S.A. 114, E8611–E8617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noel J. K., Chahine J., Leite V. B. P., Whitford P. C., Capturing transition paths and transition states for conformational rearrangements in the ribosome. Biophys. J. 107, 2881–2890 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song H., Parsons M. R., Rowsell S., Leonard G., Phillips S. E., Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution. J. Mol. Biol. 285, 1245–1256 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Steitz T. A., A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 9, 242–253 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Liu C. C., Jewett M. C., Chin J. W., Voigt C. A., Toward an orthogonal central dogma. Nat. Chem. Biol. 14, 103–106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilson D. N., Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12, 35–48 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Vale R. D., Switches, latches, and amplifiers: Common themes of G proteins and molecular motors. J. Cell Biol. 135, 291–302 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pavlov M. Y., et al. , Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. U.S.A. 106, 50–54 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seip B., Innis C. A., How widespread is metabolite sensing by ribosome-arresting nascent peptides? J. Mol. Biol. 428, 2217–2227 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Murugan A., Huse D. A., Leibler S., Speed, dissipation, and error in kinetic proofreading. Proc. Natl. Acad. Sci. U.S.A. 109, 12034–12039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peil L., et al. , Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc. Natl. Acad. Sci. U.S.A. 110, 15265–15270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodnina M. V., Wintermeyer W., GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc. Natl. Acad. Sci. U.S.A. 92, 1945–1949 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bilgin N., Ehrenberg M., Stoichiometry for the elongation factor Tu.aminoacyl-tRNA complex switches with temperature. Biochemistry 34, 715–719 (1995). [DOI] [PubMed] [Google Scholar]
- 75.Ehrenberg M., Rojas A. M., Weiser J., Kurland C. G., How many EF-Tu molecules participate in aminoacyl-tRNA binding and peptide bond formation in Escherichia coli translation? J. Mol. Biol. 211, 739–749 (1990). [DOI] [PubMed] [Google Scholar]
- 76.Ferguson A., et al. , Functional dynamics within the human ribosome regulate the rate of active protein synthesis. Mol. Cell 60, 475–486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi J., Puglisi J. D., Three tRNAs on the ribosome slow translation elongation. Proc. Natl. Acad. Sci. U.S.A. 114, 13691–13696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Traxler M. F., et al. , The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dave R., Terry D. S., Munro J. B., Blanchard S. C., Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 96, 2371–2381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aitken C. E., Marshall R. A., Puglisi J. D., An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All experimental data, materials and software are available upon request.