Significance
Ebolaviruses pose a significant threat to the global community, particularly in areas of equatorial and West Africa. While Ebola virus has caused the greatest number of human cases, Sudan virus is responsible for numerous outbreaks with equally lethal consequences. Antibody-based immunotherapies against ebolaviruses have gained prominence in recent years, resulting in the development of several Ebola virus-specific antibody cocktails, yet no Sudan virus-specific cocktails exist. In collaboration with industry partners, we generated a library of human chimeric monoclonal antibodies (mAbs) specific for the glycoprotein of Sudan virus. Through a series of in vitro assays and rodent efficacy studies, we down-selected a lead mAb cocktail that demonstrated protective efficacy in a nonhuman primate model of Sudan virus infection.
Keywords: ebolavirus, Sudan virus, monoclonal antibody, therapy, cocktail
Abstract
Antibody-based therapies are a promising treatment option for managing ebolavirus infections. Several Ebola virus (EBOV)-specific and, more recently, pan-ebolavirus antibody cocktails have been described. Here, we report the development and assessment of a Sudan virus (SUDV)-specific antibody cocktail. We produced a panel of SUDV glycoprotein (GP)-specific human chimeric monoclonal antibodies (mAbs) using both plant and mammalian expression systems and completed head-to-head in vitro and in vivo evaluations. Neutralizing activity, competitive binding groups, and epitope specificity of SUDV mAbs were defined before assessing protective efficacy of individual mAbs using a mouse model of SUDV infection. Of the mAbs tested, GP base-binding mAbs were more potent neutralizers and more protective than glycan cap- or mucin-like domain-binding mAbs. No significant difference was observed between plant and mammalian mAbs in any of our in vitro or in vivo evaluations. Based on in vitro and rodent testing, a combination of two SUDV-specific mAbs, one base binding (16F6) and one glycan cap binding (X10H2), was down-selected for assessment in a macaque model of SUDV infection. This cocktail, RIID F6-H2, provided protection from SUDV infection in rhesus macaques when administered at 50 mg/kg on days 4 and 6 postinfection. RIID F6-H2 is an effective postexposure SUDV therapy and provides a potential treatment option for managing human SUDV infection.
The 2013–2016 Ebola virus (EBOV) epidemic in western Africa and the ongoing outbreak in the Democratic Republic of the Congo clearly illustrates the devastating impact ebolaviruses can have on human health. The Ebolavirus genus, of the Filoviridae family, consists of six antigenically distinct species: Zaire ebolavirus (type member Ebola virus), Sudan ebolavirus (Sudan virus [SUDV]), Bundibugyo ebolavirus (Bundibugyo virus), Tai Forest ebolavirus, Reston ebolavirus (1), and the recently discovered Bombali ebolavirus (2). Although EBOV was the causative agent of the 2013–2016 outbreak that developed into the largest ebolavirus epidemic in recorded history (3), a related but antigenically divergent virus, SUDV, also presents a significant health threat. Since its discovery in 1976, SUDV has caused eight confirmed outbreaks in equatorial Africa and infected 779 people, 412 of whom died (4).
Currently, no US Food and Drug Administration (FDA) approved countermeasures for ebolaviruses exist, although several vaccines and therapeutics are in advanced stages of development (5–10). Until recently, postexposure antibody immunotherapy for the treatment of ebolavirus disease (EVD) was largely ignored due to several failed attempts to protect nonhuman primates (NHPs) from EBOV challenge (11, 12). Dye et al. (13) were the first to report protection of macaques from EBOV infection using antibody-based immunotherapies. Subsequently, several independent groups reported the postexposure efficacy of monoclonal antibody (mAb)-based immunotherapies against EVD in macaques when administered as mixtures of two or more mAbs (14–17). These landmark studies demonstrated that combination mAb-based immunotherapies are a viable treatment option for EVD and spurred therapeutic antibody development for filoviruses. ZMapp, a cocktail of three EBOV glycoprotein (GP)-specific mAbs, was the only therapy tested in a randomized controlled trial during the West Africa outbreak where it showed strong evidence of efficacy prior to the trial ending prematurely, due to lack of new cases and the end of the outbreak (6, 18). ZMapp and two additional mAb products, mAb114 (19) and REGN-EB3 (20), are currently being evaluated in a randomized trial being conducted in response to the current EBOV outbreak in the Democratic Republic of the Congo. Preliminary reports from this trial indicate that mAb treatment significantly reduces mortality rates relative to untreated individuals (21). While these are promising developments, the efficacy of these three products are limited to EBOV alone, leaving the public health sector with no approved treatment options for other ebolaviruses known to cause disease in humans, including SUDV. In fact, few SUDV therapies have been developed thus far. Thi et al. (22) reported the in vivo efficacy of lipid encapsulated siRNA targeting SUDV VP35 in rhesus macaques when treatment was initiated as late as day 4 postexposure. In addition, favipiravir was reported to provide protection against SUDV exposure in a guinea pig model of disease when delivered as late as day 5 postexposure (23). Finally, recent advancements in antibody discovery technologies (24) and refined screening methods have recently yielded several cross-reactive mAbs and the development of pan-ebolavirus (25, 26) and pan-filovirus (27) cocktails with broad-spectrum activity against multiple filovirus species.
A number of large-scale antibody manufacturing methods are available, chief among them being mammalian cell-based expression systems (28). With the introduction of transgenic plants capable of glycosylating with mammalian N-glycans, and recent advancements in plant expression technology, plant-based manufacturing has the potential to become an alternative manufacturing system (29, 30). EBOV-specific mAb material, utilized in studies referenced above, was derived from various sources, including plants and mammalian cells. Pros and cons of each production method are still a topic of debate, but deliberations typically focus on aspects of regulatory affairs, safety, scalability, or immunogenicity, and rarely on antibody performance. Previously reported evaluations of EBOV antibody therapies employed mAbs produced using disparate platforms, including hybridoma, mammalian cell and plant production, making direct comparisons challenging.
Here, we describe the development of a protective SUDV-specific mAb cocktail while directly comparing plant- and mammalian-produced mAbs.
Results
Chimerization and Production of SUDV-Specific mAbs.
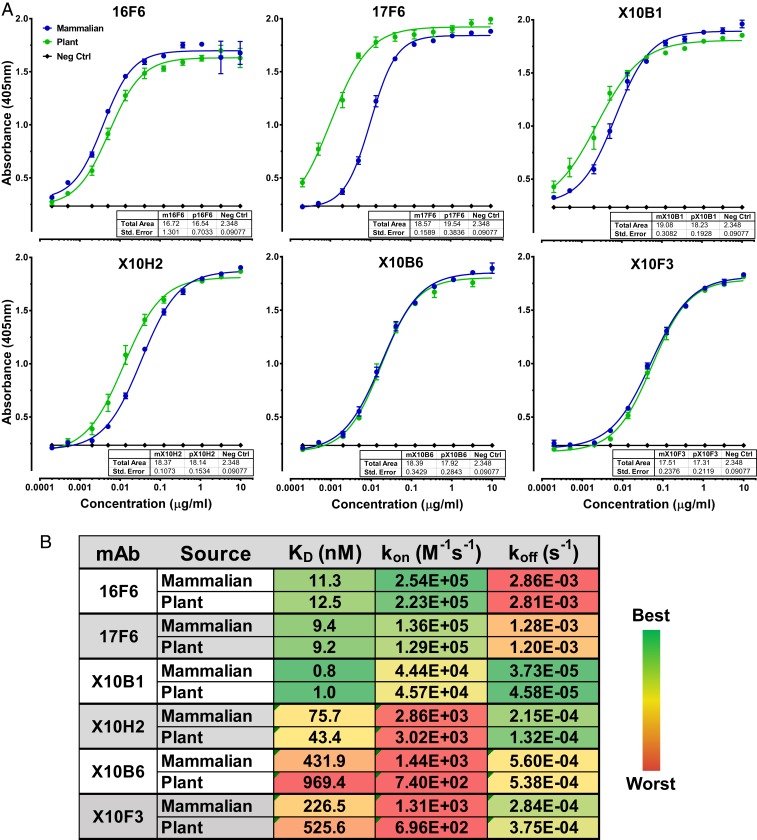
To develop SUDV-specific mAbs with the potential for human use, we leveraged two existing libraries of antibodies specific for SUDV GP. The first library consisted of monoclonal antibodies derived from BALB/c mice immunized with Venezuelan equine encephalitis virus replicon particle (VRP) vaccine expressing SUDV-Boniface (SUDV-Bon) GP, as described previously (31). The second library was comprised of novel single chain variable fragment (scFv) antibodies isolated from macaques immunized with VRP vaccine expressing SUDV-Bon GP (32). We generated human IgG1 chimeric mAbs by cloning variable heavy (VH) and variable light (VL) chains from murine and macaque antibodies into human IgG1 constant heavy and constant light chain expression vectors (SI Appendix, Fig. S1). Nucleotide sequences encoding each chimeric mAb were codon optimized for expression in either Nicotiana benthamiana plants or mammalian NS0 cells. Human IgG1 chimeric heavy and light chains were coexpressed either transiently in plants or stably in mammalian cells, and IgGs were purified using protein A affinity resin. The binding specificity of each humanized mAb was evaluated using ELISA plates coated with recombinant vesicular stomatitis virus (rVSV) expressing SUDV-Bon GP (rVSV-SUDV GP) (Fig. 1A). Overall, binding profiles of plant- and mammalian-derived mAbs were equivalent with the exception of 17F6 where plant 17F6 bound more efficiently at lower concentration with a slightly higher total area under the curve (AUC), though not statistically significant. We also evaluated cross-reactivity of each mAb against recombinant EBOV GP or Bundibugyo virus (BDBV) GP in a biolayer interferometry (BLI) assay using the Octet platform and none of the mAbs were reactive against either ebolavirus GP (SI Appendix, Fig. S2).
Fig. 1.
Antigen specificity and affinity of humanized SUDV-specific mAbs. (A) Mammalian-derived (blue) and plant-derived (green) mAbs were serially diluted and added to plates coated with rVSV-SUDV-Bon GP. Following incubation with anti-human IgG-HRP, ABTS substrate was added, and absorbance was measured. Embedded table reports the total AUC calculated for each mAb. Data points represent the mean ± SEM of two replicate assays, each having two technical replicates. (B) Antibody affinities for recombinant SUDV-Bon GP were determined using bilayer interferometry on the Octet platform. AHC biosensors coated with plant or mammalian mAbs were sequentially dipped into assay buffer containing recombinant SUDV-Bon GP to measure GP on rate (kon), then into assay buffer alone to measure GP off rate (koff). Affinity (KD) was calculated using ForteBio data analysis software. Data represent the mean of two replicate assays, each having two technical replicates.
In Vitro Characterization of SUDV-Specific mAbs.
Next, we determined affinities of each mAb to recombinant SUDV GP using the Octet platform. Biosensors loaded with chimeric mAbs were dipped into assay buffer containing serially diluted recombinant SUDV GP. SUDV GP association and dissociation was determined and KD was calculated using the ForteBio data analysis software. X10B1 demonstrated low nanomolar affinity for SUDV-Bon GP, the lowest KD of all of the antibodies tested, and had a particularly slow off rate (Fig. 1B). mAbs 17F6 and 16F6 had the next lowest KD, with the most rapid on rates, but also the most rapid off rates. The remaining antibodies, X10H2, X10B6, and X10F3, demonstrated high nanomolar affinities with the slowest on rates. Affinity of each mAb for SUDV-Gulu GP was also evaluated and each mAb had equivalent or lower KD for SUDV-Gulu GP relative to SUDV-Bon GP (SI Appendix, Table S1).
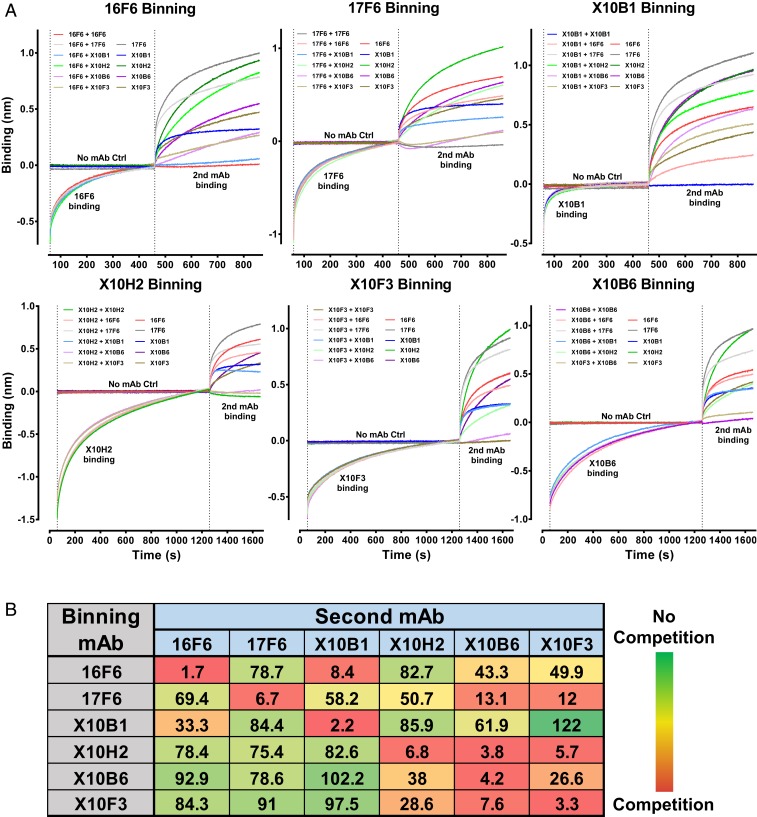
Competition groups for the panel of SUDV-specific mAbs were defined using the Octet platform and competitive ELISA. For BLI assays, biosensors loaded with recombinant SUDV GP were first dipped into assay buffer containing indicated binning mAb or assay buffer alone and binning mAb association was allowed to reach saturation. Biosensors were then dipped into assay buffer containing a mixture of the binning mAb and indicated second mAb. Data were normalized to the starting time point of the second association step. Association of the second mAb, in the presence of saturation binning mAb, was compared to binding activity of the second mAb alone (Fig. 2A). AUC for each binning mAb plus second mAb condition was compared to the AUC for each second mAb alone and reported as the percent of the second mAb alone (Fig. 2B). Two competition groups were identified by BLI analysis, mAbs competing with 16F6 and mAbs competing with X10H2. mAbs 16F6 and X10B1 significantly inhibited binding of one another, though somewhat directionally with residual 16F6 binding in the presence of X10B1 (Fig. 2). X10H2, X10B6, and X10F3 competed with each other, also somewhat directionally and in correlation with antibody affinity (Fig. 2). mAb 17F6 was found to compete only directionally with X10B6 and X10F3 (Fig. 2). These results were confirmed by competitive ELISA, in which chimeric mAbs were incubated with SUDV GP-coated plates prior to the addition of the murine version of each mAb. Binding of murine mAb alone was compared to observed binding levels in the presence of the chimeric mAbs (SI Appendix, Fig. S3).
Fig. 2.
Competitive binding of humanized SUDV-specific mAbs. (A) Competitive binding of individual mAbs was determined using bilayer interferometry on the Octet platform. HIS1K biosensors coated with recombinant SUDV GP were sequentially dipped into assay buffer alone or assay buffer containing the binning mAb, then into assay buffer containing the binning mAb and a second mAb. Association of the second mAb in the presence of saturating amounts of the binning mAb was compared to association of the second mAb alone. Data presented are from a single assay that is representative of three replicate assays, each having two technical replicates. (B) AUC was determined for all second mAb binding curves. AUC for second mAb in the presence of the binning mAb was normalized to the AUC for the second mAb alone. Data are reported as the percent of second mAb alone AUC.
We then attempted to determine binding footprints for each SUDV-specific mAb. The binding epitope for 16F6 was previously determined by X-ray crystallography to be located at the base of the GP trimer, overlapping GP1 and GP2 (24). Based on our competition data, we assigned both 16F6 and X10B1 as GP base binders. We employed SPOT membranes coated with 13-amino acid-long (13 mer) linear peptides of SUDV GP from Boniface and Gulu isolates to identify mAbs with linear epitope specificity. 17F6 was the only mAb that recognized a linear epitope, a mucin-like domain (MLD) amino acid sequence 353LH(V/I)PEGETT361 (SI Appendix, Fig. S4). To map the binding site of X10H2 and competing antibodies, we selected viral escape variants from antibody neutralization by serially passaging rVSV-SUDV GP in the presence of X10H2 until resistance to neutralization was observed. Following passage four, the viral population was plaque purified and sequenced to identify the mutations in SUDV GP that engender viral neutralization escape. We identified a single mutation in eight of eight clones sequenced, substitution of glutamine (Q) with lysine (K) at a residue located in the glycan cap of GP (Fig. 3A). Consistent with other glycan cap binding antibodies, X10H2 was unable to neutralize cleaved SUDV GP, further corroborating glycan cap binding specificity of X10H2 and competing mAbs X10F3 and X10B6 (Fig. 3A). The SUDV GP structure with defined structural elements is depicted in Fig. 3B. Amino acid residues of the 16F6 epitope, located near the base of the GP trimer, are colored purple and are presumed to overlap or be in close proximity to residues of the X10B1 epitope. The residue critical for X10H2 neutralization, is colored red with a 10A zone around the residue colored yellow to approximate the outline of the putative epitope. The binding residues for X10H2 and the two competing mAbs, X10F3 and X10B6, are likely to be located within the 10A zone.
Fig. 3.
In vitro characterization of SUDV-specific mAbs. (A) rVSV-SUDV GP escape mutants to X10H2 were selected by growing the virus in the presence of IC90 concentrations of X10H2. Escape variants were then sequenced to identify GP mutations. Neutralizing activity of X10H2, X10B6, and X10F3 against wild-type rVSV-SUDV GP (GPWT) and escape mutant rVSV-SUDV GP (GPmut) was evaluated. Neutralizing activity of X10H2 against cleaved rVSV-SUDV GP (GPCL) was also evaluated. Data points represent the mean ± SEM of two replicate assays, each having two technical replicates. (B) SUDV GP structure with labeled subunits, functional elements, and known antibody epitopes. (C) Neutralizing activity of plant (p, dotted line) and mammalian (m, solid line) mAbs were evaluated by microneutralization assay. Serially diluted mAbs were mixed with SUDV prior to adding to Vero cells at an MOI of 0.5. Cells were fixed 48 h after infection and immunostained with SUDV-specific antibody and fluorescently labeled secondary antibody. Cells were imaged and the percent of infected cells was determined using an Operetta and Harmonia Software. Data are presented as percent inhibition relative to untreated, infected control cells and represent the mean ± SEM of two replicate assays, each having two technical replicates.
Neutralizing activity of each mAb against authentic SUDV-Bon was evaluated using a microneutralization assay (26). Antibodies 16F6 and X10B1 were the most potent neutralizers of SUDV-Bon with subnanomolar (nM) half-maximal inhibitory concentrations (IC50) (Fig. 3C). Competing antibodies X10H2, X10F3, and X10B6 were all weak neutralizers with comparable IC50 values between 32 and 67 nM (Fig. 3C). Antibody 17F6 failed to neutralize SUDV-Bon at the highest concentration (∼70 nM) tested (Fig. 3C). No significant difference in neutralizing activity was observed between plant-derived and mammalian-derived mAbs.
Protective Efficacy of SUDV-Specific mAbs in Mice.
To evaluate the protective efficacy of SUDV-specific mAbs, we used the IFN-alpha/beta receptor knockout (IFNAR−/−) mouse model of SUDV infection (33). We first compared the efficacy of plant- and mammalian-derived mAbs using 5- to 8-wk-old IFNAR−/− mice, for which the primary metric of protection is reduced weight loss following SUDV-Bon challenge. Here, groups (n = 10) of age-matched IFNAR−/− mice were treated intraperitoneally (I.P.) with 200 µg of plant- or mammalian-derived mAbs on days 1 and 4 postexposure to 1,000 plaque forming units (pfu) of SUDV-Bon via I.P. inoculation, and weight loss was monitored. Base binding mAbs 16F6 and X10B1 reduced weight loss relative to vehicle control animals with 16F6 performing slightly better than X10B1 (Fig. 4A). Mucin domain binding mAb 17F6 afforded no protective benefit against SUDV-Bon challenge and was dropped from further consideration (Fig. 4B). Of the glycan cap binding mAbs, X10H2 appeared to be the most protective, followed by X10F3, then X10B6 (Fig. 4B). In all cases, no significant difference in weight loss was observed between mice treated with plant- or mammalian-derived mAbs.
Fig. 4.
In vivo efficacy of SUDV-specific mAbs in rodent model of SUDV disease. (A and B) Protective efficacy of mammalian- and plant-derived SUDV-specific mAbs were compared directly in a weight loss mouse model of SUDV disease. Groups (n = 10) of IFNAR−/− mice (5 to 8 wk old) were infected with 1,000 pfu of SUDV and treated I.P. with 200 µg of (A) base binding mAbs or (B) mucin/cap binding mAbs on days 1 and 4 postexposure. Vehicle control mice were treated I.P. with an equal volume of PBS on days 1 and 4 postexposure. Percent weight change was calculated using the daily average mouse weight (group weight/N) relative to starting average mouse weight. (C and D) SUDV-specific mAbs from each competition group were down-selected using a lethal mouse model of SUDV disease. Groups (n = 10 per study) of IFNAR−/− mice (4 wk old) were infected with 1,000 pfu of SUDV and treated I.P. with indicated mAb on days 1 and 4 postexposure. (C) Percent weight change was calculated using the daily average mouse weight (group weight/N) relative to starting average mouse weight and data points represent the mean ± SEM of two replicate studies (except X10B6). (D) Overall percent survival calculated using survival data from both studies combined (n = 20 mice per group except X10B6 where n = 10) (**P < 0.01, ***P < 0.001).
We next focused on down-selecting lead mAb candidates from the base binding and glycan cap binding groups. For these studies, we used 4-wk-old IFNAR−/− mice, which is a lethal model of disease. As before, groups (n = 10 to 20) of IFNAR−/− mice were treated with 200 µg of mAbs on days 1 and 4 postexposure to 1,000 pfu of SUDV-Bon, and weight loss and survival were monitored. In this model, base binders 16F6 and X10B1 significantly (P = 0.0007 and 0.0147, respectively) reduced weight loss relative to vehicle control-treated mice (Fig. 4C) and provided 90 to 100% protection (P < 0.001) (Fig. 4D). Glycan cap binding mAbs X10H2, X10F3, and X10B6 also provided 90 to 100% protection (P < 0.01) (Fig. 4D) and X10H2 and X10F3 limited weight loss significantly (P = 0.05 and 0.008, respectively) (Fig. 4C). mAbs 16F6 and X10H2 were selected for further development, given there slightly better and more consistent reduction of overall weight loss relative to other mAbs within the same competition group.
Protective Efficacy of SUDV-Specific Cocktail in Rhesus Macaques.
The protective efficacy of the SUDV-specific antibody cocktail comprised of 16F6 and X10H2, hereafter RIID F6-H2, was evaluated in nonhuman primates using the rhesus macaque model of EVD (34, 35). Eight adult rhesus macaques were randomized into either RIID F6-H2 treatment (n = 4) or control treatment (n = 4) groups and blinded to study personnel for the duration of the study. Macaques were exposed to a target dose of 1,000 pfu of SUDV-Bon via intramuscular injection on day 0 of the study. On days 4 and 6 postexposure, four experimental macaques were treated I.V. with plant-derived RIID F6-H2 at 25 mg/kg per mAb (50 mg/kg total per dose). Plant-produced mAbs were used in this study due to availability of product at the required quantity and the previously established equivalent protective efficacy (Fig. 4). In the interest of animal welfare, control-treated macaques were divided into two treatment regimens to align with regimens of two studies being run simultaneously. Control (Ctrl) macaques 1 and 2 were treated I.V. with an equivalent volume to weight ratio of diluent on days 4 and 6, whereas control macaques 3 and 4 were treated with an equivalent volume to weight ratio of diluent on day 5 only. Macaques were observed daily to assess overall health, behavior, and appetite. In addition, physical examinations and blood collections were completed approximately every 3 to 4 d to assess clinical disease progression. All RIID F6-H2–treated macaques survived SUDV-Bon exposure with little to no clinical signs of disease (Fig. 5 A and B and SI Appendix, Table S2). By contrast, 50% of control macaques succumbed to SUDV-Bon infection and all displayed clinical signs of EVD (Fig. 5 A and B and SI Appendix, Table S2). All control macaques were PCR positive for SUDV by day 4 postexposure (Fig. 5C); by day 6, all control macaques had detectable viremia by plaque assay (Fig. 5D). Viral titers in the control macaques peaked by day 8 postexposure between 4 and 5 log10/mL in 75% of the animals. Peak viral titer for Ctrl 3 was approximately one log10 lower at just over 3.0 log10/mL, which may explain less severe disease symptoms observed in this macaque. All experimental macaques were PCR positive for SUDV-Bon prior to receiving the first antibody treatment on day 4 postexposure (Fig. 5C), and viremia was detected by plaque assay in two of four experimental macaques on day 4 (Fig. 5D). Following RIID F6-H2 treatment on day 4, viral titers decreased rapidly in all four experimental macaques and remained below the limit of plaque assay detection through the end of the study. All four experimental macaques were PCR negative by day 8 postexposure, 2 d after the last antibody treatment. As anticipated, SUDV GP-specific IgG titers increased rapidly following antibody delivery and remained elevated through day 28 (Fig. 5E). A de novo SUDV GP-specific IgG response was detectable in control macaques as early as day 8 postexposure; by day 21 postexposure, surviving control macaques had similar titers relative to treated macaques (Fig. 5E). All control macaques had elevated serum levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP) (Fig. 6 A–C), and thrombocytopenia (Fig. 6D) during the acute phase of disease. Elevated levels of blood urea nitrogen (BUN), gamma-glutamyl transferase (GGT), and creatinine (CRE), and reduced levels of amylase (AMY) were also observed in serum of control animals that succumbed (SI Appendix, Fig. S5). With the exception of elevated AMY in experimental animal 4 (Exp4), RIID F6-H2–treated macaques were free of abnormal serum chemistries or hematology associated with EVD (Fig. 6 and SI Appendix, Fig. S5).
Fig. 5.
In vivo efficacy of SUDV-specific mAbs in macaque model of SUDV disease. Rhesus macaques were exposed intramuscularly to 1,750 pfu of SUDV. Groups (n = 4) were treated I.V. with either 50 mg/kg of SUDV-specific mAb mixture 16F6/X10H2 and/or equal volume-to-weight ratio of vehicle (0.9% NaCl) on days 4 and 6 postexposure (Ctrl 3 and Ctrl 4 treated on day 5 only). ↓ indicates mAb treatment days. (A and B) Macaques were monitored daily for (A) survival and (B) clinical signs of disease. (C–E) Blood was collected on indicated days postexposure to evaluate viremia by (C) RT-PCR (****P < 0.0001) or (D) plaque assay (*P < 0.05) and (E) SUDV GP-specific IgG titers by end titer ELISA (****P < 0.0001). Individual data points represent the mean from two replicate assays, each having at least two technical replicates. Lines represent group means.
Fig. 6.
Serum chemistries and complete blood counts (CBCs) of rhesus macaques following SUDV exposure. Rhesus macaques were exposed intramuscularly to 1,750 plaque forming units of SUDV. Groups (n = 4) were treated I.V. with either 50 mg/kg of SUDV-specific mAb mixture 16F6/X10H2 and/or equal volume-to-weight ratio of vehicle (0.9% NaCl) on days 4 and 6 postexposure (Ctrl 3 and Ctrl 4 treated on day 5 only). (A–D) Serum and whole blood was collected on indicated days postexposure to evaluate serum enzymes (A) ALT, (B) AST, and (C) ALP by Piccolo and (D) platelet counts by CBC. Individual data points are from a single assay using instruments validated daily with appropriate instrument controls. Lines represent group means. *P < 0.05, **P < 0.01, ****P < 0.0001.
Discussion
Our efforts to develop an antibody-based therapy for SUDV have yielded a cocktail of two mAbs capable of protecting macaques from SUDV exposure when delivered after animals are PCR positive, a critical clinical diagnosis and trigger-to-treat milestone. Antibody down-selection criteria focused on identifying neutralizing antibodies that targeted noncompeting epitopes of SUDV GP. Cross-reactivity against other ebolaviruses was also considered, but none of the mAbs recognized GPs of EBOV or BDBV. Out of a small panel of six antibodies, three competition groups were identified and only two groups (encompassing five mAbs in total) had measurable neutralizing activity against SUDV. The mAbs 16F6 and X10B1, the most potent neutralizing and protective mAbs, bind at the base of the GP trimer, similar to KZ52 and ZMapp components 2G4 and 4G7 (30, 31). The 16F6 epitope consists of residues located within both GP1 and GP2 (24) and 16F6/X10B1 potency may be attributed to its ability to either “lock” GP in a prefusion conformation and impede necessary structural rearrangements required for membrane fusion or block cathepsin cleavage of GP, as is reported for KZ52 and 2G4 (32). The other neutralizing and protective mAbs, X10H2, X10F3, and X10B6, bind an epitope located within the glycan cap, an epitope similar to that targeted by ZMapp component 13C6 (30, 31). While it was not our intention to develop a ZMapp-like cocktail, our down-selection strategy was similar to that used to develop ZMapp, which may have skewed our selection unintentionally toward ZMapp-like mAbs. In addition, both cocktails originated from a relatively small panel of mAbs derived from animals vaccinated with either EBOV GP or SUDV GP (31, 36, 37). Similarities in epitope specificity between ZMapp and RIID F6-H2 suggest that structural features of GP vulnerability to antibody therapies may be conserved across ebolavirus species, as proposed previously (31).
Based on published studies describing EBOV-specific mAb treatment of infected macaques (12–16), we elected to treat SUDV-infected macaques twice with 50 mg/kg of the antibody cocktail for our first proof-of-concept study. Given the rapid decrease in viremia following treatment and few clinical signs of disease observed in treated macaques, we are hopeful that less aggressive dosing strategies may also be protective. Recently published data do in fact support the idea that less frequent and lower mAb dosing can be protective in NHPs, though efficacy is expected to largely depend on antibody cocktail potency (25). A full understanding of the pharmacokinetic/pharmacodynamic (PK/PD) properties for RIID F6-H2 will help to further optimize dosing regimens and to determine manufacturing objectives as this cocktail transitions to advanced development. Future preclinical studies will focus on defining the minimal protective dose and the therapeutic window for treating SUDV infection in macaque models.
In parallel to developing a SUDV-specific antibody cocktail, we also completed a head-to-head comparison of antibodies produced in plants and mammalian expression systems. Our comparison was limited to antibody performance, specifically antigen binding, virus neutralization, and in vivo protection. Our in vitro assessment did not illustrate any significant differences between the plant- and mammal-derived mAbs studied here. For individual mAbs, plant and mammalian products had similar antigen binding affinities and virus neutralization activity. It is important to note that our in vitro assays are a measure of and dependent on Fab activity; none of our in vitro assays evaluated Fc function directly. Our in vivo assessment also did not identify any significant differences between plant- and mammal-derived mAbs. Both performed equally well in mice exposed to SUDV. Significant differences in Fc function, if they exist, could potentially have been observed by in vivo testing, although the mouse model may not be suitable to elucidate small but significant differences in the Fc effector functions of plant and mammalian mAbs.
Poor cross-reactivity of ZMapp, mAb114, REGN-EB3, and RIID F6-H2 is due to limited GP sequence homology across ebolaviruses (31–33). Many of the conserved epitopes are protected from neutralizing antibodies owing to the large, bulky mucin-like domain and the glycan cap (32). Recent antibody development efforts have focused on expanding the breadth of antibody reactivity using multiple strategies. Bispecific antibody engineering, combining EBOV and SUDV GP-specific mAbs, or the “Trojan horse” approach of targeting conserved intramolecular epitopes has yielded broadly active therapeutic candidates with in vivo efficacy in rodents against both viruses (26, 34). Heterologous prime-boost vaccination strategies to elicit broadly reactive mAbs targeting conserved epitopes have also been successful (33, 35, 36). Others have identified naturally developed, broadly reactive antibodies from human survivors of EBOV or Bundibugyo virus infections (32, 37). Given their breadth of activity, new pan-ebolavirus antibody therapies incorporating these mAbs may provide a single treatment option for managing ebolavirus outbreaks caused by any known ebolavirus species or potentially novel ebolaviruses yet to emerge. However, the utility of many of these pan-ebolavirus antibody therapies has yet to be fully realized as many are still in early stages of development. The Trojan horse strategy remains untested in animal models higher than mice. The pan-ebolavirus mAb mixture MBP134 has demonstrated broad spectrum efficacy in macaques against EBOV, SUDV, and BDBV infection (25) while a pan-filovirus mixture consisting of broadly reactive ebolavirus-specific mAbs FVM04 and CA45 and Marburg virus (MARV)-specific mAb MR191 has demonstrated protective efficacy against EBOV, SUDV, and MARV in macaque models (27). As more antibody cocktails transition to the clinic, viruses will encounter selective pressures to mutate away from these treatments in order to survive, potentially leading to the emergence of mAb therapy-resistant variants. The availability of numerous antibody therapeutics with divergent GP epitope specificities and mechanisms of action may be critical for addressing this possibility. Synergy between antibodies and small molecule or siRNA therapies may also mitigate resistance. As such, concurrent development of multiple ebolavirus mAb cocktails or combination therapies could offer an insurance policy of sorts should therapies be rendered ineffective, just as multiple antibiotics afford protection against antimicrobial resistance.
Materials and Methods
Cells and Viruses.
Vero E6 cells (ATCC) were maintained in Eagle’s minimal essential medium (EMEM) supplemented with 5% heat-inactivated fetal bovine serum (ΔFBS) and gentamicin (50 µg/mL) at 37 °C, 5% CO2, and 80% humidity. Sudan virus/Homo sapiens-gp-tc/SDN/1976/Boniface-The US Army Medical Research Institute of Infectious Disease (USAMRIID)111808 (SUDV/Bon-USAMRIID111808; “SUDV-Boniface 1976”) (38) was used for all in vitro and in vivo studies requiring live virus.
Enzyme-Linked Immunosorbent Assay.
SUDV GP reactivity or SUDV GP-specific IgG titers were determined by enzyme-linked immunosorbent assay (ELISA) using rVSV-SUDV-Bon GP or recombinant SUDV-Bon GP (National Cancer Institute, Protein Expression Lab), respectively, as previously described (13). Briefly, polyvinyl chloride ELISA plates (Dynatech Laboratories) were coated with rVSV-SUDV-Bon GP or recombinant GP, diluted in phosphate-buffered saline (PBS), overnight at 4 °C before blocking with 5% milk protein in PBS/0.02% Tween-20 at room temperature. Monoclonal antibodies or serum samples were serially diluted in blocking buffer and added to antigen-coated plates for 2 h at room temperature. Plates were washed with wash buffer (PBS/0.02% Tween-20) before adding horseradish peroxidase (HRP)-conjugated goat anti-human IgG (Rockland) for 1 h at room temperature. Following a final wash, 2,2′-Azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt (ABTS) substrate (Kirkegaard and Perry Laboratories, Inc.) was added and absorbance values were read at 405 nm using a Spectramax plate reader after 30 min (Molecular Devices, LLC). For serum sample antibody end titers, cutoff values for each dilution were defined using prechallenge serum samples from each individual macaque. Cutoff values for each dilution were calculated using absorbance values of prechallenge serum for that dilution and the following formula: average prevaccination serum absorbance + 3× SD. End-point titers for each serum sample are expressed as the last dilution to exceed the cutoff value for a given dilution.
Antibody Affinity Determination.
Binding affinity of each mAb to recombinant GPs was determined by BLI using the Octet Red96 system (ForteBio, Molecular Devices). Antibodies were loaded onto anti-human Fc (AHC) biosensors (ForteBio, Molecular Devices) at 3 μg/mL diluted in kinetic buffer (Tris buffered saline/50 μg/mL BSA/0.0001% Tween-20). Association of recombinant SUDV-Bon GP, SUDV-Gulu GP, EBOV-Kik GP, or BDBV GP proceeded across threefold serial dilutions in kinetic buffer, 10, 3.3, and 1.1 μg/mL for 16F6, 17F6, and X10B1 and 300, 100, and 33.3 μg/mL for X10B6, X10F3, and X10H2. Per manufacturer's instructions, baseline and dissociation steps were allowed to proceed in the same kinetic buffer. Binding kinetics were determined using a 1:1 binding model, though this binding interaction is likely to be more complex, given the bivalency of mAbs and trivalent configuration of the recombinant GPs.
Binning Assays by BLI.
Binning antibodies into competition groups was completed by BLI using the Octet Red96 system (ForteBio, Molecular Devices). Histidine-labeled recombinant SUDV-Bon GP was loaded onto Anti-Penta-HIS (HIS1K) biosensors (ForteBio, Molecular Devices) at 25 ug/mL diluted in kinetic buffer (Tris buffered saline/50 μg/mL BSA/0.0001% Tween-20). Association of the binning mAb at 50 μg/mL in kinetic buffer was allowed to proceed to near saturation followed by association of the second mAb at 50 μg/mL in kinetic buffer containing 25 μg/mL of the binning mAb, to maintain saturation of the binning mAb epitope. Association of the second mAb at 50 μg/mL in the absence of the binning mAb was also assessed within the same assay. AUC was determined for association of the second mAb in the presence and absence of the binning mAb. The AUC for the second mAb plus binning mAb was normalized to the AUC for the second mAb alone and expressed as a percent of second mAb alone. Antibodies were considered competitive if binding was <30%, partially competitive if binding was 30 to 70%, and noncompetitive if binding was ≥70% in both directions.
Competitive ELISA.
Polyvinyl chloride ELISA plates (Dynatech Laboratories) were coated with SUDV-Bon GP, diluted in PBS, overnight at 4 °C before blocking with 5% milk protein in PBS/0.02% Tween-20 at room temperature. Chimeric human IgGs of each mAbs were diluted to 0.1 mg/mL in PBS and added to antigen-coated plates for 20 min at room temperature. Murine IgGs of indicated mAbs were diluted to 0.1 mg/mL in PBS and added to wells containing chimeric human IgGs or PBS alone for 20 min at room temperature. Plates were washed with wash buffer (PBS/0.02% Tween-20) before adding horseradish peroxidase-conjugated goat anti-mouse IgG (Rockland) for 1 h at room temperature. Following a final wash, ABTS substrate (Kirkegaard and Perry Laboratories, Inc.) was added and absorbance values were read at 405 nm using a Spectramax plate reader after 30 min (Molecular Devices, LLC).
SPOT Membrane Assay.
SPOT membrane assays were completed using Genosys SPOT (Sigma-Aldrich) membranes in accordance with the manufacturer’s instructions. Briefly, SPOT membranes, coated with 13-mer overlapping linear peptides (95% purity) from SUDV-Bon and SUDV-Gulu were incubated sequentially with SUDV-specific mAbs and β-galactosidase conjugated anti-murine IgG secondary antibody. Signal development solution was added to the membrane before washing with PBS and imaging with a ChemiDoc XRS+ imaging system (Bio-Rad Laboratories, Inc.).
Plaque Assay.
Serial log dilutions of serum or virus were prepared in EMEM supplemented with 5% ΔFBS, 2 mM l-glutamine, and 1% gentamicin. A total of 200 µL of each dilution was inoculated onto six-well plates containing confluent monolayers of Vero E6 cells. After adsorption for 1 h at 37 °C, 5% CO2, and 80% humidity, monolayers were overlaid with a mixture of one part 1% agarose (Seakem) and one part 2× Eagle’s basal medium (EBME), 30 mM Hepes buffer, and 5% ΔFBS. After incubation at 37 °C, 5% CO2, and 80% humidity for 8 d, a second overlay, supplemented with 5% neutral red, was added. Plaques were counted the following day and titers were expressed as plaque forming units per milliliter.
Microneutralization Assay.
Neutralizing activity of antibodies was assessed by microneutralization assay as previously described (39). Antibodies were diluted to indicated concentrations in culture media and incubated with SUDV-Bon for 1 h. Vero E6 cells were exposed to antibody/virus inoculum at a multiplicity of infection (MOI) of 0.5 pfu/cell for 1 h. Antibody/virus inoculum was then removed and fresh culture medium was added. At 48 h postinfection, cells were fixed with formalin and blocked with 1% BSA. SUDV-infected cells and uninfected controls were incubated with SUDV GP-specific mAb 3C10 (USAMRIID). Cells were washed with PBS prior to incubation with goat anti-mouse IgG conjugated to Alexa 488. Cells were counterstained with Hoechst stain (Invitrogen), washed with PBS, and stored at 4 °C. Infected cells were quantitated by fluorescence microscopy and automated image analysis. Images were acquired at 20 fields/well with a 20× objective lens on an Operetta high-content device (Perkin-Elmer). Operetta images were analyzed with a customized scheme built from image analysis functions available in Harmony software.
rVSV Neutralization Assay.
rVSV expressing both enhanced green fluorescent protein (EGFP) and recombinant surface GP (rVSV-SUDV) in place of VSV G are previously described (40). rVSV neutralization assays were completed as previously described (41). Vero cells were seeded at 6.0 × 104 cells/well and cultured overnight in EMEM supplemented with 10% FBS and 100 IU/mL penicillin and 100 mg/mL streptomycin at 37 °C and 5% CO2. The next day, virus was incubated with serial threefold antibody dilutions beginning at 350 nM (∼50 μg/mL) in serum-free DMEM for 1 h at room temperature before infecting Vero cell monolayers in 96-well plates. The virus was incubated with the cells in 50% vol/vol DMEM supplemented with 2% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin at 37 °C and 5% CO2 for 14 to 16 h before the cells were fixed and the nuclei stained with Hoescht. rVSV infectivity was measured by counting EGFP-positive cells in comparison to the total number of cells indicated by nuclear staining using a Cellinsight CX5 automated microscope and accompanying software (Thermo Fisher Scientific).
Selection of Viral Neutralization Escape Mutants.
Escape mutant selections were performed as previously described (42). Breifly, rVSV-SUDV-Bon GP particles were serially passaged in the presence of the selection mAb X10H2. Serial threefold dilutions of virus were preincubated with a concentration of mAb corresponding to the IC90 value (∼100 nM) derived from neutralization assays, and then added to confluent monolayers of Vero cells in 12-well plates, in duplicate. Infection was allowed to proceed to completion (>90% cell death by eye), and supernatants were harvested from the infected wells that received the highest dilution (i.e., the least amount) of viral inoculum. Following five subsequent passages under mAb selection with virus-containing supernatants as above, supernatants from passage 4 were tested for viral neutralization escape. If resistance was evident, individual viral clones were plaque purified on Vero cells, and their GP gene sequences were determined as described previously (40).
qRT-PCR Assay.
Viral RNA copy number in blood samples collected from macaques was determined by qRT-PCR as previously described (14). Whole blood was mixed 1:3 with TRIzol LS (Thermo Fisher) and RNA was extracted using the QIAamp Viral RNA Mini Kit in accordance with manufacturer guidelines. qRT-PCR reactions were completed using SUDV-specific primer/probe pairs on an Applied Biosystems 7500 Fast Dx System. Synthetic RNA, representative of the primer target region of the SUDV genome, was used to generate an eight-point standard curve. Viral genome copy numbers for each sample were calculated by applying cycle-threshold values of an individual sample to the standard curve. The Qiagen QuantiFast Internal Control RT-PCR assay was used to monitor PCR inhibition and extraction integrity. Data are expressed as genome equivalents (ge) per milliliter of blood with a lower limit of quantitation of 8 × 104 ge/mL.
Mouse Studies.
Four- to 8-wk-old male and female type I IFNAR−/− (Jackson Labs) were exposed to a target dose of 1,000 pfu (actual dose: 234 to 910 pfu) of SUDV-Bon via I.P. injections. Mice were treated I.P. on days 1 and 4 postexposure with PBS vehicle or 200 µg of plant- or mammalian-derived mAbs diluted in 0.2 mL of PBS. Animals were observed daily for clinical signs of disease and lethality. Total weight of all animals in a given group was determined daily and the average weight was determined by dividing the total weight by the number of remaining mice in the group. The percent weight change was calculated daily by dividing the daily average weight by average weight on day 0. Daily observations were increased to a minimum of twice daily while mice were exhibiting signs of disease. Moribund mice were humanely euthanized on the basis of institutional animal care and use committee (IACUC)-approved criteria.
Rhesus Macaque Study.
Male and female rhesus macaques (4 to 7.8 kg) were randomly assigned to treatment groups, with even distribution of male and female macaques, and study personnel remained blinded to all treatment groups. Macaques were inoculated intramuscularly (I.M.) with a target dose of 1,000 pfu (actual dose: 1,750 pfu) of SUDV-Bon. Experimental macaques (n = 4) were treated I.V. with a mixture of plant-produced 16F6 and X10H2, diluted in 0.9% NaCl solution, at 50 mg/kg total (25 mg/kg per antibody) on days 4 and 6 postexposure. Control macaques 1 and 2 received an equal volume to weight ratio of vehicle (0.9% NaCl solution) I.V. on days 4 and 6 postexposure. In the interest of animal welfare and to conserve animal life, two additional control macaques from a concurrently running study were used. Control macaques 3 and 4 received an equal volume-to-weight ratio of vehicle I.V. on day 5 postexposure. Blood samples were collected from experimental macaques and control macaques 1 and 2 on days 0, 4, 6, 8, 11, 14, 21, and 28 postexposure and from control macaques 3 and 4 on days 0, 3, 5, 8, 11, 14, 21, and 28 postexposure. Blood samples were used to evaluate viremia, blood chemistries, and hematology. Physical examinations were also completed on blood collection days to assess the overall physical condition of the macaque and disease progression. Cage side observations were completed at least once daily through day 28 to assess the general disposition of each animal and disease progression. Clinical scores for each macaque were derived from cage side observations of awake animals and physical observations completed on anesthetized animals. Animals were assessed and given a cumulative score based on severity of several behavioral and physical parameters, including responsiveness to stimuli, petechial rash, temperature change, weight change, gastrointestinal abnormalities, renal output, bleeding, anorexia, labored or agonal breathing, dehydration, and edema. Higher clinical scores indicate more severe signs of disease. Macaques determined to be moribund based on either responsiveness score and/or secondary euthanasia criteria were promptly euthanized in an attempt to minimize pain and distress. Responsiveness score was based on a scale of 0 to 4 with normal animals scoring 0. Animals with slightly diminished general activity but responded normally to external stimuli were scored 1. Animals appearing withdrawn, mildly unresponsive to external stimuli, and/or having hunch posture or laying down scored 2. Animals moderately or significantly unresponsive to external stimuli and/or prostrate but able to rise in response to external stimuli scored 3. Animals severely or completely unresponsive to external stimuli, persistently prostrate, and/or displaying signs of respiratory distress scored 4. Animals with a responsiveness score of 4 were deemed moribund and euthanized immediately. Animals with a responsiveness score of 3 were further evaluated using secondary euthanasia criteria. If an animal with a responsiveness score of 3 also had a body temperature below 34 °C or any two of the following serum chemistry thresholds were met, the animal was deemed moribund and euthanized: BUN ≥68 mg/dL; Ca ≤6.8 mg/dL; CRE ≥2.8 mg/dL; GGT ≥391.
Hematology and Blood Chemistry Analysis.
Phlebotomy was performed while the animals were anesthetized, and blood was collected from the femoral vein using a venous blood collection system (Becton Dickinson). Hematological values for blood samples collected in tubes containing EDTA were determined using a Coulter Ac-T Diff analyzer (Beckman Coulter). Serum chemistry was analyzed using Piccolo General Chemistry 13 reagent discs and a Piccolo Xpress point-of-care blood analyzer (Abaxis).
Animal Welfare Statement.
Murine and macaque challenge studies were conducted under IACUC-approved protocols in compliance with the Animal Welfare Act, Public Health Service Policy, and other applicable federal statutes and regulations relating to animals and experiments involving animals. The facility where these studies was conducted (USAMRIID) is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, International and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011.
Statistical Analysis.
Antibody binding curves were evaluated using a four-parameter nonlinear regression analysis under assumption of normality. Dose–response neutralization curves were evaluated using nonlinear regression analysis (normalized response-variable slope) to determine IC50 concentrations under assumption of normality. Analysis of mouse weight change was completed by two-way ANOVA for each time point relative to control-treated animals. Significances reported were lowest P values for any time point for given antibody treatment group. Mouse survival curves were evaluated using Fisher’s exact test. Analysis of viremia and antibody titer data from the NHP efficacy study was completed by two-way ANOVA for each time point having paired data in both treatment groups. Alpha was 0.05 for all statistical tests. All analyses were carried out using GraphPad Prism.
Data Availability.
All data will be available upon request to the corresponding author.
Supplementary Material
Acknowledgments
We thank USAMRIID’s Veterinary Medicine Division for technical assistance with animal studies and USAMRIID’s Diagnostics Systems Division for technical assistance with RT-PCR analysis. We thank The Geneva Foundation, specifically Audra Earl and Andrea Masterson, for grants management and administrative support. We thank Hong Bi, Shireen Shaikh, Andrew Jenner, Stephen Tottey, and Annamalai Padmanaban (Fraunhofer USA) for technical assistance and Caitlin Morris (Fraunhofer USA) for manuscript review. Funding: This work was supported by the National Institutes of Health grant R01AI111516 and the Defense Threat Reduction Agency grant CB4088 (to J.M.D.), and the National Institutes of Health grant R01AI132256 (to K.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Opinions, conclusions, interpretations, and recommendations are those of the authors and are not necessarily endorsed by the US Army. The mention of trade names or commercial products does not constitute endorsement or recommendation for use by the Department of the Army or the Department of Defense.
Footnotes
Competing interest statement: The US Army Medical Research Institute of Infectious Disease (USAMRIID) has filed a patent application on the murine mAb 16F6 (WO2011071574 A2) entitled “Monoclonal Antibodies against Glycoprotein of Ebola Sudan Boniface Virus” with J.M.D. as a co-inventor. An invention disclosure entitled “Cross Reactive Antibodies to Ebola and Sudan Virus” has been filed for macaque mAbs X10B1, X10H2, X10F3, and X10B6 with A.S.H., J.W.F., and J.M.D. as co-inventors. N.B., O.B., D.H.K., M.H.P., and J.V. are employees of Mapp Bioparmaceutical, Inc. K.J.W. and L.Z. are employees and owners of Mapp Biopharmaceutical, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914985117/-/DCSupplemental.
References
- 1.Kuhn J. H., et al. , Virus nomenclature below the species level: A standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch. Virol. 158, 301–311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein T., et al. , The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat. Microbiol. 3, 1084–1089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC , Ebola outbreak in West Africa–Case counts. (2014). https://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/case-counts.html. Accessed 7 July 2019.
- 4.CDC , Outbreaks chronology: Ebola virus disease. https://www.cdc.gov/vhf/ebola/outbreaks/history/chronology.html. Accessed 7 July 2019.
- 5.Agnandji S. T., et al. , Phase 1 trials of rVSV Ebola vaccine in Africa and Europe. N. Engl. J. Med. 374, 1647–1660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davey R. T., Jr, et al. ; PREVAIL II Writing Group; Multi-National PREVAIL II Study Team , A randomized, controlled trial of ZMapp for Ebola virus infection. N. Engl. J. Med. 375, 1448–1456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huttner A., et al. ; VSV-Ebola Consortium , The effect of dose on the safety and immunogenicity of the VSV Ebola candidate vaccine: A randomised double-blind, placebo-controlled phase 1/2 trial. Lancet Infect. Dis. 15, 1156–1166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ledgerwood J. E., et al. ; VRC 207 Study Team , Chimpanzee adenovirus vector Ebola vaccine. N. Engl. J. Med. 376, 928–938 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Regules J. A., et al. ; rVSVΔG-ZEBOV-GP Study Group , A recombinant vesicular stomatitis virus Ebola vaccine. N. Engl. J. Med. 376, 330–341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu F. C., et al. , Safety and immunogenicity of a recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in Sierra Leone: A single-centre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 389, 621–628 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Oswald W. B., et al. , Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog. 3, e9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jahrling P. B., Geisbert J. B., Swearengen J. R., Larsen T., Geisbert T. W., Ebola hemorrhagic fever: Evaluation of passive immunotherapy in nonhuman primates. J. Infect. Dis. 196 (suppl. 2), S400–S403 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Dye J. M., et al. , Postexposure antibody prophylaxis protects nonhuman primates from filovirus disease. Proc. Natl. Acad. Sci. U.S.A. 109, 5034–5039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olinger G. G., Jr, et al. , Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc. Natl. Acad. Sci. U.S.A. 109, 18030–18035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu X., et al. , Successful treatment of ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci. Transl. Med. 4, 138ra81 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Marzi A., et al. , Protective efficacy of neutralizing monoclonal antibodies in a nonhuman primate model of Ebola hemorrhagic fever. PLoS One 7, e36192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu X., et al. , Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyon G. M., et al. ; Emory Serious Communicable Diseases Unit , Clinical care of two patients with Ebola virus disease in the United States. N. Engl. J. Med. 371, 2402–2409 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Corti D., et al. , Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 351, 1339–1342 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Pascal K. E., et al. , Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in nonhuman primates. J. Infect. Dis. 218 (suppl. 5), S612–S626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dyer O., Two Ebola treatments halve deaths in trial in DRC outbreak. BMJ 366, l5140 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Thi E. P., et al. , Rescue of non-human primates from advanced Sudan ebolavirus infection with lipid encapsulated siRNA. Nat. Microbiol. 1, 16142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahim M. N., et al. , Postexposure protective efficacy of T-705 (Favipiravir) against Sudan virus infection in Guinea pigs. J. Infect. Dis. 218 (suppl. 5), S649–S657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bornholdt Z. A., et al. , Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science 351, 1078–1083 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bornholdt Z. A., et al. , A two-antibody pan-ebolavirus cocktail confers broad therapeutic protection in ferrets and nonhuman primates. Cell Host Microbe 25, 49–58.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wec A. Z., et al. , Development of a human antibody cocktail that deploys multiple functions to confer pan-ebolavirus protection. Cell Host Microbe 25, 39–48.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brannan J. M., et al. , Post-exposure immunotherapy for two ebolaviruses and Marburg virus in nonhuman primates. Nat. Commun. 10, 105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frenzel A., Hust M., Schirrmann T., Expression of recombinant antibodies. Front. Immunol. 4, 217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q., Davis K. R., The potential of plants as a system for the development and production of human biologics. F1000Res. 5, F1000 Faculty Rev-912 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strasser R., et al. , Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol. J. 6, 392–402 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Dias J. M., et al. , A shared structural solution for neutralizing ebolaviruses. Nat. Struct. Mol. Biol. 18, 1424–1427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Froude J. W., et al. , Post-Exposure protection in mice against Sudan virus by a two antibody cocktail. Viruses 10, E286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brannan J. M., et al. , Interferon α/β receptor-deficient mice as a model for ebola virus disease. J. Infect. Dis. 212 (suppl. 2), S282–S294 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Ellis D. S., Bowen E. T., Simpson D. I., Stamford S., Ebola virus: A comparison, at ultrastructural level, of the behaviour of the Sudan and Zaire strains in monkeys. Br. J. Exp. Pathol. 59, 584–593 (1978). [PMC free article] [PubMed] [Google Scholar]
- 35.Geisbert T. W., et al. , Recombinant vesicular stomatitis virus vector mediates postexposure protection against Sudan Ebola hemorrhagic fever in nonhuman primates. J. Virol. 82, 5664–5668 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson J. A., et al. , Epitopes involved in antibody-mediated protection from Ebola virus. Science 287, 1664–1666 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Qiu X., et al. , Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clin. Immunol. 141, 218–227 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Anonymous , Ebola haemorrhagic fever in Sudan, 1976. Report of a WHO/International Study Team. Bull. World Health Organ. 56, 247–270 (1978). [PMC free article] [PubMed] [Google Scholar]
- 39.Wec A. Z., et al. , A “Trojan horse” bispecific-antibody strategy for broad protection against ebolaviruses. Science 354, 350–354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong A. C., Sandesara R. G., Mulherkar N., Whelan S. P., Chandran K., A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol. 84, 163–175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.West B. R., et al. , Structural basis of broad ebolavirus neutralization by a human survivor antibody. Nat. Struct. Mol. Biol. 26, 204–212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wec A. Z., et al. , Antibodies from a human survivor define sites of vulnerability for broad protection against ebolaviruses. Cell 169, 878–890.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data will be available upon request to the corresponding author.