Significance
The highly attenuated vaccine vector MVA is an approved smallpox vaccine and is undergoing clinical trials as a vaccine vector for other infectious diseases. Although an inability to replicate in human cells contributes to the safety of MVA, the basis for its host restriction is not understood. Here we identify a host-range gene C16L/B22R, which is present as two copies in most orthopoxviruses but is inactivated in MVA. Repair of this gene allows replication of MVA in several tested human cell lines and, in combination with a second missing host-range gene C12L, restores full replication. Furthermore, a human cell line expressing both proteins is permissive for MVA. This knowledge may contribute to further engineering of MVA vaccines.
Keywords: vaccine vector, vaccinia virus, poxvirus, host range
Abstract
Modified vaccinia virus Ankara (MVA), a widely used vaccine vector for expression of genes of unrelated pathogens, is safe, immunogenic, and can incorporate large amounts of added DNA. MVA was derived by extensively passaging the chorioallantois vaccinia virus Ankara (CVA) vaccine strain in chicken embryo fibroblasts during which numerous mutations and deletions occurred with loss of replicative ability in most mammalian cells. Restoration of the deleted C12L gene, encoding serine protease inhibitor 1, enhances replication of MVA in human MRC-5 cells but only slightly in other human cells. Here we show that repair of the inactivated C16L/B22R gene of MVA enhances replication in numerous human cell lines. This previously uncharacterized gene is present at both ends of the genome of many orthopoxviruses and is highly conserved in most, including smallpox and monkeypox viruses. The C16L/B22R gene is expressed early in infection from the second methionine of the previously annotated Copenhagen strain open reading frame (ORF) as a 17.4-kDa protein. The C16/B22 and C12 proteins together promote MVA replication in human cells to levels that are comparable to titers in chicken embryo fibroblasts. Both proteins enhance virion assembly, but C16/B22 increases proteolytic processing of core proteins in A549 cells consistent with higher infectious virus titers. Furthermore, human A549 cells expressing codon-optimized C16L/B22R and C12L genes support higher levels of MVA replication than cell lines expressing neither or either alone. Identification of the genes responsible for the host-range defect of MVA may allow more rational engineering of vaccines for efficacy, safety, and propagation.
Vaccinia virus (VACV) and the closely related cowpox virus (CPXV) were the first vaccines used to prevent an infectious disease, namely, smallpox (1). The protection afforded by VACV and CPXV is due to their close sequence relationship to variola virus, the causative agent of smallpox. Since the eradication of smallpox, VACV and other poxviruses have taken on new roles as vectors to express immunogens that can prevent diseases caused by unrelated agents and as cancer therapeutics (2, 3). Modified VACV Ankara (MVA), a vaccine strain that was developed by extensive passaging of the parent chorioallantois vaccinia virus (CVA) strain in chicken embryo fibroblasts (CEF), is widely used as a vector because of its safety and immunogenicity (4, 5). The safety of MVA depends on its inability to replicate in human cells and the loss of many immune defense genes. Despite the licensing of MVA as a smallpox vaccine and ongoing clinical trials of MVA-vectored vaccines, the basis for its host-range defect is unknown. An early study indicated that the defect occurred late during infection of human HeLa cells resulting in arrest of morphogenesis (6). A comparison of MVA with its parent CVA reveals 71 orthologous open reading frames (ORFs) predicted to encode identical proteins, whereas the remaining 124 ORFs encode gene products with amino acid changes, insertions, or deletions, complicating the efforts to identify the missing host-range gene(s) (7). The approach of making deletions in a fully replication-competent strain is further confounded by functional redundancies (8). After removal of DNA corresponding to the six large deletions totaling 25 kbp, both the CVA and Lister vaccine strains remained competent to replicate in human cells (9, 10). To simplify the task of identifying the host-range genes, Wyatt and coworkers (11) used marker rescue with overlapping 40-kbp cosmids, which narrowed the region of interest to the left side of the MVA genome. Sequencing of the rescued viruses further narrowed the critical region to an ∼6-kbp deletion in the MVA genome, which contained C12L*, a known host-range gene encoding serine protease inhibitor 1 (SPI-1) (12). The insertion of C12L into MVA partially restores replication in human MRC-5 cells but only slightly in other human cell lines, suggesting the existence of one or more additional host-range genes. Here we identify C16L/B22R, which is duplicated at the two ends of most orthopoxviruses, as a second host-range gene. In MVA, C16L, located at the left end of the genome, is severely truncated, whereas B22R, at the right end of the genome, has a 45-bp deletion. Restoration of a single intact copy of C16L/B22R enables replication of MVA in human cells and in combination with C12L to similar levels as in permissive CEF. In addition, a human cell line expressing both C12L and C16L/B22R supports MVA replication.
Results
Identification of C16L/B22R as a Human Host-Range Gene.
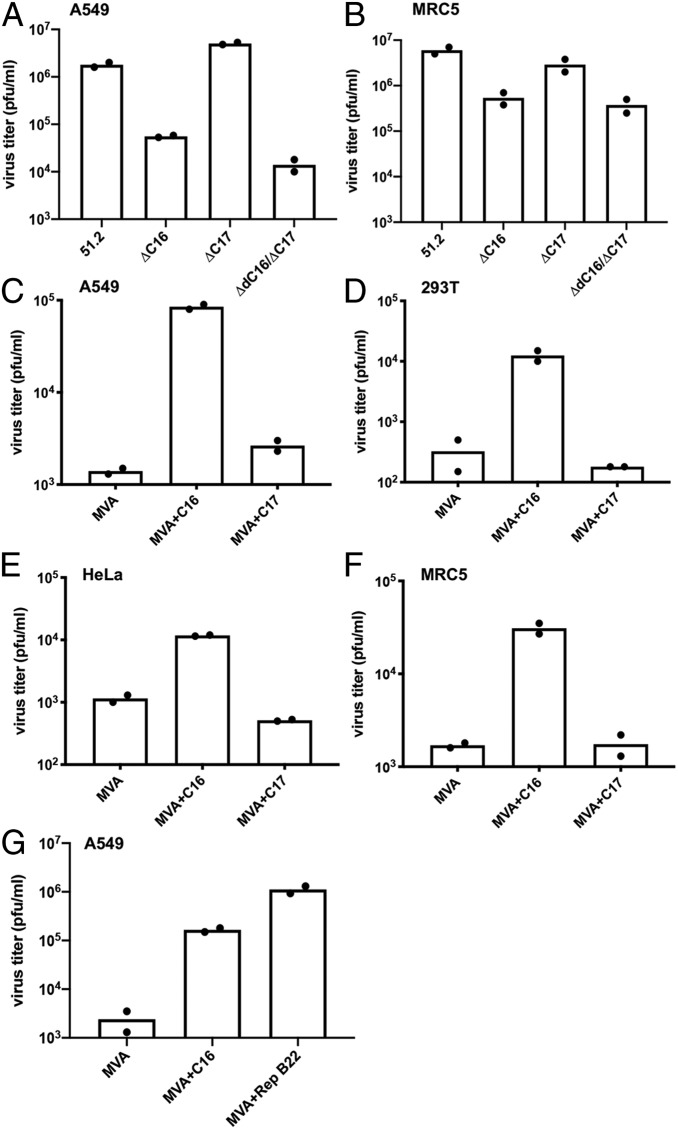
The ability of v51.2, a recombinant host-range extended MVA, to replicate in human cells is attributed to the insertion of DNA that repaired a large deletion near the left end of the MVA genome (11). Addition of C12L, one of the genes missing from MVA but present in v51.2, increases MVA replication in human MRC-5 cells but has little benefit in other human cell lines (12). As v51.2 replicates in many human cell lines, additional host-range genes must be present in the latter virus. To identify other putative host-range genes of v51.2, we deleted individual or multiple ORFs from the inserted region. To delete genes, CEF were infected with v51.2 and transfected with DNA encoding a green fluorescent protein (GFP) or mCherry expression cassette flanked by sequences adjacent to the two ends of the targeted gene to replace the latter by homologous recombination. The progeny was screened by plaque assay, and virus from fluorescent plaques was clonally purified. Since some ORFs at the left end of the genome are duplicated at the right end, PCR was used to ensure that both copies were deleted. The cloned recombinant viruses were then assessed for their ability to replicate in human A549 and MRC-5 cells. Initially, we found that a mutant with deletions of the neighboring C16L and C17L ORFs and their duplicates B22R and B23R, respectively, near the right end of the genome exhibited a replication defect. However, when specific gene deletion mutants were made, loss of C16L/B22R but not C17L/B23R diminished replication in both cell lines (Fig. 1 A and B and Table 1).
Fig. 1.
Effect of deletion and insertion of C16L/B22R on replication of MVA in human cells. A549, MRC-5, 293T, and HeLa cells were infected with v51.2 and mutants in which C16L/B22R or C17L/B23R or both were deleted (A and B), or MVA mutants in which the C16L or C17L ORFs were added (C–F), or in which the C16L was added or the deletion in B22R was repaired (G). In A and B, v51.2ΔC16/ΔB22 and v51.2ΔC17/ΔB23 are abbreviated ΔC16 and ΔC17, respectively. Cells were infected in duplicate with 0.01 pfu per cell of virus for 48 h, and the titers from each were determined by plaque assay on CEF. Virus titers from each infection are shown as dots, and the bar represents the mean value.
Table 1.
Recombinant Viruses
| Virus name | C17L | C16L | C12 | B22R | B23R | Insert | MRC-5* | A549† |
| v51.2 | + | + | + | + | + | none | +++ | +++ |
| V51.2ΔC17/B23 | mCherry¶ | + | + | + | mCherry | none | +++ | +++ |
| V51.2ΔC16/B22 | + | GFP | + | GFP | + | none | ++ | + |
| V51.2ΔC17ΔC16 | mCherry | mCherry | + | mCherry | mCherry | none | ++ | + |
| V51.2ΔC12 | + | + | GFP | + | + | none | + | ++ |
| MVA | FS‡ | truncated | − | Δ45 bp | FS | none | − | − |
| MVA+C17 | FS | truncated | − | Δ45 bp | FS | C17L | − | − |
| MVA+C16 | FS | truncated | − | Δ45 bp | FS | C16L | + | ++ |
| MVA+B22§ | FS | truncated | − | repaired | FS | none | + | ++ |
| MVA+C16/C17 | FS | truncated | − | Δ45 bp | FS | C16L+C17L | + | ++ |
| MVA+C12 | FS | truncated | − | Δ45 bp | FS | C12L | ++ | + |
| MVA+C12/C16 | FS | truncated | − | Δ45 bp | FS | C16L+C12L | +++ | +++ |
Replication in MRC-5 cells. −, +, ++, and +++ indicate no, low, moderate, and high replication, respectively.
Replication in A549 cells. −, +, ++, and +++ indicate no, low, moderate, and high replication, respectively.
Frame-shift.
B22R repaired by homologous recombination.
mCherry or GFP replaced indicated ORF.
To further compare their roles, an intact C16L or C17L ORF including its natural promoter copied by PCR from v51.2 was inserted between ORFs 069 and 070 of MVA by homologous recombination. The mCherry ORF, which was regulated by a separate VACV promoter, was simultaneously inserted downstream to facilitate plaque isolation and cloning. Sequencing revealed that the original defective C16L/B22R and C17L/B23R ORFs of MVA were not corrected by homologous recombination so that MVA+C16L and MVA+C17L had only single intact copies of these genes in a new location (Table 1). Addition of C16L but not C17L increased MVA replication in A549, 293T, HeLa, and MRC-5 cells (Fig. 1 C–F). Together, these data indicated that C16L/B22R is a previously unrecognized human host-range gene.
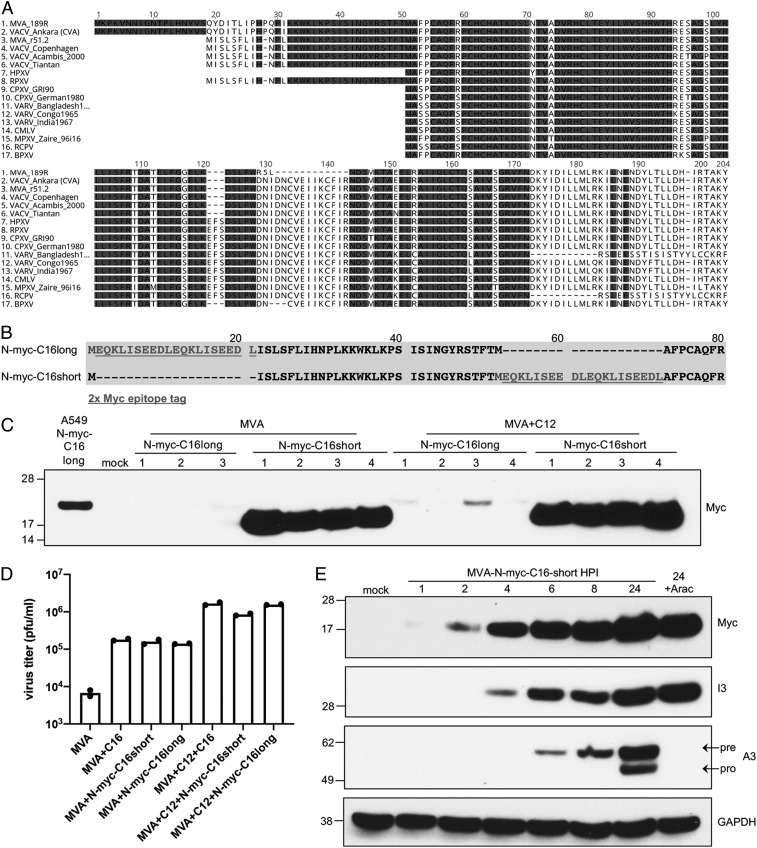
The C16L and B22R ORFs are identical in v51.2, whereas in MVA the C16L ORF has a large N-terminal truncation and the B22R copy appeared to be intact (12). However, when the B22R ORF was aligned with the C16L/B22R genes of other orthopoxviruses including v51.2 and the MVA parent CVA, it became apparent that MVA B22R (labeled 189R in Fig. 2A) has a deletion resulting in loss of 15 amino acids. Aside from this small deletion, the sequence of the MVA B22R is identical to that of other orthopoxviruses (Fig. 2A). The importance of this short sequence was confirmed by demonstrating that correction of the deletion of the MVA B22R ORF by homologous recombination was sufficient to increase replication of MVA in A549 cells (Fig. 1G). Apparently, the protein with the internal deletion is less stable or poorly expressed as quantitative mass spectrometry analysis using tandem mass tag labeling of trypsin-digested total extracts revealed 17- to 33-fold more C16L/B22 from A549 cells infected with v51.2 compared to MVA.
Fig. 2.
Sequence, expression, and activity of C16/B22 protein. (A) Multiple sequence alignment of C16L/B22R coding sequences from the indicated poxviruses. Only the B22R (189R) ORF of MVA is shown. For other orthopoxviruses, the two copies of the gene are identical, or only one copy is present. One hundred percent conserved residues are shaded. (B) Diagram showing placement of myc tag (underlined) before the first (N-myc-C16long) or second (N-myc-C16short) methionine. (C) A549 cells were mock-infected or infected with 5 pfu per cell of MVA+N-myc-C16long, MVA+N-myc-C16short, or the corresponding viruses that also express C12. C16long and C16short refer to placement of the Myc-tags after the Met at number 19 or 51 respectively, of the v51.2 C16 ORF in A. After 24 h, the cells were lysed and the proteins resolved by polyacrylamide gel electrophoresis. The C16 protein from A549 cells stably expressing codon-optimized C16 with a N-terminal Myc-tag after the first methionine regulated by the CMV promoter is shown in the first lane. Anti-myc-HRP was used for protein detection on Western blots. Numbers indicate individual clones of recombinant viruses. (D) A549 cells were infected with the indicated virus in duplicate at 0.01 pfu per cell for 48 h. Virus titers from each infection are shown as dots, and the bar represents the mean value. (E) A549 cells were mock-infected or infected with MVA-N-myc-C16short at 3 pfu per cell and after 1, 2, 4, 6, 8, and 24 h postinfection (hpi), analyzed by Western blotting. Numbers on the left are the masses in kDa and positions of marker proteins. The names on the right indicate the proteins recognized by antibodies. Pre and pro refer to precursor and processed proteins, respectively.
Further comparisons of the C16L/B22R ORFs of orthopoxviruses indicated variability in the length and sequence of the N-terminal region (Fig. 2A). The longest ORFs were in CVA and MVA; shorter ones were in the VACV strains v51.2, Copenhagen, Acambis, Tiantan, and rabbitpox; and still shorter ones were in horsepox, cowpox, variola, camelpox, monkeypox, racoonpox, and buffalopox viruses (Fig. 2A). However, in each of the longer C16L/B22R ORFs the second in-frame methionine corresponds to the start codon of each of the shorter C16L/B22R ORFs. Therefore, it seemed possible that the second methionine is also the start codon of the longer ORFs. To evaluate this hypothesis, recombinant MVAs were constructed by inserting the C16L ORF with a 2xMyc epitope tag after the first or second methionine to produce MVA+N-mycC16long and MVA+N-mycC16short, respectively (Fig. 2B). The mCherry ORF regulated by a separate VACV promoter was simultaneously inserted downstream of the C16 ORF to facilitate the isolation of recombinant viruses. Corresponding MVA+C12 viruses with long and short N-mycC16 were produced similarly. As a control, an A549 cell line was made in which a myc tag was placed after the first methionine of a codon-optimized C16 that was regulated by a cytomegalovirus (CMV) promoter. Western blots of lysates of the cell line and several independent recombinant MVA clones probed with anti-myc antibody are shown in Fig. 2C. The myc-tagged C16 expressed by the cell line migrated as a 23-kDa protein. A protein of the same size was barely detected by anti-myc antibody when MVA expressed C16 with the tag at the first methionine. In contrast, there was an intense slightly faster migrating band when the tag was placed after the second methionine suggesting that the latter is used as the start codon in recombinant MVA. Consistent with this interpretation, an A+T-rich sequence (ATTAAAAAAATGGAAATTAA) similar to the consensus early promoter motif (13, 14) is located between the first and second methionines. Regardless of where the Myc-tag was inserted, the MVA+C16 constructs enabled replication in A549 cells (Fig. 2D).
Since the Myc-tagged construct had the natural promoter, an experiment was carried out to determine the time of synthesis. The early I3 and late A3 proteins were analyzed simultaneously for comparison. The C16 protein was detected at 2 h after infection and was synthesized in the presence of AraC, an inhibitor of DNA replication, consistent with early regulation (Fig. 2E).
Together C16/B22 and C12 Enhance Replication More than Either Alone.
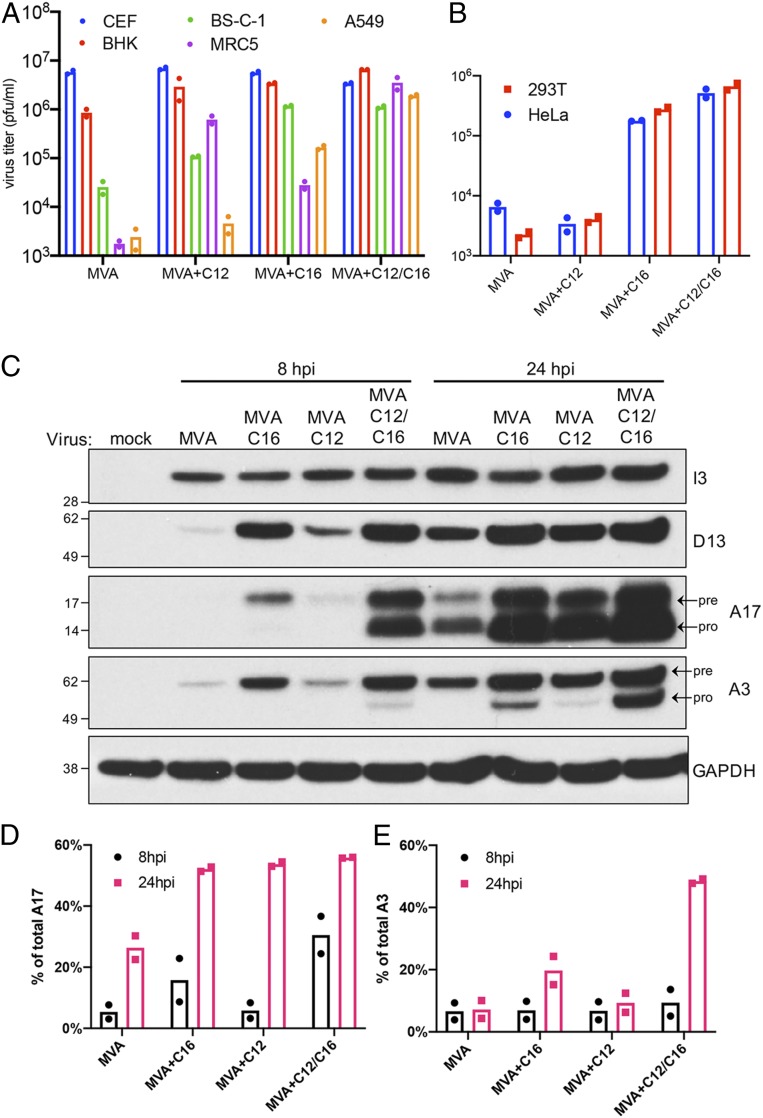
As previously shown, MVA replicates to high titers in CEF and BHK-21 cells, to a low level in BS-C-1 cells, and not appreciably in MRC-5 or A549 cells (Fig. 3A). Expression of C12 alone enhanced MVA replication in human MRC-5 cells and BS-C-1 cells but had only a slight effect in A549 cells (Fig. 3A). In contrast, expression of C16 increased replication of MVA more in A549 and BS-C-1 cells than in MRC-5 cells (Fig. 3A). A recombinant MVA that expresses both C16 and C12 further increased replication in both A549 and MRC-5 cells to levels similar to that in CEF (Fig. 3A). C16 expression also increased replication of MVA in human HeLa and 293T cells, whereas C12 did not (Fig. 3B). Both C16L and C12L are true host-range genes as they did not increase replication of MVA in CEF.
Fig. 3.
Expressions of C12 and C16/B22 enhance infection more than either alone. CEF, BHK-21, BS-C-1, MRC-5, A549 (A) and 293T and HeLa (B) cells were infected in duplicate with indicated viruses at 0.01 pfu per cell for 48 h. Virus titers from each infection determined by plaque assay on CEF are shown as dots, and the bar represents the mean. (C) Duplicate A549 cell monolayers were mock-infected or infected with 5 pfu per cell of the indicated viruses. Cells were lysed at 8 and 24 hpi and analyzed by Western blotting. Numbers on the left are the masses in kDa and positions of marker proteins. Names on the right indicate the proteins recognized by antibodies. Pre and pro refer to precursor and processed proteins, respectively. The percentages of total A17 (D) and A3 (E) processed in duplicate infections were calculated by ImageJ software based on band density. Dots are ratios from individual infections, and the bar is the mean.
C12 and C16 Overcome Assembly Defects at Different Stages.
The assembly of poxvirus virions begins in discrete areas of the cytoplasm known as factories. The first discernible structures are crescent membranes and spherical immature virions enclosing dense amorphous material bounded by a spicule-coated single-membrane bilayer. The next stage involves proteolytic processing of the A17 membrane protein and release of the D13 spicule layer followed by condensation of the core and cleavage of A3 and other core proteins. Processing of A17 and A3 are indicators of early and late stages of morphogenesis, respectively.
The synthesis and processing of viral proteins expressed by MVA, MVA+C16, MVA+C12, and MVA+C12/C16 in A549 cells were investigated by Western blotting with specific antibodies (Fig. 3 C–E). Early proteins represented by I3 were abundant at 8 h and increased at 24 h in cells infected with each of the viruses. At 8 h, the intensities of the D13 intermediate and the A17 and A3 late proteins were MVA+C12/C16 > MVA+C16 > MVA+C12 > MVA. In cells infected with MVA+C12/C16 at this time, appreciable processing of the A17 membrane protein and slight processing of the A3 core protein was discerned by the relative intensities of the upper and lower bands. At 24 h, synthesis of the early, intermediate, and late proteins was increased in each case, although the order of abundances remained the same as at 8 h. Processing of the A17 membrane protein occurred during each infection regardless of the abundance of the protein. In contrast, there was no detectable processing of the A3 core protein in cells infected with MVA and only slight processing in cells infected with MVA+C12, whereas extensive processing occurred in cells infected with MVA+C16 and MVA+C12/C16.
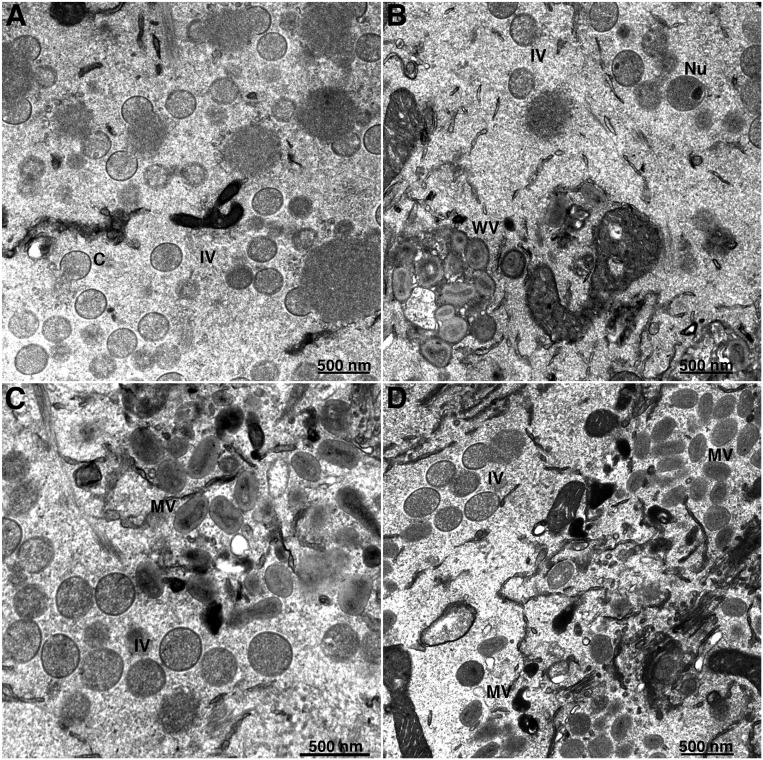
Electron microscopy was used to further analyze the roles of C16/B22 and C12 in overcoming the host-range defect of MVA. A previous study had shown accumulation of immature virions with very few mature virions in HeLa cells infected with MVA (6, 15). We found a similar block in morphogenesis in MVA-infected A549 cells (Fig. 4A). In contrast, mature-looking and wrapped virus particles formed in A549 cells infected with MVA expressing C12L, C16L, or both (Fig. 4 B–D). Manual counting of particles in 50 randomly chosen cell sections confirmed the difference in numbers of all three types of mature particles (mature, wrapped, and enveloped virions) in cells infected with MVA recombinants expressing C12, C16, or both compared to MVA (Table 2). Despite the greater cleavage of the A3 core protein in cells infected with MVA+C16 compared to MVA+C12, morphological differences in the mature particles were not apparent by transmission electron microscopy.
Fig. 4.
Electron microscopy of cells infected with MVA and recombinant MVAs. A549 cells were infected with MVA (A), MVA+C12 (B), MVA+C16 (C), and MVA+C12/C16 (D) for 20 h, and samples were fixed and analyzed by transmission electron microscopy. Abbreviations: MV, mature virion; WV, wrapped virion; C, crescent; IV, immature virion; Nu, IV that contains a nucleoid. Magnification shown below each panel.
Table 2.
Immature and Mature Virions*
| Virus stage | MVA | MVA+C12 | MVA+C16 | MV+C12/C16 |
| Crescents | 1562 | 536 | 432 | 255 |
| IVs† | 238 (3) | 510 (19) | 800 (39) | 1025 (46) |
| DVs‡ | 1 | 5 | 7 | 12 |
| MVs§ | 0 | 78 | 82 | 106 |
| WVs¶ | 0 | 31 | 22 | 57 |
| EVs# | 0 | 55 | 65 | 153 |
| Total mature|| | 0 | 164 | 169 | 316 |
Total number of virus forms in 50 A549 cell sections at 20 h after infection.
Total immature virions and those with nucleoids in parentheses.
Dense spherical virions.
Mature virions.
Wrapped virions.
Enveloped virions.
Total MVs, WVs, and EVs.
Complementation of MVA by Cells Expressing C12 and C16/B22.
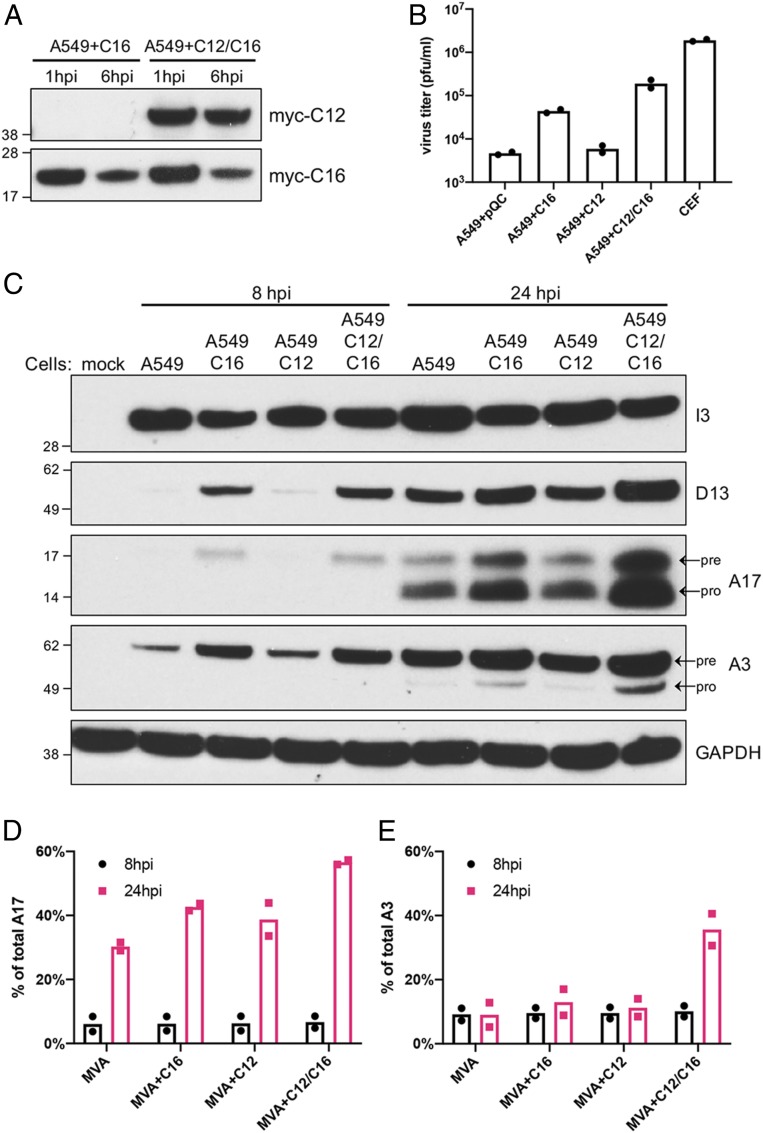
Thus far, we have shown that repair of C12L and C16L/B22R allowed MVA to replicate in monkey and human cell lines. We also investigated whether ectopic expression of the proteins in cells would complement replication of MVA. Codon-optimized N-myc–tagged C12, C16, or C12 and C16 regulated by CMV promoters were cloned into retrovirus vectors containing a puromycin-resistant marker; then the retroviruses containing the transgenes were packaged in 293T cells and used to transduce A549 cells. Stable cell lines A549+C16, A549+C12, and A549+C12/C16 expressing the proteins were maintained in the presence of antibiotic selection. Expression of the myc-tagged proteins was detected by Western blotting (Fig. 5A). When the cells were infected with MVA, C16 diminished between 1 and 6 h after infection likely resulting from turnover accompanying the virus-induced shutdown of cellular protein synthesis. As anticipated, C16 expression increased MVA replication more than C12, and the combination was better than C16 alone (Fig. 5B). However, replication of MVA in A549+C12/C16 cells was lower than in CEF, suggesting that further optimization of C12 and C16 expression may be helpful for MVA propagation.
Fig. 5.
A549 cells stably expressing C12 and C16 are permissive for MVA. (A) A549 cell lines stably expressing myc-tagged C16 or C12/C16 were infected in duplicate with 3 pfu per cell of MVA. Total protein was collected at 1 and 6 hpi for Western blotting with anti-myc antibodies. (B) Duplicate wells of CEF and of A549 cells stably expressing C12, C16, C12/16, or none (pQC) were infected with 0.01 pfu per cell of MVA. After 48 h, virus yields were determined by plaque assay on CEF. (C) Indicated cells were mock-infected or infected with 5 pfu per cell of MVA in duplicate. Cells were lysed at 8 and 24 hpi and analyzed by Western blotting. Numbers on the left are the masses in kDa and positions of marker proteins. The names on the right indicate the proteins recognized by antibodies. Pre and pro refer to precursor and processed proteins, respectively. The percent of total A17 (D) and A3 (E) that was processed in each infection was calculated by ImageJ software based on band density. Dots are ratios from individual infections, and the bar is the mean value.
Western blotting was carried out at 8 and 24 h after MVA infection to compare the synthesis and processing of viral proteins in the modified A549 cells (Fig. 5 C–E). Expression of the early I3 protein was similar in each of the cell lines. At 8 h, expression of D13, A17, and A3 was higher in A549+C16 and A549+C12/C16-expressing cell lines than in unmodified A549 and A549+C12 cells. There was increased accumulation of D13, A17, and A3 at 24 h, but the relative order of expression remained the same in the different cell lines. The abundance of the lower A17 band indicated good processing of this membrane protein in each of the cell lines at 24 h. However, processing of A3 was greatest in the A549+C12/C16 cells, next in the A549+C16 cells, and barely detectable in the A549+C12 and control A549 cells transduced with vector only. These results mimicked those obtained with the recombinant MVAs, although the extent of A3 processing was lower.
Discussion
MVA has 124 ORFs that encode gene products with amino acid changes, insertions, or deletions relative to its replication-competent parent CVA (7); nevertheless, replacement or repair of only C12L and C16L/B22R was sufficient to abrogate the host-range restriction of MVA for human cells. Because of their contiguity, it is possible that C12L and the N-terminal part of C16L were lost in a single deletion event during passaging of CVA. However, the small 45-bp deletion in the B22R copy must have occurred independently. Whether the mutations affecting both C16L and B22R are neutral or provide a slight selective advantage on serial passage of MVA in CEF is uncertain. Both C16L and B22R are intact in v51.2, which replicates well in CEF and human cells. Since the cosmid used for marker rescue of MVA to form v51.2 contained only the C16L copy, B22R must have been repaired by homologous recombination. Our finding that B22R as well as C16L of MVA is defective provides an explanation as to why previous studies failed to identify C16L/B22R as important for the host-range restriction of MVA (9, 10). In the latter studies, C16L but not B22R was deleted from the replication-competent CVA and Lister strains of VACV. The significance of C16L/B22R was also not appreciated in a recent study that did not analyze deletion of these genes alone from v51.2 or their addition to MVA (12).
The conservation of C16L and B22R in most orthopoxviruses suggests an important role. In fact, each of the replication-competent viruses in Fig. 2A, except for VARV, CMLV, and MPXV, have two identical copies, one in each of the inverted terminal repeats near each end of the genome. Nevertheless, the gene is absent from the replication-competent Western Reserve (WR) strain of VACV, suggesting the presence of another gene with a redundant function. Bioinformatic analysis using the HHpred server suggests that the C16/B22 protein has significant similarities (>99% probability) to poxvirus B cell lymphoma 2 (BCL-2) proteins B15, A52, K7, A46, and N1, which function as immunomodulators (16, 17). Interestingly, MVA is missing A52R and another BCL-2–like gene, C1L. In addition, there are sequence differences between the C6L, B15R, and N1R ORFs of MVA and CVA, whereas K7R and A46R are identical in the two viruses. Whether any of these BCL-2–like proteins can substitute for C16/B22 in overcoming the host-range defect of MVA is an interesting question that we are pursuing.
The higher yield of infectious virus when both C12 and C16 are expressed compared to either alone suggests that they enable different steps in MVA replication. Although C12 or C16 expression partly overcomes the block in morphogenesis in A549 cells as seen by increased numbers of mature-looking virus particles, less proteolytic processing of core proteins occurs when C12 is expressed compared to C16, corresponding to the difference in production of infectious virus. Since neither C12 nor C16 are necessary for MVA replication in CEF, the roles of C12 and C16 on morphogenesis in human cells are unlikely to be direct. Moreover, the majority of virion proteins are expressed following DNA replication, whereas C12 and C16 are synthesized early in infection. The presence of C12 (SPI-1) was reported in one mass spectrometry analysis of purified virions from the Copenhagen strain (18) but not in three other analyses of VACV WR virions (19–21). C16/B22 protein was not detected in purified Copenhagen virions, although it is encoded by that strain but not by WR. The host-range effect and early expression make it likely that C12 and C16/B22 enhance morphogenesis indirectly.
MVA+C12 replicates better in MRC-5 cells than MVA+16, whereas the reverse is true for A549 cells suggesting that different host factors inhibit MVA in the two cell lines. A human genome-wide RNAi screen implicated three genes (IRF2, FAM111A, and RFC3) in the restriction of rabbitpox and VACV WR C12L deletion mutants in human cells (22). Further experiments are needed to determine whether the latter host factors also restrict MVA in MRC-5 cells and whether additional host factors inhibit MVA in A549 cells.
In summary, the inability of MVA to replicate in human cells can be explained by the inactivation of only two viral genes C12L and C16L/B22R. The incorporation of two other host-range genes, K1L and C7L, with partially redundant functions into the attenuated VACV strain NYVAC enhances replication in vitro and immunogenicity in vivo without an apparent increase in virulence (23, 24). Whether the addition of C12L and C16L/B22R to MVA will enhance immunogenicity or decrease safety are important questions to consider for vaccine design.
Materials and Methods
Cell Culture and Stable Cell Lines.
Methods were previously described with minor changes (12). A549 cells (ATCC CCL-185) were maintained in Dulbecco’s Modified Eagle Medium (DMEM)/F-12 (Life Technologies) with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 units of penicillin, and 100 µg of streptomycin per ml (Quality Biologicals, Inc.). HeLa cells (ATCC CCL-2) and 293T cells (ATCC CRL-3216) were propagated in DMEM supplemented as above. Primary CEF from 10-d-old fertile eggs (Charles River) and BS-C-1 (ATCC CCL-26) and MRC-5 (ATCC CCL-171) cells were grown in minimum essential medium with Eagle’s balanced salts (EMEM) supplement as above.
The construction of an A549 cell line expressing C12 by retroviral transduction was previously described (12), and similar methods were used to stably express C16 alone and in combination with C12. Eukaryotic codon-optimized C12 and C16 ORFs with an N-terminal 2xMyc tag were inserted into pQC-XIP (Takara) to make pQC-XIP-2xMyc-C12 and pQC-XIP-2xMyc-C16. Retroviruses were packaged in 293T cells, and virus particles were collected and filtered (0.22 µm) to remove cell debris. A549 cells were transduced in the presence of 5 µg/mL of polybrene for 48 h. Cells were passaged three times in complete medium supplemented with 1 µg/mL of puromycin (Sigma-Aldrich). To generate C12/C16 double-expressing cells, 2xMyc-C12 was inserted into a pQC-XIN vector (Takara), and retrovirus particles were generated as described above to transduce A549+C16 cells. The cells were maintained in the presence of 1 µg/mL puromycin and 500 µg/mL geneticin. The expression of C12 and C16 was determined by Western blotting using an anti-Myc antibody (Santa Cruz, 9E10).
Antibodies.
Rabbit antibody to VACV WR strain was described previously (25); anti-c-Myc antibody (9E10) conjugated to horse radish peroxidase (HRP) was purchased from Santa Cruz Biotechnology; rabbit anti-GAPDH antibody was purchased from Cell Signaling Technologies (#5174); rabbit anti-A3, anti-A17 (26), and anti-D13 (27) and mouse anti-I3 (28) were described previously. HRP-conjugated secondary antibodies, anti-rabbit IgG (#7074) and anti-mouse IgG (#7076), were purchased from Cell Signaling Technologies.
Viruses.
Human replication-competent recombinant virus v51.2 was described previously (11). Additional recombinant viruses were constructed by homologous recombination using fluorescent reporter genes (mCherry or GFP) for plaque selection. Generation of MVA+C12 was described previously (12). Briefly, DNA containing the C12L ORF regulated by the mH5 promoter (29) followed by the GFP ORF regulated by the P11 promoter (30) was flanked by sequences around the deletion III locus of MVA. To generate MVA+C16, the C16L DNA segment along with the upstream 339 bp presumed to contain the natural promoter was copied by PCR from the genome of v51.2 and inserted into a cassette that contained the mCherry ORF regulated by the P1 1promoter with flanking sequences comprised of ORFs 069 and 070. To make MVA+C12L/C16L, the same cassette was inserted into MVA+C12. Homologous recombination was performed by infecting CEF cells at multiplicity of 3 plaque-forming units (pfu) per cell for 2 h, followed by transfection of assembled and purified PCR products using Lipofectamine 2000 (Thermo Fisher Scientific). Cells were harvested after 24 h and lysed by three freeze–thaw cycles. The lysates were serially diluted and were used to infect CEF cells for plaque selection. Virus from fluorescent plaques was picked and replaqued five times. Viral DNA was isolated from lysed cells with the DNeasy Blood & Tissue kit from Qiagen and was PCR-amplified and sequenced by the Sanger method to confirm successful recombination. All recombinant viruses were propagated in CEF.
Virus Yield Determination and Plaque Assay.
Procedures were carried out as previously described with minor changes (12). Cell monolayers in 24-well plates were infected in duplicate with viruses at a multiplicity of 0.01 pfu per cell in EMEM containing 2.5% FBS for 2 h. Cells were then washed twice with phosphate buffered saline (PBS) and incubated for 48 h. Cells were harvested and lysed by three free–thaw cycles. Duplicate CEF monolayers were infected with serially diluted viruses for 2 h, and then the inoculum was aspirated and replaced with medium containing 2.5% FBS and 0.5% methylcellulose. After 48 h, infected cells were fixed with methanol–acetone (1:1. vol/vol), washed with distilled water, and incubated with anti-VACV WR antibody diluted 1:1,000 in 3% FBS-PBS for 1 h. Cells were then washed twice with distilled water and incubated with a 1:3,000 dilution of protein A-HRP (Thermo Fisher Scientific). After 1 h, protein A-HRP was aspirated and replaced with ethanol-saturated dianisidine, diluted 1:50 in PBS containing 0.3% H2O2. The dianisidine solution was removed after 10 min of incubation at room temperature, and cells were washed with distilled water. Plaques were counted manually, and duplicates were averaged.
Transmission Electron Microscopy.
Infected A549 cells were fixed, dehydrated, and embedded in Embed 812 resin as described previously (31). Specimens were viewed with an FEI Tecnai Spirit transmission electron microscope (FEI).
Western Blotting.
Procedures were carried out as previously described with minor changes (12). Cells were washed once with ice-cold PBS and lysed in wells with 1x lithium dodecyl sulfate buffer supplemented with 0.5 M DTT and 1x proteinase/phosphatase inhibitor mixtures (Thermo Fisher Scientific). Cell lysates were sonicated for 1 min to reduce viscosity. Proteins were resolved on 4–12% NuPAGE Bis-Tris gels (Thermo Fisher Scientific) and transferred to nitrocellulose membranes in iBlot2 machines (Thermo Fisher Scientific). Membranes were blocked with 5% nonfat milk in Tris-buffered saline (TBS) for 1 h and then incubated with primary antibodies diluted in 5% nonfat milk in TBS with 0.1% Tween 20 (TBST) overnight at 4 °C. The membranes were washed three times with TBST and incubated with secondary antibody conjugated with horseradish peroxidase in TBST containing 5% nonfat milk for 1 h at room temperature. The bound proteins were detected with SuperSignal West Dura substrates (Thermo Fisher Scientific).
Availability Statement.
Materials and additional details regarding methods and protocols may be obtained by contacting Bernard Moss at bmoss@nih.gov.
Acknowledgments
We thank Andrea Weisberg for electron microscopy, Ruikang Liu for materials and discussions, and Catherine Cotter for cell culture. Research was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Competing interest statement: C.P. and B.M. are listed as inventors on a patent filed by the US government that contains findings from this study.
*The Copenhagen nomenclature is used for gene or ORF names. The number refers to the position in a HindIII endonuclease restriction fragment, and L or R refers to the direction of transcription. Two names separated by a slash indicate the presence of duplicate copies, one within each inverted terminal repeat. Protein names correspond to gene names, except for the absence of L and R.
See online for related content such as Commentaries.
References
- 1.Fenner F., Henderson D. A., Arita I., Jezek Z., Ladnyi I. D., Smallpox and Its Eradication (World Health Organization, Geneva, ed. 1, 1988), 1460 pp. [Google Scholar]
- 2.Moss B., Genetically engineered poxviruses for recombinant gene expression, vaccination, and safety. Proc. Natl. Acad. Sci. U.S.A. 93, 11341–11348 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paoletti E., Applications of pox virus vectors to vaccination: An update. Proc. Natl. Acad. Sci. U.S.A. 93, 11349–11353 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volz A., Sutter G., “Modified vaccinia virus Ankara: History, value in basic research, and current perspectives for vaccine development” in Advances in Virus Research, Kielian M., Mettenleiter T. C., Roossinck M. J., Eds. (Elsevier, Amsterdam, 2017), vol. 97, pp. 187–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gómez C. E., Perdiguero B., García-Arriaza J., Esteban M., Clinical applications of attenuated MVA poxvirus strain. Expert Rev. Vaccines 12, 1395–1416 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Sutter G., Moss B., Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc. Natl. Acad. Sci. U.S.A. 89, 10847–10851 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meisinger-Henschel C., et al. , Genomic sequence of chorioallantois vaccinia virus Ankara, the ancestor of modified vaccinia virus Ankara. J. Gen. Virol. 88, 3249–3259 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Dobson B. M., Tscharke D. C., Redundancy complicates the definition of essential genes for vaccinia virus. J. Gen. Virol. 96, 3326–3337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meisinger-Henschel C., et al. , Introduction of the six major genomic deletions of modified vaccinia virus Ankara (MVA) into the parental vaccinia virus is not sufficient to reproduce an MVA-like phenotype in cell culture and in mice. J. Virol. 84, 9907–9919 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimier J., et al. , Deletion of major nonessential genomic regions in the vaccinia virus Lister strain enhances attenuation without altering vaccine efficacy in mice. J. Virol. 85, 5016–5026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyatt L. S., et al. , Marker rescue of the host range restriction defects of modified vaccinia virus Ankara. Virology 251, 334–342 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Liu R., et al. , SPI-1 is a missing host-range factor required for replication of the attenuated modified vaccinia Ankara (MVA) vaccine vector in human cells. PLoS Pathog. 15, e1007710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davison A. J., Moss B., Structure of vaccinia virus early promoters. J. Mol. Biol. 210, 749–769 (1989). [DOI] [PubMed] [Google Scholar]
- 14.Yang Z., Bruno D. P., Martens C. A., Porcella S. F., Moss B., Genome-wide analysis of the 5′ and 3′ ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts. J. Virol. 85, 5897–5909 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sancho M. C., Schleich S., Griffiths G., Krijnse-Locker J., The block in assembly of modified vaccinia virus Ankara in HeLa cells reveals new insights into vaccinia virus morphogenesis. J. Virol. 76, 8318–8334 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González J. M., Esteban M., A poxvirus Bcl-2-like gene family involved in regulation of host immune response: sequence similarity and evolutionary history. Virol. J. 7, 59 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres A. A., Albarnaz J. D., Bonjardim C. A., Smith G. L., Multiple Bcl-2 family immunomodulators from vaccinia virus regulate MAPK/AP-1 activation. J. Gen. Virol. 97, 2346–2351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoder J. D., et al. , Pox proteomics: Mass spectrometry analysis and identification of Vaccinia virion proteins. Virol. J. 3, 10 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung C. S., et al. , Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 80, 2127–2140 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resch W., Hixson K. K., Moore R. J., Lipton M. S., Moss B., Protein composition of the vaccinia virus mature virion. Virology 358, 233–247 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Ngo T., Mirzakhanyan Y., Moussatche N., Gershon P. D., Protein primary structure of the Vaccinia Virion at increased resolution. J. Virol. 90, 9905–9919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panda D., Fernandez D. J., Lal M., Buehler E., Moss B., Triad of human cellular proteins, IRF2, FAM111A, and RFC3, restrict replication of orthopoxvirus SPI-1 host-range mutants. Proc. Natl. Acad. Sci. U.S.A. 114, 3720–3725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kibler K. V., et al. , Improved NYVAC-based vaccine vectors. PLoS One 6, e25674 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kibler K. V., et al. , Replication-competent NYVAC-KC Yields improved immunogenicity to HIV-1 antigens in rhesus macaques compared to nonreplicating NYVAC. J. Virol. 93, e01513-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies D. H., et al. , Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus Ankara is comparable to that of Dryvax. J. Virol. 82, 652–663 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betakova T., Wolffe E. J., Moss B., Regulation of vaccinia virus morphogenesis: Phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J. Virol. 73, 3534–3543 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sodeik B., Griffiths G., Ericsson M., Moss B., Doms R. W., Assembly of vaccinia virus: Effects of rifampin on the intracellular distribution of viral protein p65. J. Virol. 68, 1103–1114 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y. C., et al. , Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly. J. Virol. 82, 5922–5932 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyatt L. S., Shors S. T., Murphy B. R., Moss B., Development of a replication-deficient recombinant vaccinia virus vaccine effective against parainfluenza virus 3 infection in an animal model. Vaccine 14, 1451–1458 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Bertholet C., Stocco P., Van Meir E., Wittek R., Functional analysis of the 5′ flanking sequence of a vaccinia virus late gene. EMBO J. 5, 1951–1957 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maruri-Avidal L., Domi A., Weisberg A. S., Moss B., Participation of vaccinia virus l2 protein in the formation of crescent membranes and immature virions. J. Virol. 85, 2504–2511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]