In Kessler et al. (1), the small molecule BI-2852 is shown to bind—at nanomolar affinity—to KRAS between switch I and II, inhibiting interactions with effectors. Here, we identify an alternative explanation for the inhibitory activity. While investigating the BI-2852 KRAS complex (Protein Data Bank [PDB] code 6GJ8), we noticed, by revealing the molecules in the neighboring unit cell, BI-2852 induces a KRAS dimer with rotational symmetry. This dimer complex consists of four molecules (two of BI-2852 and two of KRAS; Fig. 1A).
Fig. 1.
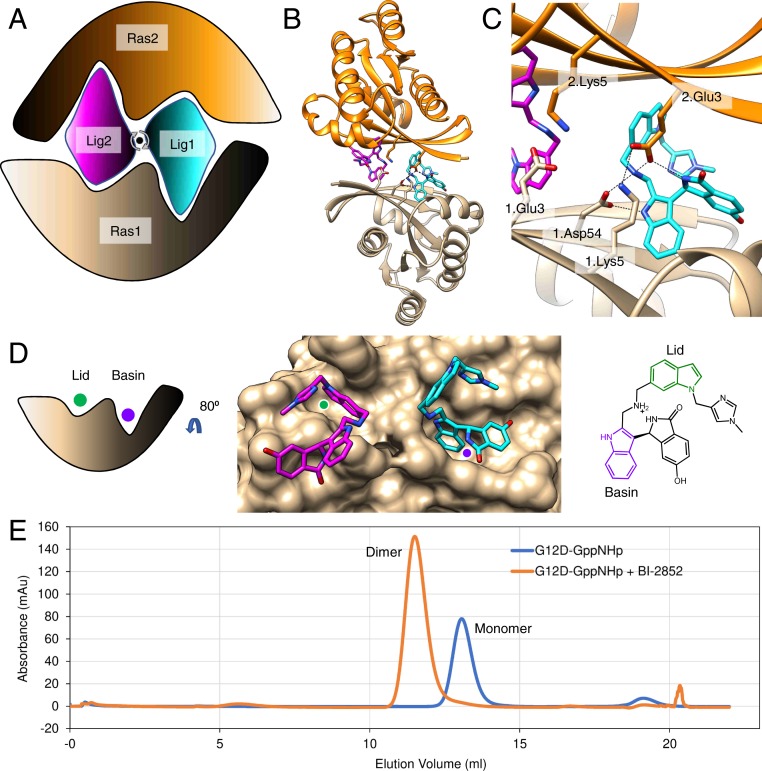
Dimer of KRAS with BI-2852. (A) Cartoon of BI-2852 KRAS dimer. Lig1 (cyan) and Lig2 (magenta) interact with both Ras1 (beige) and Ras2 (orange). (B) KRAS (ribbons) and key side chains are shown. (C) Zoom-in of the binding site: salt–bridge interactions shown (dashed lines). (D) Lid (green) and Basin (purple) are indicated on Ras1. Both Lig1 and Lig2 are shown on Ras1. The indole rings, colored green and purple, engage the Lid and Basin, respectively. (E) Absorbance A280 plotted as a function of elution volume: KRAS G12D-GppNHp with BI-2852 (orange) and apo KRAS G12D-GppNHp (blue).
To show BI-2852 induces dimers of KRAS, we performed the following: analysis of the crystal structures to characterize the dimer; performance of size exclusion column chromatography (SEC) experiments using protein with and without BI-2852, followed by analysis using native mass spectrometry; and measurement of binding affinity using isothermal titration calorimetry (ITC) and surface plasmon resonance (SPR).
Through structural analysis of PDB code 6GJ8, we identify critical interactions that stabilize the dimer (Fig. 1). Focusing on the pocket formed about Lig1 (one of two copies of BI-2852), the pocket is formed by two KRAS molecules—with one KRAS forming a larger pocket (Basin) and a second KRAS molecule forming a smaller pocket (Lid) (Fig. 1 A–D). There are two salt–bridge interactions about Lig1: Lys5 of Ras1 interacts with Glu3 of Ras2, Asp54 of Ras1 interacts with the amine of Lig1, and the amine of Lig1 also interacts with Glu3 of Ras2 (Fig. 1C and Figshare [https://doi.org/10.6084/m9.figshare.11425995, section 3.1; ref. 2]). We observe symmetric interactions about Lig2; thus, there are four salt–bridge interactions at the interface.
In the SEC experiment, we observe the formation of a KRAS dimer in solution when BI-2852 is present and a monomer when ligand is absent (Fig. 1E), and by native mass spectrometry, we confirm their molecular weight (Figshare [https://doi.org/10.6084/m9.figshare.11425995, section 3.3; ref. 2]). From SPR, where KRAS is a monomer on the sensor surface, we measure an affinity of 22 μM. Moreover, the response is over double the theoretical maximal binding predicted for a 1:1 interaction, implying there are two binding sites (Figshare [https://doi.org/10.6084/m9.figshare.11425995, section 3.4.2; ref. 2]). Using ITC, we reproduced the affinity values of 720 nM for KRAS G12D-GppHHp with BI-2852, consistent with Kessler et al. This increased affinity over that of the SPR data is likely due to the avidity of the two compound binding sites and stabilization of the dimer. We surmise the ligand binds to both the Basin (switch I/II pocket) and the Lid (Fig. 1D). These experimental observations are consistent with a dimer of KRAS stabilized by two molecules of BI-2852.
The biological effects of this compound, at least in part, are due to the formation of the KRAS dimer, resulting in enhanced binding affinity (22 μM to 720 nM). Moreover, the dimer occludes the binding footprint of the RAF1 RBD (Figshare [https://doi.org/10.6084/m9.figshare.11425995, Fig. S2; ref. 2]). To our knowledge, such a structure of KRAS as a dimer has never been observed; albeit, this likely nonfunctional dimer is distinct from a biologically relevant KRAS multimer. The formation of a nonfunctional KRAS dimer seen in structures reported by Kessler et al. reveals a strategy of directly targeting KRAS.
Acknowledgments
We thank Boehringer Ingelheim for providing us with the compound and releasing their work in an intriguing publication and Muyan Zhou for preliminary structural analysis. This project was funded in whole or in part with federal funds from National Cancer Institute–NIH Contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and the mention of trade names, commercial products, or organizations does not imply endorsement by the US Government.
Footnotes
Competing interest statement: F.M. is a consultant for the following companies: AduroBiotech; Amgen; Daiichi, Ltd.; PellePharm; Pfizer, Inc.; PMV Pharma; and Portola Pharmaceuticals. F.M. is Scientific Director of the National Cancer Institute RAS Initiative at Frederick National Laboratory for Cancer Research, Leidos Biomedical Research, Inc. F.M. is a consultant for and cofounder of the following companies: Avidity, BridgeBio, KGen, and Quartz.
Data deposition: This article contains supporting information online at Figshare (https://doi.org/10.6084/m9.figshare.11425995).
References
- 1.Kessler D., et al. , Drugging an undruggable pocket on KRAS. Proc. Natl. Acad. Sci. U.S.A. 116, 15823–15829 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran T. H., et al. , Data from “SI. The small molecule BI-2852 induces a non-functional dimer of KRAS.” Figshare. 10.6084/m9.figshare.11425995. Deposited 30 December 2019. [DOI]