Significance
Immunotherapy clinical trials have shown promising results only in some glioblastoma patients, warranting further research on glioblastoma-induced immune suppression. Cancer-driven changes in glycosylation are beginning to be appreciated as key modulators of cancer immune evasion, since antigen-presenting cells express glycan-recognizing receptors that trigger immune-suppressive signaling upon interaction with their ligands. Here we identify an O-linked glycan signature, characterized as a ligand of the tolerogenic C-type lectin receptor MGL, as modulator of immune suppression in glioblastoma. An in vivo model showed that overexpression of the MGL ligand in glioblastoma led to increased infiltration of PD-L1+ macrophages, which could also be found in patient-derived glioblastoma tissues. An improved understanding of the glioblastoma glyco-code could lead to new prognostic and therapeutic clinical applications.
Keywords: glioblastoma, immunosuppression, macrophage galactose lectin, macrophages, O-linked glycosylation
Abstract
Glioblastoma is the most aggressive brain malignancy, for which immunotherapy has failed to prolong survival. Glioblastoma-associated immune infiltrates are dominated by tumor-associated macrophages and microglia (TAMs), which are key mediators of immune suppression and resistance to immunotherapy. We and others demonstrated aberrant expression of glycans in different cancer types. These tumor-associated glycans trigger inhibitory signaling in TAMs through glycan-binding receptors. We investigated the glioblastoma glycocalyx as a tumor-intrinsic immune suppressor. We detected increased expression of both tumor-associated truncated O-linked glycans and their receptor, macrophage galactose-type lectin (MGL), on CD163+ TAMs in glioblastoma patient-derived tumor tissues. In an immunocompetent orthotopic glioma mouse model overexpressing truncated O-linked glycans (MGL ligands), high-dimensional mass cytometry revealed a wide heterogeneity of infiltrating myeloid cells with increased infiltration of PD-L1+ TAMs as well as distant alterations in the bone marrow (BM). Our results demonstrate that glioblastomas exploit cell surface O-linked glycans for local and distant immune modulation.
Glioblastoma (World Health Organization grade IV [WHO IV] glioma) is the most common and aggressive brain malignancy, with a median overall survival of less than 15 mo (1), for which immunotherapy has, to date, failed to achieve major advances. Sustainable responses remain limited to a number of individual patients benefiting from early-phase clinical trials (2, 3). Because some of these patients show impressive results (3, 4), the future for immunotherapy in glioblastoma is promising, but the brain is an immunologically challenging environment, and the complexity of immune suppression in glioblastoma is not yet fully elucidated.
The surface of mammalian cells is highly glycosylated, and the composition of this glycan coating depends on the activation status, environmental stimulation, aging, and malignant transformation of the cell (5, 6). Immune cells express a variety of glycan-binding receptors (lectins), which could modulate the immune response through interactions with tumor-associated glycans (5, 7). Therefore, glycans and their glycan-binding receptors could play a crucial role in glioblastoma immune escape.
Glioblastoma immune infiltrates are dominated by tumor-associated macrophages (TAMs) and microglia (8, 9), which contribute to immunotherapy resistance (10). TAMs express glycan-binding receptors including C-type lectin receptors (CLRs) and sialic acid-binding Ig-like lectins (siglecs) (6). These receptors specifically recognize glycan structures, often resulting in internalization and immune inhibitory signaling (5, 6, 11, 12). Several studies have demonstrated the involvement of glycan-binding receptors such as siglecs, dendritic cell (DC)-specific ICAM-3-grabbing nonintegrin 1 (DC-SIGN) and macrophage-galactose lectin (MGL) in immune suppression (6, 13–16). MGL belongs to the family of CLRs which specifically recognize glycans on pathogens and glycoproteins and trigger phagocytosis and signaling, thereby contributing to modulation of immune responses (17). MGL is typically expressed by immature DCs (18) and alternatively activated macrophages (19), conferring immune-suppressive (tolerogenic) signaling (20, 21). Binding of MGL to its ligands contributes to immune suppression by increasing IL-10 secretion and induction of T cell apoptosis (6, 22, 23).
Glycosylation is one of the major and most complex posttranslational modifications of proteins. The two major groups of glycans, formed with the aid of a variety of glycosyltransferases and glycosidases, are N-linked (Asn-linked) and O-linked (Ser/Thr-linked) glycans. Malignant transformation is accompanied with differential expression of multiple components of this glycosylation machinery (24, 25). Typical tumor-associated glycans arise from incomplete glycosylation leading to the formation of truncated glycans, or de novo glycosylation, such as sialylated Lewis antigens (24). Aberrant protein glycosylation is deeply enrooted in malignant transformation and contributes to aggressive tumor behavior due to altered protein function and cell−cell interactions. Well-known pathological processes that promote tumor progression via glycosylation include modulation of growth factor activity, impaired cell−cell adhesion due to aberrant glycoforms of E-cadherin promoting tumor cell invasion, and metastasis formation via tumor-associated N-linked glycans (Lewis antigens) that form the ligands for selectins (15, 24). Recent evidence demonstrates that cancer-associated glycan changes also contribute to cancer aggressiveness by triggering antiinflammatory signaling circuits in tumor-infiltrating immune cells (6, 15, 16, 22–24, 26–28). Tumor-associated glycans that are relevant for immune modulation include sialic acids, (sialylated) Lewis antigens, galectin ligands, and truncated O-linked glycans, such as the Tn antigen or its sialylated version (sTn), and we recently have collectively coined them as the cancer “glyco-code” (6).
In this study, we sought to identify glycan-based regulatory circuits in glioblastoma. We investigated whether the glioblastoma glyco-code could act as a tumor-intrinsic immune suppressor. First, we detected increased expression of tumor-associated truncated O-linked glycans in brain tumor tissues derived from patients with a glioblastoma compared to those from lower-grade diffuse gliomas (LGG) or epilepsy. Next, we evaluated the expression of MGL, a known immunosuppressive CLR on macrophages and DCs that interacts with truncated O-linked glycans. Stimulation of MGL by its ligands resulted in increased tumor infiltration by immune cells with an immune-suppressive phenotype. By overexpressing truncated O-linked glycans in a murine model, we studied the influence of these glioblastoma-associated glycans on immune composition using high-dimensional characterization of the tumor infiltrate, splenocytes, and bone marrow (BM) by mass cytometry.
Results
Glioblastomas Overexpress Tumor-Associated O-Linked Glycans.
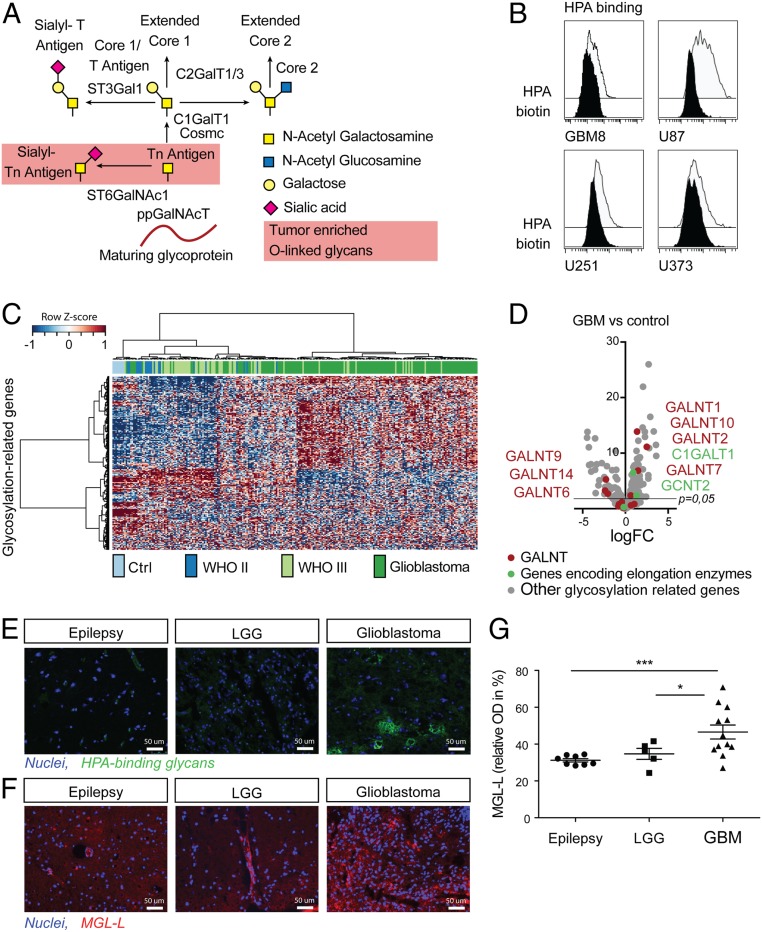
Using a broad panel of lectins, we initially profiled several glioblastoma cell lines (U78, U251, U373, and GBM8) for their glycan composition (SI Appendix, Fig. S1) and found reactivity against the α-GalNAc-specific snail lectin Helix pomatia agglutinin (HPA), which recognizes, among others, the cancer-associated Tn antigen (29, 30) (Fig. 1 A and B). Tn antigen is neoexpressed or overexpressed in many types of cancer, including gastrointestinal cancers, lung cancer, and breast cancer (6, 31). High expression of Tn antigen has been reported in human glioblastoma cell lines (32). Although Tn antigen has never been reported in healthy human brain tissue, syndecan-3−associated Tn antigen has been described as a transiently expressed antigen during the development of the mouse brain (33, 34). To further investigate the potential relevance of Tn antigen in glioma (WHO II to IV) patients, we assessed the relative expression of 270 genes involved in the glycosylation machinery (SI Appendix, Table S1) in a publicly available microarray dataset (Fig. 1C) (35). We observed different messenger RNA expression profiles in control brain tissues, LGG, and glioblastomas, suggesting an association between gliomagenesis and aberrant regulation of the glycosylation machinery. Among 42 glycosylation-related genes that were differentially expressed in glioblastomas versus healthy control tissues, we found 10 genes encoding for glycosyltransferases involved in the O-linked glycosylation pathway (Fig. 1D), suggesting an alteration of O-glycosylation in glioblastomas. Among the genes involved in the synthesis of the Tn antigen, GALNT1, GALNT2, GALNT7, and GALNT10 were up-regulated in glioblastoma (Fig. 1D). Altered GALNT expression in glioblastoma has not been described in the literature, but increased GALNT1, GALNT2, and GALNT10 have been associated with other malignancies (36–38). O-linked glycan elongation enzymes C1GalT and GCNT2 were also increased in glioblastoma versus healthy control brain tissues (Fig. 1D). In agreement with our observation in the GBM cell lines (Fig. 1B), the observed reprogramming in the O-glycosylation machinery could indicate, according to known biosynthetic pathways, that the glioblastoma’s glyco-code may be characterized by a high Tn antigen score. In addition, mucins known to carry the Tn antigen, such as MUC1 or MUC4, have been previously reported to be expressed in glioblastoma (39–41). Thus, we hypothesized that aberrant glycosyltransferase expression in patients with glioblastomas points to a glyco-code characterized by the prevalence of immature O-glycans such as the Tn antigen. To investigate this, we investigated the presence of HPA-reactive glycans in freshly collected surgically resected brain tissues from patients. HPA binding to tumor tissues derived from patients with glioblastoma confirmed the expression of terminal α-GalNAc−containing glycans, such as Tn, the most extensively characterized ligand for MGL (21), in these tumors (Fig. 1E and SI Appendix, Fig. S2). MGL is a CLR typically expressed by immature DCs and macrophages conferring immunosuppressive (tolerogenic) signaling (20, 21) via the modulation of IL-10 secretion (6, 22, 23). To evaluate the expression of MGL ligands (MGL-Ls), we used our recombinant MGL/mouseFc (42) on immunofluorescence microscopy (Fig. 1F and SI Appendix, Fig. S2) and used ELISA on tissue lysates (Fig. 1G). As shown in Fig. 1 F and G, tumor tissues derived from patients with glioblastoma expressed significantly more MGL-Ls compared to resected tissues derived from patients with epilepsy (P = 0.005) or LGG (P = 0.04). Thus, glioblastomas overexpress HPA-reactive glycans which are also ligands of MGL, such as the Tn antigen (6, 24).
Fig. 1.
Glioblastomas overexpress immature O-linked glycans. (A) Schematic representation of the O-glycosylation pathway. More than 20 different GALNTs have been characterized in humans that have overlapping activity but differ in peptide substrate specificities and expression pattern. GalNAc transferases catalyze the synthesis of GalNAcα1-O-serine/threonine (also known as the Tn antigen) as the first step in O-glycosylation (38). The Tn antigen is further elongated by the concerted action of several glycosyltransferases. (B) HPA binding to four glioblastoma cell lines (U87, U251, U373, and GBM8) measured by flow cytometry. (C) Heatmap of publicly available microarray data showing expression of genes encoding for glycosyltransferases and glycosidases in healthy brain tissue samples and 240 glioma WHO II (astrocytomas and oligodendrogliomas, n = 21), WHO III (astrocytomas and oligodendrogliomas, n = 60), and WHO IV (n = 159) samples. (D) Volcano plot showing 270 glycosylation-specific genes (SI Appendix, Table S1) with O-linked glycosylation related genes highlighted. C and D include a reanalysis of raw data obtained from Gravendeel et al. (35), including, for D, healthy brain tissue (n = 8), astrocytoma WHO II (n = 13), astrocytoma WHO III (n = 60), and glioblastoma (WHO IV, n = 159) samples. (E) Seven-micrometer cryosections of surgical epilepsy, LGG, and glioblastoma (GBM) samples, representative for five patients per condition, stained with HPA (green) and Hoechst as nuclear counterstain (blue). (F) Seven-micrometer cryosections of surgical epilepsy, LGG, and glioblastoma samples, representative for five patients per condition, stained with MGL-mouseFc (red) and Hoechst as nuclear counterstain (blue). (G) Human MGL-binding ELISA with lysates of glioblastoma (n = 12), LGG (n = 5), and epilepsy (n = 8) tissues, showing a significant difference (*P < 0.01, ***P = 0.005) between glioblastoma, and epilepsy samples (OD 450 nm).
TAMs in the Glioblastoma Microenvironment Express More MGL Compared to Myeloid Cells in Patient-Derived Lower-Grade Glioma and Epilepsy Tissues.
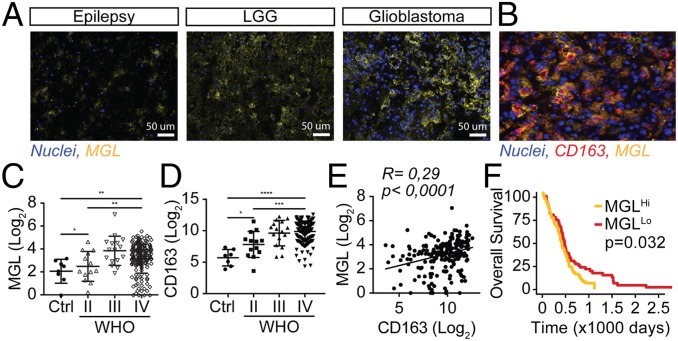
Given the prominent infiltration of suppressive myeloid cells in glioblastomas (9, 43, 44) and the abundance of MGL-L, we investigated the presence of MGL+ myeloid cells within the tumors. Patient-derived glioblastoma tissues were highly infiltrated with MGL+ cells (Fig. 2A and SI Appendix, Fig. S2) that were also CD163+ (Fig. 2B), a phenotype that is typically associated with immune-suppressive TAMs. In silico analysis of a publicly available dataset (35) also showed higher expression of MGL (Fig. 2C, P = 0.017) and CD163 (Fig. 2D, P = 0.0002) in glioblastoma, as well as a significant moderate correlation between the two (Fig. 2E, R = 0.29, P < 0.0001). This finding further supports our discovery that MGL is expressed by immune-suppressive CD163+ macrophages, but also by other cells in the glioblastoma microenvironment. The Cancer Genome Atlas data show a survival benefit for patients with lower expression levels of MGL (Fig. 2F, P = 0.032), further supporting our hypothesis that triggering of the MGL/MGL-L axis could represent a mechanism by which the tumor glycocalyx contributes to immune suppression in the glioblastoma microenvironment.
Fig. 2.
TAMs in the glioblastoma microenvironment express MGL. (A) Human glioblastoma, LGG, and epilepsy tissues, representative for five patients per condition, stained for the MGL receptor (yellow) and nuclei (blue). (B) Human glioblastoma tissue stained for the MGL receptor (yellow), CD163 (red), and nuclei (blue). (C) MGL expression in glioblastoma, astrocytoma grades II and III, and control tissues. (D) CD163 expression in glioblastoma, astrocytoma grades II and III, and control tissues. (E) Scatterplot with expression of CD163 on the x axis and MGL on the y axis showing a moderate correlation in glioblastoma tissue (Pearson’s R = 0.29, P ≤ 0.0001). C−E include a reanalysis of raw data obtained from Gravendeel et al. (35), including, for C and D, healthy brain tissue (n = 8), astrocytoma WHO II (n = 13), astrocytoma WHO III (n = 60), and glioblastoma (WHO IV, n = 159) samples. (F) Kaplan−Meier curve showing survival benefit for patients with lower expression of MGL (n = 84) versus patients with higher expression of MGL (n = 85, P = 0.032, with median expression value of 3.25 as cutoff). *P < 0.05, **P < 0.01, ***P < 0.001.
High-Dimensional Characterization of the Immune System in a Murine Glioma Model.
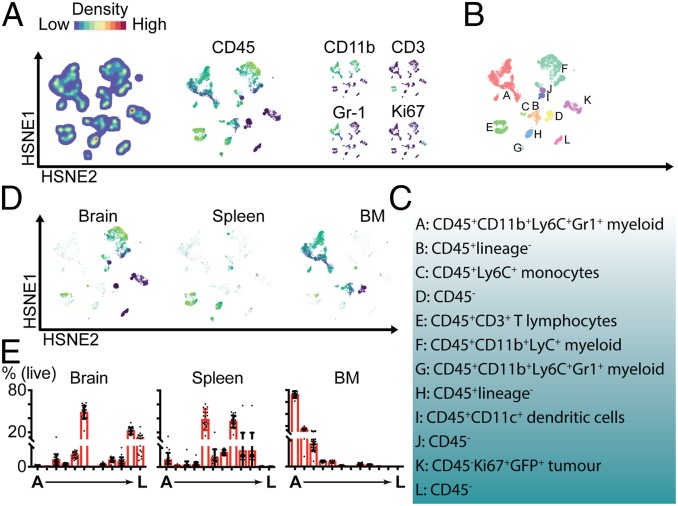
In order to recapitulate in vivo the MGL-LHi phenotype observed in human glioblastoma tissues, we knocked out the C1galt1c1 gene (also known as Cosmc) in the mouse glioma cell line GL261 using CRISPR/Cas9. Cosmc is a chaperone necessary for the synthesis of T antigen using the Tn antigen as substrate (Fig. 1A). Loss of Cosmc activity results in abrogation of extended Core 1 and Core 2 O-linked glycans and accumulation of (s)Tn-rich glycoproteins on the surface of the Cosmc−/− cells (45). Indeed, GL261 lacking Cosmc expression appeared to have a TnHi (SI Appendix, Fig. S3A), MGL-LHi (SI Appendix, Fig. S3 B and C) phenotype, even in established tumors 14 d after intracranial injection. To study whether the overexpression of MGL-Ls on glioblastoma cells triggered a tolerogenic phenotype on macrophages, we tested, in vitro, the impact on the expression of PD-L1, CD206, CD200R, and CCR2. The results, both on macrophages and microglia, clearly demonstrated that the glioma MGL-LHi cells induced higher expression of tolerogenic markers on macrophages and expression of IL-10 on microglia (SI Appendix, Fig. S4). In order to address the effects of MGL-L expression on the local intratumor and peripheral immune system, we performed high-dimensional analysis using cytometry by time-of-flight mass spectrometry (CyTOF) (46). We characterized tumor infiltrates (brain) and systemic (BM and spleen) immune composition, comparing mice with established GL261-MGL-LHi tumors to GL261-wild-type (WT) tumors 14 d after orthotopic implantation. Our antibody panel included 37 metal-conjugated antibodies (SI Appendix, Table S2) and enabled the identification of immune cell lineages, complemented with functional markers for subclassification. Our workflow (SI Appendix, Fig. S5) was based on clustering and visualization using Hierarchical Stochastic Neighbor Embedding (HSNE), a derivation of t-distributed (t)SNE in which hierarchies of nonlinear similarities are built that can be interactively explored in a stepwise manner (47) within the Cytosplore (48) environment. First, all cells from the brains, BM, and spleens of 20 mice (10 mice with GL261-MGL-LHi tumors and 10 mice with GL261-WT tumors) were classified into main cell clusters at the overview level according to similarity in the multidimensional space. HSNE built a hierarchical representation of the complete dataset (∼12 × 106 cells), without down-sampling, and allowed for subsequent clustering of the cells into consecutive levels. Across the different tissue samples, we identified 12 major overview-level cell clusters (A through L) with distinct phenotypes. Based on their relative marker expression, color-coded onto the HSNE maps, clusters from the lymphoid and myeloid cell lineages located to different areas of the HSNE map (Fig. 3 A–C and SI Appendix, Fig. S6). T lymphocytes localized together in cluster E (CD45+CD3+), whereas myeloid populations were spread over clusters A, F, and G (CD45+CD11b+Ly6C+). Clusters A and G also expressed the granulocyte marker Gr-1. Ki67+ proliferating cells were mainly located in cluster K and part of F. Clusters D, J, K, and L represented four types of CD45−CD11b−CD3−Gr1− nonimmune cells, and, due to the expression of Ki67 (Fig. 3A) and GFP (SI Appendix, Fig. S6) in cluster K and their essentially exclusive localization in the brain (Fig. 3 D and E), these cells were identified as GFP+ GL261 tumor cells. Cells derived from the tumor-bearing brain located predominantly in clusters F (myeloid), K (tumor), and L (CD45−; Fig. 3 D and E). Therefore, cluster F represented a large group of CD45+CD11b+ myeloid cells in the brain. Compared to this large number of tumor-infiltrating myeloid cells, tumor-infiltrating T lymphocytes (CD45+CD3+TCRβ+, cluster E) were largely underrepresented, which corresponds to recent literature describing poor lymphocyte infiltration of glioblastomas (2, 49). Clusters E (T lymphocytes), G (Gr1+), H (CD45+, lineage−), I (CD45+CD11c+), and J (CD45−) were mostly represented in the spleen (Fig. 3 D and E), where cluster I was positive for the DC marker CD11c. The BM was mainly populated by cells that were assigned to clusters A (CD45+CD11b+ myeloid cells), B (CD45+, lineage−), I, C (CD45+Ly6C+), and D (CD45−). HSNE maps color-coded for all markers are provided in SI Appendix, Fig. S6. Since cluster A represented the most dominant cluster of myeloid cells in the BM, E represented T lymphocytes which could be differentially activated in the context of glioma glycoforms, and F represented the dominant cluster of (myeloid) immune cells in the brain, we choose to compare these clusters between mice with GL261-MGL-LHi tumors and mice with GL261-WT tumors in subsequent analyses.
Fig. 3.
High-dimensional mass cytometric analysis of a murine model for glioblastoma overexpressing immature O-linked glycans. (A) Overview level HSNE clustering of 12-mln cells pooled from brains, BM, and spleens of the 20 mice (10 MGL-LHI and 10 WT tumors), showing cell density within different clusters, and expression of some key markers for cell lineage identification. (B) HSNE map, color-coded by cluster as identified in the first-level HSNE clustering. (C) Summary of cluster phenotypes. (D) HSNE maps selectively showing cells originating from brain, spleen, or BM. (E) Percentages of cells per cluster, relative to the total number of cells per organ (brain, spleen, or BM).
Glioblastoma-Associated MGL-Ls Attract PD-L1+ TAMs.
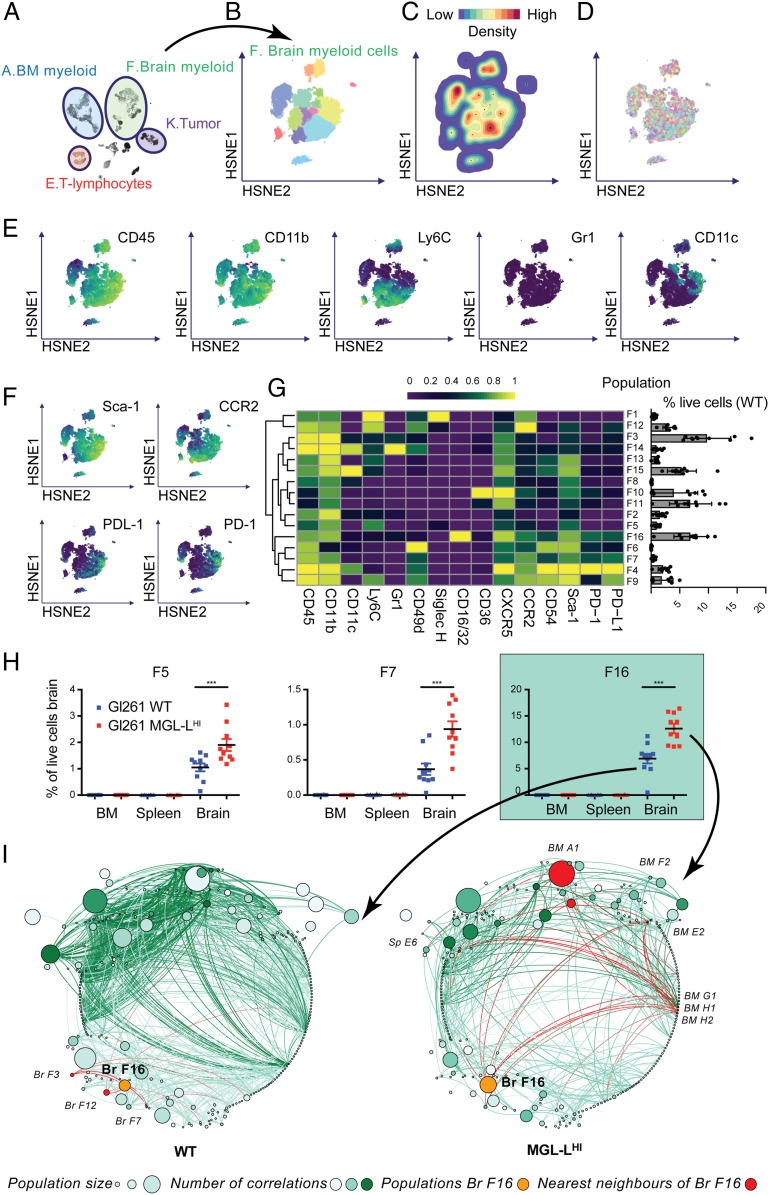
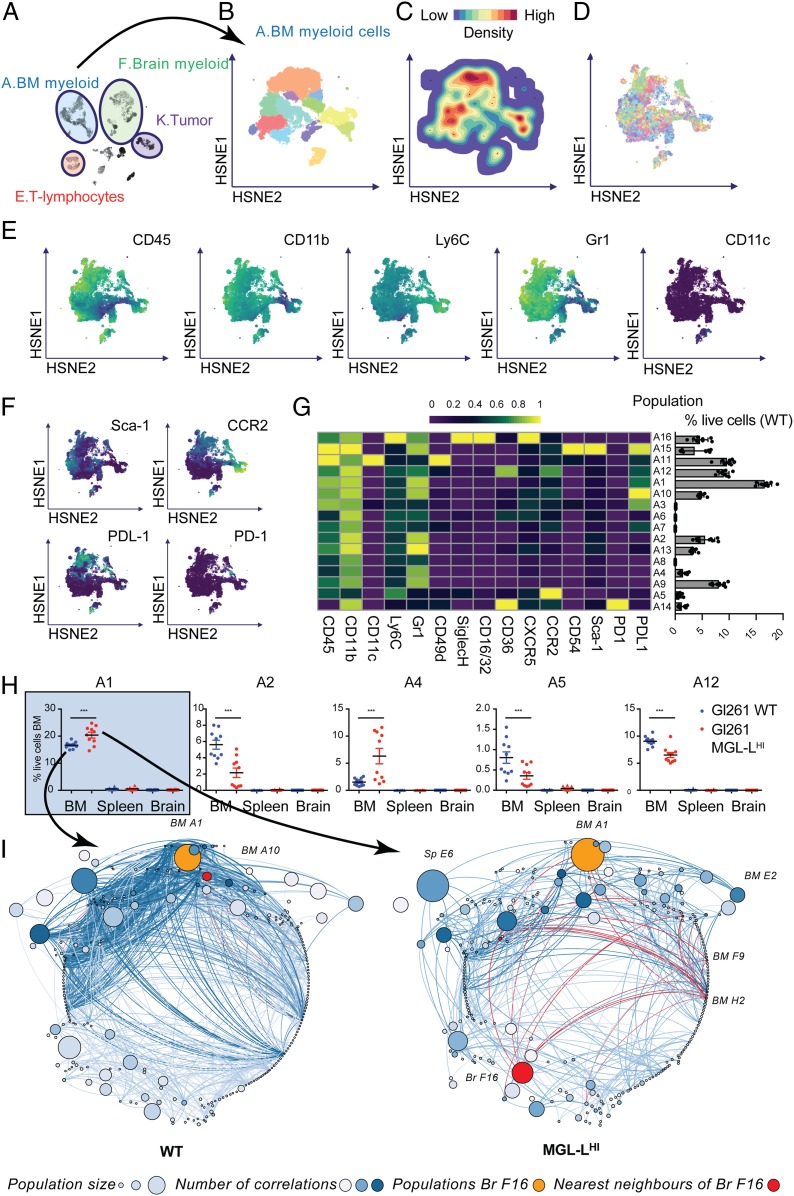
Since cells from the myeloid lineage formed the largest group of immune cells infiltrating the tumor-implanted mouse brains, we explored the phenotypes of brain-derived myeloid cells before we compared them between the two study groups. We zoomed into the HSNE embedding of cluster F (brain myeloid, Fig. 4A) and identified 16 distinct subclusters (Fig. 4B). The cell density (Fig. 4C), mouse sample identity (ID, showing absence of sample bias; Fig. 4D), and the expression levels of myeloid markers (CD45, CD11b, Ly6C, Gr1, and CD11c) were overlaid on the HSNE map (Fig. 4E). The phenotypes of the 16 subclusters of brain-infiltrating myeloid cells were characterized by lineage myeloid markers, activation markers such as Sca-1 and PD-1, and migration markers such as CCR2 (Fig. 4 F and G). The size of the clusters (percentage of live cells in the brain samples) was quantified in the bar graph lining the right side of the heatmap in Fig. 4G. Given the combined expression of CD45, CD11b, Ly6C, and CCR2 on most of the subsets (F1, F3, F5, F9, F12, F13, F14, and F15), these tumors were likely highly infiltrated with monocyte-derived macrophages in different stages of differentiation (44). Some of the immature macrophage subclusters could also be classified as monocytic myeloid-derived suppressor cells (MDSC) based on their expression of CD11b, Gr-1, and Ly6C (F1, F5, F9, and F12) (50). Clusters F14 and F15 could also be monocyte-derived DCs due to the expression of CD11c. Low expression levels of Ly6C in some of the other CD45hiCD11b+ subsets may indicate a more mature differentiated type of macrophages (subclusters F2, F6, F7, F8, and F16) or DCs (F4), although most of these subclusters still expressed high levels of CCR2 (subclusters F4, F6, F7, and F16). This could contribute to retention of the cells in the CCL2+ tumor microenvironment (51, 52). The phenotypes of subclusters that are CD11b+CD45dimCD49d− such as F10 and F11 correspond mostly to the classical definition of microglia. These cells were clearly outnumbered by the high number of immature and mature CD45hi myeloid cells. Microglia-like subcluster F10 displayed activation and proliferation markers, given the expression of Ki-67, Sca-1, CCR2, and CD54. Also, clusters F5 and F8 showed relatively low expression of CD45 and hierarchically clustered close to F10 and F11, implying a possible microglia-like phenotype. Looking more into the expression profile of other markers, F5 is Ly6C+ and F8 is CD11c+, which could be signs of activation or plasticity. The brain-derived myeloid subclusters were mostly positive for activation markers Sca-1 and CD54. In summary, the immune infiltrate in the GL261 (both WT and MGL-LHI)-harboring mouse brains was dominated by cells from the myeloid lineage with a wide heterogeneity of phenotypes that corresponded to classical definitions of microglia, monocyte-derived macrophages, and MDSC, but also show variations to the classically described phenotypes. We next explored the frequency (percent live cells) of myeloid subclusters as percentage of total live brain-derived cells and discovered three subclusters that were significantly larger in the MGL-LHi tumors (Fig. 4H): subcluster F5, F7, and F16. All three clusters were positive for CD45, CD11b, CXCR5, and Sca-1, but showed slightly different phenotypes: F5 was Ly6C+; subcluster F7 was CD49d+CCR2+CD54+PD1+PD-L1+, and the largest subcluster (F16) was CD16/32+CCR2+CD54+PD1+PD-L1+. Frequencies of all of the clusters with significant differences in any of the organs are summarized in SI Appendix, Fig. S7. Since the immune system shows features of a complex network (53, 54), we further looked into the size of all of the HSNE subclusters spread over brain, spleen, and BM, and generated a robust correlation matrix for cluster sizes in MGL-LHi tumors and WT tumors (Fig. 4I). Statistically significant correlations were visualized in a correlation network with edges (lines) indicating a correlation between two subclusters, and nodes representing the subclusters coded by size for mean subcluster size and coded by color for the number of correlations (edges) with other subclusters. Subcluster F16 turned out to be particularly interesting, not only because it was the largest subcluster of myeloid cells with a significant increase of 7% of live cells in the brains with WT tumors to 12% of live cells in the brains with MGL-LHi tumors (Fig. 4H) but also because of the gain of systemic correlations in the MGL-LHi tumors, especially with subcluster A1 (CD45+CD11b+Ly6C+Gr1+CCR2+). In the networks, F16 is shown as the orange cluster, with its nearest neighbors in red (Fig. 4I). Thus, in MGL-LHi tumors, increased numbers of myeloid cells accumulated, especially from subcluster F16, which represents a phenotype that corresponds to the description of PD-L1+ monocyte-derived macrophages (9, 44, 52, 55, 56). In mice with MGL-LHI tumors, this population (F16) gained a strong correlation with subcluster A1 in the BM as well as some smaller BM clusters. An equivalent of these cells characterized by the expression of CD163, PD-L1, and CCR2 can also be found in tissue derived from glioblastoma patients (SI Appendix, Fig. S8).
Fig. 4.
Glioblastoma-associated MGL-Ls attract CCR2+PD-L1+ macrophages. (A) HSNE map with the main clusters of interest highlighted. (B) HSNE map, colored by 16 subclusters as identified in the second-level HSNE subclustering of cluster F: tumor-infiltrating myeloid cells. (C) Density features depicting the local probability density of cells in cluster F. (D) Sample distribution. Each color represents a mouse, indicating a lack of sample bias in our data. (E) HSNE maps representing relative expression of the classifying myeloid markers (CD45, CD11b, Ly6C, Gr1, and CD11c). (F) HSNE maps representing relative expression of functional markers (Sca-1, CCR2, PDL-1, and PD-1). (G) Heatmap with relative ArcSinH(5) transformed expression of shown markers in the second-level HSNE of cluster F (Left) with a bar graph showing the percentages of total live cells in the brain per cluster (Right). (H) The three subclusters (F5, F7, and F16) with statistically significant differences in size between MGL-LHi and WT tumors. (I) Networks of subclusters from WT (Left) and MGL-LHi (Right) tumors with nodes visualizing all of the level-two cell subclusters in the experiment and edges representing correlation coefficients for relationships between subclusters. The size of the nodes represents the size of the population, the color intensity of the nodes represents the number of connections to other nodes, and the color intensity of the edges represents the correlation coefficient. The orange nodes highlight population F16, and the red nodes highlight the nearest neighbors of population F16 in both MGL-LHi- and WT tumors. ***P < 0.001.
Glioblastoma-Associated MGL-Ls Affect the Myeloid Composition of the BM.
From the correlation networks in Fig. 4I, BM subcluster A1 (CD45+CD11b+Ly6C+Gr1+CCR2+) stands out as a large subcluster with a significant correlation with the brain tumor macrophage subcluster F16. Therefore, we selected HSNE overview-level cluster A to look in further detail into the subclusters of myeloid cells in the BM. We embedded cluster A at the next HSNE level (Fig. 5 A and B), and visualized cell density (Fig. 5C), mouse ID (Fig. 5D), and the expression levels of classifying myeloid markers overlaid on the HSNE maps (Fig. 5E). The second HSNE level of cluster A resulted in 16 BM myeloid subclusters (Fig. 5 F and G). Subclusters A2 (CD45+CD11b+Ly6C+Gr1+Sca-1+), A5 (CD45+CD11b+Ly6C+Siglec-H+CXCR5+CCR2hi), and A12 (CD45+CD11b+Ly6C+Gr1+Sca-1+CCR2+) were significantly decreased in the BM of mice with MGL-LHi tumors (Fig. 4H), which could result from either a decline in BM production of these cells or an increased recruitment to the tumor. Subclusters A1 and A4 (CD45+CD11b+Ly6C+Gr1+) showed a significant increase in size in the BM of mice implanted with MGL-LHi tumors, possibly caused by tumor-induced increased production (Fig. 5H). Cross-talk of glioblastoma cells with BM-derived MDSCs has been described (8) and could be regulated by soluble proteins, and possibly also via extracellular vesicles and blood platelets (9). In the network visualization of our correlation analysis, the nearest neighbors of population A1 (shown in orange with nearest neighbors in red), indeed lacked correlations between subcluster A1 in BM and subclusters in the brains in WT tumors, whereas significant correlations were present in the MGL-LHi tumors (Fig. 5I). So, the altered GL261 glycocalyx influenced not only the myeloid composition of the local tumor infiltrate but also the immune cell composition of the BM.
Fig. 5.
Glioblastoma-associated MGL-Ls affect the myeloid composition of the BM. (A) HSNE map with the main clusters of interest highlighted. (B) HSNE map, colored by cluster as identified in the second-level HSNE clustering of cluster A: BM-derived myeloid cells. (C) Cell density. (D) Sample distribution. (E) HSNE maps representing relative expression of the classifying myeloid markers. (F) HSNE maps representing relative expression of functional markers. (G) Heatmap with relative Arcsinh transformed expression of shown markers in the second-level HSNE of population A (Left) with a bar graph showing the percentages of total live cells in the BM that the subclusters contain (Right). (H) The five subpopulations with statistically significant differences in relative frequency between mock transfected tumors and Cosmc KO tumors. (I) Networks of subclusters from WT (Left) and MGL-LHi (Right) tumors with nodes representing all of the subclusters in the experiment and edges representing correlation coefficients for relationships between the subclusters. The size of the nodes represents the size of the population, the color intensity of the nodes represents the degree (number of connections to other nodes), and the color intensity of the edges represents the correlation coefficient. The orange nodes highlight population A1, and the red nodes highlight the nearest neighbors of population A1 in both WT and Cosmc KO tumors. ***P < 0.001.
Discussion
In the present study, we evaluated the glioblastoma glyco-code as tumor-intrinsic modulator of immune suppression. We found that an α-GalNAc−terminal glycan, possibly the Tn antigen, is highly expressed on glioblastoma cell lines and in patient-derived glioblastoma tissues, as well as at lower levels in lower grade gliomas. In concert, we detected a high infiltration of immune-suppressive CD163+ TAMs expressing MGL, an immune-suppressive receptor that binds Tn antigen. In an in vivo murine model recapitulating high expression of Tn antigen (MGL-L) on glioblastomas, we profiled infiltrating immune cells with a wide heterogeneity of phenotypes that corresponded to classical definitions of microglia and monocyte-derived macrophages, but also showed variable expression of activation and migration markers. Our data demonstrate that overexpression of O-linked glycans increases the frequency of immune-suppressive PD-L1+ macrophages in murine MGL-Lhi tumors as well as inducing distant alterations in immune cell frequencies in the BM.
O-linked glycosylation is initiated at the Golgi apparatus by a family of N-acetylgalactosaminyltransferases, which mediate the addition of N-acetylgalactosamine (GalNAc) to the amino acids serine or threonine (38). The initial addition of GalNAc to serine or threonine forms the Tn antigen, which can then be further extended to form elongated O-linked glycans (38). T synthase is the key enzyme in the first step of O-glycan elongation, and its function depends on the chaperone Cosmc (C1GalT1C1) (38, 45). In healthy tissues, Tn antigen is covered by other carbohydrates, due to O-linked glycan elongation, but, due to incomplete glycosylation, it becomes exposed in cancer tissues. After Tn antigen was first described in breast cancer (57), other studies on the expression of Tn antigen in cancer followed (6, 14, 24, 31, 58–62). Tn antigen has been reported as a prognostic factor in lung cancer (63), breast cancer (64), and colorectal cancer (14, 65). The expression of Tn antigen has been described in human glioblastoma cell lines (32) and the developing mouse brain (33, 34), but not in healthy human brain tissues. Here, we report overexpression of Tn antigen in glioblastomas compared to control epilepsy tissues. In microarray data on bulk glioblastoma and lower-grade glioma tissues, we detected differential regulation of O-glycosylation−related genes. Future studies should be conducted to investigate the proteins expressing Tn antigen and evaluate the prognostic value of Tn antigen in glioblastoma. We hypothesized that the expression of Tn antigen could play a role in modulating the immune response via MGL on macrophages.
Glioblastoma cells exploit many strategies, such as recruitment of TAMs, to evade the immune system. TAMs comprise at least one-third of the cells in the glioblastoma microenvironment and facilitate tumor progression by creating a supportive stroma for tumor cell proliferation and migration (44). TAMs differentiate into a phenotype characterized by impaired phagocytic function and the expression of immunosuppressive cytokines and immune checkpoint ligands, such as PD-L1, correlated with prognosis (66, 67). Here we show that human glioblastoma-infiltrating TAMs express high levels of MGL, a CLR with immune suppressive properties.
Recapitulating the glyco-code that we identified in human glioblastomas, we developed a murine model overexpressing the truncated O-linked glycan Tn antigen. We created a mouse glioblastoma cell line (GL261) with increased exposure of Tn antigen by knocking out the essential chaperone for O-linked glycan elongation Cosmc (68). The resulting cell line was able to trigger an M2-like phenotype on macrophages and microglia in vitro. Using high-dimensional characterization of the tumor infiltrate, splenocytes, and BM by mass cytometry, we demonstrate a wide heterogeneity of tumor-infiltrating myeloid cells with increased infiltration of PD-L1+ TAMs in the MGL-LHI tumors, as well as alterations of immune cell frequencies in the BM. In the spleens, we did not find significant or biologically relevant differences. Future research should focus on the expected role of MGL in this glycan-dependent mechanism of immune modulation. Although humans have one type of MGL, mice have two different MGL genes: mgl1 and mgl2 (69). Based on the glycan specificity, the mouse homolog of human MGL is MGL2 (70). A diphtheria toxin receptor knockin mouse targeting the Mgl2 gene is commercially available, but, given the high expression of MGL and the pronounced role of macrophages in glioblastoma, ideally, loss of MGL should be studied while maintaining the TAMs. Future experimental evaluation of mgl2−/− mice (69, 70) should be performed to further assess the role of truncated O-linked glycan in immune modulation.
Glioblastoma is an incurable tumor deeply invading the parenchyma of the brain. Glioblastoma cells prefer to travel through the brain via the subarachnoid space and existing highways such as white matter tracts (71) or blood vessels (72) with oxygen and nutrient supply. Presentation of Tn antigen along blood vessels to infiltrating alternatively activated MGL+ macrophages could contribute to the immune-suppressive tumor microenvironment by activating regulatory pathways (22). In the time of precision medicine, particularly important for immunotherapy, the selection of glioblastoma patients is challenging. The increased infiltration of MGL-LHI glioblastomas by PD-L1+ TAMs could have implications for patient selection for immune checkpoint blockade against PD1 or PD-L1. Therefore, future evaluation of the clinical correlation between Tn antigen expression and PD-L1 expression in glioblastomas would be valuable. Given the high abundance of MGL ligands in glioblastomas, it would be interesting to explore whether Tn-specific CAR-T cells could be used in conjunction with anti−PD-L1 therapy for a more effective immunotherapy. Finally, the interaction between MGL and its ligands is susceptible to competitive inhibition using the nontoxic carbohydrate α-GalNAc, a strategy that could be used in combination with the current gold standard and/or the abovementioned proposed immunotherapy regime.
Altogether, we conclude that the Tn antigen-dominated glyco-code in glioblastomas has major consequences for both local and systemic immune responses, providing possible diagnostic/therapeutic avenues. We hypothesize that Tn antigen is of prognostic value in glioblastoma, as it is in several other types of cancer, and that this glycan may modulate the anti-glioblastoma immune response both locally and systemically. Follow-up studies should focus on the prognostic potential of Tn antigen and its potential for selection of patients for immune checkpoint inhibition, and investigate patient blood signatures to evaluate systemic immune modulation.
Materials and Methods
Cell Culture.
Human glioblastoma U87, U251, U373, and GBM8 cell lines and mouse GL261 cell lines (Developmental Therapeutics Program/National Cancer Institute Tumor Repository, Charles River Laboratories, Inc.) were cultured under standard conditions with 5% CO2 and 37 °C in a humidified incubator. U87, U251, and U373 cells were maintained in Dulbecco’s Modified Eagle Medium (Thermo Fisher) supplemented with 100 U/mL penicillin/streptomycin (Lonza), 2 mM l-glutamine (Lonza), and 10% fetal calf serum (FCS) (Biowest). GBM8 cells were maintained in Neurobasal medium (Invitrogen) supplemented with 100 U/mL penicillin/streptomycin (Lonza), 2 mM l-glutamine (Lonza), 1× B27 supplement (Life technologies), 0.5× N2 supplement (Life Technologies), 20 nM recombinant human fibroblast growth factor (Pepro Tech), 20 nM recombinant human epidermal growth factor (Pepro Tech), and 20 μg/mL heparin (Sigma). Mouse glioma GL261 cells grow in Roswell Park Memorial Institute medium (RPMI; Thermo Fisher) supplemented with 100 U/mL penicillin/streptomycin (Lonza), 2 mM l-glutamine (Lonza), and 10% FCS (Biowest). To introduce reporter molecules, the mouse glioma Gl261 cell line was stably transduced using a CSCW2 lentiviral vector (73) encoding for firefly luciferase and GFP separated by an internal ribosome entry site (IRES) domain (GL261.Fluc.IRES.GFP).
Generation of GL261-MGL-LHi Cells.
GL261.Fluc-IRES.GFP cells were transfected with a CRISPR/Cas9 construct targeting Cosmc (C1GalT1C1) to generate the MGL-LHi phenotype or an empty CRISPR/Cas9 construct to generate the GL261-WT (Mock) phenotype. The CRISPR/Cas9 construct was constituted according to the protocol as described by Ran et al. (74) using the pSpCas9(BB)-2A-Puro plasmid (a gift from Feng Zhang, Broad Institute of Massachusetts Institute of Technology, Addgene #62988). Cosmc (C1GalT1C1) was targeted by the following guide RNA strands: top CACCGGTTTTCTTACCTCCAAA; bottom CCAAAAGAATGGAGGTTTCAAA.
Lipofectamine LTX with PLUS reagent (ThermoFisher Scientific) was used for transfection and applied according to manufacturer’s protocol. One day after transfection, puromycin was added for selection. Mutated cells were sorted based on a Tn antigen high phenotype using the Tn antigen-specific snail lectin HPA (Sigma). Cells were incubated with biotinylated HPA (5 μg/mL; Vector Laboratories) for 30 min at 4 °C and, after a washing step, incubated with Streptavidin-AF647 (Jackson Immunolaboratories) for 30 min at 4 °C before they were sorted on a FACSAria Fusion (BD). Cells were sorted two times and frozen in aliquots in order to implant low passage cells into mice.
Surgical Samples.
Surgical samples of patients with epilepsy, LGG (Astrocytoma WHO II and III), glioblastoma, and intracranial metastasis were collected and frozen en block or in optimal temperature cutting (OCT)-Compound (Tissue Tek). All samples were collected at the Amsterdam UMC, location VU University Medical Center (Amsterdam, The Netherlands) after approval by The Medical Ethics Review Committee of the Amsterdam UMC and written informed consent. All patients were diagnosed by clinical presentation at the Department of Neurology and confirmed by histopathological analysis at the Department of Pathology of our hospital. Patients with epilepsy underwent surgery when resistant to medical treatments; patients with LGG and glioblastoma underwent surgery as primary treatment followed by radiotherapy and/or chemotherapy according to treatment standards. Patient samples were grouped according to the diagnoses assigned by a pathologist in the Amsterdam UMC, location VU University Medical Center, and handled according to national and institutional medical ethical standards regarding the use of human tissue. All patient samples were pseudoanonymized prior to data analysis.
Syngeneic Mouse Model.
Mouse studies were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee and performed according to their safety and animal well-being guidelines. Mice were maintained under a 12-h light/dark cycle with access to water and food. Adult (10 to 12 wk old), male C57/BL/6 mice (Charles River Laboratories) were maintained under specific-pathogen-free conditions. After anesthetizing mice with isofluorane, 2 µL containing 1 × 105 Gl261.FG-Cosmc-KO or Gl261.FG-Mock cells were injected into the left striatum (from Bregma: 2 mm anterior, 0.5 mm left, 2.5 mm deep) using a stereotactic frame. After 2 wk, tumor growth was comparable between groups, and mice were deeply anesthetized with a mixture of ketamine (17.5 mg/mL) and xylazine (2.5 mg/mL) followed by transcardial perfusion with 50 mL of Dulbecco’s phosphate-buffered saline (PBS) without Mg2+ and Ca2+ (DPBS) using a perfusion pump (Minipump Variable Flow; Fisher Scientific). The brain was then removed and frozen in OCT-Compound (Tissue Tek) for immunofluorescence, and prepared for mass cytometry staining or fluorescence-activated cell sorting (FACS).
In Vitro Coculture Assays.
Peritoneal macrophages were obtained by peritoneal lavage of C57BL/6 mice with 8 mL of PBS supplemented with 10 mM (ethylenedinitrilo)tetraacetic acid (EDTA). Cells were plated on 24-well plates and allowed to adhere for 24 h. Upon resting, cells were exposed to Gl261.FG-Mock or Gl261.FG-Cosmc-KO (ratio 1:5, macrophage:Gl261) for ∼48 h, stained for CD11b, CD206, CD200R, PD-L1, CD86, CD80, MHC-II, CCR2, CD163, and CD11b, and measured by flow cytometry. Alternatively, the microglia cell line BV-2 was used in an identical setting, and the expression of PD-L1 was measured by flow cytometry or the secretion of IL-10 was determined by ELISA. Where indicated, the incubation included 500 nM of the MGL inhibitor αGalNAc (Sigma).
Lectin Binding (FACS).
Glioblastoma cell lines were plated in V-bottom 96-well plates at a density of 50 to 200 × 103 cells per well and washed with Hank’s Balanced Salt Solution (HBSS; Gibco) supplemented with 0.5% bovine serum albumin (BSA). After blocking with HBSS 0,5% BSA (HBA) for 30 min at 4 °C, cells were incubated with biotinylated HPA (Vector Laboratories) or soluble recombinant human MGL-Fc, fused to a human IgG1 fragment, in the absence or presence of the calcium chelator ethylene glycol bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid. HPA and MGL-Fc binding to glioblastoma cells was detected using Streptavidin-AF488/AF647 (Jackson Immunolaboratories) or goat-anti-human-IgG- fluorescein isothiocyanate (FITC)/allophycocyanin (APC) (Jackson). After 10 min of incubation in 7AAD/eFluor780/eFluor510 (1:1,000; Jackson Immunolaboratories) in HBA for exclusion of dead cells, fluorescence signal was measured by flow cytometry (BD Fortessa).
MGL-Binding ELISA.
Surgical samples were gently homogenized and incubated in lysis buffer (5 mM EDTA, 1% Nonidet P-40, 0,5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 0.15 M NaCl, 50 mM Tris⋅HCl, 50 mM Tris) supplemented with protease inhibitors at 4 °C under rotation for 1 to 2 d, depending on the sample volume. The lysates were centrifuged at 10,000 × g for 15 min and stored at −80 °C. Protein concentrations were determined by bicinchoninic acid (BCA) assay (Micro BCA Protein Assay Kit, Thermo Fischer) with BSA as standard, and diluted until equal protein concentrations were reached for all samples. Epilepsy and tumor lysates were coated (50 μg/mL) onto Nunc maxisorb 96-well plates (Nunc) in ELISA coating buffer (0,2 mM Tris⋅HCl, pH 9,2) overnight at room temperature (RT). Then 1% BSA in coating buffer was coated as negative control. Plates were washed in Tris-sodium-magnesium (TSM) buffer (20 mM Tris⋅HCl, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, pH 8.0) and blocked for 30 min at 37 °C with TSM buffer supplemented with 1% BSA. Next, recombinant human MGL-Fc, the MGL carbohydrate recognition domain fused to a mouse IgG1 fragment, was added and incubated for 2 h at RT, mildly shaking, followed by detection by 30 min of incubation at RT with peroxidase-conjugated goat-anti-mouse-IgG (Jackson). The reaction was started using a substrate buffer containing 0,1 M Sodium Acetate (Sigma), 0.1 M citric acid (Sigma), pH 4, 0.006% H2O2, and 0.1 mg/mL tetramethylbenzidine (Sigma) and stopped with a stop buffer containing 0.2 M H2SO4. Signal was measured using a microplate absorbance spectrophotometer at a wavelength of 450 nm (Bio-Rad).
Immunofluorescence Microscopy Imaging.
Cryosections (7 to 20 μm) of Tissue-Tek embedded surgical samples or mouse brains were fixed in 2% paraformaldehyde for 10 min, followed by blocking with either 0.5% BSA in TSM buffer or HBSS (Gibco) for lectin staining, or 10% serum (species of secondary antibody) in PBS for antibody staining. Tissue sections were incubated with primary lectins or antibodies, which were diluted in 1% BSA in TSM or PBS, respectively, for 2 h at 37 °C or overnight at 4 °C. Antibody binding was detected with fluorochrome-conjugated secondary antibodies, and incubated for 60 min at RT followed by thorough washing with TSM or PBS. Sections were counterstained with Hoechst or TO-PRO3 and imaged on a wide-field fluorescence microscope (Leica DM6000 or Nikon eclipse TE2000-U). Alternatively, fluorescence images were acquired at either 0.5 or 0.25 µm per pixel resolution, using a Vectra Polaris scanner (Akoya Biosciences) with DAPI, FITC, TexRed and CY5 filters. Immunopositive cells were identified by DAPI nuclear staining by applying constant thresholds for light intensity and object size maintained across all sections with ImageJ software (NIH).
Mouse Brain Single-Cell Preparation.
After perfusing the mice with ice-cold DPBS, the brains were removed and processed into single-cell suspensions for mass cytometry staining, or FACS using the GentleMacs Dissociator and Neural Tissue dissociation Kit P (Miltenyi Biotech) according to manufacturer’s instructions. The tumor-containing hemispheres were cut into small pieces and placed into a GentleMacs C-tube (Miltenyi Biotech) with 1,900 µL of Buffer X. When all brains were harvested, 50 µL of enzyme P was added, and GentleMacs program Brain_1 was run two times followed by incubation at 37 °C for 15 min. After running GentleMacs program Brain_2 two times, 30 µL of enzyme A solution (20 µL of Buffer Y + 10 µL of Enzyme A) was added, followed by a second incubation at 37 °C for 10 min. After a third round of mechanical dissociation using the Brain_3 program and 10 min of incubation at 37 °C, single-cell suspensions were passed through a 70-μm filter (Fisher Scientific). After a washing step, myelin was removed using myelin removal beads (Miltenyi Biotec), and red blood cells (RBC) were lysed during 2 min of incubation with Mouse RBC lysis buffer (Boston Bioproducts). The final single-cell suspensions were washed and stained for FACS sorting or mass cytometry.
Mouse Splenocyte and BM Isolation.
After perfusing the mice with ice-cold PBS, the spleens and BM were removed for preparation of single-cell suspensions for mass cytometry staining. To minimize differences due to enzyme use or other aspects of the digestion protocol, isolation of splenocytes was performed following the same protocol as used for the single-cell preparation of mouse brains. For the isolation of BM, the lower limbs were removed and cleaned from all surrounding tissue, followed by a cleaning step in 70% ethanol. After flushing the bones with HBSS, cells were passed through a 70-μm filter (Fisher Scientific), and incubated with the Neural Tissue Dissociation Kit P (Miltenyi Biotech) enzymes according to the protocol for isolation of splenocytes and brain-derived single cells, but without further mechanical dissociation. RBC were lysed during 2 min of incubation with mouse RBC lysis buffer (Boston Bioproducts). Due to low yield of splenocytes in mouse MGL1 and MGL6, we excluded the spleen data of these mice from further analysis.
Live/Death Staining and Barcoding.
Single-cell suspensions were washed in RPMI (no FCS) and incubated with Cisplatin (5 µM; Fluidigm) for 5 min at 37 °C, followed by a second wash with RPMI 10% FCS to quench the Cisplatin. To ensure homogenous staining, up to 3 × 106 cells per sample were barcoded as previously described (75) using a 20-plex Pd Barcoding kit (Fluidigm). After fixation and permeabilization according to manufacturer’s directions, all samples were barcoded, assuring matching barcodes per mouse. All brain samples were then pooled together, as were all spleen samples and all BM samples, and frozen in FCS-dimethyl sulfoxide (80:20) at −80 °C.
Antibodies.
For each antibody used in our mass cytometry panel, the clone, metal tag, and provider are listed in SI Appendix, Table S2. Most of the antibodies we obtained from Fluidigm included metal label. Some antibodies, however, were labeled in house according to manufacturer’s instructions using the Maxpar Antibody Labeling Kit (Fluidigm). After metal conjugation, antibody concentrations were determined using nanodrop measurements and diluted to appropriate concentrations. Antibodies used in this study were titrated for optimal concentrations, and epitope availability was tested after samples were treated with enzymes (76).
Antibody Staining.
Pooled cells were thawed, washed with Cell Staining Buffer (CSB; Fluidigm), and incubated with FcR blocking reagent (FcBlock Trustain fcX; Biolegend) for 10 min at RT. Next, cells were incubated with a mixture of metal-conjugated antibodies against surface markers at a concentration of 20 × 106 cells per mL for 30 min at RT. Following two washing steps with CSB, cells were fixed with Fix-1 buffer (Fluidigm) for 45 min and permeabilized by washing with permeabilization buffer (Fluidigm) for 45 min. Then, cells were incubated with antibodies against the intracellular markers FoxP3, T-bet, CTLA-4, and Ki-67 for 30 min at RT. After two washing steps with CSB, cells were then incubated overnight with 191/193Ir DNA intercalator (1:4,000) diluted in Fix-1 buffer. The next day, cells were washed once with PBS and two times with H2O before measurement.
Mass Cytometry Data Acquisition.
After washing in H2O, samples were diluted in 0.1× EQ Four Element Calibration Beads and were acquired on a Helios mass cytometer at a rate of 200 to 250 events per s. Samples were split into aliquots of ∼0.8 million cells/mL to reduce sample deterioration in H2O. After acquisition of two sample aliquots, fluidics tubing was washed with H2O, and instrument tuning was repeated. Data were normalized during acquisition, using bead normalization (77). Deconvolution of pooled samples was performed by processing FCS files the standard single-cell debarcoding algorithm for CyTOF data [single-cell debarcoding algorithm for CyTOF data (75)].
Phenotype Specification.
Debarcoded samples were then processed in FlowJo to gate for stable flow, exclude beads, single events, and live cells. The resulting events were then exported into FCS3.1 files and opened in Cytosplore (47) with an arcsinh (x/5) transformation, and HSNE was computed based on CD3, CD4, CD5, CD8, CD11b, CD11c, CD16/32, CD36, CD44, CD45, CD49d, CD54, F4/80, GFP, Ki67, Ly-6C, Ly-6G.C, and TCRb. We used the standard recommended parameter settings for the hierarchical construction: random walks for landmark selection, n = 100; Random walk length, L = 15; random walks for influence computation, n = 15. We first overclustered the embedding at the overview level and merged clusters in a supervised manner. Decisions were made based on (near-)identical expression of phenotyping markers and visual inspection of the accompanying heatmap. We then proceeded to explore successive HSNE levels of each of the overview-level clusters until we could not find further fragmentation into clearly recognizable subpopulations. In each new embedding, we also started by overclustering to later merge identical clusters based on the integrated descriptive marker statistics and heatmap visualization. The resulting cell clusters were exported as FCS3.1 files, as well as the HSNE maps color-coded for sample, cluster, and all markers. Marker expression profiles per cell cluster were visualized in heatmaps based on relative Arcsinh (x/5) values using the ggplot2 package in R. Frequencies per cell clusters were extracted using FlowJo, and statistical differences between groups were investigated using nonparametric means comparison methods in SPSS.
Statistical Analysis.
For the reanalysis of microarray data obtained from Gravendeel et al. (35), the dataset was downloaded from the publicly accessible data platform R2. Data were log2-transformed, and patients with grade I glioma or a grade II or III histological diagnosis of oligo-astrocytoma were excluded from analysis, resulting in an analysis cohort consisting of n = 21 patients with grade II, n = 60 patients with grade III, and n = 159 patients with grade IV glioma, and n = 8 control tissue samples. Selected glycosylation related genes (SI Appendix, Table S1) were filtered and visualized in a heatmap. Comparisons of optical density (OD) values or gene expression data between the different glioma WHO grades were tested using a one-way ANOVA with post hoc analysis (Tukey’s multiple comparisons test). Overall survival was displayed in Kaplan−Meier curves. Differences in cell abundance of clusters between mice with a WT or MGL-Lhi tumor were tested using a one-way ANOVA with post hoc analysis. Correlations among the resulting cell clusters were calculated using Spearman correlation coefficients. To ensure robust measurements and avoid the influence of outliers, we only considered for further analysis Spearman Correlation coefficients that remained significant (P < 0.05) after 10 sequential iterations in which one sample was artificially removed from the dataset. Robust correlations were ranked and visualized as a circular network using Gephi (78), with nodes representing immune cell types, size-coded for mean frequency for that cell type in either WT tumor-injected mice or Cosmc knockout (KO) tumor-injected mice. Nodes were color-coded for degree, representing the number of correlations with other cell types. Edges in the network represent correlation coefficients, filtered for significant correlations. Populations and their nearest neighbors were highlighted in orange and red.
Data Availability.
The CyTOF dataset reported in this paper is publicly available via Cytobank, https://community.cytobank.org/cytobank/experiments# (no. 86623 “Dusoswa et al. Glioblastomas exploit O-glycosylation – PNAS”).
Supplementary Material
Acknowledgments
We thank Tommy van Solinge for assistance in intracranial injections, workup of mouse tissues, and fueling our creativity in discussions. We are thankful for the help from the O2Flow Core facility in setting up the CyTOF experiments. This work was supported by grants from the Dutch Institute for Chemical Immunology (S.A.D.), The Cancer Center Amsterdam (J.J.G.-V.), the Amsterdam Infection and Immunity Institute (S.A.D.), 642870-Immunoshape (E.R.), ERC339977 (Y.v.K.), CA069246 (X.O.B.), CA232103 (X.O.B.), Agencia Nacional de Promoción Científica y Técnica (G.A.R., PICT 2014-3687), Richard Lounsbery Foundation (G.A.R.), and Sales Foundation (G.A.R.).
Footnotes
The authors declare no competing interest.
Data deposition: The CyTOF dataset reported in this paper is publicly available via Cytobank, https://community.cytobank.org/cytobank/experiments# (no. 86623 “Dusoswa et al. Glioblastomas exploit O-glycosylation – PNAS”).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907921117/-/DCSupplemental.
References
- 1.Stupp R., et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group , Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Lim M., Xia Y., Bettegowda C., Weller M., Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 15, 422–442 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Brown C. E., et al. , Regression of glioblastoma after chimeric antigen receptor T-cell therapy. N. Engl. J. Med. 375, 2561–2569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schalper K. A., et al. , Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat. Med. 25, 470–476 (2019). [DOI] [PubMed] [Google Scholar]
- 5.van Kooyk Y., Rabinovich G. A., Protein-glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 9, 593–601 (2008). [DOI] [PubMed] [Google Scholar]
- 6.RodrÍguez E., Schetters S. T. T., van Kooyk Y., The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 18, 204–211 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Geijtenbeek T. B. H., Gringhuis S. I., Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 9, 465–479 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parney I. F., Waldron J. S., Parsa A. T., Flow cytometry and in vitro analysis of human glioma-associated macrophages. Laboratory investigation. J. Neurosurg. 110, 572–582 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wurdinger T., Deumelandt K., van der Vliet H. J., Wesseling P., de Gruijl T. D., Mechanisms of intimate and long-distance cross-talk between glioma and myeloid cells: How to break a vicious cycle. Biochim. Biophys. Acta 1846, 560–575 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Komohara Y., et al. , Importance of direct macrophage-tumor cell interaction on progression of human glioma. Cancer Sci. 103, 2165–2172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crocker P. R., Paulson J. C., Varki A., Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Lübbers J., Rodríguez E., van Kooyk Y., Modulation of immune tolerance via Siglec-sialic acid interactions. Front. Immunol. 9, 2807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perdicchio M., et al. , Sialic acid-modified antigens impose tolerance via inhibition of T-cell proliferation and de novo induction of regulatory T cells. Proc. Natl. Acad. Sci. U.S.A. 113, 3329–3334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenos K., et al. , MGL ligand expression is correlated to BRAF mutation and associated with poor survival of stage III colon cancer patients. Oncotarget 6, 26278–26290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfert M. A., Boons G. J., Adaptive immune activation: Glycosylation does matter. Nat. Chem. Biol. 9, 776–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J. Y., Oswald D. M., Oliva K. D., Kreisman L. S. C., Cobb B. A., The glycoscience of immunity. Trends Immunol. 39, 523–535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehres C. M., Garcia-Vallejo J. J., Unger W. W. J., van Kooyk Y., Skin-resident antigen-presenting cells: Instruction manual for vaccine development. Front. Immunol. 4, 157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higashi N., et al. , The macrophage C-type lectin specific for galactose/N-acetylgalactosamine is an endocytic receptor expressed on monocyte-derived immature dendritic cells. J. Biol. Chem. 277, 20686–20693 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Raes G., et al. , Macrophage galactose-type C-type lectins as novel markers for alternatively activated macrophages elicited by parasitic infections and allergic airway inflammation. J. Leukoc. Biol. 77, 321–327 (2005). [DOI] [PubMed] [Google Scholar]
- 20.van Vliet S. J., van Liempt E., Geijtenbeek T. B. H., van Kooyk Y., Differential regulation of C-type lectin expression on tolerogenic dendritic cell subsets. Immunobiology 211, 577–585 (2006). [DOI] [PubMed] [Google Scholar]
- 21.van Vliet S. J., Saeland E., van Kooyk Y., Sweet preferences of MGL: Carbohydrate specificity and function. Trends Immunol. 29, 83–90 (2008). [DOI] [PubMed] [Google Scholar]
- 22.van Vliet S. J., et al. , MGL signaling augments TLR2-mediated responses for enhanced IL-10 and TNF-α secretion. J. Leukoc. Biol. 94, 315–323 (2013). [DOI] [PubMed] [Google Scholar]
- 23.van Vliet S. J., Gringhuis S. I., Geijtenbeek T. B. H., van Kooyk Y., Regulation of effector T cells by antigen-presenting cells via interaction of the C-type lectin MGL with CD45. Nat. Immunol. 7, 1200–1208 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Pinho S. S., Reis C. A., Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Ashkani J., Naidoo K. J., Glycosyltransferase gene expression profiles classify cancer types and propose prognostic subtypes. Sci. Rep. 6, 26451 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinho S. S., et al. , Gastric cancer: Adding glycosylation to the equation. Trends Mol. Med. 19, 664–676 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Jandus C., et al. , Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Invest. 124, 1810–1820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perdicchio M., et al. , Tumor sialylation impedes T cell mediated anti-tumor responses while promoting tumor associated-regulatory T cells. Oncotarget 7, 8771–8782 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez J. F., et al. , Biochemical and structural analysis of Helix pomatia agglutinin. A hexameric lectin with a novel fold. J. Biol. Chem. 281, 20171–20180 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Rambaruth N. D. S., Greenwell P., Dwek M. V., The lectin Helix pomatia agglutinin recognizes O-GlcNAc containing glycoproteins in human breast cancer. Glycobiology 22, 839–848 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Fu C., et al. , Tumor-associated antigens: Tn antigen, sTn antigen, and T antigen. HLA 88, 275–286 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Lou Y.-W., et al. , Stage-specific embryonic antigen-4 as a potential therapeutic target in glioblastoma multiforme and other cancers. Proc. Natl. Acad. Sci. U.S.A. 111, 2482–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akita K., et al. , Developmental expression of a unique carbohydrate antigen, Tn antigen, in mouse central nervous tissues. J. Neurosci. Res. 65, 595–603 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Akita K., et al. , Identification of the core protein carrying the Tn antigen in mouse brain: Specific expression on syndecan-3. Cell Struct. Funct. 26, 271–278 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Gravendeel L. A. M., et al. , Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 69, 9065–9072 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Nguyen A. T., et al. , Organelle specific O-glycosylation drives MMP14 activation, tumor growth, and metastasis. Cancer Cell 32, 639–653.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Wu Q., et al. , Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J. Biol. Chem. 290, 1170–1185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennett E. P., et al. , Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry A., Wesseling P., Histologic classification of gliomas. Handb. Clin. Neurol. 134, 71–95 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez F. J., Scheithauer B. W., Giannini C., Bryant S. C., Jenkins R. B., Epithelial and pseudoepithelial differentiation in glioblastoma and gliosarcoma: A comparative morphologic and molecular genetic study. Cancer 113, 2779–2789 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W., et al. , MUC4 modulates human glioblastoma cell proliferation and invasion by upregulating EGFR expression. Neurosci. Lett. 566, 82–87 (2014). [DOI] [PubMed] [Google Scholar]
- 42.van Vliet S. J., Paessens L. C., Broks-van den Berg V. C. M., Geijtenbeek T. B. H., van Kooyk Y., The C-type lectin macrophage galactose-type lectin impedes migration of immature APCs. J. Immunol. 181, 3148–3155 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Komohora Y., Ohnishi K., Kuratsu J., Takeya M., Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 220, 114–125 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Hambardzumyan D., Gutmann D. H., Kettenmann H., The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 19, 20–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., et al. , Cosmc is an essential chaperone for correct protein O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 107, 9228–9233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bendall S. C., et al. , Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Unen V., et al. , Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat. Commun. 8, 1740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Höllt T., et al. , Cytosplore: Interactive immune cell phenotyping for large single-cell datasets. Comput. Graph. Forum 35, 171–180 (2016). [Google Scholar]
- 49.Chongsathidkiet P., et al. , Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat. Med. 24, 1459–1468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bronte V., et al. , Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 7, 12150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alieva M., et al. , Preventing inflammation inhibits biopsy-mediated changes in tumor cell behavior. Sci. Rep. 7, 7529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z., et al. , Cellular and molecular identity of tumor-associated macrophages in glioblastoma. Cancer Res. 77, 2266–2278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bergthaler A., Menche J., The immune system as a social network. Nat. Immunol. 18, 481–482 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Rieckmann J. C., et al. , Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol. 18, 583–593 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Pyonteck S. M., et al. , CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 19, 1264–1272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ajami B., et al. , Single-cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nat. Neurosci. 21, 541–551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Springer G. F., Desai P. R., Banatwala I., Blood group MN specific substances and precursors in normal and malignant human breast tissues. Naturwissenschaften 61, 457–458 (1974). [DOI] [PubMed] [Google Scholar]
- 58.Springer G. F., T and Tn, general carcinoma autoantigens. Science 224, 1198–1206 (1984). [DOI] [PubMed] [Google Scholar]
- 59.Springer G. F., Immunoreactive T and Tn epitopes in cancer diagnosis, prognosis, and immunotherapy. J. Mol. Med. (Berl.) 75, 594–602 (1997). [DOI] [PubMed] [Google Scholar]
- 60.Desai P. R., Immunoreactive T and Tn antigens in malignancy: Role in carcinoma diagnosis, prognosis, and immunotherapy. Transfus. Med. Rev. 14, 312–325 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Bapu D., Runions J., Kadhim M., Brooks S. A., N-acetylgalactosamine glycans function in cancer cell adhesion to endothelial cells: A role for truncated O-glycans in metastatic mechanisms. Cancer Lett. 375, 367–374 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Julien S., Videira P. A., Delannoy P., Sialyl-tn in cancer: (how) did we miss the target? Biomolecules 2, 435–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laack E., et al. , Lectin histochemistry of resected adenocarcinoma of the lung: Helix pomatia agglutinin binding is an independent prognostic factor. Am. J. Pathol. 160, 1001–1008 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Welinder C., Baldetorp B., Blixt O., Grabau D., Jansson B., Primary breast cancer tumours contain high amounts of IgA1 immunoglobulin: An immunohistochemical analysis of a possible carrier of the tumour-associated Tn antigen. PLoS One 8, e61749 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Konno A., Hoshino Y., Terashima S., Motoki R., Kawaguchi T., Carbohydrate expression profile of colorectal cancer cells is relevant to metastatic pattern and prognosis. Clin. Exp. Metastasis 19, 61–70 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Heiland D. H., et al. , Comprehensive analysis of PD-L1 expression in glioblastoma multiforme. Oncotarget 8, 42214–42225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nduom E. K., et al. , PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 18, 195–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ju T., Aryal R. P., Kudelka M. R., Wang Y., Cummings R. D., The Cosmc connection to the Tn antigen in cancer. Cancer Biomark. 14, 63–81 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsuiji M., et al. , Molecular cloning and characterization of a novel mouse macrophage C-type lectin, mMGL2, which has a distinct carbohydrate specificity from mMGL1. J. Biol. Chem. 277, 28892–28901 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Singh S. K., et al. , Characterization of murine MGL1 and MGL2 C-type lectins: Distinct glycan specificities and tumor binding properties. Mol. Immunol. 46, 1240–1249 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Wang J., et al. , Invasion of white matter tracts by glioma stem cells is regulated by a NOTCH1-SOX2 positive-feedback loop. Nat. Neurosci. 22, 91–105 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Lagerweij T., et al. , Optical clearing and fluorescence deep-tissue imaging for 3D quantitative analysis of the brain tumor microenvironment. Angiogenesis 20, 533–546 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sena-Esteves M., Tebbets J. C., Steffens S., Crombleholme T., Flake A. W.. Optimized large-scale production of high titer lentivirus vector pseudotypes. J. Virol. Methods 122, 131–139 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zunder E. R., et al. , Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 10, 316–333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dusoswa S. A., Verhoeff J., Garcia-Vallejo J. J., OMIP 054: Broad immune phenotyping of innate and adaptive leukocytes in the brain, spleen, and bone marrow of an orthotopic murine glioblastoma model by mass cytometry. Cytom. Part A 95, 422–426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finck R., et al. , Normalization of mass cytometry data with bead standards. Cytom. Part A 83, 483–494 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacomy M., Venturini T., Heymann S., Bastian M., ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi Software. PLoS ONE 9, e98679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The CyTOF dataset reported in this paper is publicly available via Cytobank, https://community.cytobank.org/cytobank/experiments# (no. 86623 “Dusoswa et al. Glioblastomas exploit O-glycosylation – PNAS”).