Significance
In HIV-infected patients on combination antiretroviral therapy (cART), greater than 95% of proviruses in the peripheral blood are “defective.” Historically, these defective proviruses have been thought to be dead-end products with no real pathophysiological significance, as they do not encode replication-competent viruses. Contrary to this view, we have identified cells in tissue culture and from cART-treated patients that harbor defective proviruses and produce viral proteins. Features found in these translationally competent yet defective proviruses suggest that HIV-1 infection results in modification of the CD4+ T cell genome analogous to human endogenous retroviruses. We propose that these defective HIV-1 proviruses are biologically significant, despite being “replication incompetent,” have the potential to elicit immune activation, and may serve as a barrier to HIV-1 cure.
Keywords: HIV, provirus, immune activation
Abstract
HIV-1 proviruses persist in the CD4+ T cells of HIV-infected individuals despite years of combination antiretroviral therapy (cART) with suppression of HIV-1 RNA levels <40 copies/mL. Greater than 95% of these proviruses detected in circulating peripheral blood mononuclear cells (PBMCs) are referred to as “defective” by virtue of having large internal deletions and lethal genetic mutations. As these defective proviruses are unable to encode intact and replication-competent viruses, they have long been thought of as biologically irrelevant “graveyard” of viruses with little significance to HIV-1 pathogenesis. Contrary to this notion, we have recently demonstrated that these defective proviruses are not silent, are capable of transcribing novel unspliced forms of HIV-RNA transcripts with competent open reading frames (ORFs), and can be found in the peripheral blood CD4+ T cells of patients at all stages of HIV-1 infection. In the present study, by an approach of combining serial dilutions of CD4+ T cells and T cell–cloning technologies, we are able to demonstrate that defective proviruses that persist in HIV-infected individuals during suppressive cART are translationally competent and produce the HIV-1 Gag and Nef proteins. The HIV-RNA transcripts expressed from these defective proviruses may trigger an element of innate immunity. Likewise, the viral proteins coded in the defective proviruses may form extracellular virus-like particles and may trigger immune responses. The persistent production of HIV-1 proteins in the absence of viral replication helps explain persistent immune activation despite HIV-1 levels below detection, and also presents new challenges to HIV-1 eradication.
The presence and the persistence of “defective” proviruses in HIV-infected patients has been well appreciated from the early years of HIV/AIDS research (1–14). The pool of cells harboring defective proviruses is initially established early during infection (6). Cells harboring these defective proviruses are able to clonally expand and can persist for greater than 10 years in vivo (10). As these defective proviruses are unable to encode intact viruses, they have long been thought of as biologically irrelevant “graveyard” of viruses with little significance to HIV-1 pathogenesis. Contrary to this notion, we have recently demonstrated that HIV-1 defective proviruses lead to the transcription of novel HIV-RNA species. The long-term persistence of antibodies to various proteins of HIV-1 in patients on combination antiretroviral therapy (cART) (5, 15) and the demonstration that cytotoxic T lymphocytes (CTLs) may recognize cells transfected with DNA constructs carrying the sequences from defective proviruses (16) provide indirect evidences that the defective proviruses may be associated with protein production. However, there has been no report of direct demonstration of HIV-1 protein expression from defective proviruses thus far.
The long-term production of foreign (HIV-1) proteins in patients on cART may have a deleterious effect on the host and may be reflected in the observation that chronic HIV-1 infection is associated with persistent immune activation. Of note, persistent inflammation and ongoing immune activation can be observed in HIV-infected individuals who have achieved prolonged suppression of plasma viremia and may be associated with an increase in all-cause mortality, liver disease, renal disease, and cancer (17–19). A puzzling observation, under such circumstances, is the persistence of immune responses, including seropositivity to HIV-1 (5, 15), in the setting of “undetectable” levels of virus in plasma. In the present study, we studied the ability of naturally occurring defective HIV-1 proviruses to encode viral proteins through two approaches: one, using single-cell clones derived from the chronically infected T cell line H9MN (20, 21); and, two, isolating and characterizing a CD4+ T cell clone isolated from an HIV-infected individual on cART.
Results
Expression of HIV-1 Proteins in H9MN T Cell Clones Harboring Defective Proviruses.
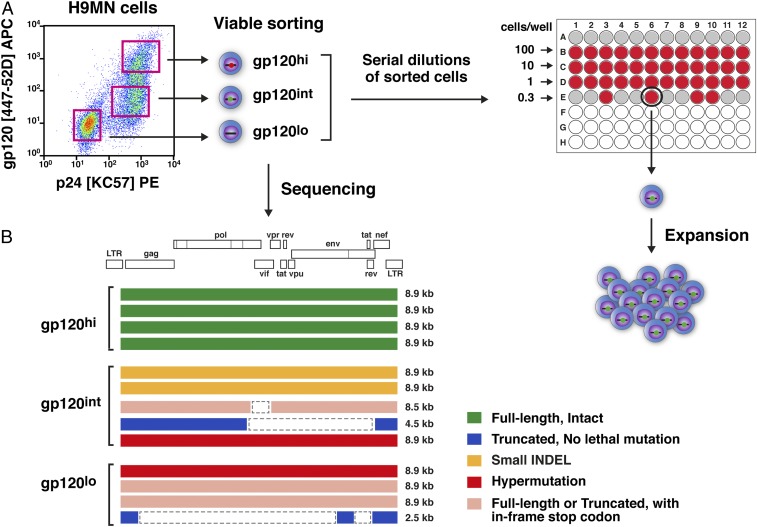
In order to assess viral protein expression from defective proviruses, we first turned to an in vitro cell culture system. H9MN is a T cell line that was established from H9 cells (Hut78 derivative cells) chronically infected with the MN strain of HIV-1 (20, 21). The original uncloned H9MN cell line was found to contain a mixture of cells with different levels of HIV-1 Gag p24, and Env gp120 protein expression (Fig. 1A). These cells were viably sorted into gp120hi, gp120int, and gp120lo fractions. Sequence analysis of the proviruses present in these protein-expressing cells demonstrated an extreme degree of sequence and size heterogeneity, ranging from 2.5 to 8.9 kb in length (Fig. 1B). Clones were obtained from these fractions by limiting dilution in 96-well plates (Fig. 1A).
Fig. 1.
Isolation of single-cell clones from the H9MN cell line. (A) Schematic diagrams of the strategies used to isolate H9MN single-cell clones harboring defective proviruses. Flow cytometry plot of HIV-1 MN cells showing the expression of intracellular Gag p24 protein (clone: KC-57) and cell surface Env gp120 (clone: 447–52D) protein. The H9MN cells were flow-sorted into three populations based on cell surface expression of Env gp120 protein and CD3: gp120hi, gp120int, and gp120lo. The flow-sorted cells were serially diluted in 96-well plates and left to grow to a confluent status (2 weeks). Cells from the wells exhibiting growth (visually inspected by microscopy) were transferred to 48-well plates. A small aliquot of cells from the wells in the 48-well plates was taken to detect the presence of HIV-DNA by 5′LTR-to-3′LTR PCR. Cells from wells positive for HIV-DNA were further expanded in 24-well plates for an additional week. Upon completion of the expansion culture, one portion (1 × 106) of the cells was used for simultaneous extraction of DNA and RNA (after cDNA synthesis), followed by 5′LTR-to-3′LTR single-genome amplification and direct amplicon sequencing; the other portion (2 × 106) of the cells was used for Western blot and confocal microscopy (1 × 106 cells) for detection of HIV-1 proteins. (B) Proviral sequences present in the three sorted fractions of H9MN cells.
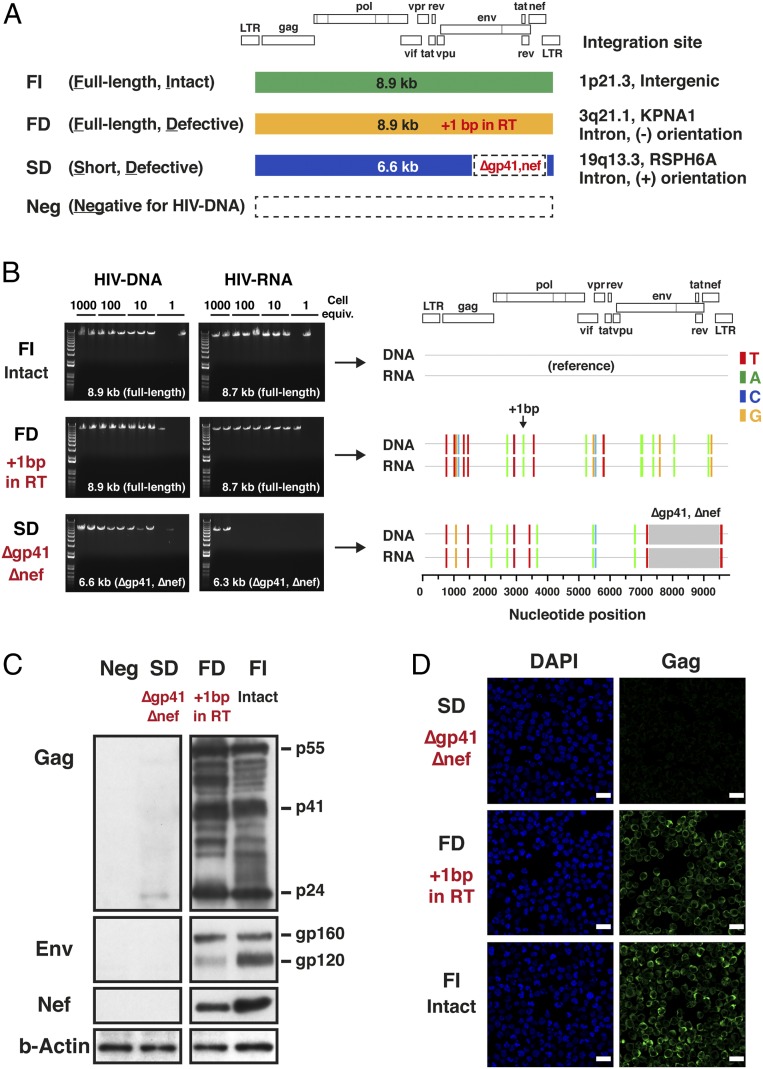
Four distinct single-cell clones were isolated from the H9MN cells (Fig. 2A). The FI clone harbored an intact full-length (8.9-kb) HIV-1 provirus encoding all open reading frames (ORFs) of HIV-1. The FD clone contained an 8.9-kb full-length defective provirus with a 1-bp frameshift insertion that resulted in total abolishment of reverse transcriptase (RT) and integrase with intact expression of Gag and Env (SI Appendix, Fig. S1). The SD clone possessed a short defective provirus with a large (2.4-kb) internal deletion, affecting the gp41 and Nef coding regions. The Neg clone did not contain HIV-DNA and served as a negative control for the subsequent Western blot analyses. All four single-cell clones were negative for human T cell lymphotropic virus type 1 (HTLV-1) by quantitative PCR (SI Appendix, Table S1).
Fig. 2.
Expression of HIV-DNA, HIV-RNA, and HIV-1 proteins in H9MN subclones harboring defective proviruses. (A) A total of four distinct single-cell clones were isolated: FI, a subclone containing an 8.9-kb full-length intact provirus; FD, a subclone containing an 8.9-kb full-length defective provirus with a 1-bp frameshift lethal mutation in the RT gene (HXB2 coordinate 3204); SD, a subclone containing a short defective provirus with a large (∼2.3-kb) internal deletion affecting the gp41 and nef coding regions; and Neg, a subclone negative for HIV-DNA. (B) Agarose gel pictures depicting sizes of HIV-DNA and HIV-RNA PCR fragments generated for FI (full-length intact provirus), FD (full-length defective provirus), and SD (short defective provirus) single-cell clones using 5′LTR-to-3′LTR PCR. The highlighter analysis was performed using the provirus sequence derived from the FI clone as a reference. The nucleotide positions that differ from the HIV-DNA sequence in the FI clone are indicated by color-coded bars. The gray areas indicate gaps. HIV-RNA sequences corresponded precisely to the HIV-DNA sequences for each clone. The integration sites of the proviruses present in the FI, FD, and SD clones were analyzed by restriction enzyme digestion of genomic DNA by BclI and inverse PCR. (C) Expression of HIV-1 proteins in three distinct H9MN single-cell clones by Western blot: SD, FD, and FI. A mouse monoclonal antibody for HIV-1 p24 (clone: 39/5.4A), a goat polyclonal antibody for HIV-1 Env, and a mouse monoclonal antibody for HIV-1 Nef (clone: EH1) were used. The SD clone expressed no Env or Nef and only a low level of p24 Gag. The FD and FI clones expressed Gag p55/p24 and Env gp160/gp120 proteins with molecular weights predicted by the HIV-RNA sequences. (D) Confocal microscopy analysis of the intracellular expression of Gag p24 in the three distinct single-cell clones. A mouse monoclonal antibody for HIV-1 p24 (clone: 39/5.4A) was used. Nuclei were visualized by DAPI (blue). Original magnification was ×63. (Scale bars, 20 µm.)
All of the clones harboring HIV-1 proviruses were found to express HIV-RNA of a length and sequence similar to those of their corresponding provirus (Fig. 2B). An analysis of integration sites for the three provirus-containing clones revealed three different chromosomal sites of integration: 1p21.3 (intergenic region) in FI; 3q21.1 (KPNA1, intron, [−] orientation) in FD; and 19q13.3 (RSPH6A, intron, [+] orientation) in SD (Fig. 2A). No associations were noted between the proviral integration site, the orientation of integration relative to the host gene, and the level of HIV-RNA transcription. We next examined whether any of these clones expressed HIV-1 proteins.
As predicted from the sequence data and their level of RNA transcripts, the SD clone expressed neither Env nor Nef and only a low level of p24 Gag by Western blot and confocal immunofluorescence microscopy (Fig. 2 C and D). The provirus and RNA sequences in both the FI and FD clones predicted that these clones could produce Gag, Env, and Nef proteins. This was found to be the case by Western blot and confocal microscopy (Fig. 2 C and D). Of note, the +1-bp frameshift mutation in the RT gene present in the provirus in the FD clone led to complete abolishment of the RT and integrase expression in these cells (SI Appendix, Fig. S1), demonstrating that this defective provirus in the FD clone, while defective from the perspective of encoding an intact virus, encodes intact Gag, Env, and Nef.
CD4+ T Cell Clones Harboring Defective Proviruses from a Patient with HIV-1 Infection Express HIV-1 Proteins In Vivo.
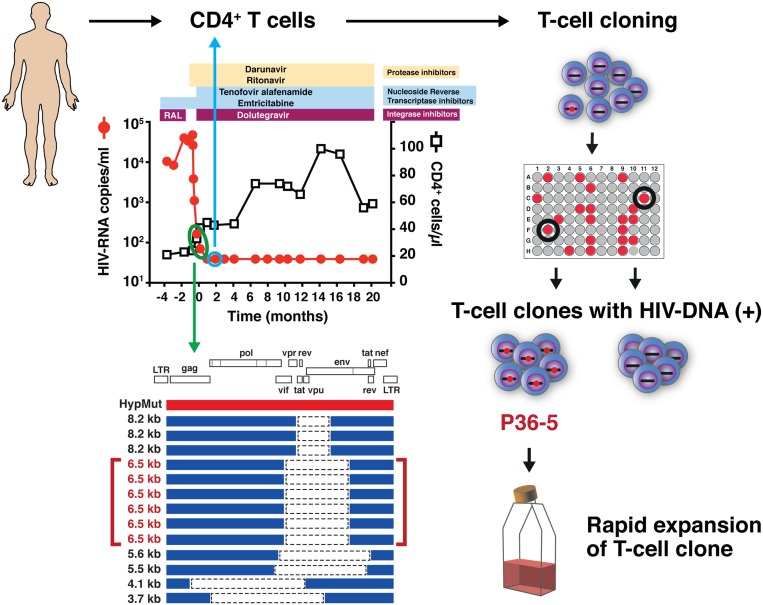
Having determined that defective proviruses can lead to the production of HIV-1 proteins following in vitro infection, we next sought to determine whether or not this was occurring in vivo in a patient with HIV-1 RNA levels <40 copies/mL on cART. We obtained frozen cell samples from an HIV-infected individual who had recently been placed on suppressive cART (Fig. 3). We found that the cells from this patient were highly enriched for cells harboring HIV-DNA with an estimated frequency of 1 in 100 CD4+ T cells. All of the provirus species found in this patient’s cells were defective, mostly represented by truncated proviruses with large internal deletions. To generate CD4+ T cell clones from this subject, we chose a time point ∼2 months after achieving a plasma viral load (pVL) <40 copies/mL We carefully chose this time point, as we thought that by the time of 2 months after achieving pVL <40 copies/mL, most of the cells harboring intact full-length proviruses encoding replication-competent viruses would have been cleared from the blood circulation. From a total of 6,000 cells initially plated, we were able to isolate two CD4+ T cell clones harboring the same defective provirus, called P36-5 (Fig. 3). Of note, this provirus with a 2.4-kb internal deletion was the predominant species of HIV-1 provirus present in the patient’s cells in vivo (Fig. 3). In addition, the HIV-RNA transcript matching the defective provirus in the P36-5 clone was detected in CD4+ T cells freshly isolated from the study subject (SI Appendix, Fig. S3).
Fig. 3.
Isolation of CD4+ T cell clones harboring defective proviruses from an individual with advanced HIV-1 infection. Schematic diagram of the strategy used to isolate CD4+ T cell clones harboring defective proviruses from a patient with HIV-1 infection. CD4+ T cells were obtained from a time point 2 months after achieving a pVL <40 copies/mL (blue circle). Out of 6,000 cells initially plated, two T cell clones harboring the same defective provirus, P36-5, were isolated. This provirus, with a 2.4-kb internal deletion, was the predominant species present in the patient’s cells in vivo (green circle). By isolating T cell clones, the capacity to transcribe HIV-RNA and produce viral proteins at the single-cell level can be assessed.
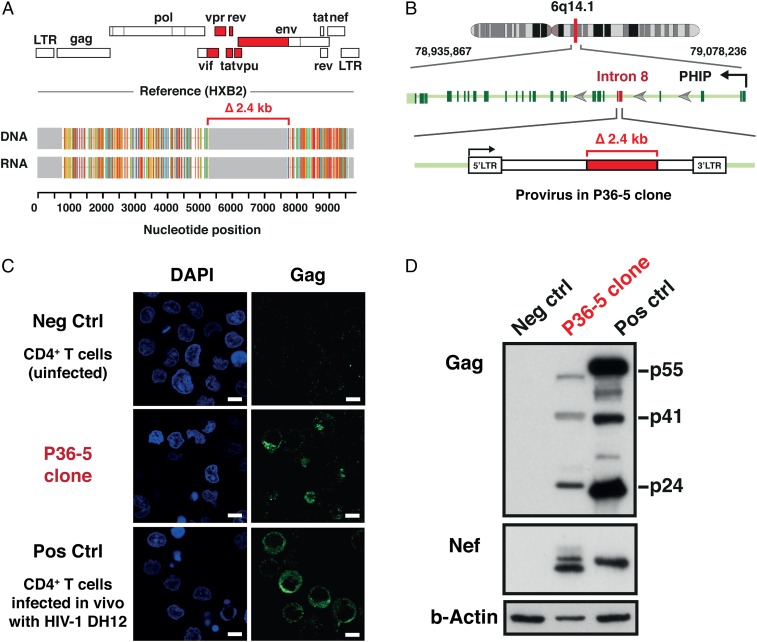
As in the case of the cells generated in vitro, the CD4+ T cell clones derived from the patient harbored HIV-RNA transcripts identical in size and sequence to those of the proviruses (Fig. 4A). The 5’ long terminal repeat (LTR)-to-3’ LTR PCR yielded uniform PCR products of ∼6.5 kb in length at the limit of detection for the assay (100% detection sensitivity is ∼10 copies per reaction). The highlighter analysis confirmed the sequence identity between the 6.5-kb provirus and the HIV-RNA transcripts in the P36-5 clone (Fig. 4A). It also confirmed the presence of a 2.4-kb internal deletion affecting the region encoding the HIV-1 accessory proteins (including the first exon of tat and rev) and the gp120 portion of the Env protein in the provirus genome (Fig. 4A). The mechanism(s) leading to initiation of transcription of HIV-RNA in the absence of an intact Tat is currently unknown. Recruitment of host transcription factors to the HIV-1 LTR independent of the viral transactivator protein HIV-1 Tat is one plausible explanation. In addition, readthrough transcription from the adjacent host gene may play some role (22–25). The whole-genome sequencing of the P36-5 cells revealed the presence of two intact LTRs and the proviral integration site in intron 8 of the PHIP (pleckstrin homology domain interacting protein) gene in the orientation opposite to that of the host gene (Fig. 4B), making readthrough transcription less likely.
Fig. 4.
Expression of HIV-DNA, HIV-RNA, and HIV-1 proteins in the CD4+ T cell clone harboring a defective provirus derived from an HIV-infected individual during cART. (A) HIV-RNA sequences corresponded precisely to the HIV-DNA sequences for the P36-5 clone. Analysis of the genome structure revealed a 2.4-kb internal deletion affecting the region encoding the HIV-1 accessory proteins (tat, rev, vpu, and others) and the gp120 portion of the Env protein (red). The Gag, Pol, and Nef regions remained intact in the provirus present in the P36-5 clone. (B) Whole-genome sequencing confirmed the presence of two intact LTRs and revealed the provirus to be integrated in intron 8 of the PHIP gene in the opposite orientation to the PHIP gene. (C) Confocal microscopy analysis of the intracellular expression of the Gag p24 protein. A mouse monoclonal antibody for HIV-1 p24 (clone: 39/5.4A) was used. Nuclei were visualized by DAPI (blue). Original magnification was ×63. (Scale bars, 5 µm.) (D) Expression of HIV-1 proteins by Western blot. Negative control (Neg Ctrl), uninfected CD4+ T cells; and positive control (Pos Ctrl), CD4+ T cells infected in vitro with the DH12 strain of HIV-1. A mouse monoclonal antibody for HIV-1 p24 (clone: 39/5.4A) and a mouse monoclonal antibody for HIV-1 Nef (clone: 3A2) were used. The predicted sizes of the Nef proteins, based on the provirus sequences for DH12 and P36-5, were 23.4 kDa and 22.6 kDa, respectively. The higher molecular weight band seen for Nef in P36-5 likely represents posttranslational modifications such as myristoylation.
The capacity of the transcription-competent defective provirus present in the P36-5 clone to produce HIV-1 proteins was assessed by both Western blot and confocal immunofluorescence microscopy (Fig. 4 C and D). Despite having a large internal deletion, the defective provirus in the P36-5 clone retained intact ORFs for the gag, pol, and env coding regions. As predicted from the sequence, Gag and Nef proteins with the predicted molecular weights were clearly detected in the P36-5 clone by Western blot. The higher molecular weight band seen for Nef in the P36-5 clone likely represents Nef proteins with posttranslational modifications such as myristoylation (Fig. 4D) (26, 27). In addition, the intracellular production of Gag could be demonstrated by confocal microscopy, providing additional direct evidence of viral protein expression in cells harboring a defective provirus. Unfortunately, we were not able to identify an antibody that would allow us to detect gp41 by Western blot.
Discussion
In the present study, we have provided direct in vitro and in vivo evidence that cells harboring defective HIV-1 proviruses are capable of producing HIV-1 proteins. In order to assess the ability of defective proviruses to express viral proteins in vivo, we isolated single-cell CD4+ T cell clones carrying distinct individual defective proviruses from a patient with HIV-1 infection and HIV-1 RNA levels <40 copies/mL This T cell–cloning strategy also provided a means of enriching viable CD4+ T cells harboring defective HIV-1 provirus without fixation and permeabilization. In order to increase our chance of isolating a CD4+ T cell clone harboring a defective provirus, we chose a time point 2 months following virologic suppression; at that time, the precursor frequency of HIV-DNA positive cells was 1%. We are currently examining the long-term kinetics of this and related populations and their potential correlation with clinically validated markers of immune activation.
The reverse transcription process for HIV-1 is extremely error prone (28–30). Because of this, defective proviruses are frequently found in the CD4+ T cells of patients with HIV-1 infection (4–6). The frequent presence of defective proviruses is a common feature observed for all viruses belonging to the family of retroviruses. Human endogenous retroviruses (HERVs) are proviral remnants of past retroviral infections of the germ line (31–34). HERV-derived proteins and particles have been shown to elicit immune responses (31, 34). A similar scenario may be occurring with a modern-day exogenous retrovirus, HIV-1.
The in vivo fate of the pool of cells harboring replication-incompetent yet viral protein-producing “zombie” defective proviruses is currently unknown. Under normal circumstances, any cells expressing foreign viral proteins would be eliminated by the cytotoxic T lymphocyte response of the host immune system. In the setting of chronic HIV-1 infection, the cells harboring defective proviruses may not be recognized by the host as foreign due to inadequate presentation by the major histocompatibility complex (MHC) (35–40). This might serve as an explanation for the clonal expansion and persistence observed in vivo for the cells harboring defective proviruses. Interestingly, the HIV-1 Nef protein can continue to be detected in plasma or peripheral blood mononuclear cells (PBMCs) of HIV-infected patients long after suppression of plasma viremia to HIV-RNA <50 copies/mL on cART is achieved (41, 42). It is plausible that the Nef protein production seen in the setting of absence of active virus replication, at least in part, is explained by the pool of cells harboring translation-competent defective proviruses.
While the major focus of therapy for HIV-1 infection is stopping the replication of the virus, these data indicate that defective proviruses may be biologically active and play a role in HIV-1 pathogenesis. The HIV-RNA transcripts and resulting proteins expressed from the defective proviruses may trigger elements of both innate and adaptive immunity leading to a state of persistent immune activation with long-term clinical consequences. It is possible that the viral proteins coded in the defective proviruses are able to form extracellular viruslike particles that trigger immune responses. In addition to providing a mechanism for persistent immune activation, defective proviruses may also present an obstacle to eradication of HIV. Recombination occurs in many RNA viruses, and it is theoretically possible for these defective proviruses to recombine to form full-length intact HIV-1 viruses that are replication competent. Recombination of defective proviruses may help explain the delayed viral rebound in certain rare cases following discontinuation of cART (43).* Given that cART does not eliminate these cells, consideration should be given to strategies that target cells harboring defective proviruses.
Methods
Study Samples.
All samples were collected following National Institute of Allergy and Infectious Diseases Institutional Review Board–approved HIV-1 clinical research protocols. Study participants provided written informed consent before enrollment.
Simultaneous Isolation of HIV-1 DNA and HIV-1 RNA and cDNA Synthesis.
Isolation of HIV-1 DNA and RNA and cDNA synthesis procedures can be found in SI Appendix.
Amplification of Near Full-Length HIV-1 DNA and Unspliced HIV-1 RNA.
PCR conditions used for amplification of near full-length HIV-1 DNA and RNA can be found in SI Appendix.
Sequencing and Sequence Analyses.
Sequencing condition and sequence analysis procedures can be found in SI Appendix.
Analysis of HIV-1 Integration Sites.
HIV-1 integration site analysis procedures can be found in SI Appendix.
Western Blot.
Whole cell lysates were prepared with radioimmunoprecipitation assay (RIPA) lysis buffer containing 50 mM Tris⋅hydrochloride (HCl) pH7.5, 150 mM NaCl, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate (SDS), and 0.5% sodium deoxycholate (Boston BioProducts) supplemented with a protease inhibitor (Pierce) before use. Ten micrograms of protein were loaded per lane, separated on 4% to 12% NuPAGE Novex Bis-Tris Gel (ThermoFisher Scientific), and transferred to a 0.2-µm nitrocellulose membrane. The detection of target proteins was performed with the use of an alkaline phosphatase-based iBind chemiluminescent kit (ThermoFisher Scientific). After detection of the target proteins, the membranes were stripped and reprobed with rabbit anti-beta-Actin antibody (Abcam, ab1801). The following antibodies were used in the Western blot assays: mouse monoclonal anti-p24 antibody (clone: 39/5.4A, Abcam, ab9071); goat polyclonal anti-Env antibody (Abcam, ab21179); and mouse monoclonal anti-Nef antibody (clone: EH1, AIDS Reagent Program, 3689).
Confocal Immunofluorescence Microscopy.
Cells for microscopy were resuspended in 4% formaldehyde (Polysciences), mounted on a Poly-L-Lysin–coated 8-chamber glass-bottom slide (ibidi), centrifuged at 300 × g for 5 min to enhance the attachment of the cells to the glass slide, incubated for 10 min at 37 °C, and washed twice with Hank’s Balanced Salt Solution (HBSS) buffer (ThermoFisher Scientific). Following fixation, the cells were washed three times with HBSS buffer and permeabilized with 0.2% Tween 20 (Roche Diagnostics) for 5 min at room temperature. The cells were washed three times with HBSS buffer and treated with a blocking buffer (3% BSA in HBSS) for 45 min at room temperature. The cells were incubated with a mouse monoclonal anti-p24 antibody (clone: 39/5.4A, Abcam, ab9071) at a final concentration of 10 µg/mL in the blocking buffer for 18 to 20 h at 4 °C. After washing twice with the blocking buffer, the cells were incubated in the dark for 1 h at room temperature with a secondary antibody conjugated to a fluorescent tag (Alexa Fluor 488 goat anti-mouse IgG, Molecular Probes) at a final concentration of 10 µg/mL. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (ThermoFisher Scientific). Images were acquired using a Zeiss LSM 800 with a ×63 oil immersion objective lens, and captured by the Airyscan detector (Zeiss) with a deconvolution module, the ZEN imaging software (Zeiss). The images were processed using the Fiji image-processing package (44).
Cloning of H9MN Cells.
A stock culture of H9MN (H9/HTLV-IIIMN NIH 1984, AIDS reagent program, 402) was maintained in RPMI 1640 supplemented with 10% FBS, 5 mM Hepes, and 10 µg/mL gentamicin. H9MN cells from the stock culture were stained with CD3 FITC (fluorescein isothiocyanate) (clone: SK7, BD Bioscience) and a human monoclonal antibody for HIV-1 Env (clone: 447–52D, AIDS Reagent Program, 12123) at a final concentration of 10 µg/mL, followed by staining with secondary Alexa Fluor 647 conjugated donkey antihuman IgG heavy and light (H+L) chains (Jackson ImmunoResearch Laboratories) at a final concentration of 3.75 ng/µl. Cells were viably sorted with BD FACSAria II SORP (BD Biosciences) based on the cell surface expression of CD3 and HIV-1 Env into three fractions: gp120hi, gp120int, and gp120lo (Fig. 1A). Cells from each sorted fraction were plated into 96-well round-bottom plates at cell densities of 100, 10, 1, and 0.3 cells per well. Irradiated (30 Gy) PBMCs from a healthy volunteer were added as feeder cells (5 × 104 cells per well). The overall cloning procedure is summarized in Fig. 1A. Cells in 96-well plates were left to grow to a confluent status for 2 weeks in the complete medium used for the H9MN stock culture. Wells exhibiting cell growth were visually identified by microscopy. Cells from the wells exhibiting growth (visually inspected by microscopy) were transferred to 48-well plates and allowed to expand for an additional week. Small aliquots of cells from the wells positive for cell growth were used to detect the presence of HIV-DNA by 5′LTR-to-3′LTR PCR. Cells from the wells positive for HIV-DNA were further expanded in 24-well plates for one additional week. T cell cloning was considered successful if the following criteria were met: 1) The nested PCR for detection of HIV-DNA achieved single-copy sensitivity (positive at the limit of detection of 10 copies per reaction); 2) HIV-1 proviral DNAs detected by 5′LTR-to-3′LTR PCR were uniform in size and sequence; and 3) a single proviral integration site was found. To rule out the possibility of viral protein production from contaminant full-length proviruses in putative T cell clones harboring truncated defective proviruses with large internal deletions, the absence of full-length proviruses was confirmed by a nested PCR that specifically amplified the env region of the HIV-1 genome, a region that was frequently deleted in the truncated defective proviruses (SI Appendix, Fig. S2). Of 15 HIV-1–positive individual H9MN clones isolated, 4 were selected as single-cell clones for further analyses.
Isolation of CD4+ T Cell Clones from an HIV-Infected Individual.
Cells used for T cell cloning were obtained from an HIV-infected 56-y-old man who was known to have been HIV-1 positive since 1994. He was admitted to the National Institute of Allergy and Infectious Diseases (NIAID) inpatient service to receive an antiretroviral regimen designed for treatment of multidrug-resistant HIV-1 and was started on dolutegravir, ritonavir-boosted darunavir, tenofovir, and emtricitabine 12 weeks prior to obtaining the blood sample used for T cell cloning. At that time, his plasma HIV-RNA levels had been suppressed to below 40 copies/mL for 8 weeks. Total CD4+ T cell counts ranged between 43 and 100 cells/µL during this time. CD4+ T cells were obtained by negative selection using the CD4+ T Cell Isolation Kit II (Miltenyi Biotec) of cryopreserved PBMCs. The CD4+ T cells were plated in 96-well plates at cell densities of 100 and 1,000 cells per well in X-VIVO 15 medium (Lonza) supplemented with 50 U/mL recombinant human IL-2 (Roche Diagnostics), 30 ng/mL anti-CD3 antibody (clone: UCHT1, BD Pharmingen), and 10 nM dolutegravir (Selleckchem.com) (to prevent any possible outgrowth of replication-competent HIV-1). Irradiated (30 Gy) autologous PBMCs were added as feeder cells (5 × 104 cells per well). Cells were initially cultured for 10 to 14 days. Cells from the wells exhibiting growth (visually inspected by microscopy) were transferred to 48-well plates and monitored for an additional 10 to 14 days. Half the volume of medium was replaced with fresh medium (without anti-CD3 antibody) whenever the medium turned a yellow-orange color. Small aliquots of cells from the wells positive for cell growth were tested for the presence of HIV-DNA by 5′LTR-to-3′LTR PCR. In order to obtain enough cells for the subsequent Western blot and confocal immunofluorescence microscopy analyses, CD4+ T cells expressing defective proviruses were further expanded by modification of a rapid expansion protocol (described in ref. 45). Briefly, cells from the wells positive for HIV-DNA (4 × 104 cells) were cocultured with a 625-fold excess of irradiated (30 Gy) autologous PBMC feeder cells obtained from the donor 16 months after he had achieved a pVL <40 copies/mL. The cells were cultured in standing T25 flasks in 25 mL complete medium consisting of X-VIVO 15 medium (Lonza) supplemented with 50 U/mL recombinant human IL-2 (Roche Diagnostics), 50 ng/mL anti-CD3 antibody (clone: UCHT1, BD Pharmingen), and 10 nM dolutegravir (Selleckchem.com). The success of T cell cloning from the HIV-infected individual was determined using the same criteria used for the H9MN cloning.
Data Availability.
All data are included in the manuscript and SI Appendix. Sequence information has been provided to GenBank (accession nos. MN887608–MN887638).
Supplementary Material
Acknowledgments
We thank R. Dewar, H. Highbarger, and A. Shah for running the Abbott HIV-1 assay; M. Bosche for coordinating whole-genome sequencing; T. Zhai for help with Sanger sequencing; and U. O’Doherty, S. Migueles, F. Maldarelli, and S. Hughes for helpful discussions. This work was funded in part through the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, NIH, and in part with federal funds from the National Cancer Institute, NIH, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
The authors declare no competing interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. MN887608–MN887638).
*A. Violari, TUPDB0106LB, 9th IAS Conference on HIV Science, Paris, 2017.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917876117/-/DCSupplemental.
References
- 1.Imai H., et al. , A defective proviral DNA with a 2.6-kb deletion of human immunodeficiency virus type 1 (HIV-1) in a persistently HIV-1 infected cell clone. Virus Genes 5, 81–88 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Li Y., et al. , Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: Identification of replication-competent and -defective viral genomes. J. Virol. 65, 3973–3985 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez G., Xu X., Chermann J. C., Hirsch I., Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J. Virol. 71, 2233–2240 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ho Y. C., et al. , Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imamichi H., et al. , Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A. 113, 8783–8788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruner K. M., et al. , Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 22, 1043–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzon M. J., et al. , Long-term antiretroviral treatment initiated at primary HIV-1 infection affects the size, composition, and decay kinetics of the reservoir of HIV-1-infected CD4 T cells. J. Virol. 88, 10056–10065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Josefsson L., et al. , The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc. Natl. Acad. Sci. U.S.A. 110, E4987–E4996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray J. M., et al. ; PINT Study Team , HIV DNA subspecies persist in both activated and resting memory CD4+ T cells during antiretroviral therapy. J. Virol. 88, 3516–3526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imamichi H., et al. , Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS 28, 1091–1099 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Kearney M. F., et al. , Origin of rebound plasma HIV includes cells with identical proviruses that are transcriptionally active before stopping of antiretroviral therapy. J. Virol. 90, 1369–1376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finzi D., Plaeger S. F., Dieffenbach C. W., Defective virus drives human immunodeficiency virus infection, persistence, and pathogenesis. Clin. Vaccine Immunol. 13, 715–721 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Besson G. J., et al. , HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin. Infect. Dis. 59, 1312–1321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golob J. L., et al. , HIV DNA levels and decay in a cohort of 111 long-term virally suppressed patients. AIDS 32, 2113–2118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendoza D., et al. , Comprehensive analysis of unique cases with extraordinary control over HIV replication. Blood 119, 4645–4655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollack R. A., et al. , Defective HIV-1 proviruses are expressed and can be recognized by cytotoxic T lymphocytes, which shape the proviral landscape. Cell Host Microbe 21, 494–506.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulware D. R., et al. ; INSIGHT Study Group , Higher levels of CRP, D-dimer, IL-6, and hyaluronic acid before initiation of antiretroviral therapy (ART) are associated with increased risk of AIDS or death. J. Infect. Dis. 203, 1637–1646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuller L. H., et al. ; INSIGHT SMART Study Group , Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. 5, e203 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverberg M. J., et al. , Risk of cancers during interrupted antiretroviral therapy in the SMART study. AIDS 21, 1957–1963 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Gallo R. C., et al. , Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224, 500–503 (1984). [DOI] [PubMed] [Google Scholar]
- 21.Shaw G. M., et al. , Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science 226, 1165–1171 (1984). [DOI] [PubMed] [Google Scholar]
- 22.Takebe Y., Telesnitsky A., Evidence for the acquisition of multi-drug resistance in an HIV-1 clinical isolate via human sequence transduction. Virology 351, 1–6 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Y., et al. , Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4, 134–146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bullen C. K., Laird G. M., Durand C. M., Siliciano J. D., Siliciano R. F., New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 20, 425–429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasternak A. O., et al. , Minor contribution of chimeric host-HIV readthrough transcripts to the level of HIV cell-associated gag RNA. J. Virol. 90, 1148–1151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glück J. M., Hoffmann S., Koenig B. W., Willbold D., Single vector system for efficient N-myristoylation of recombinant proteins in E. coli. PLoS One 5, e10081 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan C. R., Miglionico B. V., Engen J. R., Effects of HIV-1 Nef on human N-myristoyltransferase 1. Biochemistry 50, 3394–3403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts J. D., Bebenek K., Kunkel T. A., The accuracy of reverse transcriptase from HIV-1. Science 242, 1171–1173 (1988). [DOI] [PubMed] [Google Scholar]
- 29.Preston B. D., Poiesz B. J., Loeb L. A., Fidelity of HIV-1 reverse transcriptase. Science 242, 1168–1171 (1988). [DOI] [PubMed] [Google Scholar]
- 30.Hu W. S., Hughes S. H., HIV-1 reverse transcription. Cold Spring Harb. Perspect. Med. 2, a006882 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson P. N., et al. , Demystified. Human endogenous retroviruses. Mol. Pathol. 56, 11–18 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurth R., Bannert N., Beneficial and detrimental effects of human endogenous retroviruses. Int. J. Cancer 126, 306–314 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Hohn O., Hanke K., Bannert N., HERV-K(HML-2), the best preserved family of HERVs: Endogenization, expression, and implications in health and disease. Front. Oncol. 3, 246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young G. R., Stoye J. P., Kassiotis G., Are human endogenous retroviruses pathogenic? An approach to testing the hypothesis. BioEssays 35, 794–803 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins K. L., Baltimore D., HIV’s evasion of the cellular immune response. Immunol. Rev. 168, 65–74 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Mann J. K., et al. , Nef-mediated down-regulation of CD4 and HLA class I in HIV-1 subtype C infection: Association with disease progression and influence of immune pressure. Virology 468–470, 214–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pawlak E. N., Dikeakos J. D., HIV-1 Nef: A master manipulator of the membrane trafficking machinery mediating immune evasion. Biochim. Biophys. Acta 1850, 733–741 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Dirk B. S., et al. , HIV-1 Nef sequesters MHC-I intracellularly by targeting early stages of endocytosis and recycling. Sci. Rep. 6, 37021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howcroft T. K., Strebel K., Martin M. A., Singer D. S., Repression of MHC class I gene promoter activity by two-exon Tat of HIV. Science 260, 1320–1322 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Kamp W., Berk M. B., Visser C. J., Nottet H. S., Mechanisms of HIV-1 to escape from the host immune surveillance. Eur. J. Clin. Invest. 30, 740–746 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Wang T., et al. , Intracellular Nef detected in peripheral blood mononuclear cells from HIV patients. AIDS Res. Hum. Retroviruses 31, 217–220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferdin J., et al. , Viral protein Nef is detected in plasma of half of HIV-infected adults with undetectable plasma HIV RNA. PLoS One 13, e0191613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persaud D., et al. , Absence of detectable HIV-1 viremia after treatment cessation in an infant. N. Engl. J. Med. 369, 1828–1835 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xufré C., et al. , Low frequency of GITR+ T cells in ex vivo and in vitro expanded Treg cells from type 1 diabetic patients. Int. Immunol. 25, 563–574 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and SI Appendix. Sequence information has been provided to GenBank (accession nos. MN887608–MN887638).