Significance
5-Methylcytosine (m5C) is one of the most abundant RNA modifications, but its function in adult stem cell development remains poorly defined. This study shows that Ypsilon schachtel (YPS) promotes germ line stem cell (GSC) maintenance, proliferation, and differentiation in the adult Drosophila ovary by preferentially binding to m5C-containing RNAs. Highly conserved cold-shock domains (CSDs) of YPS and its human homolog Y box binding protein 1 (YBX1) exhibit preferential binding to m5C-containing RNAs through hydrophobic interactions. Human YBX1 can functionally replace YPS to promote GSC development in the Drosophila ovary, and overexpressing RNA-binding–defective YBX1 and YPS mutant proteins disrupts normal GSC development. Thus, this study provides insight into the importance of m5C RNA modification in adult stem cell development.
Keywords: m5C, GSC, RNA methylation, YPS, YBX
Abstract
5-Methylcytosine (m5C) is a RNA modification that exists in tRNAs and rRNAs and was recently found in mRNAs. Although it has been suggested to regulate diverse biological functions, whether m5C RNA modification influences adult stem cell development remains undetermined. In this study, we show that Ypsilon schachtel (YPS), a homolog of human Y box binding protein 1 (YBX1), promotes germ line stem cell (GSC) maintenance, proliferation, and differentiation in the Drosophila ovary by preferentially binding to m5C-containing RNAs. YPS is genetically demonstrated to function intrinsically for GSC maintenance, proliferation, and progeny differentiation in the Drosophila ovary, and human YBX1 can functionally replace YPS to support normal GSC development. Highly conserved cold-shock domains (CSDs) of YPS and YBX1 preferentially bind to m5C RNA in vitro. Moreover, YPS also preferentially binds to m5C-containing RNAs, including mRNAs, in germ cells. The crystal structure of the YBX1 CSD-RNA complex reveals that both hydrophobic stacking and hydrogen bonds are critical for m5C binding. Overexpression of RNA-binding–defective YPS and YBX1 proteins disrupts GSC development. Taken together, our findings show that m5C RNA modification plays an important role in adult stem cell development.
5-Methylcytosine (m5C) is one of the most abundant RNA modifications in eukaryotic cells (1). Such an RNA modification was once thought to be only common on rRNAs and tRNAs (2, 3), but with the development of high-throughput and next-generation sequencing technologies, the transcriptome-wide mapping of m5C on RNAs has shown that the majority of m5C sites are localized on mRNAs of animal and plant cells (4–9). Interestingly, m5C is situated mainly in the coding region and the vicinity of the translational start site in vertebrate and mammalian cells (4–6, 8, 9). The deposition of m5C in mRNAs is catalyzed primarily by the methyltransferase Misu/NSUN2 (10). ALYREF is the protein that recognizes m5C sites in mRNAs for their cytoplasmic export (6). Although Misu/NSUN2 was previously reported to be required for embryonic stem cell differentiation (11, 12), little is known about the functions of m5C RNA modification in adult stem cell development.
In Drosophila, Ypsilon schachtel (YPS) is the closely related homolog of human Y box binding protein 1 (YBX1) (13) (SI Appendix, Fig. S1). YBX1 has been shown to be important for late mouse embryonic development and tumorigenesis in humans (14, 15). YPS has been shown to bind to osk mRNA and regulate its localization and translation during late Drosophila oogenesis (16); however, whether YPS also regulates germ line stem cell (GSC) development in the Drosophila ovary has been unclear. In this study, we show that YPS promotes GSC maintenance, proliferation, and differentiation in the Drosophila ovary, and that its human homolog YBX1 can replace YPS function in supporting GSC development. More importantly, the cold-shock domains (CSDs) of YBX1 and YPS can preferentially bind to m5C-containing RNA in vitro using hydrophobic stacking interactions, and YPS also preferentially binds to m5C-containing RNAs in germ cells, including those mRNAs encoding known GSC self-renewal and differentiation factors. Therefore, our results show that the m5C modification in mRNAs plays an important role in GSC development.
Results
YPS Promotes GSC Maintenance, Proliferation, and Differentiation in the Drosophila Ovary.
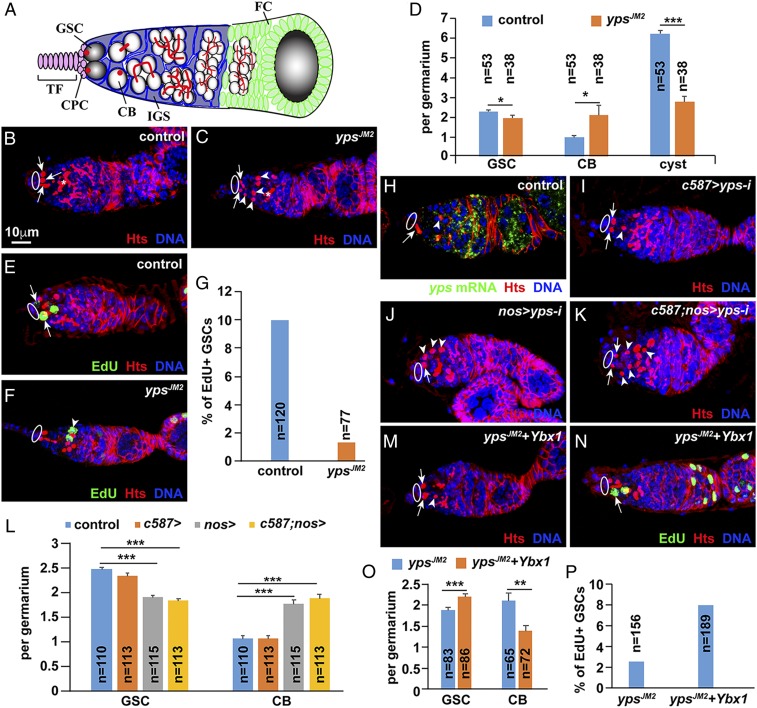
To determine whether YPS regulates GSC development in the Drosophila ovary, we immune-stained control and ypsJM2 mutant ovaries for Hts, a fusome component, to quantify GSCs and cystoblasts (CBs). ypsJM2 is a null mutation deleting the initiation ATG and CSD (16). In the ovary, two or three GSCs at the tip of the germarium continuously divide to generate CBs and cysts (17) (Fig. 1A). GSCs and CBs carry a spherical fusome known as the spectrosome, but only GSCs directly contact cap cells, whereas cysts contain a branched fusome (Fig. 1A). Interestingly, 2-wk-old yps mutant germaria contain significantly fewer GSCs and accumulate significantly more CBs than the control germaria (Fig. 1 B–D). The CB accumulation is indicative of the delayed differentiation into cysts (18, 19). In addition, 5-ethynyl-2′-deoxyuridine (EdU) labeling results show that yps mutant GSCs proliferate slower than normal (Fig. 1 E–G). Taken together, our results indicate that YPS is important for promoting GSC maintenance, proliferation, and progeny differentiation.
Fig. 1.
YPS is required intrinsically to promote GSC maintenance, proliferation, and differentiation in the Drosophila ovary. (A) Schematic diagram of the germarium: spectrosome (red dot) and fusome (red line). TF, terminal filament; CPC, cap cells; IGS, inner germarial sheath cells; FC, follicle cell. Ovals and arrows indicate cap cells and spectrosomes in GSCs, respectively, whereas arrowheads and asterisks denote spectrosomes in CBs and fusomes in the cysts, respectively. (B–D) Control germarium carries three GSCs and no CBs (B), while yps mutant germaria contains two GSCs and four CBs (C). (D) Quantification results. (E–G) yps mutant germaria contain fewer EdU-positive GSCs than controls. G shows quantification results. Arrows in E indicate EdU+ GSCs; arrowhead in F indicates EdU+ CB. (H) mRNA in situ hybridization results show that yps mRNA (green) is expressed primarily in GSCs, CBs, and cysts. (I–L) Germ line-specific, but not niche-specific, knockdown significantly decreases GSCs and increases CBs. L presents quantification results. (M–P) Ubiquitous expression of human Ybx1 can rescue the defects in GSC maintenance, proliferation, and differentiation in the yps mutant.
Our mRNA in situ hybridization results show that yps mRNA is expressed primarily in germ cells, including GSCs and CBs (Fig. 1H). We used c587-Gal4 and nos-gal4 to express a shRNA against yps to knock down its expression in niche cells and germ cells, respectively (20, 21), and found that germ line-specific, but not niche-specific, knockdown of yps can recapitulate the yps mutant GSC phenotypes (Fig. 1 I–L). These results indicate that YPS is required intrinsically for promoting GSC maintenance, proliferation, and differentiation.
Human YBX1 Can Functionally Replace YPS to Support GSC Development.
Drosophila YPS and human YBX1 share high homology in the CSD domain (SI Appendix, Fig. S1). We used Actin5c-gal4 to ubiquitously express human Ybx1 in the yps mutant ovary to explore whether YBX1 can functionally replace YPS. Interestingly, although it does not cause any discernible phenotypes in wild-type ovaries, ubiquitous Ybx1 expression significantly and fully rescues the GSC maintenance, proliferation, and differentiation defects in yps mutant ovaries (Fig. 1 M–P). These results indicate that human YBX1 can functionally replace YPS to support GSC development in the Drosophila ovary.
YPS Preferentially Binds to m5C-Containing RNAs in Germ Cells.
YBX1 and m5C methylase NSUN2 are required for loading tRNAs and rRNAs into exosomes, raising the possibility that YBX1 might bind to m5C-containing RNAs (22). To test this possibility, we used an anti-m5C antibody and germ line-expressed Flag-YPS to immunoprecipitate m5C-containing RNAs and YPS-bound mRNAs from adult ovaries, respectively (SI Appendix, Fig. S2). Interestingly, Flag-YPS and m5C antibody brought down 5,887 and 4,933 RNAs, respectively, which share 4,283 RNA species, suggesting that YPS binds to m5C-containing RNAs (Fig. 2A). In addition to rRNAs and tRNAs known to carry m5C modifications, other noncoding RNA species, such as long noncoding RNAs (lncRNAs), microRNA precursors (premiRNAs), antisense RNAs (asRNAs), small nucleolar RNAs (snoRNAs), and small nuclear RNAs (snRNAs), also carry m5C modifications based on Flag-YPS and anti-m5C pulldown results (Fig. 2 B–G). Interestingly, >85% of the m5C-containing RNAs are mRNAs, indicating that most of m5C modification sites are on mRNAs (Fig. 2H). These results indicate that YPS preferentially bind to m5C-containing RNAs in Drosophila.
Fig. 2.
Drosophila YPS preferentially bind to m5C-containing RNAs in germ cells. (A–H) Germ line-specific Flag-YPS and anti-m5C antibody pull down mostly overlapped total RNAs, rRNAs, tRNAs, LncRNAs, premiRNAs, asRNAs, snoRNAs, snRNAs, and mRNAs (numbers represent different RNAs). (I and J′) YPS-binding sites are closely correlated with m5C sites in mad and eIF4A mRNAs (I and I′) and in mei-P26 and sxl mRNAs (J and J″). (K and L) m5C sites and YPS-binding sites are more prevalent in the CDS and intron than in the 5′ UTR and the 3′ UTR. (M) Top 15 significantly overrepresented pathways for the mRNAs sharing YPS-binding sites and m5C sites based on KEGG pathway analysis.
Since RNAs were fragmented before the pulldown experiments, each peak likely represents one m5C site (SI Appendix, Fig. S2). Consistently, the RNA peaks pulled down by YPS and anti-m5C are almost perfectly correlated on individual mRNAs (Fig. 2 I and J′). For the mRNAs encoding known GSC self-renewal factors, mad carries m5C modifications in the coding sequence (CDS) only, while eIf4A has m5C modifications in the 3′ UTR and CDS (Fig. 2 I and I′). For the mRNAs encoding known differentiation factors, mei-P26 carries m5C sites in the 5′ UTR, whereas sxl contains m5C sites in the intron, CDS, and 3′ UTR (Fig. 2 J and J′). The majority of m5C sites on mRNAs are located in the CDS, and the remaining sites reside in the intron, 5′ UTR, and 3′ UTR (Fig. 2 K and L). Based on KEGG pathway analysis, the m5C-containing mRNAs are significantly enriched for those encoding the proteins involved in protein synthesis, mRNA splicing, endocytosis, and other functions (Fig. 2M). These results indicate that m5C sites are abundantly located in the CDS and intron, suggesting m5C’s potential importance in translation and splicing.
YBX1 CSD Exhibits Preferential Binding toward the m5C-Containing RNA Primarily through Hydrophobic Stacking Interactions.
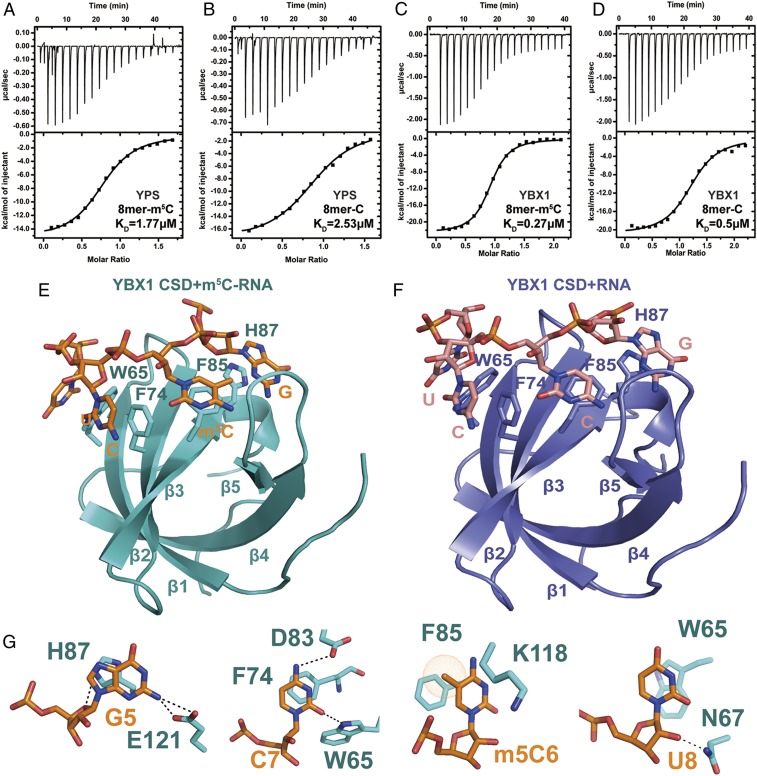
Since highly conserved CSDs in YBX family proteins are known to bind RNAs (23), we used isothermal titration calorimetry (ITC) to determine the binding affinity of YBX1 and YPS CSDs for m5C-containing and unmethylated RNAs. The 8-mer m5C and unmethylated RNA oligos 5′-A1C2C3A4G5m5C6C7U8-3′ and 5′-A1C2C3A4G5C6C7U8-3′ were chosen based on the previously reported YBX1 target sequence (22). YPS and YBX1 CSDs exhibited ∼1.5-fold and 2-fold higher affinity, respectively, for the m5C-containing RNA than for the unmethylated RNA (Fig. 3 A–D). Along with the in vivo YPS results, these results indicate that YBX1/YPS CSDs function as the reader for m5C RNA.
Fig. 3.
The CSDs of Drosophila YPS and human YBX1 preferentially bind to the m5C-containing RNA. (A and B) ITC analysis of Drosophila YPS binding to m5C-containing and RNA oligos. (C and D) ITC analysis of human YBX1 binding to m5C-containing and RNA oligos. (E and F) Overall structure of the human YBX1 CSD domain in complex with to m5C-containing and RNA oligos. The m5C RNA is shown in sticks, while the YBX1 CSD is shown in cartoon. The aromatic residues that interact with the RNA bases are indicated. (G) Detailed presentation of the interactions between YBX1 CSD domain aromatic residues and individual m5C RNA bases. The residues in the CSD domain and the m5C RNA bases are shown in cyan and orange, respectively.
Since the YPS CSD is highly similar to the YBX1 CSD (SI Appendix, Fig. S1), we solved the crystal structure of the YBX1 CSD in complex with the 8-mer methylated or unmethylated RNA oligo at a resolution of 2.0 Å and 1.4 Å, respectively. Both of the overall structures belong to the P62 space group and adopt a canonical five-stranded antiparallel β-barrel CSD fold, which is similar to the previously determined bacterial CSD-RNA crystal structure (24). As the RNA wraps around the CSD, four bases of either the methylated or unmethylated RNA, (G5, C6, C7, and U8) establish π-stacking interactions with the aromatic side chains of CSD, including Trp65, Phe74, Phe85, and His87, which are conserved in the Drosophila YPS (Fig. 3 E and F and SI Appendix, Fig. S1). In addition, multiple hydrogen bonds were detected between the RNA and the CSD (Fig. 3G). Interestingly, Phe85 stacks with the base of m5C6, and a weak cation–π interaction between residue Lys118 and m5C6 also exists. Therefore, our structural results suggest that the methyl group in m5C likely strengthens hydrophobic interactions with the CSD, thereby enhancing YBX1/YPS binding to m5C-containing RNAs.
We next determined whether Trp65, Phe74, Phe85, His87, and Lys118 in the YBX1 CSD are important for m5C-containing RNA binding by mutating them to alanine. The ITC binding data show that the YBX1 CSD carrying a mutation in W65, F74, F85, and H87 has diminished binding affinity to both the m5C-containing and unmethylated RNAs, suggesting that they contribute to general RNA binding (SI Appendix, Fig. S3 A and B). Interestingly, the K118A mutant decreases RNA binding only slightly, suggesting that Lys118 contributes less to RNA binding (SI Appendix, Fig. S3 A and B). Consistently, the W68A and F88A mutant YPS CSDs (equivalent to W65A and F85A, respectively, in YBX1) also lose RNA binding (SI Appendix, Fig. S3 C and D′). Taken together, these results further confirm the importance of the aromatic amino acid residues in the CSD for RNA binding.
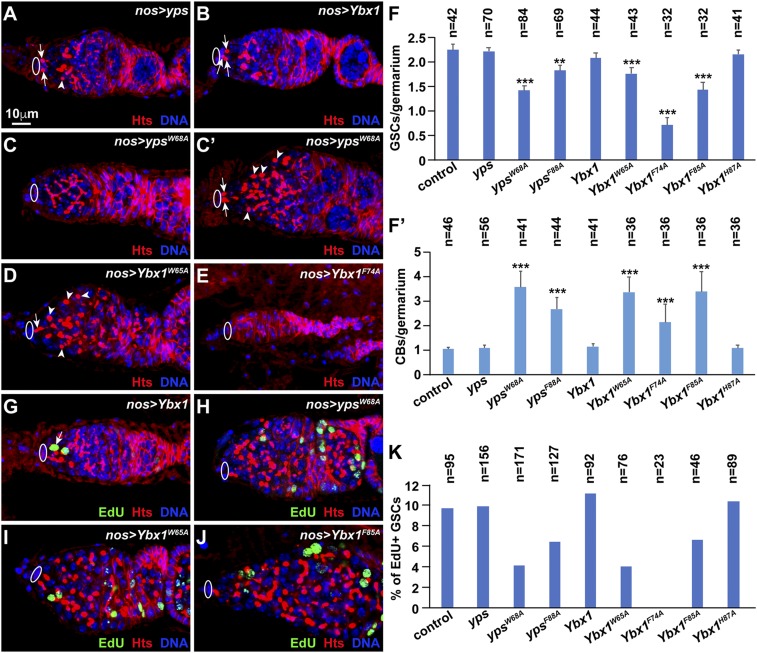
Overexpression of RNA-Binding–Defective Mutant YBX1 and YPS Proteins Severely Disrupts GSC Development.
We next used c587-Gal4 and nos-gal4 to test the functions of RNA-binding–defective yps and Ybx1 mutants by overexpressing them in niche cells and germ cells, respectively. Overexpressed wild-type yps and Ybx1 in either germ cells or niche cells as well as overexpressed Ybx1W65A, Ybx1F74A, and Ybx1F85A in niche cells behaved like the controls (Fig. 4 A and B and SI Appendix, Fig. S4). In contrast, germ line-specific overexpression of ypsW68A, ypsF88A, Ybx1W65A, Ybx1F74A, and Ybx1F85A caused a significant loss of GSC, a decrease in EdU-positive GSCs, and an increase in CBs, indicating that these mutants can dominantly disrupt GSC maintenance, proliferation, and differentiation (Fig. 4 C–K). Germ line-expressed Flag-YPS, Flag-YPSW68A, and Flag-YPSF88A could still bind similar protein partners in germ cells, suggesting that the GSC developmental defects likely arise from titrating out the interacting partners from endogenous wild-type YPS protein (SI Appendix, Fig. S5). Surprisingly, overexpression of YbxH87A in germ cells did not produce any obvious GSC developmental defects, suggesting that His87 does not contribute to the function of YBX1 in vivo, even though it is essential for RNA binding in vitro (Fig. 4 F, F′, and K). Taken together, our results indicate that Trp65, Phe74, and Phe85 in YBX1 and YPS are critical for their functions in vivo.
Fig. 4.
Germ line-specific expression of RNA binding-defective yps and Ybx1 mutants severely compromises GSC development. Ovals indicate cap cells, whereas arrows and arrowheads point to spectrosomes in GSCs and CBs, respectively. (A–F′) Germ line-specific expression of ypsW68A, Ybx1w65A, and Ybx1F74A, but not of Ybx1H87A, significantly decreases GSCs and increases CBs compared with overexpression of wild-type yps and Ybx1. F and F′ show results. Some yps and Ybx1 mutant-expressing germaria contain no GSCs (C and E) or one GSC (D), whereas other germaria accumulate many CBs (C′ and D). (G–K) Germ line-specific expression of yps or Ybx1 mutants except Ybx1H87A decreases EdU+ GSCs compared with controls. In G, the arrow indicates EdU+ GSC. K shows quantification results.
Discussion
The m5C RNA modification has recently been shown to be widespread in plant and human cells (4–9). This modification is involved in regulating various biological processes, including embryonic development, maternal-to-zygotic transition, and tumorigenesis, by modulating mRNA stability and translation (4–6, 8); however, whether it regulates adult stem cell development has been unclear. In this study, we show that YPS promotes GSC maintenance, proliferation, and differentiation in the Drosophila ovary by preferentially binding to m5C-containing RNAs. Human YBX1 can functionally replace YPS to support GSC development in the Drosophila ovary. In germ cells, YPS preferentially binds to m5C-containing RNAs, most of which are mRNAs, including those encoding known GSC self-renewal and differentiation factors.
In addition, we used crystal structures to show that the YBX1 CSD binds to the RNA primarily through hydrophobic stacking interactions, and that m5C helps strengthen such hydrophobic interactions. Finally, forced expression of RNA binding-defective YPS or YBX1 mutants in germ cells sufficiently dismantles GSC development in a dominant-negative manner, indicating that m5C binding is important for YPS to regulate GSC development. Therefore, our findings provide insight into the importance of YBX family protein YPS in adult GSC development by preferentially recognizing m5C-containing RNAs.
This study provides important structural information on how YPS and YBX1 proteins preferentially recognize m5C-containing RNAs. First, our RNA-immunoprecipitation results show that YPS preferentially binds to m5C-containing RNAs, including mRNAs. Approximately 88% of mRNAs and 75% of noncoding RNAs bound by YPS are m5C-containing RNAs, indicating that YPS preferentially binds to m5C-containing RNA in vivo. This finding is consistent with the two studies on YBX1 in zebrafish and humans, which were published during the revision of this current study (4, 5) and challenges the previous notion that YBX family proteins are low-specificity RNA chaperones (25). Second, our ITC results show that both YBX1 and YPS CSDs preferentially bind to m5C-containing RNAs over unmethylated RNAs. Third, crystal structures show that four aromatic amino acid residues form hydrophobic stacking interactions with four RNA bases, and that m5C can further strengthen the stacking interactions. Consistently, mutating one of the four amino acid residues involved in hydrophobic stacking interactions (Trp65, Phe74, Phe85, or His87) completely abolishes the RNA binding of the YBX1 CSD. Finally, YBX1 mutants changing Trp65, Phe74, or Phe85 into alanine behave dominant-negatively as the corresponding YPS mutants interfere with YPS function in the Drosophila ovary. Since YBX family CSDs are highly conserved, our results can help explain how YBX family proteins bind to m5C-containing RNAs.
This study also provides important insight into how YPS regulates adult GSC development. YPS has previously been shown to bind to osk mRNA and regulate its localization and translation in the oocyte (16). Our findings reveal that YPS is required intrinsically to promote GSC maintenance, proliferation, and differentiation in the Drosophila ovary, since yps null mutant germaria have significantly fewer GSCs and EdU-positive GSCs and more CBs compared with control germaria. In addition, cell type-specific shRNA-mediated knockdown indicates that YPS is required intrinsically to support GSC development. Moreover, human YBX1 can functionally replace YPS to support GSC development in the ovary, since ubiquitous expression of human Ybx1 in the yps mutant can fully rescue all the GSC developmental defects. In addition, germ line-specific expression of RNA binding-defective YPS and YBX1 mutant proteins can similarly disrupt GSC development in the ovary. Interestingly, YBX2 and YBX3 are required for mouse spermatogenesis, suggesting that the functions of YBX proteins in germ cell development are conserved (26).
Another important finding is that YPS promotes GSC development through binding to m5C-containing RNAs, including mRNAs and noncoding RNAs. Some of YPS-bound mRNAs encode known self-renewal factors (e.g., Mad, eIF4A) and differentiation-promoting factors (e.g., Mei-P26, Sxl), which could potentially explain its requirement for GSC development. Interestingly, YPS more frequently binds to the CDS and intron in mRNAs than to the 5′ UTR and 3′ UTR, suggesting that YPS preferentially regulates mRNA translation and splicing. YPS-bound noncoding RNAs include tRNAs, rRNAs, miRNA precursors, lncRNAs, snoRNAs, and snRNAs, which are known to regulate translation, RNA splicing, or transcription. Although YBX1 is proposed to bind to DNA in mammalian cells, YPS protein is present almost exclusively in the cytoplasm of Drosophila ovarian germ cells, and there is little m5C DNA modification and DNA methylase Dmnt2 protein expression in adult Drosophila females based on the published results (16, 27), suggesting that YPS regulates GSC development primarily through binding to m5C-containing RNAs. In addition, it will be interesting to investigate whether YPS binding influences m5C modification on RNAs.
Taken together, our findings suggest that YPS regulates translation, splicing, and stability by preferentially recognizing m5C-containing RNAs, thereby promoting GSC development in the Drosophila ovary. Since YPS/YBX family protein functions have been conserved from Drosophila to humans, the Drosophila ovary will be an effective system for dissecting the functions of human YBX family proteins and m5C modifications in germ cell development.
Materials and Methods
Drosophila Stocks.
Flies were maintained and crossed at room temperature on standard cornmeal/molasses/agar media unless specified otherwise. To maximize the expression levels of shRNAs and transgenes, newly eclosed flies were cultured at 29 °C for 2 wk before the analysis of ovarian phenotypes. Further details on the Drosophila mutant and transgenic strains can be found in SI Appendix, Materials and Methods.
Immunostaining, EdU Labeling and mRNA In Situ Hybridization.
Ovaries were dissected at room temperature in Grace’s medium and fixed with 4% paraformaldehyde. Immunostaining, EdU detection, and mRNA in situ hybridization were performed as described in SI Appendix, Materials and Methods.
Identification of m5C-Containing and YPS-Bound RNAs.
For each RNA pulldown experiment, 400 pairs of ovaries were dissected from the females of proper genotypes for RNA immunoprecipitation by anti-m5C and anti-Flag antibodies, RNA sequencing, and bioinformatic analysis, as described in SI Appendix, Materials and Methods.
Protein Purification, ITC Measurement, and Crystal Structure Determination.
Drosophila YPS (amino acids 58 to 132) and human YBX1 (amino acids 52 to 129) CSD domains were cloned, expressed in Escherichia coli, and purified for the ITC measurement for RNA binding and for the determination of crystal structures of human CSD complexed with methylated and unmethylated RNAs, as described in SI Appendix, Materials and Methods.
Data Availability.
X-ray structures have been deposited in the RCSB Protein Data Bank (ID codes 6KTC for YBX1-m5C RNA and 6KUG for YBX1-C RNA). Original RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE139828). Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1489.
Supplementary Material
Acknowledgments
We thank the BL19U beamline staff at the Shanghai Synchrotron Radiation Facility for assistance during data collection, and Tulle Haelrigg, Allan Spradling, the Bloomington Drosphila Stock Center, and the Developmental Studies Hybridoma Bank for reagents. This work was supported by the National Natural Science Foundation of China (Grant 31870755, to S.L.), the Guangdong Innovation Research Team Fund (Grant 2016ZT06S172, to S.L.), the Shenzhen Sci-Tech Fund (Grant KYTDPT20181011104005, to S.L.), the NIH (Grant R01 HD097664, to T.X.), and the Stowers Institute for Medical Research (T.X.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. H.L. is a guest editor invited by the Editorial Board.
Data deposition: X-ray structures have been deposited in the RCSB Protein Data Bank (ID codes 6KTC for YBX1-m5C RNA and 6KUG for YBX1-C RNA) and original RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE139828). Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1489.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910862117/-/DCSupplemental.
References
- 1.Boccaletto P., et al. , MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubin D. T., Taylor R. H., The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 2, 1653–1668 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubin D. T., Stollar V., Methylation of Sindbis virus “26S” messenger RNA. Biochem. Biophys. Res. Commun. 66, 1373–1379 (1975). [DOI] [PubMed] [Google Scholar]
- 4.Yang Y., et al. , RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell. 75, 1188–1202.e11 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Chen X., et al. , 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21, 978–990 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Yang X., et al. , 5-methylcytosine promotes mRNA export: NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 27, 606–625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David R., et al. , Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell 29, 445–460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amort T., et al. , Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 18, 1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Squires J. E., et al. , Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brzezicha B., et al. , Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 34, 6034–6043 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanco S., et al. , The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 7, e1002403 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain S., et al. , The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol. Cell. Biol. 33, 1561–1570 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilhelm J. E., et al. , Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J. Cell Biol. 148, 427–440 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fotovati A., et al. , YB-1 bridges neural stem cells and brain tumor-initiating cells via its roles in differentiation and cell growth. Cancer Res. 71, 5569–5578 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Lu Z. H., Books J. T., Ley T. J., YB-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol. Cell. Biol. 25, 4625–4637 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mansfield J. H., Wilhelm J. E., Hazelrigg T., Ypsilon Schachtel, a Drosophila Y-box protein, acts antagonistically to Orb in the oskar mRNA localization and translation pathway. Development 129, 197–209 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Xie T., Control of germ line stem cell self-renewal and differentiation in the Drosophila ovary: Concerted actions of niche signals and intrinsic factors. Wiley Interdiscip. Rev. Dev. Biol. 2, 261–273 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Shen R., Weng C., Yu J., Xie T., eIF4A controls germ line stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc. Natl. Acad. Sci. U.S.A. 106, 11623–11628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S., et al. , Lissencephaly-1 controls germ line stem cell self-renewal through modulating bone morphogenetic protein signaling and niche adhesion. Proc. Natl. Acad. Sci. U.S.A. 107, 19939–19944 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song X., et al. , Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germ line stem cells in the Drosophila ovary. Development 131, 1353–1364 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Van Doren M., Williamson A. L., Lehmann R., Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8, 243–246 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Kossinova O. A., et al. , Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim. Biophys. Acta. Proteins Proteomics 1865, 664–673 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto K., Wolffe A. P., Gene regulation by Y-box proteins: Coupling control of transcription and translation. Trends Cell Biol. 8, 318–323 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Jiang W., Hou Y., Inouye M., CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272, 196–202 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Lindquist J. A., Mertens P. R., Cold shock proteins: From cellular mechanisms to pathophysiology and disease. Cell Commun. Signal. 16, 63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder E., et al. , Compound heterozygosity for Y box proteins causes sterility due to loss of translational repression. PLoS Genet. 11, e1005690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunert N., Marhold J., Stanke J., Stach D., Lyko F., A Dnmt2-like protein mediates DNA methylation in Drosophila. Development 130, 5083–5090 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
X-ray structures have been deposited in the RCSB Protein Data Bank (ID codes 6KTC for YBX1-m5C RNA and 6KUG for YBX1-C RNA). Original RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE139828). Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1489.