Significance
The cell of origin and the temporal order of oncogenic events in tumors play important roles for disease state. This is of particular interest for PCa due to its highly variable clinical outcome. However, these features are difficult to analyze in tumors. We established an in vitro murine PCa organoid model taking into account the cell of origin and the temporal order of events. We found that TIP5 primes luminal prostate cells for Pten-loss mediated oncogenic transformation whereas it is dispensable once the transformation is established. Cross-species transcriptomic analyses revealed a PTEN-loss gene signature that identified a set of aggressive tumors with PTEN-del, or low PTEN expression, and high-TIP5 expression. This paper provides a powerful tool to elucidate PCa mechanisms.
Keywords: prostate cancer, organoids, PTEN, TIP5, BAZ2A
Abstract
Prostate cancer (PCa) is the second leading cause of cancer death in men. Its clinical and molecular heterogeneities and the lack of in vitro models outline the complexity of PCa in the clinical and research settings. We established an in vitro mouse PCa model based on organoid technology that takes into account the cell of origin and the order of events. Primary PCa with deletion of the tumor suppressor gene PTEN (PTEN-del) can be modeled through Pten-down-regulation in mouse organoids. We used this system to elucidate the contribution of TIP5 in PCa initiation, a chromatin regulator that is implicated in aggressive PCa. High TIP5 expression correlates with primary PTEN-del PCa and this combination strongly associates with reduced prostate-specific antigen (PSA) recurrence-free survival. TIP5 is critical for the initiation of PCa of luminal origin mediated by Pten-loss whereas it is dispensable once Pten-loss mediated transformation is established. Cross-species analyses revealed a PTEN gene signature that identified a group of aggressive primary PCas characterized by PTEN-del, high-TIP5 expression, and a TIP5-regulated gene expression profile. The results highlight the modeling of PCa with organoids as a powerful tool to elucidate the role of genetic alterations found in recent studies in their time orders and cells of origin, thereby providing further optimization for tumor stratification to improve the clinical management of PCa.
PCa is the second leading cause of death from cancer in man (1). In the current era of PSA screening, nearly 90% of PCas are clinically localized at the time of their diagnosis (2). However, the clinical behavior of a localized PCa is highly variable. Immediate treatment is recommended for patients whose tumor biopsies are assigned a high Gleason score. In contrast, patients with tumor biopsies with a low Gleason score are considered indolent and monitored without therapeutic intervention to minimize overtreatment (3). The concern is, however, that active surveillance may miss the opportunity for early intervention of tumors that are seemingly low risk but actually are aggressive (4). It is now clear that the heterogeneity observed in the clinic is underpinned by a heterogeneous molecular landscape where genomic rearrangements and epigenetic alterations combine to amplify transcriptomic diversity (5–8). This complexity makes PCa still resilient to classification into molecular subtypes associated with distinct disease outcomes. At present, a major clinical challenge for PCa management is the identification of early biomarkers able to distinguish truly aggressive from indolent PCa and targets to be used in PCa resistant to conventional treatment regimens, such as castration resistant PCa (CRPC). A considerable limitation for the progress of these clinical applications is that the underlying mechanisms of PCa remain still elusive.
The identification of the cell type of the origin of cancer and the molecular drivers of tumor initiation is of fundamental importance in understanding the basis of distinct tumor subtypes as well as for tumor stratification and personalized treatment (9–11). In the case of PCa, the cell of the origin model is of particular interest since patients with low to intermediate grade primary tumors can have widely different outcomes (12). The adult prostate epithelium is composed of luminal, basal, and rare neuroendocrine cells (13). Prostate adenocarcinoma displays a luminal phenotype with loss of basal cells (12). There is now considerable evidence supporting a luminal origin for prostate cancer, whereas basal cells are thought to only give rise to tumors after differentiation into luminal cells (14–18). Furthermore, it has been suggested that tumors with luminal origin might correlate with aggressive disease and poor disease outcome (19).
Genomic analyses have started to dissect the heterogeneous molecular landscape of PCa. A recent comprehensive analysis of primary PCa revealed a molecular taxonomy in which 74% of tumors fell into one of seven subtypes defined by specific gene fusions (ERG, ETV1/4, and FLI1) or mutations (SPOP, FOXA1, and IDH1) (7). Recent analyses from a cohort of metastatic CRPCs identified genomic alterations, such as PIK3CA/B, R-spondin, BRAF/RAF1, APC, and β-catenin, offering clinically actionable information that could impact treatment decisions (8). However, these data alone cannot help to assess biomarkers of disease outcome and drug sensitivity since drugs against PCa cannot be tested effectively other than in clinical trials. Indeed, and in contrast to other cancer types, there is a lack of in vitro PCa models that recapitulate the diversity of human PCa due to the inability to reliably maintain PCa cells in culture (20).
Recently, new approaches have been developed for 3D culture of prostate epithelial cells termed “organoids” (21). This culture system uses conditions that enhance the survival and differentiation of stem/progenitor populations and exhibit spatial organ architecture with structures resembling the corresponding in vivo organs (22–24). Recent studies have shown the successful long-term culture of human PCa organoids from biopsy of metastatic and circulating tumor cells (20, 25). These PCa organoid lines recapitulated the the original patient sample and provided the basis for the generation of patient-derived PCa lines amenable to pharmacologic studies. However, the establishment of organoids from primary PCa is still technically challenging, owing to the overgrowth of nonmalignant prostate epithelial cells that are present within each sample (26).
Recently, we showed that TIP5 (BAZ2A), a subunit of the nucleolar-remodeling complex NoRC, is implicated in aggressive PCa (27). TIP5 is highly expressed in metastatic tumors and is required for cell proliferation, viability, and invasion. Furthermore, high TIP5 levels in tumors associate with poor prognosis and disease recurrence (27). TIP5 establishes epigenetic silencing at genes frequently repressed in metastatic PCa, and tumors with high TIP5 expression display a CpG island methylator phenotype.
In this paper, we showed that TIP5 regulates gene expression specifically in prostatic luminal cells. We found that TIP5 high expression correlates with primary PCa with PTEN-del, the most commonly lost tumor suppressor gene in primary PCa. Tumors with high TIP5 expression and PTEN-del are strongly associated with reduced PSA recurrence-free survival. We used organoid technology to elucidate the role of TIP5 in PCa initiation and found that TIP5 is critical for the initiation of PCa of luminal origin whereas it is dispensable once Pten-mediated transformation is established. Cross-species gene expression analyses revealed a PTEN gene signature that identified a group of primary PCas characterized by PTEN-del or low expression and high TIP5 expression and that is also regulated by TIP5. This PTEN gene signature is indicative of dedifferentiation states and aggressive disease as evidenced by the high Gleason score of the identified PCa group. These results highlight the in vitro modeling of PCa with organoids as a powerful mean to elucidate the role of oncogenic hits in their time order and cell of origin thereby improving tumor stratification and providing a potential informative value for the choice of initial therapeutic strategies in the clinical management of PCa.
Results
TIP5 Regulates Prostate Luminal Cells.
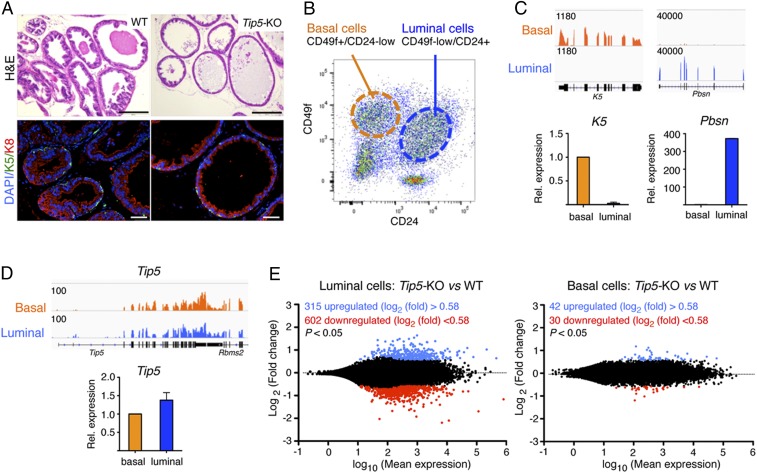
To determine whether TIP5 is implicated in prostate epithelial differentiation, we analyzed morphology and cell composition of prostate ducts in wild-type (WT) and Tip5-knockout (KO) mice at different postnatal developmental stages: before puberty (less than 6-wk old), just after reaching sexual maturity (8–10-wk old), and in mature adulthood (3–6 mo of age and more). Hematoxylin and eosin (H&E) staining and immunofluorescence (IF) analyses for basal marker keratin 5 (K5) and luminal marker keratin 8 (K8) revealed that the duct morphology and cell composition of 6-wk and 10-wk old WT and Tip5-KO mice was very similar (SI Appendix, Fig. S1A). In contrast, prostate duct structure of 6-mo-old Tip5-KO mice (two of two total) was drastically altered compared to the control group (Fig. 1A). Prostate ducts of Tip5-KO animals were dilated and had increased the diameter and thinner luminal epithelium layer, suggesting defects in prostate epithelium maintenance.
Fig. 1.
TIP5 plays a major role in luminal prostate cells. (A) TIP5 is required for prostate epithelium maintenance. H&E and IF staining of basal K5 and luminal K8 of the mouse ventral prostate from 6-mo-old WT and Tip5-KO mice. (Scale bars represent 200 μm [H&E] and 50 μm [IF].) (B) Representative FACS plots of CD49f+/CD24-low (basal) and CD49f-low/CD24+ (luminal) stained murine prostate cells. (C, Top) Integrative Genomics Viewer (IGV) browser view of RNAseq reads mapped at K5 and Pbsn loci in basal and luminal cells. (C, Bottom) Quantitative (q)RT-PCR expression analysis of K5 and Pbsn in isolated basal and luminal cells. Expression was normalized to Gapdh. Average expression value of K5 (three experiments) and Pbsn (two experiments). Error bars represent s.d. (D) TIP5 is expressed in both prostate luminal and basal cells. (D, Top) IGV view of RNAseq reads mapped at the Tip5 locus. (D, Bottom) qRT-PCR analysis of Tip5 expression. Values were normalized to Gapdh mRNA levels. Average value of three experiments. Error bars represent s.d. (E) TIP5 regulates gene expression in prostate luminal cells. MA plots of RNAseq data showing differentially expressed genes among luminal and basal WT and Tip5-KO cells.
To determine how TIP5 regulates prostate epithelial cells, we first analyzed TIP5 expression in the mouse prostate epithelium of 10-wk-old mice. Using fluorescence-activated cell sorting (FACS), we isolated basal and luminal cells from primary mouse prostate tissue based on the high expression of CD49f in basal cells and the expression of CD24 in luminal cells as previously described (24) (Fig. 1B and SI Appendix, Fig. S1B). The purity of sorted populations was verified with RNA sequencing (RNAseq) analysis and confirmed by qRT-PCR (Fig. 1C and SI Appendix, Fig. S1 C and D). Gene ontology (GO) analysis of genes significantly highly expressed in luminal vs. basal cells revealed enrichment in biological pathways linked to immune system processes and inflammatory response (SI Appendix, Fig. S1 D and E and Datasets S1 and S2). In contrast, genes highly expressed in basal cells were mainly linked to developmental processes. Similar pathways were also found in a recent gene expression analysis of human benign prostatic basal and luminal cells (28, 29). Quantification of Tip5 levels indicated similar expression in both basal and luminal cells (Fig. 1D).
To determine how TIP5 affects prostate epithelium maintenance, we performed RNAseq analysis and compared gene expression of basal (CD49f+/CD24-low) and luminal (CD49f-low/CD24+) cell compartments from 10-wk-old WT and Tip5-KO mice prostates. We did not observe significant changes in the amounts of CD49f+/CD24-low and CD49f-low/CD24+ prostate cells between WT and Tip5 KO (SI Appendix, Fig. S1B). Remarkably, loss of TIP5 induced higher differential gene expression in luminal cells than in basal cells (Fig. 1E and Dataset S3). In Tip5-KO luminal cells, 917 genes showed significant transcriptional changes whereas only 72 genes were affected in Tip5-KO basal cells. Importantly, a large fraction of luminal genes were affected by Tip5 loss in luminal cells whereas only few basal genes were affected in Tip5-KO basal cells (SI Appendix, Fig. S1F). GO analysis revealed that genes down-regulated in Tip5-KO luminal cells are strongly enriched in the immune-system process, locomotion, response to wounding, and development whereas up-regulated genes are linked to ion transport and lipid metabolic processes (SI Appendix, Fig. S1G and Datasets S3 and S4). Taken together, these results suggest that TIP5 plays a major role in prostate luminal cells.
Tip5 Deletion Affects the Capacity of Mouse Prostate Epithelium Progenitor Cells to Form Prostate Organoids.
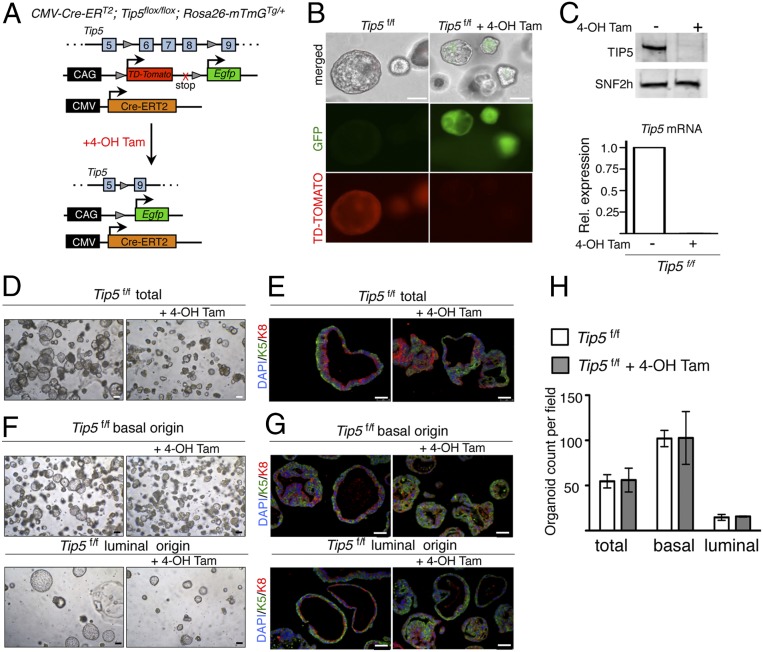
To determine how TIP5 affects the prostate epithelium, we used organoid culturing conditions (23, 24). Organoids were obtained by embedding in matrigel single cells from 10-wk-old mouse prostate tissue and culturing for 10 d according to published protocols (23, 24) (SI Appendix, Fig. S2A). Organoids generated from WT mice showed a ductal architecture resembling the prostate gland in vivo and were composed of an outer basal layer exclusively expressing basal prostate markers p63 and K5 and an inner luminal layer expressing luminal marker K8 (SI Appendix, Fig. S2 B and C). We generated a tamoxifen inducible whole-body Tip5 KO mouse line in which Tip5 exons 6–8 were flanked by loxP sites and Cre-ERT2 recombinase expression was driven by the cytomegalovirus (CMV) promoter (CMV-CreERT2; Tip5f/f) (Fig. 2A). These mice were combined with the reporter line Rosa26-mTmGTg/+ to generate the CMV-CreERT2, Tip5flox/flox, and Rosa26-mTmGTg/+ line (here after named TIP5f/f). We evaluated the efficiency of Cre recombinase activity in organoids by monitoring the tdTomato/GFP signal and TIP5 expression. Treatment of Tip5f/f prostate cells with 4-hydroxytamoxifen (4-OH Tam) was very efficient as shown by the complete loss of tdTomato+ cells, the gain of GFP+ cells, and strong reduction in Tip5 expression levels (Fig. 2 B and C and SI Appendix, Fig. S2D). We measured the ability of prostate progenitor cells to form organoids in the absence of TIP5. We generated organoids with the same number of total (not-sorted) WT or Tip5f/f prostate cells (Fig. 2 D and E and SI Appendix, Fig. S2E). Compared to Tip5f/f control organoids (not treated with 4-OH Tam) and WT control organoids treated with 4-OH Tam, Tip5-KO organoids (Tip5f/f + 4-OH Tam) displayed a smaller size and altered structures as evident by the lack of well-defined basal (K5+) and luminal (K8+) bilayered architectures, suggesting that TIP5 expression is required for a correct prostate cell differentiation. Next, we generated organoids using the same number of isolated basal and luminal cells from Tip5f/f mouse prostate tissue and cultured them in the absence or presence of 4-OH Tam (Fig. 2 F–H). Efficiency of recombination was assessed by visualization of tdTomato and GFP expressions (SI Appendix, Fig. S2F). Consistent with previous reports, organoids derived from basal and luminal cells displayed substantial differences (24, 30, 31). Basal cells generated more organoids than luminal cells (Fig. 2 F and H), indicating their higher competence as progenitors. In contrast, luminal cell-derived organoids were few and formed enlarged spheres (Fig. 2 F and H). Consistent with previous reports (23, 24), IF analysis revealed that organoids derived either from basal or from luminal cells expressed both basal and luminal markers, supporting the presence of stem or progenitor cells in both compartments (Fig. 2G). Although large organoids of luminal origin were underrepresented in IF analysis due to their extreme fragility in sample processing, smaller luminal-derived organoids displayed a clear bilayer structure, a result that is in line with previous reports (23, 24). Notably, Tip5 deletion had major effects in luminal-derived organoids (Fig. 2 F and G). Tip5-KO organoids of luminal origin were consistently smaller than control organoids whereas organoids of basal origin were not greatly affected in their size. Analysis of cell composition revealed a less clear bilayered organization in both basal- and luminal-derived Tip5-KO organoids (Fig. 2G). We did not observe significant changes in the number of organoids, indicating that TIP5 is not a prerequisite for organoids' formation, but its lack impairs proper development (Fig. 2H). These results suggest that, in the absence of TIP5, the capacity for prostate progenitors to form organoids is compromised. Moreover, the observation that the major defects were observed in luminal-derived organoids are consistent with the alteration in gene expression observed in TIP5-KO prostatic luminal cells (Fig. 1), indicating an important role of TIP5 in the prostate luminal compartment.
Fig. 2.
TIP5 is required to form prostate organoids. (A) Schematic of Cre-mediated recombination upon 4-OH Tam treatment in the CMV-Cre-ERT2, Tip5flox/flox, Rosa26-mTmGTg/+ (Tip5f/f) line. (B) Analysis showing expression of GFP protein and loss of the td-Tomato red protein signal in Tip5f/f organoids upon 4-OH Tam treatment. (Scale bars represent 150 μm.) (C, Top) Western blot analysis of organoids showing TIP5 levels in organoids derived from Tip5f/f and Tip5f/f + 4-OH Tam cells. SNF2H serves as the loading control. (C, Bottom) qRT-PCR analysis showing Tip5 mRNA levels. Values were normalized to Gapdh mRNA content. Average of two independent experiments. (D) Representative bright field images of Tip5f/f and Tip5f/f + 4-OH Tam-treated organoids derived from total prostate cells. (Scale bars represent 150 μm.) (E) IF analysis with anti-K5 and anti-K8 of organoids derived from total Tip5f/f prostate cells treated with 4-OH Tam. (Scale bars represent 50 μm.) (F) Representative bright field images of organoids derived from basal and luminal Tip5f/f cells treated with 4-OH Tam. (Scale bars represent 150 μm.) (G) IF analysis with anti-K5 and anti-K8 of organoids derived from basal and luminal Tip5f/f cells treated with 4-OH Tam. (Scale bars represent 50 μm.) (H) Quantification of organoids derived from total, basal, and luminal Tip5f/f cells treated with 4-OH Tam. Average value of three independent experiments. Error bars represent s.d.
TIP5 Is Required for the Initiation of PCa Driven by Pten-Loss in Organoids.
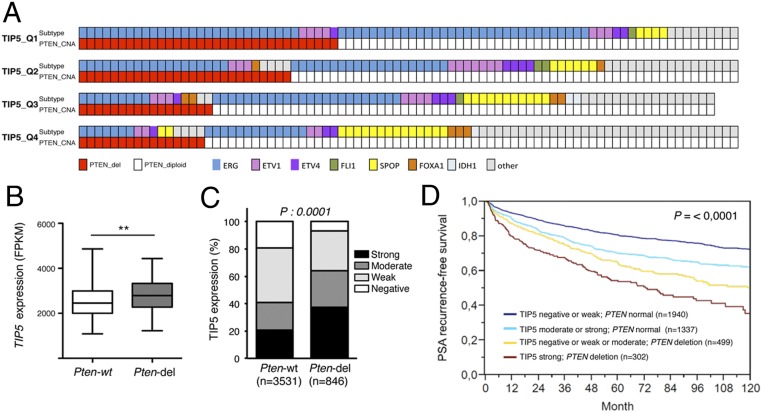
To elucidate the role of TIP5 in PCa, we analyzed TIP5 expression levels using a recent comprehensive analysis of more than 300 primary PCas (The Cancer Genome Atlas–Prostate Adenocarcinoma [TCGA–PRAD]) (7). We classified primary PCas according to TIP5 levels and found that the group with high TIP5 expression was enriched in tumors containing ERG fusions and PTEN copy number alterations (Fig. 3A and SI Appendix, Fig. S3A), supporting previous results showing that ERG gene fusions are seen early and most commonly with PTEN-loss (32). Consistent with these results, tumors with PTEN-del expressed higher TIP5 levels compared to tumors with intact PTEN (PTEN-wt) (Fig. 3B and SI Appendix, Fig. S3B). Next, we analyzed TIP5 protein levels on a large tissue microarray (TMA) where TIP5 expression and PTEN status were previously measured using immunostaining and DNA fluorescence in situ hybridization (FISH), respectively (27, 33). This earlier analysis was expanded to a total number of 6,803 samples informative for PTEN-del and 7,869 samples informative for TIP5 immunohistochemistry (IHC). Data on both TIP5 and PTEN were available from 4,377 samples. TIP5 expression was categorized as negative (17%), weak (38%), moderate (22%), or strong (24%). Some 846 out of these tumors displayed PTEN-del. The results revealed that TIP5 protein up-regulation in tumors is linked to PTEN-del (P < 0.0001, Fig. 3C). Remarkably, the time to postoperative PSA recurrence was significantly shorter in the group with strong TIP5 expression and PTEN-del (P < 0.0001) (Fig. 3D). These results indicate that TIP5 expression levels in primary PCa correlate with PTEN-del, and patients with high TIP5 levels and PTEN-del had the worst outcome.
Fig. 3.
TIP5 is highly expressed in primary tumors with PTEN-del. (A) PTEN status in PCa groups defined by TIP5 expression levels (e.g., quartile Q1: the top 25% of PCas with the highest TIP5 expression; Q4: the top 25% of PCas with the lowest TIP5 levels). The molecular subtypes of primary PCa (ERG fusions, ETV1/ETV4/FLI1 fusions, or overexpression, and SPOP/FOXA1/IDH1 mutations) defined in ref. 7 are shown. (B) TIP5 expression in primary prostate tumors with PTEN-del. Data are from ref. 7. Statistical significance was calculated using the unpaired two-tailed t test (**< 0.01). (C) TMA measurements of TIP5 expression levels in tumors with PTEN intact and deleted states. P values were obtained by the χ2 test. (D) Kaplan–Maier analysis of the time to postoperative PSA recurrence vs. TIP5 levels in all prostate tumors. P values were obtained by a log-rank test.
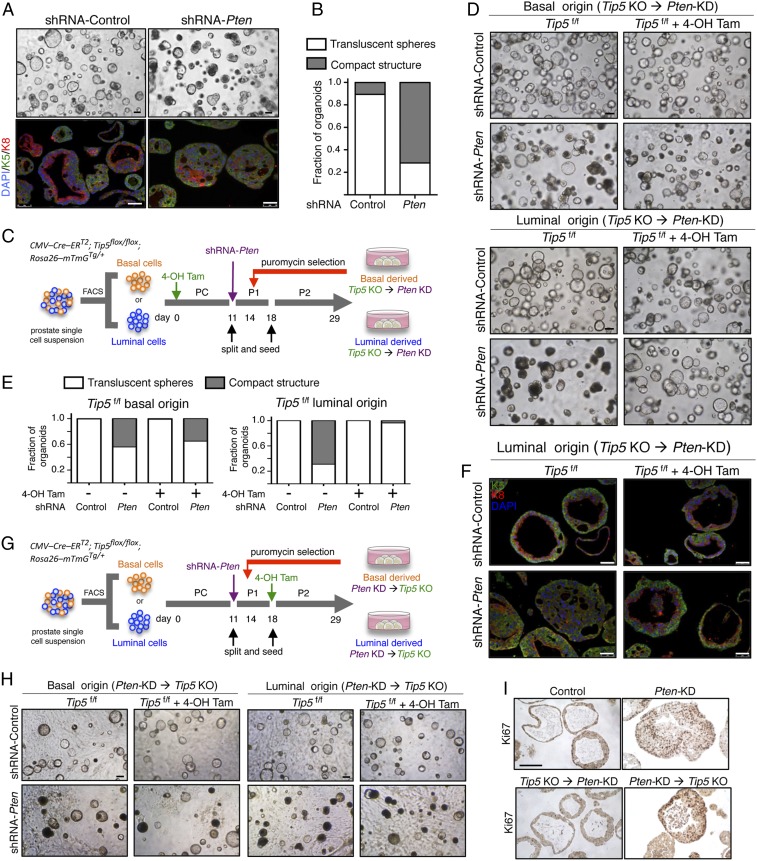
Since PTEN is one of the most commonly lost tumor suppressor genes in primary PCa (7, 34–36), we set out to down-regulate Pten expression in prostate organoids to model initiation of PCa and assess the role of TIP5 in this process. Down-regulation of Pten was achieved by infection with lentivirus encoding short hairpin RNA (shRNA) targeting Pten, which allowed >70% reduction of Pten at protein and mRNA levels (SI Appendix, Fig. S4 A and B). Consistent with previous results (24), Pten-knockdown (KD) organoids displayed altered morphology as evidenced by the solid and compact structures with a disrupted bilayered epithelium (Fig. 4 A and B). Western blot analysis revealed increased phosphorylated AKT levels, which is a known downstream effect of Pten-del in the PI3/PTEN/AKT pathway (SI Appendix, Fig. S4B). Consistent with previous studies showing reduced androgen receptor (AR) transcriptional activity in Pten-null prostate tissues (37, 38), Pten-KD organoids decreased the expression of the AR target gene Fkbp5, a result that we also observed in primary PTEN-del PCa (7) (SI Appendix, Fig. S4 C and D). These results showed that Pten down-regulation in the prostate organoid resembles cancer phenotypes observed in animal models and patient specimens and can be used to study pathways implicated in PCa.
Fig. 4.
TIP5 is required for the initiation of luminal origin PCa in organoids. (A) Representative bright field images of Pten-KD organoids derived from total prostate cells and corresponding IF analyses with anti-K5 and anti-K8 antibodies. (Scale bars represent 150 μm [bright field] and 50 μm [IF].) (B) Quantification of Pten-KD organoid morphology. Results from n > 70 organoids per condition and experiment are shown. Average of three independent experiments. (C) Schematic time line for the generation of Tip5-KO → Pten-KD organoids of basal or luminal origin. Primary culture (PC), passage 1 (P1), and passage 2 (P2). (D) Representative bright field images of Tip5-KO → Pten-KD organoids of basal or luminal origin. Data are from three independent experiments. (Scale bars represent 200 μm.) (E) Quantification of the morphology of Tip5-KO → Pten-KD organoids. Results from n > 80 organoids per condition and experiment. Average of two independent experiments. (F) IF analysis with anti-K5 and anti-K8 antibodies of Tip5-KO → Pten-KD organoids of luminal origin. (Scale bars represent 50 μm.) (G) Schematic time line for the generation of Pten-KD → Tip5-KO organoids of basal or luminal origin. (H) Representative bright field images of Pten-KD → Tip5-KO organoids. Data are from three independent experiments. (Scale bars represent 200 μm.) (I) IF analysis for Ki67 in Pten-KD luminal organoids.
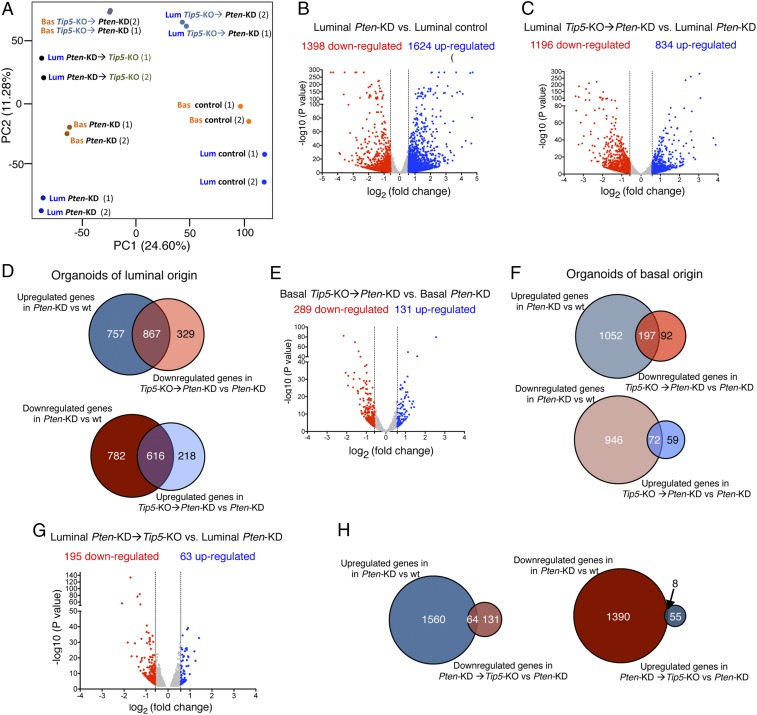
We used prostate organoids to investigate the role of TIP5 in Pten-loss PCa type taking into account the cell of origin as well as the order of Pten-KD and Tip5-KO. Pten down-regulation was performed in organoids derived from basal or luminal cells (Fig. 4 C and D) whereas Tip5 deletion was achieved before or after Pten down-regulation through treatment with 4-OH Tam. Organoids derived from shRNA-Pten luminal cells formed more solid and compact structures compared to control organoids (+shRNA control) and organoids derived from shRNA-Pten basal cells, indicating the high potential of luminal cells to undergo transformation upon Pten-depletion (Fig. 4 D and E). These results are consistent with early studies using mouse models showing that prostate luminal cells are more responsive to Pten-loss-induced mitogenic signaling whereas basal cells are resistant to direct transformation (18). Furthermore, they provide further evidence that PCa of luminal origin is more aggressive than tumors of basal origin (15, 19). Remarkably, depletion of TIP5 before induction of Pten-KD (Tip5-KO→Pten-KD) impaired the ability of organoids to undergo transformation as evident by the translucent phenotype and intact bilayer structure displayed by the majority of organoids (Fig. 4 D–F and SI Appendix, Fig. S4E). This effect was particularly evident in organoids derived from luminal cells, suggesting a major role of TIP5 in the initiation of PCa of luminal origin. Surprisingly, changing the order of the events by inducing Pten down-regulation followed by TIP5 depletion (Pten-KD → Tip5-KO) did not revert the Pten-mediated transformed phenotype (Fig. 4 G and H and SI Appendix, Fig. S4F). This switch of phenotype was not due to inefficient Tip5 deletion or Pten down-regulation as Tip5 and Pten levels were comparable between Tip5-KO → Pten-KD and Pten-KD → Tip5-KO organoids (SI Appendix, Fig. S4G). Consistent with these results, IHC staining for nuclear proliferation marker Ki67 in organoids revealed a strong positive signal in Pten-KD organoids and Pten-KD → Tip5-KO organoids compared to control organoids and Tip5-KO → Pten-KD organoids (Fig. 4I). Finally, principal component analysis (PCA) of RNAseq data from basal or luminal cells with different timings of Pten-KD and Tip5-KO revealed similar differences observed by the phenotypic analysis reported above (Fig. 5A and SI Appendix). Collectively, these results indicate that TIP5 plays a critical role for the initiation of PCa driven by Pten-loss, in particular, in luminal cells, which are considered the cell of origin of aggressive PCa. In contrast, TIP5 is dispensable once the Pten-mediated transformation is established.
Fig. 5.
TIP5 deletion affects the expression of genes regulated by PTEN during the initiation of PCa of luminal origin. (A) PCA plot of RNAseq of prostate organoids. (B) Volcano plot showing gene expression fold changes in luminal Pten-KD organoids vs. control organoids. Values of two replicates that were averaged and selected for 1.5-fold changes and P < 0.05. (C) Volcano plot showing gene expression fold changes in luminal Tip5 KO → Pten KD organoids vs. luminal Pten KD organoids. Values of two replicates. (D) Venn diagrams showing the number of PTEN-regulated genes that are affected upon Tip5-KO prior to Pten-KD in organoids of luminal origin. (E) Volcano plot showing gene expression fold changes in basal Tip5-KO → Pten-KD organoids vs. basal Pten-KD organoids. Values of two replicates. (F) Proportional Venn diagrams showing the number of PTEN-regulated genes that are affected upon Tip5-KO prior to Pten-KD in organoids of basal origin. (G) Volcano plot showing gene expression fold changes in luminal Pten-KD → Tip5-KO organoids vs. luminal Pten-KD organoids. Values of two replicates. (H) Proportional Venn diagrams showing the number of PTEN-regulated genes that are affected upon Tip5-KO after Pten-KD in organoids of luminal origin.
TIP5 Is Required for the Expression of Genes Regulated by PTEN during the Initiation of PCa of Luminal Origin.
To elucidate which pathways are regulated by TIP5 in PCa organoids, we first analyzed which genes are regulated by PTEN in both basal and luminal organoids (PTEN-regulated genes). We compared gene expression of luminal Pten-KD organoids and control organoids and found 1,398 down-regulated genes and 1,624 up-regulated genes upon PTEN depletion (Fig. 5B and Dataset S5). GO analysis revealed that up-regulated genes were enriched in pathways linked to cell cycle whereas down-regulated genes were related to epithelium and urogenital development (SI Appendix, Fig. S5A and Dataset S6). These results indicated that prostate organoids upon Pten-KD resemble cancer features, such as high cell proliferation and dedifferentiation states. Analysis of Pten-KD organoids of basal origin also showed alterations in gene expression and enrichment in pathways that were similar to the ones identified in luminal organoids (SI Appendix, Fig. S5 B and C and Datasets S5 and S6). Consistent with the PCA analysis (Fig. 5A), basal and luminal organoids differ in their gene expression profile upon Pten-KD with >40% of genes that are exclusively regulated in luminal organoids (SI Appendix, Fig. S5D and Dataset S7).
To determine which PTEN-regulated genes depend on Tip5, we compared Tip5-KO → Pten-KD and Pten-KD luminal organoids and found 1,196 down-regulated genes and 834 up-regulated genes (Fig. 5C and Dataset S5). Remarkably, the expression of a large portion of PTEN-regulated genes was altered (Fig. 5D and Dataset S7). The top 10 GO terms of genes affected by Tip5-KO anticorrelate with the pathways linked to Pten-KD in organoids (SI Appendix, Fig. S5 A and E and Dataset S6). Thus, these are genes that can be affected by Pten down-regulation in normal prostate organoids only when TIP5 is expressed. Several of these genes were previously implicated in PCa, such as Myc, Pim1, Aurka, Lif, Bub1b, Cited1, Ccnd1, Cdc25b, Id4, Bmp2, and Aox1 (39–52). We performed a similar analysis with organoids of basal origin and found a lower number of differentially regulated genes compared to luminal organoids (Fig. 5E and Dataset S5). GO analysis revealed similar biological pathways for both organoids of luminal and basal origins (SI Appendix, Fig. S5F and Dataset S6). However, only a minority of PTEN-regulated genes was affected by Tip5-loss in basal organoids (Fig. 5F and Dataset S7). These results indicate that TIP5 plays a major role in the gene expression program necessary for the initiation of PCa of luminal origin driven by Pten down-regulation whereas it is less effective in organoids of basal origin. Finally, we analyzed organoids that were Tip5 depleted after Pten down-regulation by comparing Pten-KD → Tip5-KO and Pten-KD luminal organoids (Fig. 5G and Dataset S5). As expected, the number of differentially expressed genes (195 down-regulated and 63 up-regulated genes) was lower compared to the number of genes that were affected in Tip5-KO → Pten-KD organoids. Furthermore, only few PTEN-regulated genes were affected (Fig. 5H and Dataset S7). These results are consistent with the PCA and phenotypic analyses and indicated that TIP5 is dispensable for the maintenance of the transformed state mediated by Pten down-regulation in prostate organoids.
Modeling of PCa with Organoids Identifies a Gene Signature of Primary PCa That Depends on PTEN Alterations and High TIP5 Expression.
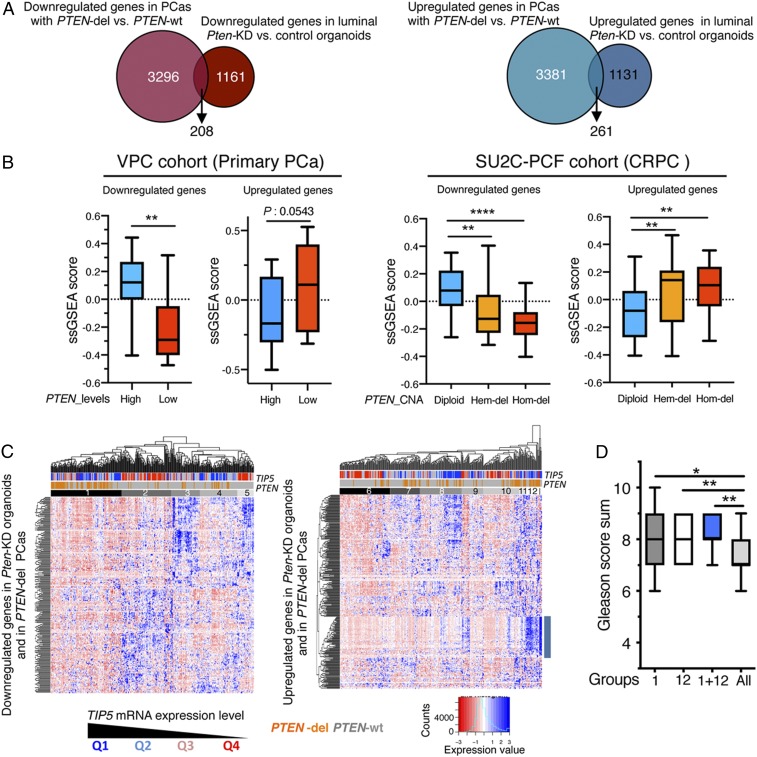
To determine whether the modeling of PCa initiation mediated by PTEN and TIP5 in murine organoids could serve to stratify PCa types, we integrated our RNAseq analyses with TCGA–PRAD data of primary PCas (7). First, we used TCGA–PRAD data to identify genes that were differentially expressed between PTEN-del and PTEN-wt PCas (3,504 down-regulated genes and 3,642 up-regulated genes, P < 0.05) (Fig. 6A and Dataset S8). We then asked how many of these differentially expressed genes were regulated by PTEN in mouse luminal prostate organoids. We found 208 and 261 genes that were down-regulated and up-regulated, respectively, in both Pten-KD luminal organoids and PTEN-del PCa types (Fig. 6A and Dataset S8). The analysis of other datasets from primary tumor samples (Vancouver Prostate Centre [VPC]) (53) and CRPCs Stand Up to Cancer (SU2C) (54) confirmed that the expression of these genes strongly correlates with PTEN status (diploid, hemizygous, and homozygous deletions) and PTEN expression levels in tumors (Fig. 6B). Interestingly, the expression of a large portion of these PTEN-regulated genes (>50%) was affected by Tip5-KO prior to Pten-KD in organoids of luminal origin (SI Appendix, Fig. S6A and Dataset S8). The dependency on PTEN status and expression levels for these TIP5-dependent genes could also be confirmed using VPC and SU2C datasets (SI Appendix, Fig. S6B). These TIP5-dependent genes were primarily associated with urogenital and epithelium developmental pathways and cell cycle processes (SI Appendix, Fig. S6A and Dataset S8). Several of these genes were previously associated with PCa, such as AOX1, ID4, RXRA, BUB1B, CITED1, CDC25C, SIX1, MCM10, and DIAPH3 (39, 40, 50, 55–60) and correlated with poor prognosis (LENG9 and GLB1L2) (61).
Fig. 6.
Expression of PTEN-regulated genes in primary PCa correlates with TIP5 levels. (A) Proportional Venn diagrams showing the number of genes that are differentially expressed in both PTEN-del PCas and Pten-KD organoids of luminal origin. (B) Single sample gene set enrichment analysis scores in primary PCa (VPC, n = 43) and CRPC (SU2C, n = 93) datasets of genes found differentially expressed in both PTEN-del PCas using the TCGA–PRAD dataset and Pten-KD organoids of luminal origin. In the analysis of the VPC dataset, measurements referred to the top 25% of PCa with the higher PTEN expression (High) and the top 25% of PCas with the lowest PTEN levels (Low). In the analysis of the SU2C dataset, measurements referred to groups of tumors with PTEN status as diploid, hemizygous (Hem-del), or homozygous (Hom-del) deletion. Statistical significance (P values) was calculated using the unpaired two-tailed t test (*< 0.05; ** <0.01; *** <0.001; **** <0.0001). (C) Unsupervised hierarchical clustering and heatmaps illustrating the expression of genes that are down-regulated or up-regulated in both PTEN-del PCas and Pten-KD organoids of luminal origin in primary PCas. The row dendrogam represents gene clusters. The column dendogram separates tumors into clusters according to gene expression. Normalized gene expression is indicated by the row Z-score value. (D) Gleason score of groups 1 and 12 and PCas common to both groups. Statistical significance (P values) was calculated using the paired two-tailed t test (*< 0.05; ** <0.01).
Next, we performed unsupervised clustering to determine whether TIP5 expression in PCa correlates with the PTEN gene signature obtained from the cross-species analysis of human PCas and murine Pten-KD organoids of luminal origin (Fig. 6C). We found that PCa patients grouped not only accordingly to PTEN alteration status (deletion or wt), but also to TIP5 expression levels. This was particularly evident in the analysis of down-regulated genes that identified a group of PCas (group 1) displaying low expression levels of these genes compared to the other PCa groups and was enriched for PTEN-del and TIP5-high PCa types (Fig. 6C and SI Appendix, Fig. S6 C and D). Remarkably, PTEN expression in tumors of group 1 with PTEN-wt status was lower compared to all other samples, further indicating that our cross-species analyses identified a group of primary PCas with low PTEN expression (SI Appendix, Fig. S6E). In the analysis of up-regulated genes, this pattern of PCas was less evident; however, we identified a group of PCas (group 12) that is also enriched in PTEN-del and TIP5-high PCa and exhibits strong up-regulation of a set of genes that are associated with the cell cycle (SI Appendix, Fig. S6 C and F and Dataset S8). Importantly, 21 out of 24 patients of group 12 belong to group 1 that was identified in the analysis of down-regulated genes. PCas common to groups 1 and 12 expressed high TIP5 levels and were enriched in PTEN-del (76%) and ERG fusions (86%) (SI Appendix, Fig. S6 C and D) Furthermore, PCas of groups 1 and 12 showed high Gleason scores compared to all of the other tumors whereas this correlation could not be found by selecting only for TIP5-high or PTEN-del PCas (Fig. 6D and SI Appendix, Fig. S6G). Therefore, the PTEN gene signature identified a subset of patients with PTEN-del and high TIP5 levels that have advanced disease.
Taken together, these results identified a class of genes whose expression in PCa correlates with both PTEN status and TIP5 expression level, and that are linked to developmental pathways and cell cycle processes. Since the transcription of these genes was altered in luminal organoids upon Pten-KD, the data suggested that their differential expression in primary PCa could directly depend on PTEN-del and that these PCas might originate from an oncogenic insult that occurred in a luminal progenitor cell. The high TIP5 expression in these PCas and the fact that >50% of these genes require TIP5 prior to Pten down-regulation in organoids of luminal origin implies a critical role of TIP5 in the establishment of this gene signature in primary PCa.
Discussion
In this paper, we showed the potential of murine organoids to model PCa heterogeneity and the feasibility of this system to study the function of different genetic and epigenetic factors that may contribute to PCa development. The possibility to engineer alterations identified in recent comprehensive analyses of primary PCas (7) using mouse prostate organoids with a defined genetic background has the advantage to facilitate the study of the very first steps of tumor initiation and the minimal requirements for neoplasia without confounding effects from other mutations.
According to the cell of the origin model, different cell types can give rise to distinct tumor subtypes, which have specific histopathological and/or molecular features that correlate with treatment response and patient prognosis (62). In this paper, we tested the possibility to model in vitro PCa initiation with organoids taking into account the cell of origin. We showed that organoids derived from luminal cells display a more severe phenotype upon Pten-loss than the one of basal origin. Furthermore, Pten-KD affected greater numbers of genes in organoids derived from luminal cells than from basal cells. Although multiple studies indicate that both prostate basal and luminal cells can serve as the cell of origin for PCa, increasing evidence has suggested that PCa of luminal origin is more aggressive (14, 18, 19, 63, 64). Our findings are consistent with studies performed with mouse models and cross-species bioinformatic analyses, revealing that luminal cells produce higher grade tumors upon loss of Pten while basal cells give rise to less aggressive PCa (15, 18, 19, 64). Thus, in vitro modeling of PCa with organoids supports previous evidence showing that the cell context in which an oncogenic event occurs plays a significant role in defining the resulting tumors.
Our results revealed that TIP5 plays a major role in prostatic luminal cells. These results were further supported by the analysis of prostate organoids derived from luminal progenitors, whose formation was compromised upon TIP5 deletion. Importantly, TIP5 plays a critical role in the initiation of PCa of luminal origin driven by Pten down-regulation whereas it is dispensable once the Pten-mediated transformed state is established. Nearly half of the genes regulated by PTEN in organoids of luminal origin require TIP5 expression to mediate initiation of oncogenic transformation, whereas gene expression after transformation was not affected by the absence of TIP5. These findings suggest that TIP5 primes luminal cells for oncogenic transformation mediated by Pten depletion. Previous work showed that TIP5 regulates gene expression in PCa through the establishment of repressive chromatin features that lead to silencing of genes linked to developmental processes and frequently repressed in metastatic PCa (27). Although chromatin alterations have been associated with all stages of tumor formation and progression, it has been reported that the abnormal epigenetic silencing occurs most frequently during the early precancerous stages of the neoplastic process through the disruption or overactivation of key developmental and cell signaling pathways (65–67). However, it is not well understood how genetic and epigenetic events act in concert to cause oncogenic states and the impact of their relative timing of occurrence. Depending on the susceptibility and second event, a precancerous cell may or may not give rise to a tumor. This concept also implies that the initiating oncogenic hit may not be required for tumor progression once epigenetic priming has already occurred (68, 69). Epigenetic priming has been recently observed in lung adenocarcinoma where chronic cigarette smoke caused progressive epigenetic alterations that sensitized for key genetic events that drive tumor development (67). Thus, tumor priming would explain the requirement of TIP5 for Pten-KD-mediated transformation, and why TIP5 is not necessary for the maintenance of already transformed organoids. Therefore, TIP5-mediated chromatin states could control key developmental pathways and tumor suppressor genes, which precede and sensitize for genetic events to drive cancer development.
The order of genetic and epigenetic oncogenic events in tumors is difficult to dissect since many of the acquired mutations are present in the same cell. Our paper showed that bottom-up cancer modeling with organoids represents a powerful tool for the stratification of tumors. Cross-species analyses using primary PCa and murine Pten-KD organoids of luminal origin revealed a PTEN gene signature that identified a group of primary PCa characterized by PTEN-del or low expression and high TIP5 expression. These results suggest that these PCas could have initiated through the deletion of PTEN specifically in a luminal cell. Genes down-regulated in this PCa group are mainly involved in developmental processes related to the prostate gland, epithelium, and urogenital system, a gene signature that is indicative of a dedifferentiation state known to mark the progression from low‐ to high‐grade cancer. Accordingly, these tumors have higher Gleason scores compared to the other PCas. Thus, the ability to convert in vitro normal prostate organoids into cancerlike organoids is a powerful system that can be used to recapitulate the PCa molecular subtypes identified in recent genomic analyses of primary tumors (7). Furthermore, it will serve to elucidate the role of these mutations in PCa development and facilitate screening for drug sensitivity that, until now, has not been possible to perform. Finally, the data highlighted the use of murine prostate organoids to classify primary PCa types taking into account the cell of origin thereby providing a potential informative value for the choice of initial therapeutic strategies in the clinical management of PCa.
Materials and Methods
Prostate Organoid Culture.
Generation of organoids was performed based on the modified procedure described by Karthaus et al. (24). Prostate cells were resuspended in cold advanced Dulbecco's modified Eagle's medium (ADMEM)/F12 5+ medium at a density of 5.000 cells per 40 μL for total organoid culture and at 4.000 cells per 40 μL for basal and luminal organoid cultures. Cell suspension was mixed with 60 μL of prethawed growth factor reduced Matrigel (Corning) and plated in a form of a drop into the middle of a well of a 24-well plate (TPP). Mixes for multiple wells of the same conditions were prepared together. Organoids were cultured in 500 μL of ADMEM/F12 supplemented with B27 (Gibco Life Technologies), Glutamax (Gibco Life Technologies), 10-mM Hepes (Gibco Life Technologies), 2% fetal bovine serum (FBS), and 100-μg/mL Primocin (ADMEM/F12 5+ medium) and contained following growth factors and components: mEGF (Gibco Life Technologies), transforming growth factor-β/Alk inhibitor A83-01 (Tocris), dihydrotestosterone (Sigma-Aldrich) with the addition of 10-μM ROCK inhibitor Y-27632 for the first week after seeding (organoid culture complete medium). For induction of Cre recombinase activity and Tip5 deletion, cells were cultured in medium supplemented with 1-μM 4-OH TAM (Sigma-Aldrich) on the day of plating (day 0). The medium was changed every 2–4 d by aspiration of the old one and addition of fresh 500 μL of culture organoid medium. A detailed summary of medium composition is listed in Dataset S9. The efficiency of organoid formation was calculated by averaging the number of organoids visible in each well after 11 d of growth using a microscope ×10 objective.
After 11 d in culture, organoids were split into single cells for the following experiments. Matrigel drops were dissolved by addition of 125-μL dispase at 5 mg/mL to each well of the 24-well plate to make the final concentration of dispase 1 mg/mL and incubation at 37 °C for 10 min. The matrigel drop was then disrupted by pipetting up and down several times, and the organoid suspension was transferred to a conical tube. Samples were centrifuged at 350 g for 5 min at room temperature. Supernatants were aspirated, pellets were washed with cold phosphate-buffered saline to remove the rest of the medium, and centrifuged at 350 g for 5 min. The 1 mL of warm TrypLE was added, and organoids were incubated at 37 °C for 5 min. To dissociate them into single cells, organoids were pipetted up and down for 30 s and transferred into a conical tube prefilled with 2-mL cold Hanks’ balanced salt solution +2% FBS to quench the reaction. Samples were centrifuged at 350 g for 5 min, and supernatants were removed. Cell pellets were resuspended in ADMEM/F12 + 2% FBS, and viable cells were counted using a hemacytometer and trypan blue.
Single cells from trypsinized organoids were cryopreserved in ADMEM/F12 5+ medium with 50% FBS and 10% dimethyl sulfoxide by gradual freezing to −80 °C and subsequent transfer into liquid nitrogen after 3 d.
Data Availability.
All raw data generated in this paper using high throughput sequencing are accessible through National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE131845).
Supplementary Material
Acknowledgments
We thank Dr. Olga Shakova for assistance with histology analyses. We thank Catherine Aquino, the Functional Genomic Center Zürich, and the flow cytometry facility of the University of Zürich for technical support. This work was supported by Krebsliga Schweiz (Grants KFS-3497-08-2014 and KLS-4527-08-2018 to R. Santoro and KLS-4248-08-2017 to J.-P.T.), the Swiss National Science Foundation (Grants 310003A-152854 and 31003A_173056 to R. Santoro and PP00P3_179072 to J.-P.T.), Sassella Stiftung (to K.P.), Krebsliga Zürich, Novartis, Fidinam Foundation, Julius Müller Stiftung, and Olga Mayenfisch Stifung.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. A.M.C. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE131845).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911673117/-/DCSupplemental.
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Penney K. L., et al. , Gleason grade progression is uncommon. Cancer Res. 73, 5163–5168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srigley J. R., et al. , One is the new six: The International Society of Urological Pathology (ISUP) patient-focused approach to Gleason grading. Can. Urol. Assoc. J. 10, 339–341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irshad S., et al. , A molecular signature predictive of indolent prostate cancer. Sci. Transl. Med. 5, 202ra122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser M., Berlin A., Bristow R. G., van der Kwast T., Genomic, pathological, and clinical heterogeneity as drivers of personalized medicine in prostate cancer. Urol. Oncol. 33, 85–94 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Boyd L. K., Mao X., Lu Y. J., The complexity of prostate cancer: Genomic alterations and heterogeneity. Nat. Rev. Urol. 9, 652–664 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network , The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson D., et al. , Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rycaj K., Tang D. G., Cell-of-Origin of cancer versus cancer stem cells: Assays and interpretations. Cancer Res. 75, 4003–4011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chua C. W., et al. , Differential requirements of androgen receptor in luminal progenitors during prostate regeneration and tumor initiation. eLife 7, e28768 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata M., Shen M. M., The roots of cancer: Stem cells and the basis for tumor heterogeneity. BioEssays 35, 253–260 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S. H., Shen M. M., Cell types of origin for prostate cancer. Curr. Opin. Cell Biol. 37, 35–41 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Toivanen R., Shen M. M., Prostate organogenesis: Tissue induction, hormonal regulation and cell type specification. Development 144, 1382–1398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., et al. , A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461, 495–500 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z. A., Toivanen R., Bergren S. K., Chambon P., Shen M. M., Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep. 8, 1339–1346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurel B., et al. , Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod. Pathol. 21, 1156–1167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meeker A. K., et al. , Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res. 62, 6405–6409 (2002). [PubMed] [Google Scholar]
- 18.Choi N., Zhang B., Zhang L., Ittmann M., Xin L., Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell 21, 253–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z. A., et al. , Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 15, 274–283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao D., et al. , Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clevers H., Modeling development and disease with organoids. Cell 165, 1586–1597 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Lancaster M. A., Knoblich J. A., Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 345, 1247125 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Chua C. W., et al. , Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell. Biol. 16, 951–961, 1–4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karthaus W. R., et al. , Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puca L., et al. , Patient derived organoids to model rare prostate cancer phenotypes. Nat. Commun. 9, 2404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S., Gao D., Chen Y., The potential of organoids in urological cancer research. Nat. Rev. Urol. 14, 401–414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu L., et al. ; ICGC Project on Early Onset Prostate Cancer , BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nat. Genet. 47, 22–30 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Zhang D., et al. , Stem cell and neurogenic gene-expression profiles link prostate basal cells to aggressive prostate cancer. Nat. Commun. 7, 10798 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang D., Zhao S., Li X., Kirk J. S., Tang D. G., Prostate luminal progenitor cells in development and cancer. Trends Cancer 4, 769–783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal S., et al. , Identification of different classes of luminal progenitor cells within prostate tumors. Cell Rep. 13, 2147–2158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drost J., et al. , Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 11, 347–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baca S. C., et al. , Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krohn A., et al. , Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am. J. Pathol. 181, 401–412 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Phin S., Moore M. W., Cotter P. D., Genomic rearrangements of PTEN in prostate cancer. Front. Oncol. 3, 240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger M. F., et al. , The genomic complexity of primary human prostate cancer. Nature 470, 214–220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimoto M., et al. , Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet. Cytogenet. 169, 128–137 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Carver B. S., et al. , Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19, 575–586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulholland D. J., et al. , Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 19, 792–804 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson V. C., et al. ; Australian Prostate Cancer BioResource , A gene signature identified using a mouse model of androgen receptor-dependent prostate cancer predicts biochemical relapse in human disease. Int. J. Cancer 131, 662–672 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Fu X., et al. , Overexpression of BUB1B contributes to progression of prostate cancer and predicts poor outcome in patients with prostate cancer. OncoTargets Ther. 9, 2211–2220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding Z., SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 470, 269–273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holder S. L., Abdulkadir S. A., PIM1 kinase as a target in prostate cancer: Roles in tumorigenesis, castration resistance, and docetaxel resistance. Curr. Cancer Drug Targets 14, 105–114 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Won H., et al. , TLR9 expression and secretion of LIF by prostate cancer cells stimulates accumulation and activity of polymorphonuclear MDSCs. J. Leukoc. Biol. 102, 423–436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ngan E. S., Hashimoto Y., Ma Z. Q., Tsai M. J., Tsai S. Y., Overexpression of Cdc25B, an androgen receptor coactivator, in prostate cancer. Oncogene 22, 734–739 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Gurel B., et al. , Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod. Pathol. 21, 1156–1167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwata T., et al. , MYC overexpression induces prostatic intraepithelial neoplasia and loss of Nkx3.1 in mouse luminal epithelial cells. PLoS One 5, e9427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kivinummi K., et al. , The expression of AURKA is androgen regulated in castration-resistant prostate cancer. Sci. Rep. 7, 17978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma P., et al. , Id4 deficiency attenuates prostate development and promotes PIN-like lesions by regulating androgen receptor activity and expression of NKX3.1 and PTEN. Mol. Cancer 12, 67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vinarskaja A., Goering W., Ingenwerth M., Schulz W. A., ID4 is frequently downregulated and partially hypermethylated in prostate cancer. World J. Urol. 30, 319–325 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Zhong C., Yang S., Huang J., Cohen M. B., Roy-Burman P., Aberration in the expression of the retinoid receptor, RXRalpha, in prostate cancer. Cancer Biol. Ther. 2, 179–184 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Møller M., et al. , Heterogeneous patterns of DNA methylation-based field effects in histologically normal prostate tissue from cancer patients. Sci. Rep. 7, 40636 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horvath L. G., et al. , Loss of BMP2, Smad8, and Smad4 expression in prostate cancer progression. Prostate 59, 234–242 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Ramnarine V. R., et al. , The long noncoding RNA landscape of neuroendocrine prostate cancer and its clinical implications. Gigascience 7, giy050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abida W., et al. , Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Towers C. G., et al. , The Six1 oncoprotein downregulates p53 via concomitant regulation of RPL26 and microRNA-27a-3p. Nat. Commun. 6, 10077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cui F., Hu J., Ning S., Tan J., Tang H., Overexpression of MCM10 promotes cell proliferation and predicts poor prognosis in prostate cancer. Prostate 78, 1299–1310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morley S., et al. , Regulation of microtubule dynamics by DIAPH3 influences amoeboid tumor cell mechanics and sensitivity to taxanes. Sci. Rep. 5, 12136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li W., et al. , Genome-wide scan identifies role for AOX1 in prostate cancer survival. Eur. Urol. 74, 710–719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komaragiri S. K., et al. , ID4 promotes AR expression and blocks tumorigenicity of PC3 prostate cancer cells. Biochem. Biophys. Res. Commun. 478, 60–66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor B. S., et al. , Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinha A., et al. , The Proteogenomic landscape of curable prostate cancer. Cancer Cell 35, 414–427.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J. J., Shen M. M., Prostate stem cells and cancer stem cells. Cold Spring Harb. Perspect. Med., a030395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldstein A. S., Huang J., Guo C., Garraway I. P., Witte O. N., Identification of a cell of origin for human prostate cancer. Science 329, 568–571 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xin L., Cells of origin for cancer: An updated view from prostate cancer. Oncogene 32, 3655–3663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feinberg A. P., Tycko B., The history of cancer epigenetics. Nat. Rev. Cancer 4, 143–153 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Yamada Y., et al. , Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 13580–13585 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vaz M., et al. , Chronic cigarette smoke-induced epigenomic changes precede sensitization of bronchial epithelial cells to single-step transformation by KRAS mutations. Cancer Cell 32, 360–376.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vicente-Dueñas C., Hauer J., Cobaleda C., Borkhardt A., Sánchez-García I., Epigenetic priming in cancer initiation. Trends Cancer 4, 408–417 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Pérez-Caro M., et al. , Cancer induction by restriction of oncogene expression to the stem cell compartment. EMBO J. 28, 8–20 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data generated in this paper using high throughput sequencing are accessible through National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE131845).