Abstract
Background
The fat mass and obesity-associated protein (FTO) was identified as a critical demethylase involved in regulating cellular mRNA stability by removing N6-methyladenosine (m6A) residues from mRNA. Emerging evidence has revealed that FTO is deeply implicated in lung cancer. However, knowledge of the function of FTO in lung adenocarcinoma (LUAC) is limited.
Methods
FTO and FTO R96Q (R96Q), an FTO missense mutant lacking demethylase activity, were ectopically overexpressed, and FTO was knocked down via siRNA in A549 and H1299 cells. The relationships between FTO with cell characteristics and mRNA m6A levels were explored. Furthermore, RNA sequencing was performed on A549 cells.
Results
FTO overexpression enhanced the proliferation, migration, and invasion ability of A549 and H1299 cells, decreased mRNA m6A levels. Interestingly, overexpression of R96Q, blunted the effects of FTO overexpression on cell proliferation and invasion. Through RNA sequencing analysis of A549 cells overexpressing FTO or R96Q and control A594 cells, 45 genes were identified as affected by m6A mRNA demethylation. Most of these genes were related to lung cancer, such as laminin γ2, thrombospondin 1, nerve growth factor inducible, integrin alpha11, and proprotein convertase subtilisin/kexin type 9. Gene ontology and Kyoto Encyclopedia of Genes and Genomes analyses suggested that these genes are fundamental to cancer development processes, such as cell migration and extracellular matrix organization.
Conclusion
Our research shows that FTO facilitates LUAC cell progression by activating cell migration through m6A demethylation; however, further research on the mechanism underlying FTO activity in LUAC is necessary.
Keywords: FTO, lung adenocarcinoma, m6A demethylase
Introduction
Lung cancer is the leading cause of cancer incidence and mortality. In 2018, 2.1 million new lung cancer cases were diagnosed, and 1.8 million deaths were predicted, accounting for nearly one in five (18.4%) cancer deaths.1 Approximately 80% of lung cancer cases are non-small-cell lung cancer (NSCLC), which includes lung adenocarcinoma (LUAC, 50%–60%) and lung squamous cell carcinoma (LUSC, 30%).2 Although targeted and anti-PD1 therapy have shown promise in lung cancer treatment, the five-year survival rate remains low.3–5 Therefore, there is an urgent need to evaluate the biological mechanism of LUAC regulation.
m6A is the most prevalent modification of mammalian RNA. Approximately 0.1%–0.4% of adenosine nucleotides in isolated mammalian RNA are chemically modified. Methyltransferase like 3 (METTL3) and METTL14 are RNA methyl transferases with the ability to mediate m6A modification.6 The FTO protein and a-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5) are demethyltransferases that can erase m6A decoration.7–9
FTO was initially identified through genome-wide association studies as a protein associated with an increased risk of obesity.10 FTO was later revealed to be an m6A demethylase. Several recent reports have indicated the potential association between aberrant m6A modification and lung cancer development. METTL3 promotes the growth, survival, and invasion of human lung cancer cells by enhancing mRNA translation.11 Thus, METTL3 could be a therapeutic target for NSCLC.12,13 High FTO expression is associated with a low survival LUSC rate.14 In addition, FTO overexpression has been indicated in various cancers, including LUSC, NSCLC, gastric cancer (GC), cervical carcinoma, pancreatic cancer, and acute myeloid leukemia.14–19 FTO is strongly associated with various cancers, but limited knowledge on the role of FTO in LUAC is available.
In the present study, the prognostic role of FTO in LUAC was evaluated. By data mining in The Cancer Genome Atlas (TCGA) database, the relationship between FTO expression and the LUAC survival rate was determined. Loss- and gain-of-function studies were performed to explore the function of FTO in A549 and H1299 cells. Finally, the underlying molecular mechanism of FTO was determined through identifying critical mRNA targets of FTO by mRNA sequencing.
Materials and Methods
Publicly Available Gene Expression Data Sets
FTO mRNA and protein expression levels in different human tissues were analyzed on the Human Protein Atlas portal (http://www.proteinatlas.org/).20 To determine the potential roles of FTO in lung cancer, the relationships between FTO expression levels and the LUSC and LUAC survival proportions in a TCGA cohort were analyzed by UALCAN (http://ualcan.path.uab.edu/index.html).21 All data are directly available online.
Clinical Samples
Archived formalin-fixed and paraffin-embedded specimens from 30 LUAC tissues and corresponding adjacent normal tissues collected in the Affiliated Tumor Hospital of Guangxi Medical University from January 2013 to December 2018 were used. The study was approved by the Ethical Committee of the Affiliated Tumor Hospital of Guangxi Medical University.
Immunohistochemistry
Paraffin-embedded sections (4 µm thick) on polylysine-coated slides were used for staining. Antigen retrieval was achieved with 10 mmol/L sodium citrate (pH 6.0). Thereafter, sections were blocked with 10% normal goat serum and then incubated with rabbit monoclonal antibody against FTO (EPR6895, Abcam, 1:1000) overnight at 4 °C. After incubation with secondary antibody (Vector Laboratories), staining was performed using the DAB Substrate Kit for Peroxidase (Vector Laboratories). FTO expression was evaluated by the immunodetection of staining intensity and the number of positive cells. Photomicrographs were imaged using an Olympus BX53 microscope with plan apochromatic objective lenses. An Olympus DP11 camera and Olympus Camedia software were used to produce the images. All cases were reviewed by two pathologists who were blinded to the study and the images were quantified using Image Pro Process software.
Cell Culture
The A549 and H1299 human LUAC epithelial cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences in 2016 July. The cells were maintained in F-12 or 1640 medium (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, NY, USA) at 37 °C under 5% CO2. Mycoplasma testing was conducted by polymerase chain reaction (PCR) every 2 months with the listed primers. Cells at passage 4 was used to generate stable FTO overexpression or knockdown cell lines. All cells used in this research were used before passage 15.
Plasmid Construction and Transfection
The FTO encoding gene (NM_001080432) was amplified by PCR using cDNA as a template and ligated into pTSB-CMV-MCS-copGFP-F2A-PuroR. The R96Q mutation indicates that the Arg at position 96 of the FTO gene was replaced by a Gln. The FTO Arg96Gln mutation resulted in the loss of FTO demethylase activity.22 The R96Q plasmid was acquired by point mutation of the FTO plasmid.
FTO small interfering RNA (siRNA) and negative control siRNA plasmids were obtained from VectorBuilder (Guangzhou, China).
Virus packaging was performed in 293T cells by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The amount of DNA packaged in the 293T cells was calculated according to the ratio of target plasmid:psPAX2:PMD2G of 5:3:2. Medium containing the retroviral supernatant was harvested 48 h after transfection, and viral titers were determined. Approximately 200 µL of viral supernatant was added to A549 and H1299 cells. The medium was replaced with fresh medium 24 h after transfection.
Total RNA Preparation, cDNA Synthesis, and Quantitative Reverse Transcription PCR (qRT-PCR)
Total RNA was extracted with E.Z.N.A.TM Total RNA Kit I (Omega). For cDNA synthesis, 2000 ng of total RNA was used for reverse transcription in a 10 µL reaction volume with a PrimeScript™ RT reagent kit (TaKaRa) following the manufacturer’s instructions. Then, qRT-PCR was performed with TB Green™ Premix Ex Taq™ II (TaKaRa) in a Roche LightCycler®96 qRT-PCR system. β-Actin was used as an endogenous control, and each reaction was run in triplicate. The qRT-PCR primers used are listed in Table S1.
Western Blot Analysis
Total protein was extracted using radioimmunoprecipitation assay lysis buffer (Sigma-Aldrich). Protein concentrations were determined using a bicinchoninic acid assay reagent kit (Beyotime, Shanghai, China). Protein lysates were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and subsequently transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were then incubated in blocking solution containing 5% (w/v) nonfat milk for 1 h at room temperature. Subsequently, the PVDF membranes were incubated with primary antibodies (anti-FTO, 1:1000, Abcam, ab124892; anti-glyceraldehyde 3-phosphate dehydrogenase, 1:1000, Abcam, ab8245) at 4 °C overnight, followed by incubation with secondary horseradish peroxidase-conjugated anti-rabbit secondary antibody for 1 h at room temperature. Bound antibodies were visualized with an ECL kit (Beyotime, China). Protein band intensities were quantified using ImageJ software (NIH, USA).
Cell Proliferation Assay
Cell proliferation was assessed by Cell Counting Kit-8 (CCK-8; Dojindo Laboratories) following the manufacturer’s instructions. The cells were briefly seeded on a 96-well plate at a density of 5000 cells/100 µL. Subsequently, CCK-8 solution (10% of total volume) was added at the indicated time points. After incubation at 37 °C for 2 h, the absorbance at 570 nm was read on a microplate reader (Thermo Varioskan™ LUX). The experiment was conducted in triplicates.
Migration Assay
Cells were seeded in six-well plates for 12 h and then scratched. Photomicrographs were obtained at different time points, and the wound-healing ratio was measured using ImageJ.
Colony Formation Assay
For the colony-formation assay, 500 cells were seeded into six-well plates. Formed colonies were counted after two weeks.
Cell Invasion Assay
For the invasion assay, 2 × 105 cells were resuspended in serum-free medium and seeded into the upper chamber of a Transwell. In addition, FBS-containing medium was added to the bottom chamber, which was separated from the upper chamber by a membrane with an 8 µm pore size coated with a thin layer of Matrigel, to serve as a chemoattractant for migration. After 24 h, migrated cells were fixed with methanol and stained with crystal violet for cell counting.
Cell Apoptosis Assay
Cells were harvested and suspended, and 5 µL of 7-AAD and 5 µL of APC Annexin V were added to the cell suspension (BD, USA). After staining for 30 min, the cells were subjected to flow cytometric analysis.
Measurement of the m6A Level
Total RNA was extracted from the cell lines using the E.Z.N.A.TM Total RNA Kit I (Omega, USA). RNA was quantified by ultraviolet-visible spectrophotometry. To detect m6A levels in the total RNA, an EpiQuikTM m6A RNA Methylation Quantification Kit (EpiGentek, USA) was used following the manufacturer’s instructions. In addition, 200 ng of RNA was seeded in each well, followed by the addition of capture and detection antibody solutions according to the manufacturer’s protocol. m6A levels were then measured colorimetrically by determining the absorbance of each well at 450 nm on a microplate reader (Thermo Varioskan™ LUX).
Processing of Transcriptome Sequencing and Bioinformatics Analysis
Total RNA was extracted from cell lines using the E.Z.N.A.TM Total RNA Kit I (Omega, USA). mRNA sequencing was performed on an Illumina HiSeq 4000 RNA sequencing platform. Base calling was adopted to convert original sequencing images to sequential data as raw reads. These raw reads were subjected to adapter trimming and low-quality filtering using the Trimmomatic program. High-quality clean reads were aligned to the human genome (hg19) by using Bowtie2. The hg19 sequence and gene annotation data were obtained from the University of California Santa Cruz Genome website (http://genome.ucsc.edu/). NOISeq with default parameters was used to profile differentially expressed genes (DEGs). Gene Ontology (GO) functional enrichment analysis was completed using the web-based GO analysis tool Web Gene Ontology Annotation Plot (WEGO) (http://wego.genomics.org.cn/cgi-bin/wego/index.pl). WEGO was used to map all DEGs and search for GO terms significantly enriched in the DEGs compared with the genomic background. Pathways were constructed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database following custom scripts. KEGG pathway enrichment analysis of DEGs compared with the transcriptome background was performed by the hypergeometric distribution test using the pHYPER function of the R software package (http://www.rproject.org/). Bonferroni correction was used to adjust the P value for each pathway. Different proteins often form complexes through complicated interactions to perform their biological functions. Therefore, protein interaction network analysis was performed by Cytoscape version 3.7.0.
Statistical Analysis
Data are presented as the means ± standard deviations. Statistical analyses were performed using SPSS 16.0 software, and graphical representations were obtained with Graph Pad Prism 5 software. Student’s t-test was carried out to analyze the significance of differences between any two groups, whereas one-way analysis of variance was used to compare differences among more than two groups. Differences with P values less than 0.05 were considered statistically significant.
Results
FTO Expression Is Correlated with Human Lung Cancer
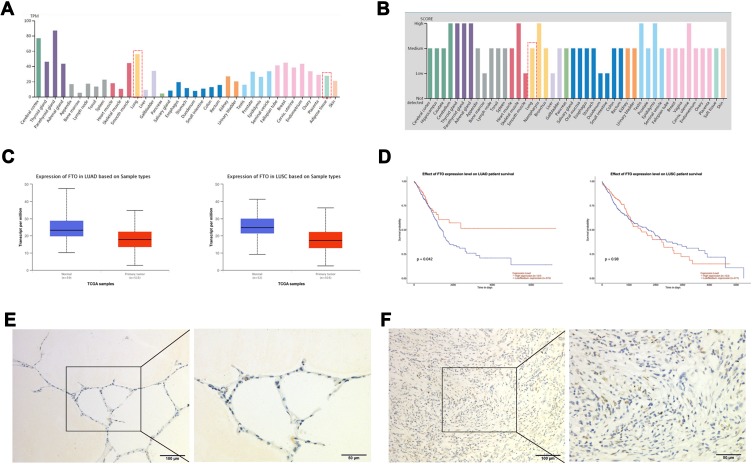
To investigate the role of FTO in lung cancer, we first investigated the mRNA expression and FTO protein levels in different tissues. Data from the Human Protein Atlas portal indicated that the FTO mRNA expression level was high in the lungs and higher than that in adipose tissue (Figure 1A). Consistently, the FTO protein was also expressed in the lungs (Figure 1B). Data from UALCAN revealed that compared with that in adjacent normal tissues, FTO expression was low in LUSC and LUAC tissues (Figure 1C), suggesting that FTO participates in lung cancer formation. Furthermore, FTO downregulation was significantly correlated with a high LUAC survival rate but not related to the LUSC survival rate (Figure 1D).
Figure 1.
FTO expression correlates with human lung cancer. (A) The mRNA level of FTO in different tissues. (B) The protein level of FTO in different tissues. (C) The expression of FTO in LUAC and LUSC tissues. (D) The effect of FTO expression level on LUAD and LUSC survival rate. (E) The FTO protein was expressed in LUAC tissues. (F) The FTO protein was expressed in normal lung tissues.
FTO Protein Expression in Human Lung Cancer Tissues
We evaluated whether FTO expression is associated with the clinicopathological features of body mass index (BMI), tumor stage (T stage), and tumor grading, in LUAC. Immunohistochemistry was used to observe FTO expression in 20 cases of LUAC tissues and matched nontumor tissues. The FTO protein was expressed in both LUAC and normal lung tissues, and FTO was mainly expressed in the nucleus (Figure 1E and F). However, no evident difference in the FTO-positive rate between LUAC and nontumor tissues was observed (Table S2). Moreover, FTO expression was not associated with age, T stage (tumor stage), LN metastasis (lymph node metastasis), TNM stage (tumor, node, metastasis stage), Ki67 expression, or BMI (Table S3).
FTO Overexpression Enhanced LUAC Cell Proliferation, Migration, and Invasion
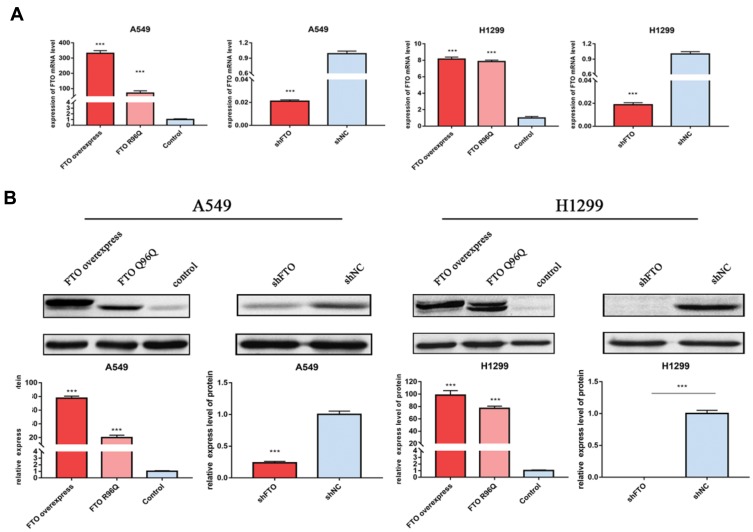
To examine the roles of FTO in LUAC, FTO was overexpressed or knocked down in A549 and H1299 cells. Successful FTO overexpression and knockdown were confirmed at the mRNA and protein levels (Figure 2A and B).
Figure 2.
Confirmation of overexpression and knockdown of FTO by qRT-PCR and Western blotting in A549 and H1299 cells. (A) The mRNA level of FTO in A549 and H1299 cells (B) The protein level of FTO in A549 and H1299 cells. Bar graphs: mean ± SD. ***P < 0.001.
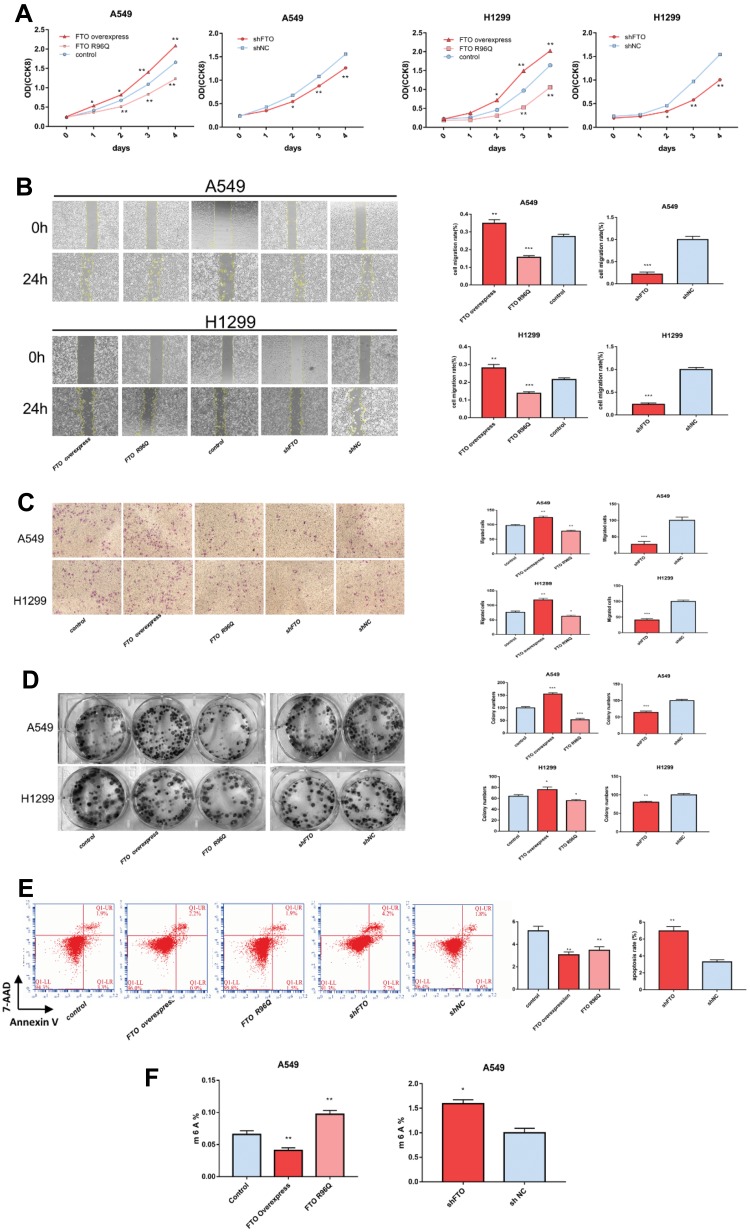
Compared with that in the control group, FTO overexpression significantly increased A549 and H1299 cell proliferation (Figure 3A). The findings of the migration assay indicated that FTO increased the migration of cells into the blank scratched area increased compared with that in the control group (Figure 3B). The results of Transwell migration and invasion assays also revealed that there were significantly more invasive cells in the FTO group than in the control group (Figure 3C). Furthermore, FTO overexpression significantly enhanced the ability of A549 and H1299 cells to form colonies (Figure 3D). Flow cytometry data revealed that FTO overexpression decreased the apoptosis rate in A594 cells (Figure 3E).
Figure 3.
FTO regulates LUAC cell characteristics and mRNA m6A levels. (A) FTO influenced LUAC cell proliferation in A549 and H1299 cells. Cell proliferation was assessed by Cell Counting Kit-8 assay. FTO influenced the following cell growth characteristics of A549 and H1299 cells: (B) migration, (C) invasion, and (D) colony-formation ability. (E) apoptosis, (F) FTO regulated the mRNA m6A levels of A549 cells. Bar graphs: mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
As expected, FTO knockdown suppressed cell proliferation, migration, colony formation, invasion, and apoptosis (Figure 3A–E). Altogether, these results suggest that FTO plays a key role in LUAC cell proliferation, metastasis, and invasion.
FTO Regulates mRNA m6A Levels of LUAC Cells
To further explore the underlying mechanisms of FTO in LUAC, m6A levels in A549 cells were determined. Compared with those in control cells, FTO overexpression decreased mRNA m6A levels, whereas FTO knockdown increased m6A levels in A549 cells (Figure 3F). Therefore, FTO plays a role in regulating the m6A modification in LUAC.
FTO Demethylase Activity Is Required for LUAC Cell Growth, Survival, and Invasion
To further verify the demethylase activity of FTO in LUAC, the FTO R96Q mutation was generated, and the mutated protein was expressed in A549 and H1299 cells. The R96Q missense mutation eliminated FTO demethylase activity. Successful R96Q overexpression was confirmed at the mRNA and protein levels (Figure 2A and B). R96Q overexpression blunted the FTO-mediated promotion of cell proliferation, migration, colony-forming ability, invasion, and apoptosis (Figure 3A–E) but had no effect on the decrease in the cellular m6A level induced by FTO (Figure 3F). Therefore, the effect of the FTO gene on LUAC is evidently due to its demethylase activity.
FTO Alters LUAC Cell Gene Expression
mRNA sequence analysis was performed on A594 cells overexpressing FTO or R96Q and control A594 cells. Compared with their expression in control cells, FTO overexpression resulted in 119 differentially expressed mRNAs with a fold change in expression of >2.0 comprising 56 upregulated and 63 downregulated genes. R96Q overexpression resulted in the upregulation of 94 genes. The genes upregulated by FTO overexpression but not R96Q overexpression were identified as gene affected by m6A mRNA demethylation. Finally, 45 genes were distinguished, and most of these genes were related to lung cancer.
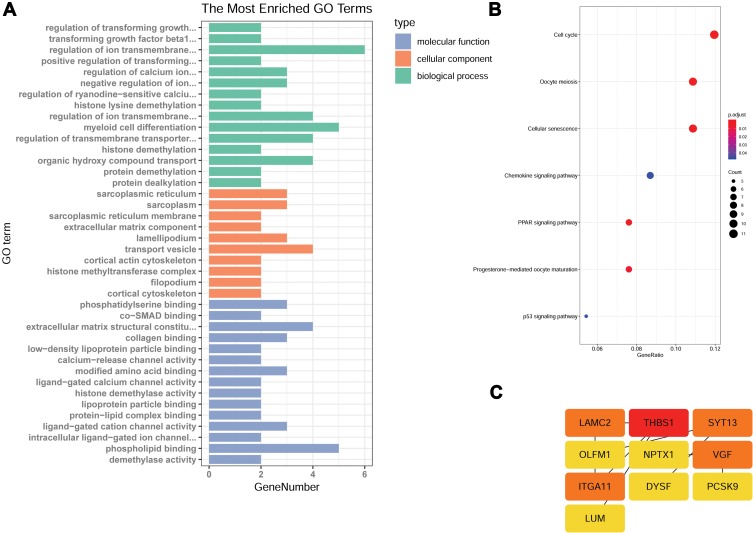
GO enrichment analysis was performed to explore the biological functions of these genes. Several genes were enriched in positive regulation of cell adhesion, extracellular matrix organization, the positive regulation of transforming growth factor beta1 production, cell migration, extracellular space and cell junction GO terms (Figure 4A).
Figure 4.
FTO affects gene expression in A549 cells. RNA-seq analysis of A594 cells overexpressing FTO or R96Q and control A594 cells identified a total of 45 genes as affected by m6A mRNA demethylation. (A) Gene ontology (GO) term categorization and distribution of differentially expressed genes. GO terms were processed and categorized under three main categories (molecular function, cellular component, and biological process). (B) Detailed figure showing KEGG pathway enrichment. (C) Protein–protein interaction networks.
To understand the biological effects of these genes, KEGG analysis was performed. A total of 40 pathways enriched in genes with significantly differential expression were identified. These pathways were mainly related to cell cycle, oocyte meiosis, cellular senescence, the chemokine signaling pathway, the peroxisome proliferator-activated receptor signaling pathway, progesterone-mediated oocyte maturation, and the p53 signaling pathway. Most of these pathways are associated with cancer development (Figure 4B).
To identify DEGs that play key regulatory roles, we applied Cytoscape version 3.7.0 to analyze the regulatory network on the basis of a large set of gene expression data. The findings revealed that the laminin γ2 (LAMC2), thrombospondin 1 (THBS1), synaptotagmin-13, nerve growth factor inducible (VGF), and integrin alpha11 (ITGA11) genes might be the master players involved in tumor cell metastasis (Figure 4C).
Discussion
According to TCGA database, FTO downregulation is significantly correlated with a high LUAC survival rate. Immunohistochemical analysis revealed that FTO expression was no different between cancerous and paracarcinoma tissues, whereas the FTO expression level exhibited no relationship with sex, tumor stage, tumor differentiation, or invasion depth. In addition, in vitro loss- and gain-of-function studies were conducted to explore whether FTO promotes the proliferation, migration, colony-forming and invasive abilities of A549 and H1299 cells. Epidemiological investigation and cell biological experiments provided conflicting results. One possible explanation for this difference is the selection biases of the database. Second, immunohistochemical studies were based on small numbers of adenocarcinoma and corresponding adjacent normal tissues; that is, only 20 pairs of cases were available. Third, FTO may play a dual role in cancer progression, and the function of FTO in cancer progression may be complicated.23
Low FTO protein expression has been associated with a short overall survival rate in GC patients.24 However, other studies drew a contrasting conclusion and indicated that FTO promotes the proliferation, migration, and invasion of GC cell lines.16 Numerous studies have indicated that high FTO expression is significantly associated with cancer development. FTO enhances LUSC proliferation and invasion by reducing m6A levels and mRNA stability in myeloid zinc finger 1 (MZF1) mRNA transcription.14 FTO mRNA and protein levels are overexpressed in human NSCLC tissues and cell lines, and FTO promotes NSCLC cell growth by increasing ubiquitin-specific-processing protease 7 expression.17 The FTO expression level is closely related to low levels of differentiation and lymph node metastasis in GC; FTO overexpression promotes the proliferation, migration, and invasion of GC cell lines.16 In cervical cancer, upregulated FTO represses the m6A modification of β-catenin and induces chemoradiotherapy resistance.18 Elevated FTO leads to a low level of m6A modification of the untranslated regions of ankyrin repeat and SOCS box protein 2 and retinoic acid receptor alpha, thus reducing the mRNA and protein levels of these anti-angiogenic genes; C21orf59, MZF1, and taxilin alpha oncogene expression levels then increase, leading to the promotion of acute myeloid leukemia.15 FTO promotes the proliferation of pancreatic cancer cells by regulating proto-oncogene MYC stability.19 In summary, FTO may have an oncogenic function that is associated with its m6A demethylation activity. Therefore, more studies should delve into the mechanisms by which FTO affects LUAC development.
m6A modification plays a central role in the epigenetic regulation of the mammalian transcriptome. The m6A modification can be reversed by the FTO and ALKBH5 proteins.15 The overexpression of R96Q, an FTO missense mutant lacking demethylase activity, did not affect the cellular m6A level.22,25 In this study, mRNA m6A levels were evaluated in A549 cells overexpressing FTO or R96Q and control A549 cells. FTO overexpression resulted in reduced m6A levels, but R96Q overexpression caused no such effect. These findings indicate FTO may promote LUAC development through its demethylase activity. This result is similar to the results of a previous study, in which FTO was found to promote the transition of 3T3-L1 preadipocytes into adipocytes with decreased m6A modification. In contrast, R96Q had no effect on the cellular m6A level and impeded differentiation.25
m6A decoration maintains mRNA stability or leads to mRNA splicing, followed by a ripple effect on mRNA expression levels. Such an effect may trigger oncoprotein expression and biological activity, thereby accelerating tumor progression. Through mRNA sequential analysis of A594 cells overexpressing FTO or R96Q and control A594 cells, 45 genes were identified as affected by m6A mRNA demethylation. Most of these genes have been shown to be related to lung cancers. For example, ITGA11 is frequently overexpressed in NSCLC, and the coimplantation of with ITGA11 -overexpressing fibroblasts in severe combined immunodeficient mice a markedly enhanced tumor growth.26 Large-scale genomic analysis demonstrated that LAMC2 and THBS1 were increased in LUAC cells.27 LAMC2 overexpression increased the traction force, migration, and invasion of A549 cells, accompanied by cell epithelial–mesenchymal transition.28 A recent analysis identified THBS1 release in LUAC cell-derived exosomes, which are involved in NSCLC cell migration; cloud-based computing was used as a source of biomarkers.29,30 GO analysis revealed that these genes are high correlated with elements of the cell division process, such as cell adhesion, extracellular matrix organization, and the positive regulation of transforming growth factor beta1 production.
Conclusion
These observations demonstrated that FTO facilitates LUAC cell progression by activating cell migration through m6A demethylation; however, further research on the mechanism underlying FTO activity in LUAC is necessary.
Funding Statement
This research was supported in part by grants from the National Natural Science Foundation of China (81760024, 81460003, 81460388 and 81760419), the Guangxi Natural Science Foundation (2018GXNSFAA138027), the Youth Science Foundation of Guangxi Medical University (GXMUYSF201603), and the Medical Excellence Award Funded by the Creative Research Development Grant from The First Affiliated Hospital of Guangxi Medical University (2017010).
Abbreviations
ALKBH5, a-ketoglutarate-dependent dioxygenase alkB homolog 5; CCK-8, Cell Counting Kit-8; FTO, fat mass and obesity-associated; GC, gastric cancer; GO, Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; LUSC, lung squamous cell carcinoma; LUAC, lung adenocarcinoma; m6A, N6-methyladenosine; METTL3, Methyltransferase like 3; NSCLC, non-small-cell lung cancer; PCR, polymerase chain reaction; TCGA, The Cancer Genome Atlas; UALCAN, http://ualcan.path.uab.edu/index.html.
Ethics Approval and Consent to Participate
The study was approved by the Ethical Committee of The Affiliated Tumor Hospital of Guangxi Medical University.
Data Sharing Statement
All data generated or analyzed during this study [and its supplementary information files] are included in this published article.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Brower V. Afatinib and chemotherapy in non-small-cell lung cancer. Lancet Oncol. 2016;17(2):e47. doi: 10.1016/S1470-2045(15)00609-9 [DOI] [PubMed] [Google Scholar]
- 3.Hussain S. Nanomedicine for treatment of lung cancer. Adv Exp Med Biol. 2016;890:137–147. [DOI] [PubMed] [Google Scholar]
- 4.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latimer KM, Mott TF. Lung cancer: diagnosis, treatment principles, and screening. Am Fam Physician. 2015;91(4):250–256. [PubMed] [Google Scholar]
- 6.Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Haim MS, Moshitch-Moshkovitz S, Rechavi G. FTO: linking m6A demethylation to adipogenesis. Cell Res. 2015;25(1):3–4. doi: 10.1038/cr.2014.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Du B. Novel positioning from obesity to cancer: FTO, an m(6)A RNA demethylase, regulates tumour progression. J Cancer Res Clin Oncol. 2019;145(1):19–29. doi: 10.1007/s00432-018-2796-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin S, Choe J, Du P, et al. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du M, Zhang Y, Mao Y, et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017;482(4):582–589. doi: 10.1016/j.bbrc.2016.11.077 [DOI] [PubMed] [Google Scholar]
- 13.Du Y, Hou G, Zhang H, et al. SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function. Nucleic Acids Res. 2018;46(10):5195–5208. doi: 10.1093/nar/gky156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Ren D, Du Z, et al. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502(4):456–464. doi: 10.1016/j.bbrc.2018.05.175 [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Weng H, Su R, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127–141. doi: 10.1016/j.ccell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu D, Shao W, Jiang Y, et al. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017;38(4):2285–2292. doi: 10.3892/or.2017.5904 [DOI] [PubMed] [Google Scholar]
- 17.Li J, Han Y, Zhang H, et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. 2019;479–485. [DOI] [PubMed] [Google Scholar]
- 18.Zhou S, Bai ZL, Xia D, et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting beta-catenin through mRNA demethylation. Mol Carcinog. 2018;57(5):590–597. doi: 10.1002/mc.22782 [DOI] [PubMed] [Google Scholar]
- 19.Tang X, Liu S, Chen D, et al. The role of the fat mass and obesity-associated protein in the proliferation of pancreatic cancer cells. Oncol Lett. 2019;17(2):2473–2478. doi: 10.3892/ol.2018.9873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 21.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Z, Niu T, Chang J, et al. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464(7292):1205–1209. doi: 10.1038/nature08921 [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Chai P, Jia R, et al. Novel insights on m(6)A RNA methylation in tumorigenesis: a double-edged sword. Mol Cancer. 2018;17(1):101. doi: 10.1186/s12943-018-0847-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Zheng D, Wang F, et al. Expression of demethylase genes, FTO and ALKBH1, is associated with prognosis of gastric cancer. Dig Dis Sci. 2019;521:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M, Zhang Y, Ma J, et al. The demethylase activity of FTO (Fat Mass and Obesity Associated Protein) is required for preadipocyte differentiation. PLoS One. 2015;10(7):e0133788. doi: 10.1371/journal.pone.0133788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu CQ, Popova SN, Brown ER, et al. Integrin alpha 11 regulates IGF2 expression in fibroblasts to enhance tumorigenicity of human non-small-cell lung cancer cells. Proc Natl Acad Sci U S A. 2007;104(28):11754–11759. doi: 10.1073/pnas.0703040104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S, Chen L, Chan DW, et al. Protein signatures of molecular pathways in non-small cell lung carcinoma (NSCLC): comparison of glycoproteomics and global proteomics. Clin Proteomics. 2017;14:31. doi: 10.1186/s12014-017-9166-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon YW, Rao G, Kim JJ, et al. LAMC2 enhances the metastatic potential of lung adenocarcinoma. Cell Death Differ. 2015;22(8):1341–1352. doi: 10.1038/cdd.2014.228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vykoukal J, Sun N, Aguilar-Bonavides C, et al. Plasma-derived extracellular vesicle proteins as a source of biomarkers for lung adenocarcinoma. Oncotarget. 2017;8(56):95466–95480. doi: 10.18632/oncotarget.v8i56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang WT, Chong IW, Chen HL, et al. Pigment epithelium-derived factor inhibits lung cancer migration and invasion by upregulating exosomal thrombospondin 1. Cancer Lett. 2019;442:287–298. doi: 10.1016/j.canlet.2018.10.031 [DOI] [PubMed] [Google Scholar]