Abstract
Background
As a key subtype of non-coding RNAs, circular RNA (circRNA) has been well documented to play a key role in the tumorigenesis of osteosarcoma (OS). circPVT1 was revealed to participate in the progression of multiple human tumors; however, the roles of circPVT1 in OS invasion and metastasis and its potential mechanisms remain elusive.
Methods
RNA expression in OS tissues and cells was examined by qRT-PCR, protein expression was measured by Western blot. circPVT1 knockdown in vitro was achieved by transfecting OS cells with specific siRNAs. OS cell proliferation was assessed via CCK-8 and colony formation assays. OS cell migration and invasion were evaluated by transwell assay. Interaction between miR-205-5p and circPVT1 or c-FLIP was validated through dual-luciferase reporter assay. Rescue experiments were performed to explore the regulatory net among circPVT1, miR-205-5p and c-FLIP in OS progression in vitro.
Results
circPVT1 and c-FLIP were highly expressed, while miR-205-5p was lowly expressed in OS tissues and cell lines. Knockdown of circPVT1 repressed cell proliferation, migration and invasion via inhibiting epithelial–mesenchymal transition (EMT) in OS. circPVT1 functioned as a sponge of miR-205-5p, and c-FLIP was targeted by miR-205-5p in OS cells. Furthermore, circPVT1 indirectly regulated c-FLIP expression through competitively binding to miR-205-5p. Inhibition of miR-205-5p or overexpression of c-FLIP abolished the effects of si-circPVT1 on cell proliferation, migration and invasion.
Conclusion
Our study demonstrated circPVT1 functions as a sponge for miR-205-5p to promote c-FLIP expression, thereby enhancing EMT and inducing OS invasion and metastasis in vitro, implying that circPVT1 might be a potential therapeutic target for further clinical therapy of OS.
Keywords: osteosarcoma, circPVT1, miR-205-5p, c-FLIP, invasion and metastasis
Introduction
Osteosarcoma (OS) is the most common primary malignant bone cancer, which mainly occurs in children and adolescents, and brings heavy economy and physiological burdens on patients.1 It predominantly arises from the metaphysis of long bones and characterized by the emergence spindle cells and osteoid.2 Although the mortality rate of OS patients has been dramatically declined in the past decades due to the tremendous improvement of medical technologies, the prognosis of OS patients needs to be further improved.3 Nowadays, the OS patients’ five-year survival rate has risen to 60–70%; however, the OS patients with distant metastasis are still lower than 20%.4 A considerable proportion of patients already exhibit metastasis at the time of diagnosis, and the most frequent target organ of metastasis is lung.5,6 The subsequent respiratory impairment is a leading cause of the death of OS patients.7 These lung-metastasis OS patients have the poorest overall prognosis, with only a 19% five-year survival rate.8 It is essential to further study the molecular mechanisms associated with OS tumorigenesis, metastasis and drug-resistance to reveal several effective therapeutic options for better management of OS.
Non-coding RNA (ncRNA), the main part of the human transcriptome, is accounting for over 98% of all transcripts.9 Up to now, the number of ncRNA that has been identified in vitro is extremely higher than that of protein-encoding genes.10 According to the structure and formative mechanisms, ncRNA could be divided into linear RNA and circular RNA (circRNA). As the most famous linear RNA, microRNA (miRNA) and long non-coding RNA (lncRNA), characterized by short and long nucleotides, respectively, have been shown a critical role in regulating tumor cell growth in multiple human cancers.11,12 Nevertheless, circRNA has recently achieved more and more attention from molecular biology. Unlike linear ncRNA, circRNA has no 5ʹ-cap and 3ʹ-poly A tail because it is formed by back-splicing of pre-mRNA or exons.13 The circular structure enables circRNA more resistant to the RNA exonucleases or RNase R, making it one of the most promising diagnostic biomarkers of cancer.14 A growing number of studies have demonstrated that circRNA participates in the tumorigenesis of numerous human cancers through multiple distinct mechanisms, including competing for endogenous RNAs (ceRNA) theory.15 ceRNA hypothesis revealed that circRNA, lncRNA and other transcripts could regulate gene expression by competitively binding to the response elements of miRNA.16 Recently, multiple circRNAs have been reported to play a role in the tumorigenesis of diverse human cancers through ceRNA mechanism,17,18 however, studies focused on the role of circRNA in OS are considerably limited.
circPVT1 is originated from a tumor susceptibility gene locus PVT1 located on chromosome 8q24.19 circPVT1 was demonstrated to function as an oncogene in multiple human cancers, such as colorectal cancer, head and neck squamous cell carcinoma and leukemia.20–22 Recently, circPVT1 was revealed to contribute to the chemo-resistance of OS cells by modulating ABCB1.23 Nevertheless, whether circPVT1 plays a role in the OS invasion and metastasis remains unknown.
The present study aims to study the function of circPVT1 in OS invasion and metastasis, attempting to understand the mechanism of circPVT1 working on OS progression and providing several therapeutic targets for OS.
Materials and Methods
OS Tissues and Cells
Our study was conducted in accordance with the principles of the Declaration of Helsinki and approved by the Ethics Committee of Xiangya Hospital of Central South University (Changsha, China). OS tissues and corresponding adjacent non-tumor soft tissues (25 pairs) were gathered from the patients who were diagnosed at the Xiangya Hospital (clinical information of patients listed in Table 1). After removed from the patients, all tissue samples were rapidly dipped into liquid nitrogen and stored at −80°C. Written informed consents for the experiment of tumor samples were obtained from each OS patients. Four OS cell lines (Saos-2, MG63, U2OS and SW1353) and a normal human osteoblast cell line (hFOB 1.19) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured with Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Life Technologies) and 100 U/mL penicillin and 100 mg/mL streptomycin (Sigma-Aldrich, St. Louis, MO, USA). OS cells were placed under the condition of 5% CO2 and 37°C, while hFOB 1.19 cells were incubated at 34°C in a humidified 5% CO2 atmosphere.
Table 1.
The Clinicopathological Features of Patients with Osteosarcoma
| Variable | Group | Cases, n |
|---|---|---|
| Gender | Male | 14 |
| Female | 11 | |
| Age, years | <20 | 8 |
| ≥20 | 17 | |
| Primary tumor diameter, cm | <8 | 15 |
| ≥8 | 10 | |
| TNM stage | I | 7 |
| II | 12 | |
| III | 6 | |
| Anatomic location | Femur/tibia | 21 |
| Elsewhere | 4 | |
| Histological type | Osteoblastic | 19 |
| Others | 6 | |
| Distant metastasis | Yes | 7 |
| No | 18 |
RNA Extraction and Quantitative Real-Time PCR Analysis (qRT-PCR)
Total RNA was extracted from OS tissues and cells lysed with TRIzol reagent (Life Technologies) following the protocols of the manufacturer. The quality of extracted RNA was examined by a NanoDrop 2000 instrument (Thermo Scientific, USA) and RNA gel electrophoresis. Next, 3 μg RNA was utilized as a template to form cDNA via a cDNA Reverse Transcription kit (Applied Biosystems, USA). The processes of RT-PCR were carried out with an ABI Prism 7700 system (PE Applied Biosystems, USA) by using a SYBR Premix Ex Taq Kit (Takara, Tokyo). GAPDH and U6 were applied as the endogenous control for mRNA/circRNA and miRNA, respectively. Fold alteration in gene expression was calculated via the 2–ΔΔCt method. Primers used in the present study are summarized in Table 2.
Table 2.
The Primer Sequences of Genes Used in qRT-PCR
| Genes | Forward Primers | Reverse Primers |
|---|---|---|
| GAPDH | 5ʹ-CCAGGTGGTCTCCTCTGA-3’ | 5ʹ-GCTGTAGCCAAATCGTTGT-3’ |
| U6 | 5ʹ-CTCGCTTCGGCAGCACA-3’ | 5ʹ-AACGCTTCACGAATTTGCGT-3’ |
| circPVT1 | 5ʹ-GGTTCCACCAGCGTTATTC-3’ | 5ʹ-CAACTTCCTTTGGGTCTCC-3’ |
| miR-205-5p | 5ʹ-CGGTCCTTCATTCCACCGG-3’ | 5ʹ-CGGCCCAGTGTTCAGACTAC-3’ |
| c-FLIP | 5ʹ-TGATGGCAGAGATTGGTGAG-3’ | 5ʹ-TCTGGGGCAACCAGATTTAG-3’ |
Cell Transfection
circPVT1-specific siRNAs (si-circPVT1-1, si-circPVT1-2 and si-circPVT1-3) and its negative control si-NC, as well as miR-205-5p mimics/inhibitor and pcDNA3.1-c-FLIP, were all designed by and obtained from Shanghai Sangon Biotech Corp. (Shanghai, China). miR-205-5p mimics/inhibitor was used to enhance or neutralize its function, respectively. Lipofectamine 3000 was used to accomplish the cell transfection following the manufacturers’ instruction. Forty-eight hrs after tansfection, OS cells were allowed for further experiments.
Cell Proliferation Analysis
For Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Kumamoto, Japan) assay, treated OS cells were harvested and re-seeded into 96-well plates at a concentration of 2 × 105 cells/well. After cultured at 37°C for the indicated time, 100 μL of CCK-8 solution was added into each well and incubated for another 2 hrs. Finally, the absorbance was determined at 450 nm using a microplate reader (Bio-Rad Laboratories, USA).
For colony formation assay, treated OS cells were collected and re-seeded into 35-mm petri dishes at a concentration of 1000 cells/per dish. After cultured at 37°C for 2 weeks, the visible colonies were fixed using 4% paraformaldehyde. Then, the colonies were subjected for staining with 0.1% crystal violet solution. After the excessive crystal violet solution was washed off, the colony number was counted manually.
Migration and Invasion Analysis
The migratory and invasive capacities of OS cells were estimated by transwell chambers coated with and without Matrigel matrix (BD Biosciences, USA), respectively. For cell migration, 100 mL treated OS cells (1 × 106 cells/mL) suspended in DMEM without serum were added into the upper room of the transwell chamber without Matrigel matrix. And 500 mL FBS (10%) contained DMEM were added into the lower room. After culture of 24 h, OS cells invaded through the membrane were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet solution. The invasive number of OS cells in five randomly selected fields were counted using a phase-contrast microscope (Olympus, Japan). For cell invasion, experiments were performed as same as migration described above with Matrigel matrix coated transwell chamber.
Western Blot Assay
After lysed in the protease inhibitors contained RIPA buffer (Sigma-Aldrich), total proteins of OS cells were isolated by high-speed centrifuge at 12,000 g/min. BCA protein assay kit (Thermo Fisher) was applied to determine the concentration of extracted proteins, and then 50 μg protein samples were loaded into and isolated by 10% SDS-PAGE (80 V, 30 min and 120 V, 45 min). Next, the isolated proteins were transferred into 0.45-μm polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA; 100 V, 60 min) followed by the incubation of 5% skimmed milk for 2 h. The membranes were incubated overnight with primary antibodies that against c-FLIP (cellular FLICE inhibitory protein, 1:800, ab8421, Abcam, UK), E-cadherin (1:500, ab15148, Abcam, UK), N-cadherin (1:1000, ab76057, Abcam, UK), Vimentin (1:2000, ab137321, Abcam, UK), Snail (1:1000, ab180714, Abcam, UK) and GAPDH (1:10,000, ab181602, Abcam, UK). GAPDH was loaded as an internal reference. After removing the excessive primary antibodies, the membranes were subjected for incubation of horseradish peroxidase conjugated secondary antibodies (IgG-HRP, Abcam, 1:2000, UK) for 2 h, and bands were detected by an enhanced chemiluminescent reagent (ECL, Germany). And the relative protein expression level was quantified using the Image J software.
Dual-Luciferase Reporter Assay
The online prediction database Starbase (http://starbase.sysu.edu.cn/) was used to predict the potential miRNA target of has-circPVT1 and downstream targets of miR-205-5p, respectively. The wild type (WT) and mutant (MUT) miR-205-5p binding sequence included fragments of circPVT1 were cloned and inserted into the pGL3 (Promega, USA) vector to form the recombinant luciferase reporter plasmids, circPVT1-WT and circPVT1-MUT. The circPVT1-WT or circPVT1-MUT and miR-205-5p or miR-NC were co-transfected into OS cells using Lipofectamine 3000. After transfection of 48 h, the luciferase intensity was of OS cell was measured by the Dual-Luciferase Reporter Assay System (Promega). The interplay between miR-205-5p and c-FLIP in OS cells was also validated following the protocols described above.
Statistical Analysis
Data presented in this study by mean ± standard deviation (SD). Student’s t-test and one-way analysis of variance were used to compare the difference between two or multi groups. Pearson’s correlation coefficient analysis was performed to assess the correlation between the two groups. P value less than 0.05 was considered significant.
Results
The Expression of circPVT1 in OS Tissues and Cell Lines
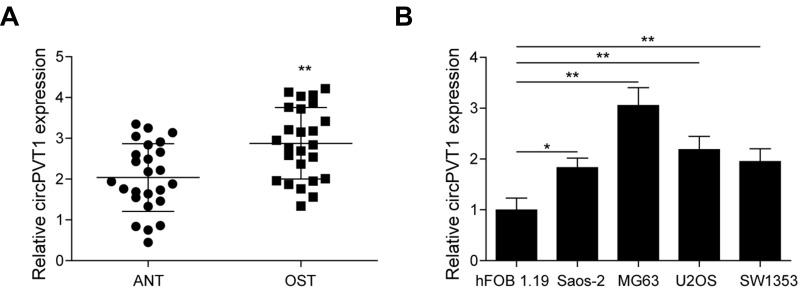
To investigate whether circPVT1 has a role in the progression of OS, we first analyzed its expression in collected 25 pairs of OS and adjacent normal tissue samples by qRT-PCR. As results presented that the circPVT1 level in OS samples was significantly higher than that in adjacent normal tissues (Figure 1A). To further confirm this result, we examined the expression of circPVT1 in four OS cell lines, including Saos-2, MG63, U2OS and SW1353. Compared to hFOB1.19 cell, the circPVT1 level was significantly increased in OS cells (Figure 1B). Among these cell lines, much higher expression levels of circPVT1 in MG63 and U2OS cells and thus were chosen for further experiments.
Figure 1.
The expression of circPVT1 in OS tissues and cell lines. (A) qRT-PCR analysis of circPVT1 in clinical OS and adjacent normal tissue samples. (B) qRT-PCR analysis of circPVT1 in hFOB1.19 cell line and four OS cell lines (Saos-2, MG63, U2OS and SW1353). The data were means ± SD of three independent assays, *P < 0.05; **P < 0.01.
circPVT1 Affects Proliferation, Migration and Invasion via Regulating EMT in OS Cells
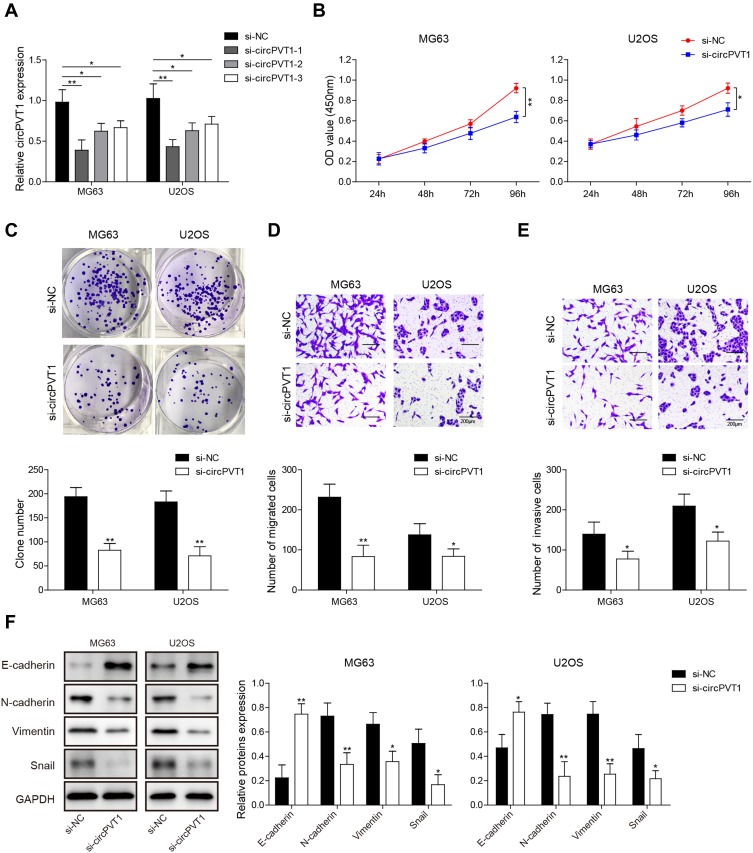
To study whether a positive function of circPVT1 working on OS in vitro, knockdown experiments were performed in MG63 and U2OS cells. Three specific siRNAs that target different parts of circPVT1 (si-circPVT1-1, si-circPVT1-2 and si-circPVT1-3) were designed and transfected into MG63 and U2OS cells. The knockdown efficiency of three siRNAs was validated by qRT-PCR, and si-circPVT1-1, exhibited the best knockdown efficiency, was selected for further study (Figure 2A). CCK-8 and colony formation assays were then adopted to analyze the influences of circPVT1 knockdown on MG63 and U2OS cell proliferation. CCK-8 results showed that the silencing of circPVT1 remarkably attenuated the MG63 and U2OS cell viability (Figure 2B). The colony number of si-circPVT1-transfected groups were markedly lower than that of si-NC transfected groups in colony formation assay (Figure 2C). Moreover, transwell assay was applied to determine the influence of si-circPVT1 on the migratory and invasive abilities of MG63 and U2OS cells. As results illustrated that circPVT1 knockdown resulted in a significant inhibition of cell migration and invasion (Figure 2D and E). In addition, Western blot analysis indicated that si-circPVT1 treatment significantly reduced the protein expression levels of N-cadherin, Vimentin and Snail, while increased E-cadherin expression in MG63 and U2OS cells (Figure 2F). Collectively, our observations demonstrated circPVT1 facilitates OS invasion and metastasis via enhancing cell epithelial–mesenchymal transition (EMT).
Figure 2.
circPVT1 affects proliferation, migration and invasion via regulating EMT in OS cells. (A) After transfected with si-NC, si-circPVT1-1, si-circPVT1-2 and si-circPVT1-3, respectively, MG63 and U2OS cells were subjected for circPVT1 detection by qRT-PCR. (B) CCK-8 and (C) colony formation assays were performed in si-circPVT1 transfected MG63 and U2OS cells to examine the effects of circPVT1 knockdown on OS cell proliferation viability. After transfected with si-circPVT1, the (D) migratory and (E) invasive capacities of MG63 and U2OS cells were evaluated by transwell assay. (F) Western blot detection of E-cadherin, N-cadherin, Vimentin, and Snail in si-circPVT1 transfected MG63 and U2OS cells. The data were means ± SD of three independent assays, *P < 0.05; **P < 0.01.
circPVT1 Functions as a Sponge for miR-205-5p
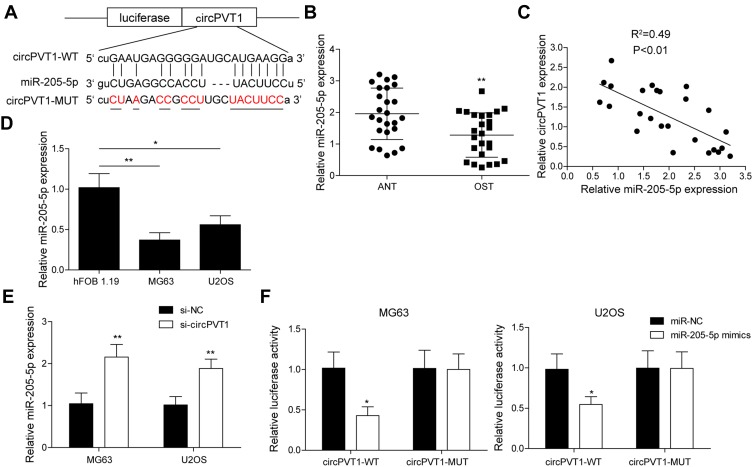
To probe the potential mechanisms of circPVT1 in OS, we hypothesized that it may serve as a sponge of miRNAs to affect the progression of OS based on ceRNA theory. By using a bioinformatics software starBase, we found that circPVT1 possessed a complementary sequence of miR-205-5p (Figure 3A). We then measured the expression of miR-205-5p in collected 25 pairs of OS and adjacent normal tissue samples via qRT-PCR. miR-205-5p expression was remarkably decreased in OS tissues compared to adjacent normal ones (Figure 3B). Pearson’s correlation analysis showed that miR-205-5p exhibited a negative correlation with circPVT1 (Figure 3C). Compared to hFOB1.19 cell, miR-205-5p expression in MG63 and U2OS cells was obviously lower (Figure 3D). Moreover, we revealed that miR-205-5p was remarkably upregulated in circPVT1-silenced MG63 and U2OS cells (Figure 3E). In addition, in dual-luciferase reporter assay, miR-205-5p mimics treatment could attenuate the luciferase activity of MG63 and U2OS driven by circPVT1-WT, but not circPVT1-MUT (Figure 3F), suggesting circPVT1 sponged miR-205-5p.
Figure 3.
circPVT1 functions as a sponge for miR-205-5p. (A) StarBase prediction showing the complementary sequence between circPVT1 and miR-205-5p. (B) qRT-PCR analysis of miR-205-5p in OS and adjacent normal tissue samples. (C) Pearson’s analysis showing the correlation between circPVT1 level and miR-205-5p level in OS sample. (D) Relative miR-205-5p expression in hFOB1.19, USOS and MG63 was detected by qRT-PCR. (E) qRT-PCR was carried out in MG63 and U2OS cells after transfected with si-circPVT1 to examine miR-205-5p level. (F) The interaction between circPVT1 and miR-205-5p was validated by dual-luciferase reporter assay in MG63 and U2OS cells. The data were means ± SD of three independent assays, *P < 0.05; **P < 0.01.
circPVT1 Acts as a ceRNA and Regulates c-FLIP Expression by Competitively Binding to miR-205-5p
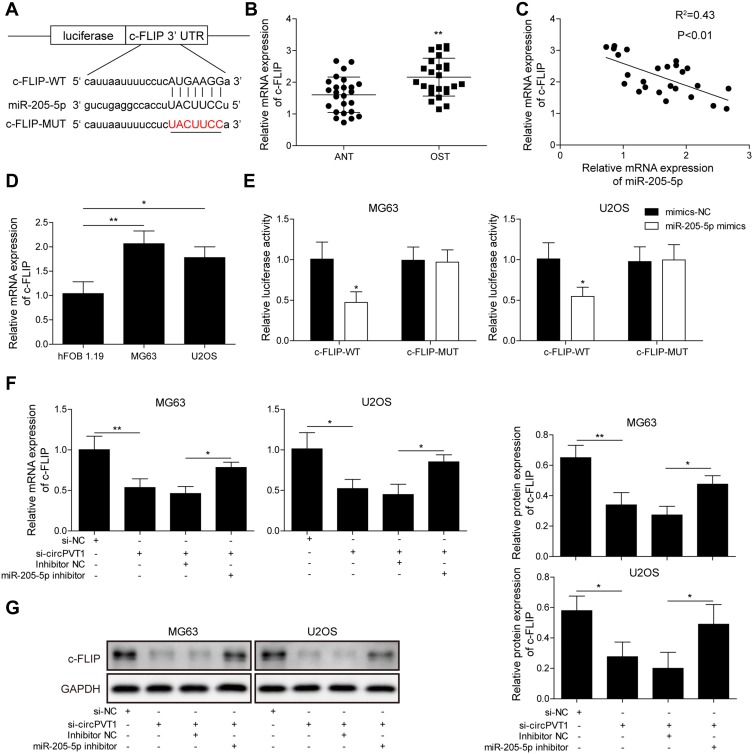
Then, we predicted the target gene of miR-205-5p by starBase. As results indicated that the 3ʹ-UTR of c-FLIP possessed the binding sequence of miR-205-5p (Figure 4A). qRT-PCR analysis showed that c-FLIP was significantly increased in OS samples compared to adjacent normal ones (Figure 4B). Pearson’s correlation analysis revealed that there was a negative correlation between the miR-205-5p level and the c-FLIP level (Figure 4C). Compared to hFOB1.19 cells, c-FLIP expression was markedly higher in U2OS and MG63 cells (Figure 4D). In the dual-luciferase reporter assay validation of c-FLIP and miR-205-5p interaction, we found that miR-205-5p, but not miR-NC, significantly reduced the luciferase intensity of MG63 and U2OS cells driven by c-FLIP-WT, but not c-FLIP-MUT (Figure 4E). To investigate whether circPVT1 could indirectly regulate c-FLIP through miR-205-5p, we examined the c-FLIP expression in MG63 and U2OS cells transfected with si-circPVT1 or miR-205-5p inhibitor by qRT-PCR and Western blot. Results from qRT-PCR indicated that circPVT1 knockdown resulted in a significant downregulation of c-FLIP in MG63 and U2OS cells, however, miR-205-5p inhibitor treatment could partially reverse this downregulation of c-FLIP caused by circPVT1 knockdown, indicating miR-205-5p could suppress the expression level of c-FLIP (Figure 4F). Western blot analysis of c-FLIP protein expression obtained consistent results with qRT-PCR (Figure 4G).
Figure 4.
circPVT1 acts as a ceRNA and regulates c-FLIP expression by competitively binding to miR-205-5p. (A) StarBase prediction showing the complementary sequence between the 3ʹ-UTR and miR-205-5p. (B) Relative c-FLIP mRNA expression of OS and adjacent normal tissue samples was tested by qRT-PCR. (C) Pearson’s analysis exhibiting the correlation between miR-205-5p level and c-FLIP level in OS sample. (D) Relative c-FLIP mRNA expression in hFOB1.19, U2OS and MG63 was measured by qRT-PCR. (E) Interaction between miR-205-5p and c-FLIP in MG63 and U2OS cells was validated via dual-luciferase reporter assay. (F, G) Expression of c-FLIP was detected in MG63 and U2OS cells treated with si-circPVT1 or miR-205-5p inhibitor by qRT-PCR and Western blot. The data were means ± SD of three independent assays, *P < 0.05; **P < 0.01.
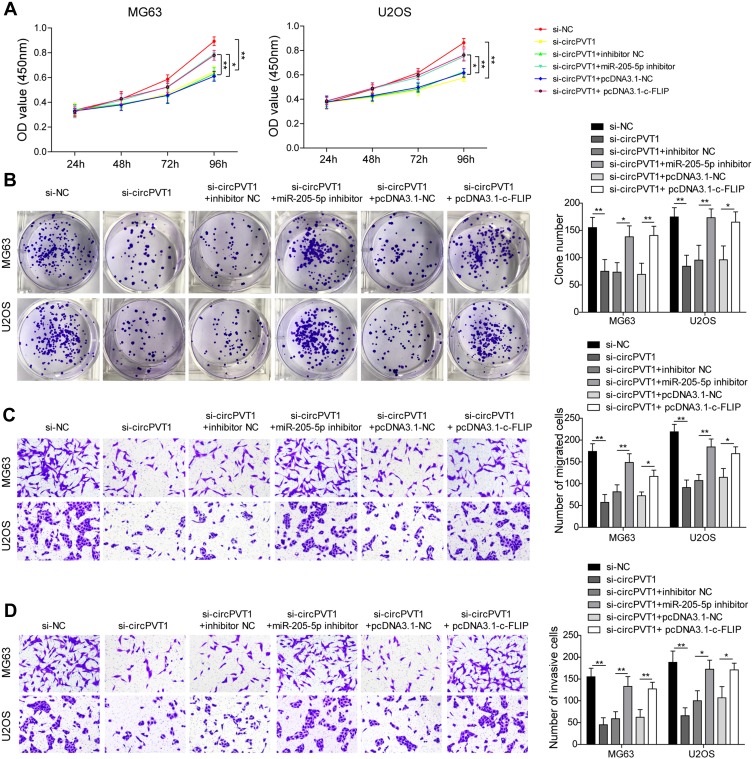
circPVT1 Regulates Invasion and Metastasis of OS Cells Through miR-205-5p/c-FLIP Axis
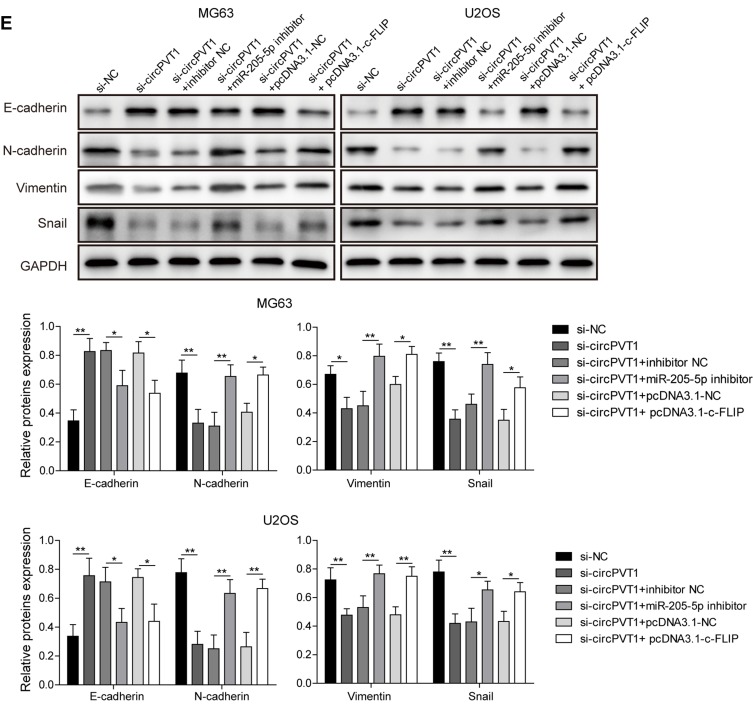
To further assess the biological function of circPVT1/miR-205-5p/c-FLIP axis in OS progression, we examined the effects of miR-205-5p inhibition and c-FLIP overexpression on the suppressive influences of si-circPVT1 on OS progression through CCK-8, colony formation and transwell assays. In the CCK-8 and colony formation assays, we revealed that si-circPVT1 transfection resulted in a significant repression on MG63 and U2OS cell proliferation, however, transfection of miR-205-5p inhibitor or c-FLIP partially abolished the effects of si-circPVT1 (Figure 5A and B). Moreover, in the transwell analysis of cell migration and invasion, we demonstrated that the suppressive roles of si-circPVT1 in MG63 and U2OS cell migration and invasion were abrogated by the treatment of miR-205-5p inhibitor or c-FLIP (Figure 5C and D). Finally, we also revealed that the inhibitory effects of si-circPVT1 on the EMT phenomenon could be blocked by miR-205-5p inhibitor or c-FLIP (Figure 5E). Taken together, circPVT1 induces cell EMT through the miR-205-5p/c-FLIP axis, thus promoting OS invasion and metastasis in vitro.
Figure 5.
circPVT1 regulates invasion and metastasis of OS cells through miR-205-5p/c-FLIP axis. (A) CCK-8 and (B) colony formation assays were utilized to examine the cell proliferation of MG63 and U2OS cells after transfection of si-circPVT1, miR-205-5p inhibitor or c-FLIP. Effects of miR-205-5p inhibitor and c-FLIP on (C) migratory and (D) invasive capacities of circPVT1 knockdown MG63 and U2OS cells were analyzed by transwell assay. (E) Western blot analysis of E-cadherin, N-cadherin, Vimentin, and Snail in si-circPVT1, miR-205-5p inhibitor or c-FLIP transfected MG63 and U2OS cells. The data were means ± SD of three independent assays, *P < 0.05; **P < 0.01.
Figure 5.
Continued.
Discussion
OS, a malignant bone tumour, commonly causes the main mortality of patients for its metastasis. Due to the limitation of OS treatment, it is urgent to explore new strategies. Recently, ncRNA has been reported to play critical roles in OS progression. In the present study, circPVT1 was revealed to be remarkably increased in OS tissues and cell lines, and knockdown of circPVT1 resulted in a repression on the processes of OS cell proliferation, migration, invasion via inhibiting EMT. In mechanism, c-FLIP was demonstrated to be indirectly regulated by circPVT1 through sponging miR-205-5p in a ceRNA manner. Finally, rescue experiments indicated that miR-205-5p inhibitor or c-FLIP transfection reversed the inhibitory effects of si-circPVT1 on OS invasion and metastasis in vitro.
The transition process from epithelium to mesenchyme is termed as EMT, in which polarized epithelium loses the ability of adhesion and acquire mesenchymal cell phenotype.24 EMT process is involved in multiple physiological and pathological events, such as embryogenesis, wound healing and tumor cell metastasis and invasion.25–27 circRNA has been proven to possess the ability of regulating the process of EMT, and thus participate in the regulation of tumor cells metastasis and invasion. For example, circ_0023642 was reported by Zhou et al in 2018 to enhance the capacities of migration and invasion of gastric cancer cells through facilitating the EMT process.28 circCDR1as was also demonstrated to promote OS cell migration through facilitating the EMT process by sponging miR-7.29 circPVT1 was firstly identified by Chen et al in gastric cancer samples using a computational algorithm (anchor alignment) in 2016.19 Up to now, the oncogenic role of circPVT1 has been confirmed in several human tumors, including OS.23,30 There is only one report involved in the effects of circPVT1 on tumor cell metastasis, and whether this effect is mediated by EMT remains undetermined.20 In our study, circPVT1 was found highly expressed in OS tissues and cell lines, which is consistent with the conclusion of the previous study.23 As we know, downregulation of E-cadherin and upregulation of N-cadherin, Vimentin are crucial events during EMT, and our results showed that knockdown of circPVT1 caused a significant increase of E-cadherin and decrease of N-cadherin, Vimentin. Taken together, our findings supported that circPVT1 might facilitate OS cell invasion and metastasis through promoting EMT phenotype.
To our knowledge, circRNA exists multiple function mechanisms, including interaction with RNA-binding proteins and sponging miRNA to regulate target mRNA. Since the discovery of ceRNA hypothesis, the functions of circRNA-miRNA-mRNA axis in human diseases have been well documented by numerous studies, including diabetes, major depressive disorder, osteoarthritis, heart failure and carcinomas.31 For example, circTCF25 was validated to be bound by miR-103a/miR-107 in bladder cancer cells and thus indirectly increased the level of CDK6 genes to promote bladder cell invasion and metastasis, implying that circTCF25-miR-103a-3p/miR-107-CDK6 axis was contributed to the progression of bladder cancer.32 circPVT1 was also revealed to facilitate the progression of non-small cell lung cancer (NSCLC) by sponging miR-125b to release E2F2, indicating a significant role of the circPVT1-miR-125b-E2F2 axis in NSCLC.33 In our study, we demonstrated miR-205-5p could bind to circPVT1 or c-FLIP, and circPVT1 could indirectly regulate c-FLIP through sponging miR-205-5p and reducing its expression. There may be some unknown mechanisms that can partially degrade binding miRNA, which is an interesting scientific topic worth exploring. Moreover, we previously study showed that miR-205-5p acted as a tumor suppressor in OS and inhibited the malignant phenotypes of OS cells.34 c-FLIP, an inhibitor of the apoptotic pathway, was significantly higher in the lung metastases than in the primary tumors and may play an important role in the metastasis of OS.35 In the present results, miR-205-5p knockdown and c-FLIP overexpression were showed to abolish the inhibitory effects of si-circPVT1 on OS cell proliferation, migration and invasion, further suggesting miR-205-5p and c-FLIP were implicated in OS invasion and metastasis progression. Accordingly, our findings supported the existence and function of circPVT1-miR-205-5p-c-FLIP axis in OS. Nevertheless, the function of circPVT1/miR-205-5p/c-FLIP axis in OS was only validated in vitro in our study, in vivo experiments need to be performed in the future research. Whether circPVT1 can interact with RNA-binding proteins to increase or decrease mRNA stability and further to regulate OS progression and drug resistance are also directions we should work on.
In conclusion, our results provided strong evidence that circPVT1 facilitates the invasion and metastasis of OS by releasing c-FLIP through the interaction with miR-205-5p. Thus, circPVT1/miR-205-5p/c-FLIP regulatory pathway might be a promising therapeutic target for OS. Nevertheless, the function of circPVT1/miR-205-5p/c-FLIP axis in OS was only validating in vitro in our study, in vivo studies are needed to further confirm in our future research.
Disclosure
The authors have no commercial or other associations that might pose a conflict of interest in this work.
References
- 1.Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. [DOI] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. [DOI] [PubMed] [Google Scholar]
- 3.Bishop MW, Janeway KA, Gorlick R. Future directions in the treatment of osteosarcoma. Curr Opin Pediatr. 2016;28(1):26–33. doi: 10.1097/MOP.0000000000000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison DJ, Geller DS, Gill JD, et al. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18(1):39–50. doi: 10.1080/14737140.2018.1413939 [DOI] [PubMed] [Google Scholar]
- 5.Khanna C, Wan X, Bose S, et al. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10(2):182–186. doi: 10.1038/nm982 [DOI] [PubMed] [Google Scholar]
- 6.Yu S, Fourman MS, Mahjoub A, et al. Lung cells support osteosarcoma cell migration and survival. BMC Cancer. 2017;17(1):78. doi: 10.1186/s12885-017-3047-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyder CL, Saltzman DA, Ferrell KL, Thompson RC, Leonard AS. A new approach to the resection of pulmonary osteosarcoma metastases. Results of aggressive metastasectomy. Clin Orthop Relat Res. 1991;270:247–253. [PubMed] [Google Scholar]
- 8.Huang X, Zhao J, Bai J, et al. Risk and clinicopathological features of osteosarcoma metastasis to the lung: a population-based study. J Bone Oncol. 2019;16:100230. doi: 10.1016/j.jbo.2019.100230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abraham KJ, Ostrowski LA, Mekhail K. Non-coding RNA molecules connect calorie restriction and lifespan. J Mol Biol. 2017;429(21):3196–3214. doi: 10.1016/j.jmb.2016.08.020 [DOI] [PubMed] [Google Scholar]
- 11.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54. doi: 10.1016/j.gpb.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429. doi: 10.2174/138920101505140828161335 [DOI] [PubMed] [Google Scholar]
- 13.He J, Xie Q, Xu H, et al. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027 [DOI] [PubMed] [Google Scholar]
- 14.Zhang HD, Jiang L-H, Sun D-W, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi: 10.1007/s12282-017-0793-9 [DOI] [PubMed] [Google Scholar]
- 15.Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu S, Liu Z, Yang X, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–309. doi: 10.1016/j.canlet.2017.11.022 [DOI] [PubMed] [Google Scholar]
- 18.He R, Liu P, Xie X, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36(1):145. doi: 10.1186/s13046-017-0614-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Su M, Xiang B, et al. Circular RNA PVT1 promotes metastasis via miR-145 sponging in CRC. Biochem Biophys Res Commun. 2019;512(4):716–722. doi: 10.1016/j.bbrc.2019.03.121 [DOI] [PubMed] [Google Scholar]
- 21.Verduci L, Ferraiuolo M, Sacconi A, et al. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017;18(1):237. doi: 10.1186/s13059-017-1368-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J, Han Q, Gu Y, et al. Circular RNA PVT1 expression and its roles in acute lymphoblastic leukemia. Epigenomics. 2018;10(6):723–732. doi: 10.2217/epi-2017-0142 [DOI] [PubMed] [Google Scholar]
- 23.Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14(3):321–330. doi: 10.7150/ijbs.24360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen T, You Y, Jiang H, et al. Epithelial-mesenchymal transition (EMT): a biological process in the development, stem cell differentiation, and tumorigenesis. J Cell Physiol. 2017;232(12):3261–3272. doi: 10.1002/jcp.v232.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gros J, Tabin CJ. Vertebrate limb bud formation is initiated by localized epithelial-to-mesenchymal transition. Science. 2014;343(6176):1253–1256. doi: 10.1126/science.1248228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee P, Venkatachalam S, Mamidi MK, et al. Vitiligo patient-derived keratinocytes exhibit characteristics of normal wound healing via epithelial to mesenchymal transition. Exp Dermatol. 2015;24(5):391–393. doi: 10.1111/exd.12671 [DOI] [PubMed] [Google Scholar]
- 27.Ruscetti M, Quach B, Dadashian EL, et al. Tracking and functional characterization of epithelial-mesenchymal transition and mesenchymal tumor cells during prostate cancer metastasis. Cancer Res. 2015;75(13):2749–2759. doi: 10.1158/0008-5472.CAN-14-3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou L-H, Yang Y-C, Zhang R-Y, et al. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22(8):2297–2303. doi: 10.26355/eurrev_201804_14818 [DOI] [PubMed] [Google Scholar]
- 29.Xu B, Yang T, Wang Z, et al. CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag Res. 2018;10:4871–4880. doi: 10.2147/CMAR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adhikary J, Chakraborty S, Dalal S, et al. Circular PVT1: an oncogenic non-coding RNA with emerging clinical importance. J Clin Pathol. 2019;72:513–519. doi: 10.1136/jclinpath-2019-205891 [DOI] [PubMed] [Google Scholar]
- 31.Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–73281. doi: 10.18632/oncotarget.v8i42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Zhang Z, Jiang H, et al. Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. Cell Physiol Biochem. 2018;51(5):2324–2340. doi: 10.1159/000495876 [DOI] [PubMed] [Google Scholar]
- 34.Zhang C, Long F, Wan J, et al. MicroRNA-205 acts as a tumor suppressor in osteosarcoma via targeting RUNX2. Oncol Rep. 2016;35(6):3275–3284. doi: 10.3892/or.2016.4700 [DOI] [PubMed] [Google Scholar]
- 35.Rao-Bindal K, Rao CK, Yu L, et al. Expression of c-FLIP in pulmonary metastases in osteosarcoma patients and human xenografts. Pediatr Blood Cancer. 2013;60(4):575–579. doi: 10.1002/pbc.v60.4 [DOI] [PMC free article] [PubMed] [Google Scholar]