Abstract
Background
Long non-coding RNAs (lncRNAs), as competing endogenous RNAs (ceRNAs), can regulate various pathophysiological processes by binding competitively to microRNAs at the post-transcription level. Our previous work demonstrated that miR-146a-5p was lowly expressed in diabetic peripheral neuropathy (DPN) rats. However, the ceRNA network in DPN mediated by lncRNAs and miR-146a-5p remains to be explored.
Methods
Two groups of rats (n=4 per group), a type 2 diabetes (T2DM) group and a DPN group, were used in this study. Sciatic nerve conduction velocity (NCV) of each rat was determined at the 6th and the 12th week. LncRNA microarray analysis was performed in the sciatic nerve of DPN and T2DM rats. Based on the TargetScan algorithm and the miRanda database, we determined the differentially expressed (DE) lncRNAs bound to miR-146a-5p. Furthermore, we verified the DE lncRNAs potentially bound to miR-146a-5p by qRT-PCR. The genes targeted by miR-146a-5p were identified by bioinformatics prediction and experimental techniques.
Results
We found 413 DE lncRNAs between DPN and T2DM rats (|log2FC| ≥ 2 and adjust P ≤ 0.05). Eight DE lncRNAs were predicted to bind to miR-146a-5p by both algorithms, of which four were verified by qRT-PCR. TRAF6, IRAK1, and SMAD4 were identified as miR-146a-5p targeted genes and were predominantly enriched in the inflammatory signaling pathway.
Conclusion
LncRNAs may contribute to the pathogenesis of DPN by regulating inflammation through functioning as ceRNAs of miR-146a-5p.
Keywords: diabetic peripheral neuropathy, long non-coding RNA, MicroRNA, inflammation
Introduction
Diabetes mellitus (DM) has emerged as one of the most common non-communicable diseases with increasing prevalence over the world.1 Diabetic peripheral neuropathy (DPN), one of the most common microvascular complications of DM, affects over 50% of diabetic patients and has emerged as a severe public health problem.2 This chronic complication causes immense financial burden and seriously decreases the life quality and expectancy of diabetic patients.3 DPN is induced by multifactorial metabolic disorders, including abnormal metabolism of glucose, lipid, and protein, vascular abnormalities, neurotrophic factor insufficiency, oxidative stress, immune damage.4,5 Current therapeutic strategies for DPN, including pathogenesis-oriented and symptomatic therapies, are of limited efficacy.6,7 This suggests that effective treatments to decrease the incidence of DPN remain to be discovered. Therefore, it is imperative to explore new mechanisms underlying DPN.
With the development of genomics, human genome transcript regions are estimated as high as 76%. However, over 98% of the transcripts are non-coding RNAs, including microRNA (miRNA), long non-coding RNAs (lncRNAs), circular RNAs, and other regulatory non-coding RNAs.8 Non-coding RNAs are demonstrated to play significant regulatory functions in various diseases, which has attracted widespread attention from researchers.9–11 LncRNAs are non-coding endogenous RNAs with a length of over 200 nucleotides and are involved in multiple biological processes, such as cell proliferation, differentiation, chromosome remodeling, epigenetic regulation, as well as transcriptional and post-transcriptional modification.12 MiRNAs, with a length of approximately 22 nucleotides, regulate gene expression through silencing or degrading of mRNA translation by base pairing at the post-transcription level.13 LncRNAs function as competing endogenous RNAs (ceRNAs) and regulate gene expression by binding competitively to miRNAs at the post-transcription level.14 These interactions between miRNA and lncRNA have been widely studied in the cytoplasm using experimental techniques. This post-transcriptional gene regulation mode is more convenient and effective in comparison to traditional gene regulation, which has attracted enormous attention in the field of epigenetic regulation.15
In recent studies, it was demonstrated that lncRNAs are significant regulators in the development of diabetic complications.16,17 Some studies have reported that several lncRNAs may be involved in the pathogenesis of DPN.18,19 However, the underlying mechanism remains unknown. We previously showed that miR-146a-5p was differentially expressed between T2DM and DPN rats and that it could suppress the inflammatory response in DPN.20,21 Therefore, in this study, we explored the potential ceRNA network targeting miR-146a-5p in a DPN rat model.
Materials and Methods
Reagents
Streptozocin (STZ) was purchased from Sigma Life sciences and biotechnology (St. Louis, MO, USA). TRIzol was obtained from Takara Technology (Kusatsu, Shiga Prefecture, Japan).
Animals
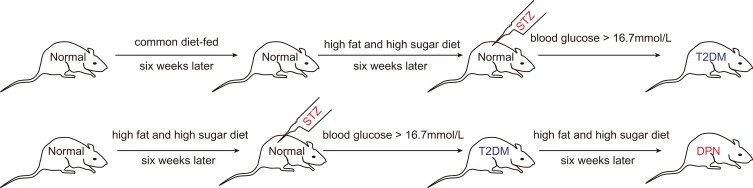
Eight male Sprague-Dawley (SD) rats were ordered from the Shanghai Slack Laboratory Animal Co., Ltd. All rats were raised according to the Shanghai Animal Management Committee regulations. The Ethics Committee of Animal Care of Jinshan Hospital approved the rat experiments (NO. Jin Medical Ethics 2016(23)). Rats use was complied with the Shanghai Animal Management Committee regulations. Rats were randomly divided into a T2DM and a DPN group, with four rats per group. The T2DM group rats were fed common feed for six weeks, and they were given a high sugar and high fat diet containing 10% lard, 20% sucrose and 70% regular feed for another 6 weeks. Then, the T2DM group rats were administered STZ intraperitoneally at a dosage of 35 mg/kg. Fasting blood glucose levels (fasted for 12 h) were measured 72 hrs after intraperitoneal injection. Rats with blood glucose levels >16.7 mmol/L were defined as T2DM rats. The DPN group rats were intraperitoneally injected with STZ after being fed the high fat and high sugar feed for 6 weeks, successfully establishing T2DM rat model. Then these T2DM rats were fed the high fat and high sugar diet for an additional 6 weeks to establish a DPN model (Figure 1).
Figure 1.
Detailed flow chart of establishing T2DM and DPN rat model.
Measurement of Nerve Conduction Velocity
The sensory nerve conduction velocity (SNCV) and motor nerve conduction velocity (MNCV) were measured at the 6th and 12th week. 10% of chloral hydrate solution was applied for anesthesia of rats at a dose of 35mg/kg. The SNCV and MNCV were determined and documented as described previously.22
Selection of Differentially Expressed (DE) lncRNAs
We extracted total RNA from the sciatic nerve of T2DM and DPN rats using the TRIzol method based on the manufacturer’s product manual. The lncRNA expression profile was determined using an Arraystar Rat LncRNA Microarray V2.0 array (Arraystar Flash RNA Labeling Kit, Arraystar, Inc., Rockville, USA). The lncRNA microarray analysis was conducted at the Shanghai KangChen Bio-tech Corporation. The experimental procedures were performed as described in detail on the KangChen Bio-tech website (http://www.aksomics.com/). The acquired array images were analyzed using Agilent Feature Extraction software (version 11.0.1.1). The GeneSpring GX v12.1 software package (Agilent Technologies) was utilized to perform quantile normalization and subsequent data processing. DE LncRNAs with statistical significance between the two groups were identified through |log2FC| ≥ 2 and adjust P < 0.05 filtering. Then, the pheatmap package of R software was used to construct a heatmap of the clustering analysis.
Identification of DE lncRNAs Bound to miR-146a-5p
As our previous study described, we found miR-146a-5p was downregulated in DPN rats as compared to T2DM rats.20 We identified DE lncRNAs that bind to miR-146a-5p according to the calculation principle of TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/) database.
Identification of mRNAs Targeted by miR-146a-5p
Moreover, we analyzed the mRNAs targeted by miR-146a-5p in the database of TargetScan, miRDB (http://www.mirdb.org/), and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/). Then we looked for intersections of these predicted genes found in the above three databases. Cytoscape software was used to build a ceRNA network.
Quantitative Real-Time PCR Analysis
Total RNA was extracted from sciatic nerve tissues using TRIzol reagent (TaKaRa, Japan). PrimeScript RT Master Mix (TaKaRa, Japan) was applied to synthesize cDNA from lncRNAs, including XR_589933, XR_351905, XR_353891, XR_357013, XR_600244, XR_595664, XR_598132 and XR_589615. SYBR Premix Ex Taq (Tli RNaseH Plus, TaKaRa, Japan) was used to determine the lncRNAs expression levels of target genes. The lncRNA expression levels were recorded using the 2−ΔΔCt system. We used β-actin as an internal quantitative control to normalize lncRNA expression levels. The primers used for the test of lncRNAs were presented in Table 1.
Table 1.
The Primers Used for the Test of lncRNAs
| Gene | Forward (5ʹ to 3ʹ) | Reverse (5ʹ to 3ʹ) |
|---|---|---|
| XR_589933 | TGGCTTCCGCTGATTCCTCACTGA | TCCTCTTGCCTCCTGCTGACTTGG |
| XR_351905 | GCCTCTGAGCGAGTTAGTGCGATA | AGAAGACGGAGGACACGACCACAT |
| XR_353891 | TGCCAAGGCTTGCTGACTGTGTTC | AAGTTCAGAGATCGCCGCCAGGAG |
| XR_357013 | GGCACAAGCCCACTGCTCCTTT | TCCCGATCCCAGCGGCACTAAA |
| XR_600244 | GCAGGAGCCACAGTAGCAGGAACA | TGAGGTCACCTTAGGTCGTGTCCA |
| XR_595664 | TTGCTTGCTTCCTGACCACCAACA | GCTCCATGCACCTCAGAACCACTC |
| XR_598132 | GCCAAACGAAATGCCTGCCCATAA | GAGAACCTGCCACCTGAAGACCAT |
| XR_589615 | TCCCAAGGCGGCTTACTGCTTT | AGGAGTTAGGCGGTGGCGGTTA |
Statistical Analysis
Experimental data were presented as the mean ± standard deviation. SPSS 25.0 (IBM Corp., Armonk, N.Y., USA) was used to conduct statistical analyses. A two-independent-sample t-test was applied to determine the differences between two independent samples. A P value < 0.05 was considered statistically significant.
Results
Measurement of NCV in the Two Groups of Rats
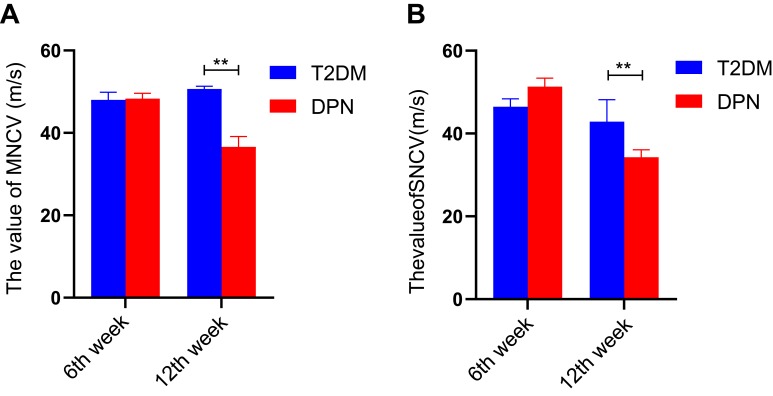
We measured the MNCV and SNCV for the two groups of rats. There existed no significant differences in either MNCV or SNCV between the T2DM and DPN groups at the 6th week (P > 0.05, Figure 2). At the 12th week, the MNCV and SNCV gradually decreased in the DPN group compared with the T2DM group (P < 0.01, Figure 2). This indicates that the DPN rat model was successfully established.
Figure 2.
NCV values for T2DM and DPN groups (m/s). (A) MNCV values for T2DM and DPN groups at assessed time points. (B) SNCV values for T2DM and DPN groups at assessed time points. ** P < 0.01.
Identification of DE lncRNAs in the Sciatic Nerve of T2DM and DPN Rats
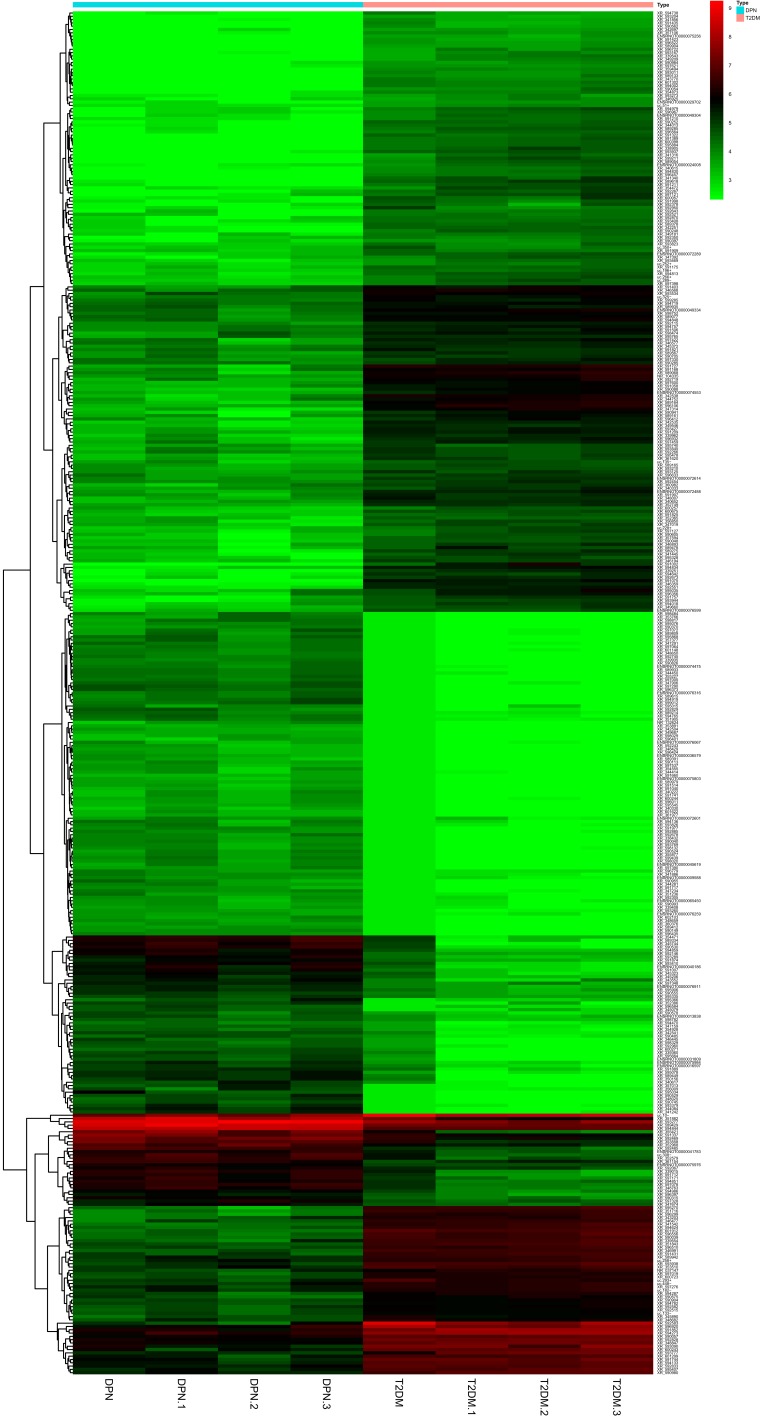
A lncRNA microarray analysis was performed to compare the lncRNA expression profile in sciatic nerves in the two groups of rats. We found 413 DE lncRNAs between the two groups of rats (|log2FC| ≥ 2 and adjust P < 0.05, Figure 3). Compared with the T2DM group, 234 lncRNAs were downregulated and 179 lncRNAs were upregulated in the DPN group. The heatmap graphically presents the DE lncRNAs. The detailed information of the lncRNAs are provided in a supplementary xlsx file.
Figure 3.
The heatmap of DE lncRNA for T2DM and DPN rats (log2FC| ≥ 2 and adjust P < 0.05). Cluster analysis of the test sample from the sciatic nerve of T2DM rats and DPN rats. These lncRNAs were selected on the basis of their differential expression (|log2FC| ≥ 2 and adjust P < 0.05). The values shown were the means of the normalized expressions from two independent group. Upregulated and downregulated lncRNAs in sciatic nerve were shown in red and green respectively.
Preliminary Screening of DE lncRNAs Bound to miR-146a-5p
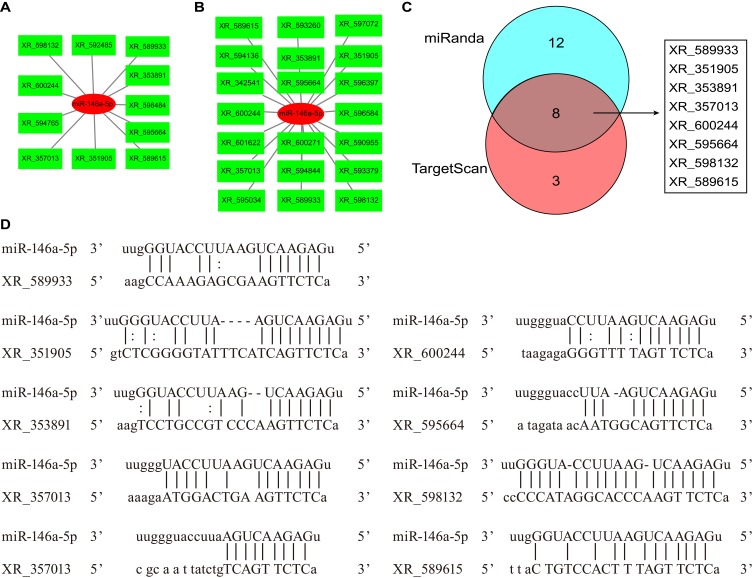
We previously established that miR-146a-5p was decreased in DPN rats compared with T2DM rats.20 In the ceRNA network, the correlation of lncRNA and mRNA was positive, while the correlation of lncRNA and miRNA was negative. Therefore, we analyzed the DE lncRNAs that were predicted to bind to miR-146a-5p among the upregulated lncRNAs. With the algorithm of the TargetScan database, 11 DE lncRNAs were found to bind to miR-146a-5p with 8mer binding sites (Figure 4A). 20 DE lncRNAs that could bind to miR-146a-5p were identified by the algorithm of the miRanda database (Figure 4B). There were eight overlapping DE lncRNAs from the two algorithms, including XR_589933, XR_351905, XR_353891, XR_357013, XR_600244, XR_595664, XR_598132, and XR_589615 (Figure 4C). Potential binding sites between miR-146a-5p and DE lncRNAs were presented (Figure 4D).
Figure 4.
DE lncRNAs predicted to bind miR-146a-5p. (A) DE lncRNAs predicted to bind miR-146a-5p by the algorithm of the TargetScan database. miR-146a-5p and lncRNAs were shown in red and green respectively. (B) DE lncRNAs predicted to bind miR-146a-5p by the algorithm of the miRanda database. miR-146a-5p and lncRNAs were shown in red and green respectively. (C) DE lncRNAs predicted to bind miR-146a-5p overlapping for the two algorithms, which was shown in gray. (D) Potential binding sites between miR-146a-5p and DE lncRNAs.
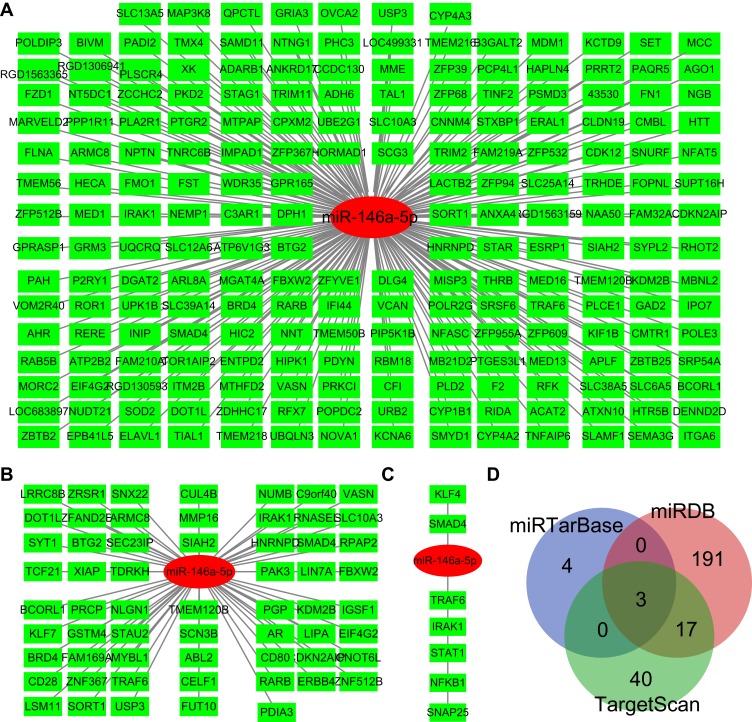
Identification of mRNAs Targeted by miR-146a-5p
To obtain miR-146a-5p targeted genes, we retrieved the TargetScan, miRDB, and miRTarBase database. In the miRDB database, we found that miR-146a-5p specifically binds to 211 genes with a target score ≥50 (Figure 5A). In the TargetScan database, there were 60 genes that may be targeted by miR-146a-5p with at least one conserved 8mer binding site (Figure 5B). Moreover, seven genes were predicted to bind with miR-146a-5p in the miRTarBase database (Figure 5C). At last, three mRNAs targeted by miR-146a-5p were found by all three databases, including tumor necrosis factor receptor-associated factor 6 (TRAF6), interleukin 1 receptor-associated kinase 1 (IRAK1), and SMAD4 (Figure 5D).
Figure 5.
mRNAs targeted by miR-146a-5p. (A) mRNAs targeted by miR-146a-5p as predicted by the miRDB database. miR-146a-5p and mRNAs were shown in red and green respectively. (B) mRNAs targeted by miR-146a-5p as predicted by the TargetScan database. miR-146a-5p and mRNAs were shown in red and green respectively. (C) mRNAs targeted by miR-146a-5p as predicted by the miRTarBase database. miR-146a-5p and mRNAs were shown in red and green respectively. (D) Overlapping mRNAs targeted by miR-146a-5p in all three databases, which was shown in gray.
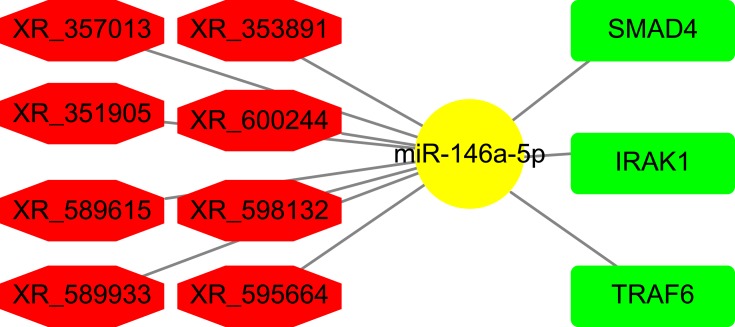
Construction of ceRNA Network
Based on the lncRNA-miRNA and miRNA-mRNA pairs, we constructed a network using the Cytoscape software. In this network, eight DE lncRNAs, miR-146a-5p, and three mRNA were included (Figure 6). The constructed network suggests that a ceRNA network is found in DPN, which may correlate with the onset of DPN.
Figure 6.
Network of ceRNAs mediated by DE lncRNAs. miR-146a-5p was shown in yellow. DE lncRNAs were shown in red. mRNAs were shown in green.
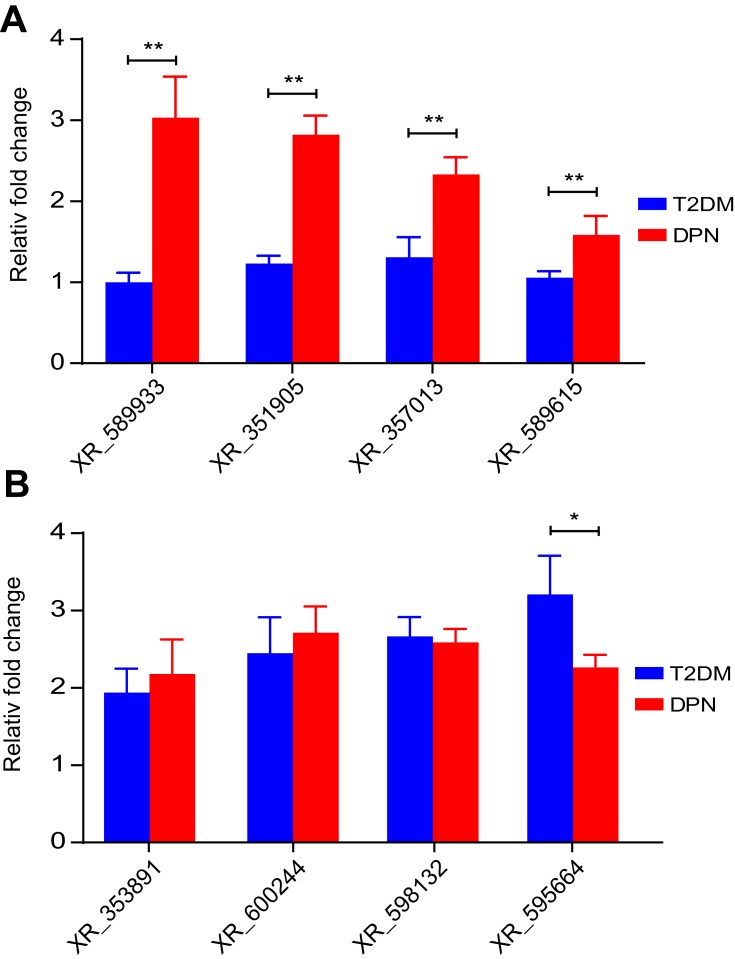
Validation of DE lncRNAs by qRT-PCR Analysis
It has previously been verified that miR-146a-5p expression in the DPN group was lower than that in the T2DM group. Therefore, we performed qRT-PCR to verify the DE lncRNAs bound to miR-146a-5p in the aforementioned ceRNA network. As shown in Figure 7A, qRT-PCR analysis confirmed that four DE lncRNAs; XR_589933, XR_351905, XR_357013, and XR_589615, were upregulated in the sciatic nerves of the DPN group in comparison to the T2DM group (P < 0.01). However, qRT-PCR analysis verified that the expression of XR_353891, XR_600244, and XR_598132 in the sciatic nerves of the DPN rats were identical to those of the T2DM group (P > 0.05, Figure 7B). The expression of XR_595664 in the sciatic nerves of the DPN rats was lower than that of the T2DM group (P < 0.05, Figure 7B). These findings indicate that these four DE lncRNAs functioning as ceRNAs may participate in the pathogenesis of DPN by absorbing miR-146a-5p.
Figure 7.
Validation of DE lncRNAs. Using qRT-PCR analysis, DE lncRNAs were measured in sciatic nerves of T2DM and DPN group rats. (A) The expression of XR_589933, XR_351905, XR_357013, and XR_589615. (B) The expression of XR_353891, XR_600244, XR_595664, and XR_598132. ** P < 0.01, * P < 0.05.
Discussion
DPN, a common microvascular complication of DM, has become the main cause of non-traumatic amputations.2 Because multiple factors are implicated in the pathogenesis of DPN, there are still no specific treatments for preventing the occurrence or development of DPN.4 It has been reported that lncRNAs are involved in the pathogenesis of DM and diabetic complications.23
LncRNAs are endogenous regulators and play vital roles in various pathophysiological processes, which can regulate gene expression through competitively binding to miRNA at the post-transcriptional level. Several studies have confirmed that lncRNAs widely contribute to the pathophysiology process of DM and diabetic complications by acting as ceNRAs.17,24 Recently, lncRNA NEAT1 was shown to regulate the mTOR signaling pathway via absorbing miR-181b like a sponge.25 In the pathogenesis of diabetic cardiomyopathy, lncRNA XIST is involved in the ceRNA regulatory network by interacting with miR‑424‑5p and miR‑497‑5p.26 It also has been reported that lncRNA-ES3 regulates the expression of Bcl-2 modifying factor and plays a role in the calcification and senescence of aorta vasculature in vitro by acting as a miR-34c-5p sponge.27 Moreover, lncRNAs are demonstrated to be associated with the pathogenesis of diabetic retinopathy by performing the function of ceRNA.28 Several microarray analyses have been conducted to reveal the potential roles of lncRNAs in DPN.18,29 However, whether ceRNA networks mediated by lncRNAs have a positive correlation with the pathophysiology of DPN still needs to be elucidated.
In this study, we identified the DE lncRNAs between DPN and T2DM rat model groups. In our previous study, we verified that the expression of miR-146a-5p was downregulated in the DPN rat as compared to the T2DM rats.20 Overexpressing miR-146a-5p significantly reduced the level of inflammatory response and ameliorated the sciatic nerve injury in DPN rats, which confirmed that miR-146a-5p played a role as an inflammation regulator in DPN.21 Accordingly, finding lncRNAs that bind to miR-146a-5p is at the core of constructing a ceRNA network in DPN. Currently, multiple databases provide functional predictions of RNA binding between lncRNAs and miRNAs, such as lncRNASNP2, miRcode, LNCediting, StarBase, and DIANA-LncBase V2. The miRcode database only contains human predictions of lncRNAs and miRNAs interactions,30 and was last updated in June 2012, which limits the probability of accurate prediction. The StarBase database was last updated in 2019. Nevertheless, the StarBase database is mainly focused on human and mouse.31 Some other databases cannot provide rat predictions about lncRNA-miRNA interaction either. The binding between lncRNA and miRNA primarily depends on microRNA recognition elements in lncRNAs, which is similar to the binding mode of miRNA and mRNA. Therefore, we predicted the interaction of lncRNA and miRNA in DPN using the algorithm applied in predicting miRNA-mRNA interactions.
As described above, the algorithms in the TargetScan and miRanda databases were used to predict the interaction between lncRNA and miRNA in rats. In the TargetScan database, the predicted targets of miRNAs are recognized by complementary combination with the seed area (nucleotides 2–8) of the miRNAs by the matched position of the 8mer, 7mer, and 6mer.32 The algorithm in miRanda is used to detect potential miRNA target sites based on various parameters, including energy threshold, alignment score threshold, scaling parameter to scale, and strict alignment in the seed region.33 TargetScan and miRanda provide the prediction of binding between lncRNA and miRNA by distinct algorithms. Therefore, we obtained lncRNA-miRNA interactions with both respective algorithms and then determined the overlap between the TargetScan and miRanda databases, which elevated the probability of an accurate prediction. Finally, we obtained eight DE lncRNAs that are predicted to bind miR-146a-5p. We further explored the ceRNA network related to miR-146a-5p. Four DE lncRNAs predicted to bind to miR-146a-5p in our microarray analysis were validated by qRT-PCR. Besides, the interaction of miRNAs and mRNAs also needs to be identified in the ceRNA network mediated by lncRNAs
Bioinformatic analysis predicted the genes targeted by miR-146a-5p to include SMAD4, TRAF6, and IRAK1.32,34,35 Then, we reviewed the literature of miR-146a-5p directly regulating related genes. In the process of hepatic stellate cell proliferation and bone regeneration, miR-146a-5p is reported to target SMAD4 as determined by a luciferase reporter assay and qRT-PCR.36,37 In the pathogenesis of LPS-induced acute lung injury, miR-146a-5p is demonstrated to target TRAF6 and IRAK1, as determined by qRT-PCR and Western blot.38 In the recovery process of spinal cord injury, TRAF6 and IRAK1 were shown to be the target gene of miR-146a-5p by luciferase reporter assay.39 Moreover, miR-146a-5p was found to bind to TRAF6 and IRAK1 in peripheral neuropathy in T2DM mice.40
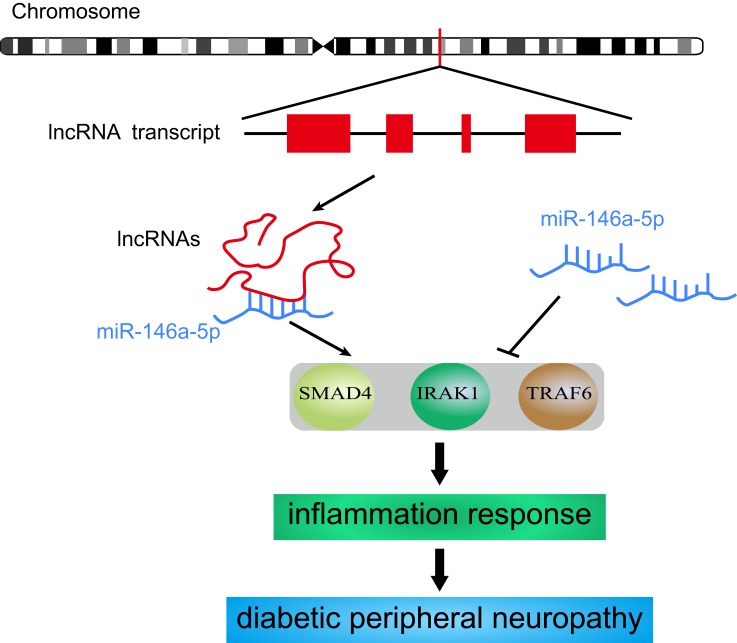
SMAD4, as a critical cytoplasmic signal cascade molecule, can regulate the transcription of TGF-β by cooperating with other transcription factors. The activation of SMAD4 is correlated with an inflammatory response. In smooth muscle cells, SMAD4 suppresses the level of inflammation via decreasing production of chemokines and preventing the recruitment of macrophages.41 IRAK1 belongs to a family of serine/threonine kinases and plays a vital role in the immune response by serving as a key molecule in the Toll-like receptor signaling pathway.42 TRAF6 is identified as an actor downstream of multiple receptor families, including the Toll-like receptor signaling pathway, and can promote the activation of the transcription factor NF-κB.43 Based on current research, SMAD4, TRAF6, and IRAK1 are all predominantly enriched in the inflammatory signaling pathway. This indicates that miR-146a-5p is involved in the inflammatory response by regulating the expression of its target genes. Therefore, four DE lncRNAs found in this study might regulate inflammation by decoying miR-146a-5p (Figure 8).
Figure 8.
The potential mechanism of DE lncRNA decoying miR-146a-5p.
Conclusion
In conclusion, our findings imply that lncRNAs functioning as ceRNAs may participate in the pathogenesis of DPN by regulating inflammation. Although the clinical significance of this work is limited at present. It does preliminary suggest that lncRNAs might play a role in DPN. Further validation of the specific lncRNAs that bind miR-146a is needed by experimental techniques. Moreover, the specific signaling pathway regulated by the ceRNA network needs to be further explored. This study provides a new perspective on the pathogenesis of DPN, which indicates lncRNA may be a potential biomarker or therapeutic target for DPN. Much more experiments need to be performed in basic and clinical fields, and promising therapeutic targets and biomarkers will be used in the future.
Acknowledgments
This work was supported by grants from the Fund of Shanghai Municipal Health Commission (201940362), the Fund of Jinshan Science and Technology Commission (2019-03-02), and Outstanding Young Talents training plan of Jinshan District Health Committee (JSYQ201903).
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Forouhi NG, Wareham NJ. Epidemiology of diabetes. Medicine. 2019;47(1):22–27. doi: 10.1016/j.mpmed.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iqbal Z, Azmi S, Yadav R, et al. Diabetic peripheral neuropathy: epidemiology, diagnosis, and pharmacotherapy. Clin Ther. 2018;40(6):828–849. doi: 10.1016/j.clinthera.2018.04.001 [DOI] [PubMed] [Google Scholar]
- 3.Hicks CW, Selvin E. Epidemiology of peripheral neuropathy and lower extremity disease in diabetes. Curr Diab Rep. 2019;19(10):86. doi: 10.1007/s11892-019-1212-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewanjee S, Das S, Das AK, et al. Molecular mechanism of diabetic neuropathy and its pharmacotherapeutic targets. Eur J Pharmacol. 2018;833:472–523. doi: 10.1016/j.ejphar.2018.06.034 [DOI] [PubMed] [Google Scholar]
- 5.Singh R, Kishore L, Kaur N. Diabetic peripheral neuropathy: current perspective and future directions. Pharmacol Res. 2014;80:21–35. doi: 10.1016/j.phrs.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 6.Callaghan BC, Cheng HT, Stables CL, Smith AL, Feldman EL. Diabetic neuropathy: clinical manifestations and current treatments. Lancet Neurol. 2012;11(6):521–534. doi: 10.1016/S1474-4422(12)70065-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selvarajah D, Kar D, Khunti K, et al. Diabetic peripheral neuropathy: advances in diagnosis and strategies for screening and early intervention. Lancet Diabetes Endocrinol. 2019;7(12):938–948. doi: 10.1016/S2213-8587(19)30081-6 [DOI] [PubMed] [Google Scholar]
- 8.Szymański M, Barciszewski J. Beyond the proteome: non-coding regulatory RNAs. Genome Biol. 2002;3(5):reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uchida S, Adams JC. Physiological roles of non-coding RNAs. Am J Physiol Cell Physiol. 2019;317(1):C1–C2. doi: 10.1152/ajpcell.00114.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75(3):467–484. doi: 10.1007/s00018-017-2626-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16(3):167–179. doi: 10.1038/nrd.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jandura A, Krause HM. The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genet. 2017;33(10):665–676. doi: 10.1016/j.tig.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 13.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 14.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas AA, Biswas S, Feng B, Chen S, Gonder J, Chakrabarti S. lncRNA H19 prevents endothelial-mesenchymal transition in diabetic retinopathy. Diabetologia. 2019;62(3):517–530. doi: 10.1007/s00125-018-4797-6 [DOI] [PubMed] [Google Scholar]
- 17.Yan B, Yao J, Liu J-Y, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116(7):1143–1156. doi: 10.1161/CIRCRESAHA.116.305510 [DOI] [PubMed] [Google Scholar]
- 18.Fachrul M, Utomo DH, Parikesit AA. lncRNA-based study of epigenetic regulations in diabetic peripheral neuropathy. In Silico Pharmacol. 2018;6(1):7. doi: 10.1007/s40203-018-0042-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu W, Zhao GQ, Cao RJ, Zhu ZH, Li K. LncRNA NONRATT021972 was associated with neuropathic pain scoring in patients with Type 2 diabetes. Behav Neurol. 2017;2017:2941297. doi: 10.1155/2017/2941297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Y, Chen L, Luo Q, Wu M, Chen Y, Shi X. Involvement of microRNA-146a in diabetic peripheral neuropathy through the regulation of inflammation. Drug Des Devel Ther. 2018;12:171–177. doi: 10.2147/DDDT [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Q, Feng Y, Xie Y, et al. Nanoparticle-microRNA-146a-5p polyplexes ameliorate diabetic peripheral neuropathy by modulating inflammation and apoptosis. Nanomedicine. 2019;17:188–197. doi: 10.1016/j.nano.2019.01.007 [DOI] [PubMed] [Google Scholar]
- 22.Shi X, Chen Y, Nadeem L, Xu G. Beneficial effect of TNF-α inhibition on diabetic peripheral neuropathy. J Neuroinflammation. 2013;10:69. doi: 10.1186/1742-2094-10-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng S-D, Yang J-H, Yao CH, et al. Potential regulatory mechanisms of lncRNA in diabetes and its complications. Biochem Cell Biol. 2017;95(3):361–367. doi: 10.1139/bcb-2016-0110 [DOI] [PubMed] [Google Scholar]
- 24.Liu S-X, Zheng F, Xie K-L, Xie M-R, Jiang L-J, Cai Y. Exercise reduces insulin resistance in Type 2 diabetes mellitus via mediating the lncRNA MALAT1/MicroRNA-382-3p/resistin axis. Mol Ther Nucleic Acids. 2019;18:34–44. doi: 10.1016/j.omtn.2019.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Z, Li X, Zhan X, et al. Construction of competitive endogenous RNA network reveals regulatory role of long non-coding RNAs in type 2 diabetes mellitus. J Cell Mol Med. 2017;21(12):3204–3213. doi: 10.1111/jcmm.2017.21.issue-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen K, Ma Y, Wu S, et al. Construction and analysis of a lncRNA‑miRNA‑mRNA network based on competitive endogenous RNA reveals functional lncRNAs in diabetic cardiomyopathy. Mol Med Rep. 2019;20(2):1393–1403. doi: 10.3892/mmr.2019.10361 [DOI] [PubMed] [Google Scholar]
- 27.Lin X, Zhan JK, Zhong JY, et al. lncRNA-ES3/miR-34c-5p/BMF axis is involved in regulating high-glucose-induced calcification/senescence of VSMCs. Aging (Albany NY). 2019;11(2):523–535. doi: 10.18632/aging.101758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Jia K, Wu H, et al. A comprehensive competitive endogenous RNA network pinpoints key molecules in diabetic retinopathy. Mol Med Rep. 2019;19(2):851–860. doi: 10.3892/mmr.2018.9715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo G, Ren S, Kang Y, et al. Microarray analyses of lncRNAs and mRNAs expression profiling associated with diabetic peripheral neuropathy in rats. J Cell Biochem. 2019;120(9):15347–15359. doi: 10.1002/jcb.v120.9 [DOI] [PubMed] [Google Scholar]
- 30.Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28(15):2062–2063. doi: 10.1093/bioinformatics/bts344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J-H, S L, Zhou H, Qu L-H, Yang J-H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–D97. doi: 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. doi: 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou C-H, Shrestha S, Yang C-D, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(1):18. doi: 10.1186/s13059-019-1629-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Y, Huang C, Sun X, Long X-R, Lv X-w LJ. MicroRNA-146a modulates TGF-beta1-induced hepatic stellate cell proliferation by targeting SMAD4. Cell Signal. 2012;24(10):1923–1930. doi: 10.1016/j.cellsig.2012.06.003 [DOI] [PubMed] [Google Scholar]
- 37.Xie Q, Wei W, Ruan J, et al. Effects of miR-146a on the osteogenesis of adipose-derived mesenchymal stem cells and bone regeneration. Sci Rep. 2017;7:42840. doi: 10.1038/srep42840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng Z, Gong H, Li Y, et al. Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp Lung Res. 2013;39(7):275–282. doi: 10.3109/01902148.2013.808285 [DOI] [PubMed] [Google Scholar]
- 39.Wei J, Wang J, Zhou Y, Yan S, Li K, Lin H. MicroRNA-146a contributes to SCI recovery via regulating and expression. Biomed Res Int. 2016;2016:4013487. doi: 10.1155/2016/4013487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu XS, Fan B, Szalad A, et al. MicroRNA-146a Mimics reduce the peripheral neuropathy in Type 2 diabetic mice. Diabetes. 2017;66(12):3111–3121. doi: 10.2337/db16-1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang P, Hou S, Chen J, et al. Smad4 deficiency in smooth muscle cells initiates the formation of aortic aneurysm. Circ Res. 2016;118(3):388–399. doi: 10.1161/CIRCRESAHA.115.308040 [DOI] [PubMed] [Google Scholar]
- 42.Wang L, Qiao Q, Ferrao R, et al. Crystal structure of human IRAK1. Proc Natl Acad Sci USA. 2017;114(51):13507–13512. doi: 10.1073/pnas.1714386114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Min Y, Kim M-J, Lee S, Chun E, Lee K-Y. Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy. 2018;14(8):1347–1358. doi: 10.1080/15548627.2018.1474995 [DOI] [PMC free article] [PubMed] [Google Scholar]