Abstract
Background
With increasing incidence, pancreatic cancer (PC) is one of the most common digestive tract tumors. However, the prognosis of PC is particularly dismal due to the highly invasive and metastatic behavior of this deadly disease. DEP domain-containing protein 1B (DEPDC1B), which is overexpressed in multiple tumors, such as breast cancer, oral cancer and non-small cell lung cancer, plays a significant role in cell movement, cell cycle and cytoskeleton reorganization. However, the function of DEPDC1B in PC remains poorly understood.
Methods
The function of DEPDC1B in the migration and invasion of PC was evaluated by wound healing and Transwell assays in vitro and PC-derived liver metastasis models in vivo. The molecular mechanisms of DEPDC1B were investigated through cell line establishment, Western blotting, qRT-PCR, immunoprecipitation, histological examination and immunohistochemistry analysis.
Results
DEPDC1B was overexpressed in PC cell lines. DEPDC1B regulated cell migration and invasion. DEPDC1B regulated the Rac1/PAK1-LIMK1-cofilin1 signaling pathway by interacting with Rac1. Rac1 inhibition suppressed DEPDC1B-induced migration and invasion in PC in vitro and DEPDC1B-induced liver metastasis in vivo.
Conclusion
DEPDC1B promoted cell migration and invasion by activating the Rac1/PAK1-LIMK1-cofilin1 signaling pathway, thus providing a potential therapeutic target against PC.
Keywords: pancreatic cancer, DEP domain-containing protein 1B, migration, invasion, Rac1/PAK1-LIMK1-cofilin1 signaling
Introduction
According to population-based cancer registration data, pancreatic cancer (PC) is one of the most malignant digestive tract tumors, ranking as the fourth leading cause of death among all cancers.1–3 Because the early symptoms of PC are obscure and because PC is prone to vascular invasion, 80–85% of patients have lost the chance for radical resection at diagnosis.4 Surgical resection is the only potential cure for PC. However, its high recurrence rate significantly impedes improvements in the postoperative prognosis of PC patients.5 Multiple signal transduction pathways, such as the VEGFR pathway, EGFR pathway, and K-Ras pathway, are reported to be dysregulated in PC.6 Accordingly, monoclonal antibodies and small molecular inhibitors have been developed for targeted therapy of PC.6 However, despite recent improvements in the comprehensive treatment of PC, the prognosis of PC remains dismal. The lack of specific biomarkers and the presence of highly invasive and metastatic features remain a major challenge for treating this deadly disease.7
DEP domain-containing protein 1B (DEPDC1B) was initially discovered by mRNA expression profiling in MDA-MB 231 human breast cancer cells.8 DEPDC1B has also been reported to be overexpressed in oral cancer,9 non-small cell lung cancer,10 prostate cancer,11 soft tissue sarcoma,12 cervical cancer13 and malignant melanoma.14 The DEPDC1B gene contains two important domains: the DEP domain and the Rho-GAP domain, which are correlated with cell proliferation, migration, invasion, cell cycle progression and cytoskeleton reorganization.15 DEPDC1B promotes cell proliferation through the Rac1-ERK1/2 signaling axis in oral cancer cell lines, and this process is mediated mainly by the Rho-GAP domain.9 In non-small cell lung cancer cells, DEPDC1B enhances cell migration and invasion via activating Wnt/β-catenin signaling.10 Moreover, DEPDC1B knockdown suppresses the viability, migration and invasion abilities of HeLa and SiHa cells through regulating EMT progression and Snai family member expression levels.13
However, the role of DEPDC1B in PC remains unknown. Our present study aimed to investigate the biological function of DEPDC1B and the associated molecular mechanisms in PC. We first evaluated the expression level of DEPDC1B in PC cell lines and normal pancreatic cell lines. Then, we studied the role of DEPDC1B in cell migration and invasion in PC using both in vivo and in vitro models. Finally, we explored the effect of DEPDC1B on the Rac1/PAK1-LIMK1-cofilin1 signaling pathway in PC.
Materials and Methods
Reagents and Antibodies
Primary antibodies against PAK1, phospho-PAK1, cofilin1, phospho-cofilin1, E-cadherin, N-cadherin, and TWIST1, horseradish peroxidase (HRP)-conjugated secondary antibodies and an Active Rac1 Detection Kit were purchased from Cell Signaling Technology (Beverly, MA, USA). Primary antibodies against Rac1, LIMK1, phospho-LIMK1 and Vimentin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). An anti-human DEPDC1B antibody was purchased from Invitrogen (Carlsbad, CA, USA). Anti-GAPDH antibody was obtained from Bioworld Technology, Inc. (St. Louis Park, MN, USA). Trizol reagent and PrimeScript RT Master Mix (Perfect Real Time) were both obtained from TaKaRa (TaKaRa Bio Inc., Naha, Japan). EHop-016, a Rac1 inhibitor, was purchased from Selleck Chemicals (Houston, TX, USA).
Cell Culture and Cell Line Establishment
All human PC cell lines and immortalized human pancreatic ductal cell lines were obtained from the American Type Culture Collection (Rockville, MD, USA) and were cultured in complete DMEM (Wisent, Inc., St-Bruno, QC, Canada) containing 10% fetal bovine serum (Wisent, Inc.) at 37°C in a 5% CO2 humidified incubator.
To create DEPDC1B-silenced cell lines, PANC-1 cells and CFPAC-1 cells were transfected with DEPDC1B-siRNA and negative control-siRNA (synthesized by Shanghai GenePharma Co., Ltd., Shanghai, China) using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, California, USA) according to the instructions. Following transfection for 48–72 h, the cells were collected for subsequent experiments. To establish stable DEPDC1B-overexpressing cell lines, SW1990 cells and BXPC3 cells were transfected with LV-DEPDC1B and LVCON319 (designed by Shanghai GenePharma Co., Ltd., Shanghai, China) according to the manufacturer’s instructions. The knockdown and overexpression efficiencies were confirmed by Western blotting. All sequences used were as follows: DEPDC1B#1-siRNA, GCAAGCAGGGAGUUGUUAUTT; DEPDC1B#2-siRNA, GCACUUCACAAGAGAACAUTT; NC-siRNA, GUACUGGGUUUGUUACAGAUU.
Western Blotting Assay
Total protein was extracted using NP40 lysis buffer (5 M NaCl, 1 M Tris–HCl [pH 7.4], 1% NP40, proteinase inhibitor, 1 mM PMSF). All of the extracted proteins were subjected to SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane. Then, the membrane was incubated with primary antibodies for approximately 12 hrs at 4°C and washed 3 times with Tris-buffered saline (TBS) containing 0.1% Tween 20. Then, the membrane was incubated with HRP-conjugated secondary antibody for 2 hrs at room temperature. After washing 3 times with TBS containing 0.1% Tween 20, the membrane was exposed to an ECL solution (Millipore, Switzerland) and imaged with a Tanon 5200 imaging system (Tanon, China). Finally, the protein bands were analyzed by ImageJ software.
Real-Time PCR (qRT-PCR)
RNA was extracted, purified and converted to cDNA using an oPlus kit and PrimeScript RT Master Mix (Takara Bio Inc., Naha, Japan) according to the manufacturer’s protocol.16 Quantitative real-time PCR was performed using a ViiA 7 real-time PCR system (Applied Biosystems, Thermo Fisher Scientific, Rockford, IL, USA) and SYBR Green PCR master mix. The primers used for real-time PCR were as follows: DEPDC1B forward 5′-AGCTACCAGGCTGTGGAATG-3′ and reverse 5′-AGCTCTTGAAACGACAGCGA-3′; GAPDH forward 5′-CCATGTTCGTCATGGGTGTGAACCA-3′ and reverse 5′-GCCAGTAGAGGCAGGGATGATGTTC-3′.
Wound Healing Assay
A total of 1.5×105 cells were seeded in each wound healing culture insert (EMD Millipore, Billerica, MA, Germany) in 24-well plates beforehand. After approximately 24 hrs, the cells reached 95% confluence, and the culture insert was then removed. The cells were washed with PBS and cultured in DMEM without serum. The wound areas were photographed at 0 and 24 hrs with a microscope. The original area and migration area were measured by ImageJ software, and the healing rates are shown according to the ratio of the migration area to the original area.
Transwell Migration and Invasion Assays
The assay was carried out with transwell inserts (EMD Millipore, Billerica, MA, Germany) coated with or without Matrigel (BD Biosciences San Jose, CA). Briefly, 2×104 cells and 300 μL serum-free DMEM were placed in the upper chamber (pore size, 8 µm; Millipore, Germany), and 1000 μL DMEM containing 10% FBS was added to the lower chamber. After 48 hrs, the cells that had migrated to or invaded the lower surface of the filter were fixed and stained, and the mean values for the cell counts in five random fields of vision under a microscope (×200) were calculated.
CCK-8 Assay
In brief, 8 × 103 cells were cultured in 96-well plates. After incubation with different concentrations of EHop-016 for 24 hrs, 10 μL Cell Counting Kit-8 (CCK-8, Bimake, USA) reagent was added to 100 μL medium and incubated for 2 hrs at 37°C. The absorbance of the samples at 450 nm was recorded by a spectrophotometer.
Co-Immunoprecipitation (Co-IP) Assay
Cell lysates from PANC-1 and CFPAC-1 cells were created using NP40 lysis buffer. Then, cell extracts (800 μg) were incubated with 5 μg RAC1 antibody at 4°C overnight with gentle rocking, and 20 μL protein A agarose (Beyotime, Shanghai, China) suspension was added for another 2 hrs. Immunoprecipitates were washed 5 times with NP40 lysis buffer, boiled for 5 mins and then subjected to Western blotting analysis.
Immunoprecipitation of RAC1-GTP
Rac1 activation was detected with an Active Rac1 Detection Kit (Kit #8815 Cell Signaling Technology, Inc.) in accordance with the manufacturer’s recommendations. To ensure that the immunoprecipitation procedures worked properly, positive control (GTPγS) and negative control (GDP) experiments were carried out before the formal experiment. In brief, cells were harvested in ice-cold 1X lysis/binding/wash buffer plus 1 mM PMSF and centrifuged at 16,000 g for 15 mins at 4°C. GST-PAK1-PBD was added to the spin cup containing glutathione resin. Immediately, the cell lysate (containing 600 µg of total protein) was transferred to the spin cup. Then, the reaction mixture was incubated at 4°C for 1 hr with gentle rocking. The beads were washed 3 times with lysis buffer, and SDS sample buffer was then added to elute the bound proteins. Western blotting was carried out as described above. A Rac1 mouse antibody was used to detect GTP-Rac1 and total RAC1 in the samples. The expression level of active Rac1-GTP was quantified relative to the total Rac1 content.
Orthotopic Tumor Implantation in Nude Mice
For the in vivo metastasis study, 24 male Balb/c nude mice (specific pathogen-free,4–6 weeks old, and weighing 18–21 g) obtained from the Model Animal Research Center of Nanjing University (Nanjing, China) were divided randomly into three groups. SW1990-LVCON319 and SW1990-LV-DEPDC1B cells were suspended at concentrations of 1 ×106/50 μL in PBS before the experiment. Subsequently, the suspended pancreatic cancer cells (1 × 106) were injected into the major pancreatic duct. Four weeks after injection, EHop-016 was dissolved in Dimethyl Sulfoxide (DMSO) and administered three times weekly (25 mg/kg) by intraperitoneal injection in the third group of mice for the following 8 weeks. Similarly, DMSO was administered to the other two groups. Finally, the mice were euthanized, and suspected liver metastatic tissues were subjected to histological examinations and immunohistochemistry. All animal protocols were reviewed and approved by the Animal Care Committee of Nanjing University in accordance with the guidelines of the Institutional Animal Care and Use Committee.
Histological Examination (HE) and Immunohistochemistry (IHC)
Tissues fixed with 4% paraformaldehyde were embedded in paraffin and cut into 5-μm-thick slices. For hematoxylin and eosin (H&E) staining, the tissue slices were dewaxed in xylene, rehydrated with decreasing concentrations of ethanol, and washed with PBS. Then, the slices were stained with hematoxylin for 30 sec with agitation and rinsed with water. After that, the slices were stained with eosin for 10–30 sec with agitation and rinsed with water. After staining, the slices were dehydrated, mounted and covered with coverslips. IHC was carried out according to a published protocol.17 Samples were incubated with a rabbit polyclonal antibody against DEPDC1B (5 µg/mL dilution, PA5-72875, Invitrogen, Carlsbad, CA, USA).
Statistical Analysis
All results are displayed as the mean ± standard deviation (SD) of three independent experiments. Student’s t-tests or ANOVA analyses were used to analyze the data, and the data were deemed to be statistically significant at P < 0.05. Statistical analysis was carried out using IBM SPSS Statistics 20 software.
Results
DEPDC1B Expression Is Upregulated in Pancreatic Cancer
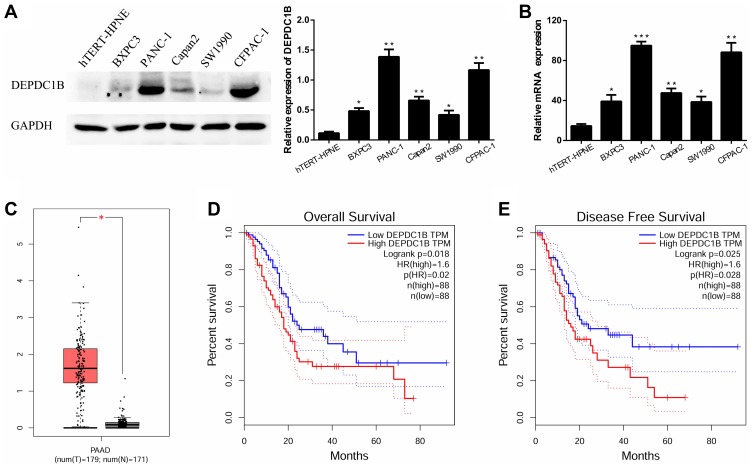
In five human PC cell lines (BXPC3, PANC-1, Capan2, SW1990 and CFPAC-1) and one normal human pancreatic ductal cell line (hTERT-HPNE), DEPDC1B protein and mRNA levels were determined by Western blotting and qRT-PCR, respectively. As displayed in Figure 1A and B, DEPDC1B expression was significantly higher in the five human PC cell lines than in hTERT-HPNE cells, and the DEPDC1B expression levels in PANC-1 and CFPAC-1 cells were the highest among all PC cells. Moreover, analysis of data from the Cancer Genome Atlas (TCGA) project (http://cancergenome.nih.gov/) and the Genotype-Tissue Expression (GTEx) project (https://gtexportal.org/home/) showed that DEPDC1B expression was higher in PC specimens than in normal pancreatic tissues (Figure 1C, p < 0.05). A Kaplan-Meier survival analysis based on DEPDC1B mRNA expression from a large cohort of 176 PC patients in the TCGA dataset revealed that patients with a high DEPDC1B transcripts per million (TPM) value had a significantly shorter overall survival (HR=1.6, p = 0.02) and disease-free survival (HR=1.6, p = 0.028) than those with a low DEPDC1B TPM value (Figure 1D and E).
Figure 1.
Expression of DEPDC1B in PC. (A, B) Protein and mRNA expression levels of DEPDC1B were analyzed in different PC cell lines and a normal human pancreatic ductal cell line. (C) DEPDC1B expression data for 179 PC tumor specimens (T) and 171 normal pancreatic tissues (N) from TCGA and GTEx. (D) DEPDC1B expression in relation to the overall survival of 176 PC patients (data from TCGA). (E) DEPDC1B expression in relation to the disease-free survival of 176 PC patients (data from TCGA). *P <0.05, **P <0.01, and ***P <0.001.
Abbreviations: DEPDC1B, DEP domain-containing protein 1B; PC, pancreatic cancer; TCGA, The Cancer Genome Atlas; GTEx, Genotype-Tissue Expression.
DEPDC1B Expression Is Correlated with Migration and Invasion in PC Cells
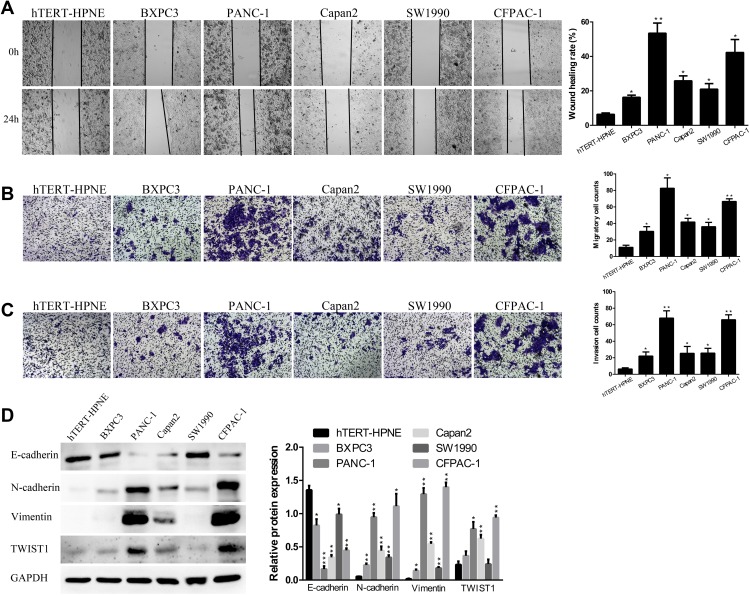
Then, in the subsequent wound healing assay, we found that five human PC cell lines migrated remarkably more quickly than hTERT-HPNE cells (Figure 2A). A transwell migration assay (without Matrigel) also confirmed that the migration ability of the five human PC cell lines was significantly higher than that of hTERT-HPNE cells (Figure 2B). Moreover, the transwell invasion assay (with Matrigel) results showed that the invasive ability of the five human PC cell lines was noticeably higher than that of hTERT-HPNE cells (Figure 2C). The expression levels of EMT-related proteins were analyzed by Western blotting; epithelial marker (E-cadherin) protein levels were lower, and mesenchymal marker (N-cadherin, Vimentin) and transcription factor (TWIST1) protein levels were higher in the five human PC cell lines than in hTERT-HPNE cells (Figure 2D). However, the expression levels of TWIST1 in BXPC3 and SW1990 cells were not significantly enhanced compared with those in hTERT-HPNE cells. These results showed that the DEPDC1B protein level may be correlated with cell migration and invasion ability as well as the expression of EMT markers in five human PC cell lines and one normal human pancreatic ductal cell line, which indicates that DEPDC1B may be a marker of tumor invasiveness and metastasis. We further examined the cellular location of E-cadherin in pancreatic cancer cells by immunofluorescence assay (see Supplemental Method). The results showed that although the expression levels of E-cadherin were consistent to protein levels in Western blotting assay, only SW1990 cells showed typical membrane location of E-cadherin (Supplementary Figure 1).
Figure 2.
The abilities of cell migration and invasion in PC. (A) Typical images of the wound healing assay results from different PC cell lines and a normal human pancreatic ductal cell line; images of cell migration to the wound were captured at 0 and 24 after wounding. (B) Typical images of the transwell migration assay (without Matrigel) and (C) transwell invasion assay (with Matrigel) showed the migration and invasion of cells on the membrane. The average migrated and invaded cell numbers from five random visual fields were determined from three independent experiments. (D) EMT-related protein levels were analyzed in different PC cell lines and a normal human pancreatic ductal cell line by Western blotting. *P <0.05, **P <0.01, and ***P <0.001.
Abbreviations: EMT, epithelial-mesenchymal transition; PC, pancreatic cancer.
Knockdown of DEPDC1B Significantly Inhibits the Migration and Invasion of PC Cells in vitro
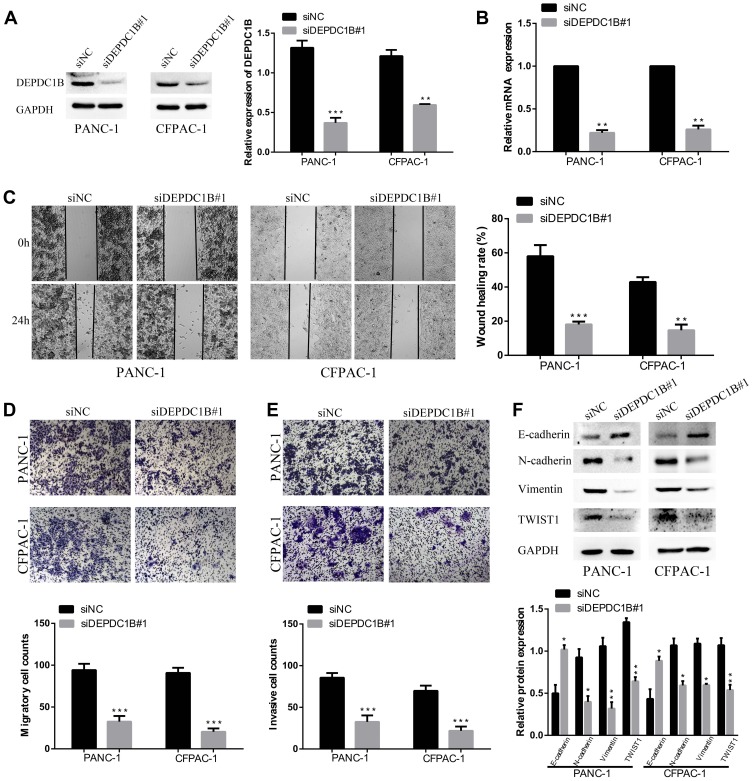
To study the effect of DEPDC1B in PC, we knocked down DEPDC1B in PANC-1 and CFPAC-1 cells by transient transfection with siNC, siDEPDC1B#1 and siDEPDC1B#2. PANC-1 and CFPAC-1 cells were selected because among the PC cells analyzed, DEPDC1B expression was highest in these. The successful knockdown of DEPDC1B in PC-siDEPDC1B#1 and PC-siDEPDC1B#2 cells was verified by Western blotting and qRT-PCR (Figure 3A and B, Supplementary Figure 2B and C). To prevent potential confounding factors brought by cell proliferation indifferent transfection conditions, we examined in detail the difference of proliferation in DEPDC1B knockdown or overexpressed cells and control cells by CCK-8 assay. Then, we found that there was no significant difference in cell proliferation between treated cells and control cells (Supplementary Figure 2A). We observed that DEPDC1B knockdown cells migrated more slowly than control cells in the wound healing assay (Figure 3C, 40% decrease in PANC-1 and 28% decrease in CFPAC-1; Supplementary Figure 2D, 24% decrease in PANC-1 and 19% decrease in CFPAC-1). Similarly, a transwell migration assay (without Matrigel) carried out in PC cells indicated that DEPDC1B knockdown contributed to a decrease in cell migration (Figure 3D, p < 0.001 in PANC-1 and CFPAC-1; Supplementary Figure 2E, p < 0.01 in PANC-1 and CFPAC-1). In addition, the transwell invasion assay (with Matrigel) results showed that cell invasion was remarkably lower in the siDEPDC1B group than in the siNC group (Figure 3E, p < 0.001 in PANC-1 and CFPAC-1; Supplementary Figure 2F, p < 0.01 in PANC-1 and CFPAC-1). Western blotting was used to confirm the expression levels of EMT marker proteins, and the results showed that knockdown of DEPDC1B in PC cells contributed to increased epithelial marker (E-cadherin) protein levels and decreased mesenchymal marker (N-cadherin, Vimentin) and transcription factor (TWIST1) protein levels (Figure 3F, Supplementary Figure 2G). All told, DEPDC1B silencing suppresses the migration and invasion of PC cells in vitro.
Figure 3.
Effect of DEPDC1B knockdown on the migration and invasion of PC cells in vitro. (A, B) The DEPDC1B interference efficiency of transient transfection with siRNAs in PC cells was examined by Western blotting and qRT-PCR. (C) Typical images of the wound healing assay for the siNC and siDEPDC1B#1 groups show cell migration to the wound; the images were captured at 0 and 24 h after wounding. (D) Typical images of the transwell migration assay (without Matrigel) and (E) transwell invasion assay (with Matrigel) showing migrated and invaded cells on the membrane. The average migrated and invaded cell numbers from five random visual fields were calculated from three independent experiments. (F) EMT-related protein levels were analyzed in the siNC group and siDEPDC1B#1 group by Western blotting. *P <0.05, **P <0.01, and ***P <0.001.
Abbreviations: DEPDC1B, DEP domain-containing protein 1B; EMT, epithelial-mesenchymal transition; PC, pancreatic cancer; qRT-PCR, quantitative real-time PCR.
Overexpression of DEPDC1B Promotes the Migration and Invasion of PC Cells in vitro
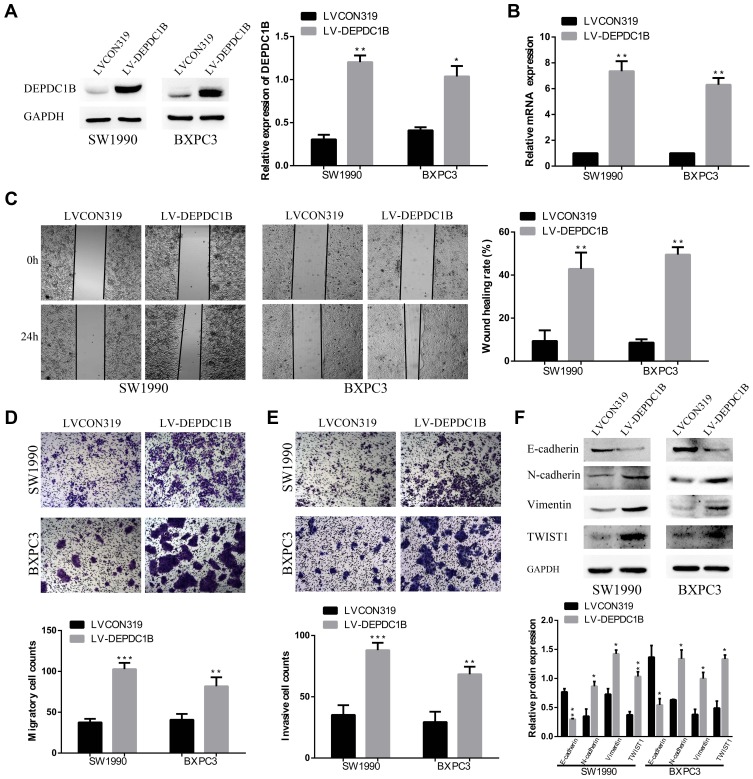
To further determine the function of DEPDC1B in PC, SW1990 and BXPC3 cells with the lowest DEPDC1B expression among all PC cells analyzed were chosen for stable transfection with LV-CON319 and LV-DEPDC1B. DEPDC1B expression was confirmed to be significantly higher in the LV-DEPDC1B group than in the LV-CON319 group by Western blotting and qRT-PCR (Figure 4A and B). In the subsequent wound healing assay, we found that DEPDC1B-overexpressing cells migrated more quickly than control cells (Figure 4C, 34% increase in SW1990 and 41% increase in BXPC3). The transwell migration assay (without Matrigel) also confirmed the finding that DEPDC1B overexpression led to enhanced migration ability (Figure 4D, p < 0.001 in SW1990; p < 0.01 in BXPC3). Moreover, the transwell invasion assay (with Matrigel) results showed that cell invasion ability was markedly higher in the LV-DEPDC1B group than in the LV-CON319 group (Figure 4E, p < 0.001 in SW1990; p < 0.01 in BXPC3). The expression levels of EMT-related proteins were analyzed by Western blotting, and we discovered that overexpression of DEPDC1B led to the decreased epithelial marker (E-cadherin) protein levels and increased mesenchymal marker (N-cadherin, Vimentin) and transcription factor (TWIST1) protein levels (Figure 4F). These results indicate that DEPDC1B overexpression promotes the migration and invasion of PC cells in vitro.
Figure 4.
Effect of DEPDC1B overexpression on the migration and invasion of PC cells in vitro. (A, B) DEPDC1B overexpression efficiency in PC cells after stable transfection with LV-CON319 and LV-DEPDC1B was examined by Western blotting and qRT-PCR. (C) Typical images of the wound healing assay of the LV-CON319 and LV-DEPDC1B groups show cell migration to the wound; the images were captured at 0 and 24 after wounding. (D) Typical images of the transwell migration assay (without Matrigel) and (E) transwell invasion assay (with Matrigel) showing the migrated and invaded cells on the membrane. The average migrated and invaded cell numbers from five random visual fields were determined from three independent experiments. (F) EMT-related protein levels were analyzed in the LV-CON319 and LV-DEPDC1B groups by Western blotting. *P <0.05, **P <0.01, and ***P <0.001.
Abbreviations: DEPDC1B, DEP domain-containing protein 1B; EMT, epithelial-mesenchymal transition; PC, pancreatic cancer; qRT-PCR, quantitative real-time PCR.
DEPDC1B Mediates the Rac1/PAK1-LIMK1-Cofilin1 Signaling Pathway via Interacting with Rac1 in PC Cells in vitro
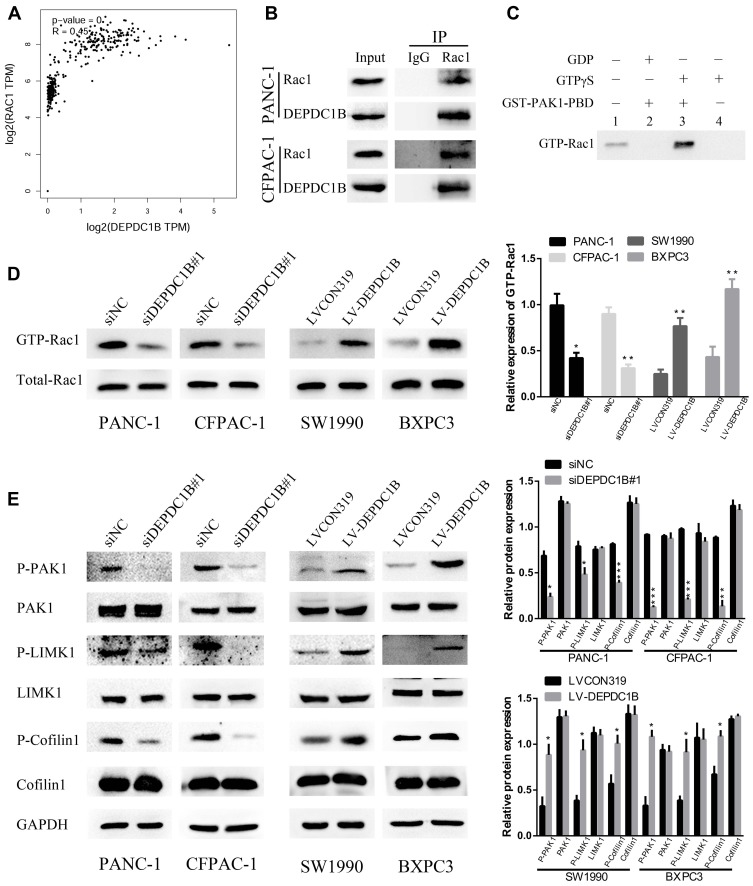
Previous studies have clearly shown that DEPDC1B induces oral cancer cell migration and cell invasion by specifically activating Rac1.9 Moreover, the Rac1/PAK1-LIMK1-cofilin1 signaling pathway could promote cancer metastasis.18 However, the correlation between DEPDC1B and Rac1 and the function of the Rac1/PAK1-LIMK1-cofilin1 signaling pathway in PC cells have not been studied. To determine whether DEPDC1B has a significant effect on PC by interacting with Rac1 to activate the downstream PAK1-LIMK1-cofilin1 signaling pathway, analysis of data from TCGA project showed that DEPDC1B expression was well related to Rac1 expression (Figure 5A, R=0.45, p < 0.001). Co-IP experiments also verified that DEPDC1B and Rac1 interacted with each other in PANC-1 and CFPAC-1 cells (Figure 5B). Subsequently, we examined the effect of DEPDC1B on the activation of Rac1 (GTP-Rac1). First, we performed experiments with GTPγS (positive control) and GDP (negative control) to ensure that the immunoprecipitation procedures worked properly (Figure 5C). The results showed that the relative protein level of GTP-Rac1 decreased when DEPDC1B was knocked down and increased when DEPDC1B was overexpressed (Figure 5D, p < 0.05 in PANC-1, p < 0.01 in CFPAC-1, p < 0.01 in SW1990 and p < 0.05 in BXPC3). Given that DEPDC1B binds to Rac1, we explored the effect of DEPDC1B on the Rac1/PAK1-LIMK1-cofilin1 signaling pathway. Strikingly, DEPDC1B silencing decreased phospho-PAK1 (P-PAK1), phospho-LIMK1 (P-LIMK1) and phospho-cofilin1 (P-cofilin1) protein levels, but the total PAK1, LIMK1 and cofilin1 protein levels were not significantly changed. Accordingly, DEPDC1B overexpression increased P-PAK1, P-LIMK1 and P-cofilin1 protein levels, but the total PAK1, LIMK1 and cofilin1 protein levels were not significantly changed (Figure 5E). Taken together, the data show that the interaction between DEPDC1B and Rac1 activates the Rac1/PAK1-LIMK1-cofilin1 pathway.
Figure 5.
DEPDC1B interacts with Rac1 and activates the Rac1/PAK1-LIMK1-cofilin1 signaling pathway. (A) A correlation between DEPDC1B expression and Rac1 expression in PC (data from TCGA). (B) Co-IP experiments using PANC-1 and CFPAC-1 cells confirmed that DEPDC1B and Rac1 can bind with each other. (C) To ensure that the immunoprecipitation procedures worked properly, positive control (GTPγS) and negative control (GDP) experiments were carried out before the formal experiment. (D) GTP-Rac1 (Rac1 activation) levels were measured by GST-PAK1-PBD pull-down assay and were analyzed by Western blotting in the siNC, siDEPDC1B#1, LVCON319 and LV-DEPDC1B groups. The expression level of GTP-Rac1 was quantified relative to the total Rac1 level. (E) The expression of related proteins in the Rac1/PAK1-LIMK1-cofilin1 signaling pathway was assayed by Western blotting in the siNC, siDEPDC1B#1, LVCON319 and LV-DEPDC1B groups. *P <0.05, **P <0.01, and ***P <0.001.
Abbreviations: DEPDC1B, DEP domain-containing protein 1B; PC, pancreatic cancer; TCGA, The Cancer Genome Atlas; Co-IP, Co-immunoprecipitation.
DEPDC1B Promotes Migration and Invasion in PC Cells via the Rac1/PAK1-LIMK1-Cofilin1 Signaling Pathway
EHop-016 Suppressed the DEPDC1B-Induced Rac1/PAK1-LIMK1-Cofilin1 Signaling Pathway
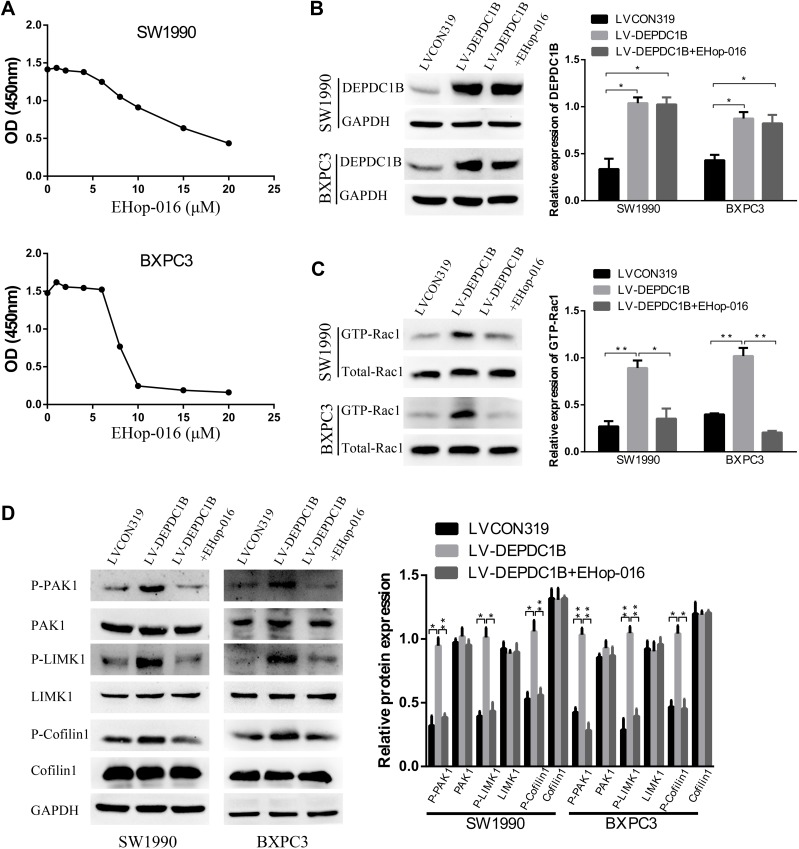
To further probe the role of the Rac1/PAK1-LIMK1-cofilin1 signaling pathway in PC cells, we studied the effect of suppressing Rac1 activation with EHop-016 (a Rac1 inhibitor) in LV-DEPDC1B stably transfected SW1990 and BXPC3 cells. A CCK-8 assay was used to observe the effect of EHop-016 on the viability of LV-DEPDC1B stably transfected PC cells, and 5 μM was chosen for the Rac1 inhibition experiment (Figure 6A). The relative expression of GTP-Rac1 was significantly lower in the EHop-016-treated LV-DEPDC1B group compared than in the LV-DEPDC1B group according to Western blotting (p < 0.05 in SW1990 and p < 0.01 in BXPC3), but DEPDC1B expression showed no significant changes (Figure 6B and C). Then, Rac1/PAK1-LIMK1-cofilin1 pathway-related protein expression was analyzed. The DEPDC1B-induced high phospho-PAK1, phospho-LIMK1 and phospho-cofilin1 expression levels were observably inhibited by EHop-016, but the total PAK1, LIMK1 and cofilin1 protein expression levels did not change in PC cells (Figure 6D). Thus, EHop-016 suppressed the DEPDC1B-induced Rac1/PAK1-LIMK1-cofilin1 signaling pathway.
Figure 6.
Effect of the Rac1 inhibitor EHop-016 on DEPDC1B-induced Rac1/PAK1-LIMK1-cofilin1 signaling activation in vitro. (A) A CCK-8 assay was used to observe the effect of EHop-016 on the viability of LV-DEPDC1B stably transfected PC cells. (B) DEPDC1B protein levels, (C) GTP-Rac1 levels and (D) related protein expression levels in the Rac1/PAK1-LIMK1-cofilin1 signaling pathway were analyzed by Western blotting in the LVCON319, LV-DEPDC1B and EHop-016 treated LV-DEPDC1B groups. *P <0.05, and **P <0.01.
Abbreviations: DEPDC1B, DEP domain-containing protein 1B; PC, pancreatic cancer.
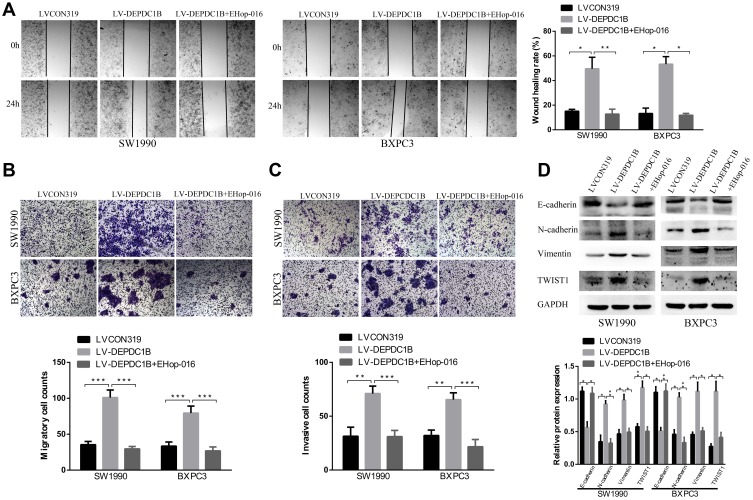
EHop-016 Suppressed DEPDC1B-Induced the Migration and Invasion of PC in vitro
Because DEPDC1B mediates the Rac1/PAK1-LIMK1-cofilin1 signaling pathway, we questioned whether DEPDC1B-induced PC cell migration and invasion could be attributed to this signaling pathway. We discovered that cells in the EHop-016-treated LV-DEPDC1B group migrated more slowly than cells in the LV-DEPDC1B group in a wound healing assay (Figure 7A, 37% decrease in SW1990 and 42% decrease in BXPC3). A transwell migration assay (without Matrigel) also confirmed this finding that EHop-016 treatment resulted in weakened cell migration ability (Figure 7B, p < 0.001 in SW1990 and BXPC-3). Moreover, transwell invasion assay (with Matrigel) results showed that cell invasion ability was obviously inhibited in the EHop-016-treated LV-DEPDC1B group compared with that in the LV-DEPDC1B group (Figure 7C, p < 0.001 in SW1990 and BXPC-3). The expression levels of EMT-related proteins were analyzed by Western blotting, and we discovered that epithelial marker (E-cadherin) protein levels increased, and mesenchymal marker (N-cadherin, Vimentin) and transcription factor (TWIST1) protein levels decreased in the EHop-016-treated LV-DEPDC1B group compared with those in the LV-DEPDC1B group (Figure 7D).
Figure 7.
Effect of the Rac1 inhibitor EHop-016 on DEPDC1B-induced migration and invasion in PC cells in vitro. (A) Typical images of the wound healing assay in the LVCON319, LV-DEPDC1B and EHop-016 treated LV-DEPDC1B groups show cell migration to the wound; the images were captured at 0 and 24 after wounding. (B) Typical images of transwell migration assays (without Matrigel) and (C) transwell invasion assays (with Matrigel) showing migrated and invaded cells on the membrane. The average migrated and invaded cell numbers from five random visual fields were determined from three independent experiments. (D) EMT-related protein levels were analyzed by Western blotting in the LVCON319, LV-DEPDC1B and EHop-016-treated LV-DEPDC1B groups. *P <0.05, **P <0.01, and ***P <0.001.
Abbreviations: DEPDC1B, DEP domain-containing protein 1B; EMT, epithelial-mesenchymal transition; PC, pancreatic cancer.
EHop-016 Suppressed the DEPDC1B-Induced Liver Metastasis of PC in vivo
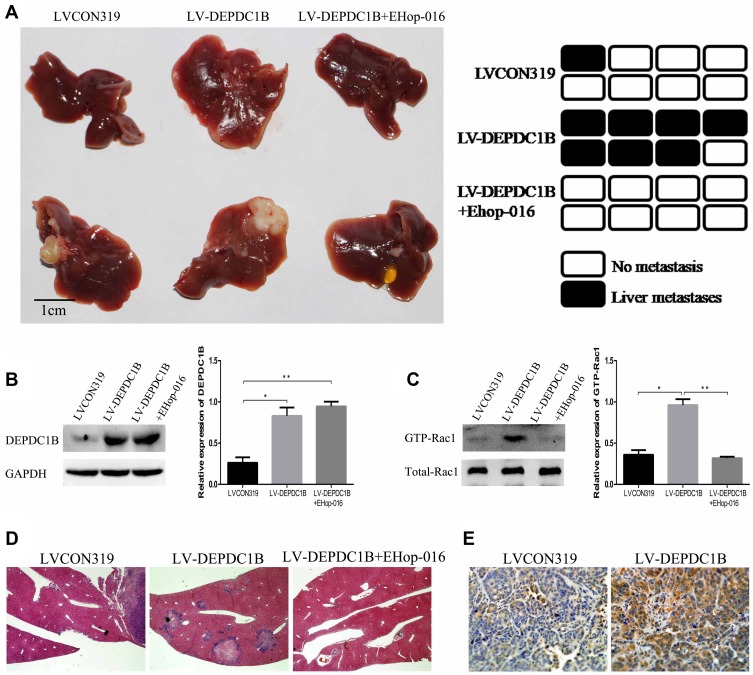
To determine whether DEPDC1B could promote PC metastasis in vivo and whether Rac1 was involved, PC-derived liver metastasis models were used. The number of mice that had liver metastatic nodules was higher in the LV-DEPDC1B group than in the LVCON319 group (Figure 8A, D); however, in the EHop-016-treated LV-DEPDC1B group, there were no mice that had liver metastatic nodules (Figure 8A, D). Moreover, compared with those in the LVCON319 group, the DEPDC1B protein levels (p < 0.05) and GTP-Rac1 levels (p < 0.05) in pancreatic orthotopical tumor tissues were significantly increased in the LV-DEPDC1B group (Figure 8B and C). IHC staining showed that liver metastatic tumors formed by SW1990-LV-DEPDC1B cells had higher DEPDC1B expression than those formed by SW1990-LVCON319 cells (Figure 8E). However, compared with those in the LV-DEPDC1B group, GTP-Rac1 levels in pancreatic orthotopical tumor tissues were significantly decreased in the EHop-016-treated LV-DEPDC1B group.
Figure 8.
DEPDC1B promotes the metastasis of PC cells in vivo. (A) PC-derived liver metastasis models were built by injecting SW1990-LVCON319 and SW1990-LV-DEPDC1B cells orthotopically into the major pancreatic duct of nude mice. Four weeks after cell injection, half of the mice in the LV-DEPDC1B group were injected intraperitoneally with EHop-016 three times weekly (25 mg/kg) for 8 weeks. Finally, the mice were euthanized, and liver metastatic tissues were subjected to HE and IHC. Liver metastatic nodules were evaluated approximately 3 months post-injection (n = 8 per group). (B, C) Pancreatic orthotopical tumor tissues were used to analyze DEPDC1B protein levels and GTP-Rac1 levels in the three groups by Western blotting. (D) Representative HE staining results from metastatic nodules in the livers from the LVCON319 group, LV-DEPDC1B group and EHop-016-treated LV-DEPDC1B group. (E) Metastatic nodules in the livers from the LVCON319 group and LV-DEPDC1B group were stained with an anti-DEPDC1B antibody for IHC. *P <0.05, **P <0.01.
Abbreviations: DEPDC1B, DEP domain-containing protein 1B; PC, pancreatic cancer; HE, histological examination; IHC, immunohistochemistry.
Discussion
The DEPDC1B gene, located at chromosome 5 (5q12.1), contains two important domains: the DEP domain and the Rho-GAP domain.15,19–21 The DEP domain enables the protein to interact with G protein-coupled receptors and mediates the GPCR signaling pathway.20 The Rho-GAP domain is related to Rho GTPase signaling.15 Previous results have proven that DEPDC1B plays a crucial role in cell growth, movement, differentiation, cell cycle progression and cytoskeleton reorganization.15 Subsequent studies demonstrated that it is also overexpressed in other types of cancer,8–14 and it is used as a prognostic factor that predicts outcomes in patients with non-small cell lung cancer10 and prostate cancer.11 In our present study, we show that the expression of DEPDC1B was obviously higher in PC cell lines than in normal human pancreatic ductal cell lines, which is similar to the results in non-small cell lung cancer10 and cervical cancer13 cell lines. In addition, DEPDC1B protein expression may be correlated with cell migration and invasion ability as well as the levels of EMT markers in five human PC cell lines and one normal human pancreatic ductal cell line in vitro. The cellular distribution of E-cadherin, which is a typical marker of epithelial phenotype, was further examined by immunofluorescence assay. The results showed that the expression levels of E-cadherin were consistent with that found in Western blotting assay. However, only in SW1990 cells, E-Cadherin demonstrated typical membrane location, indicating functional cellular junction. The detailed mechanism of this result remains unclear and whether it plays a role in the differences of migration/invasion abilities of pancreatic cancer cell lines is unknown, which should be considered as a limitation of this study. Further investigations are warranted to elucidate this finding. Since potential correlation between DEPDC1B expression and malignant behaviors such as cellular migration/invasion was observed, whether the expression of DEPDC1B correlates with pancreatic cancer differentiation becomes an interesting topic. Previous data have shown that PANC-1 and SW1990 were established from poorly-differentiated pancreatic adenocarcinoma; BXPC3 was moderate-differentiated; Capan-2 and CFPAC-1 were well-differentiated.22,23 In our present study, we found that the expression of DEPDC1B in PANC-1 was relatively high while the expression levels were relatively low in BXPC3 and Capan-2 cells, indicating that the expression levels of DEPDC1B could be correlated with the differentiation levels of the cell lines. However, the expression of DEPDC1B in SW1990 and CFPAC-1 did not rigorously correlated with the level of differentiation. Therefore, the expression of DEPDC1B showed possible but not rigid correlation with the differentiation levels of PC cells.
Previous studies have shown that silencing DEPDC1B in NSCLC suppressed cell migration and invasion in both NSCLC and cervical cancer cells.10,13 Consistent with previous reports, DEPDC1B knockdown in PANC-1 and CFPAC-1 cells significantly inhibited cell migration and invasion. Yang et al demonstrated that ectopic expression of DEPDC1B in NSCLC enhanced cell migration and invasion.10 In addition, DEPDC1B induced both cell migration in a cultured embryonic fibroblast cell line and cell invasion in oral cancer cell lines.9 It is likely that the overexpression of DEPDC1B in SW1990 and BXPC3 cells accelerated cell migration and invasion.
It has been shown that RhoGEFs could form Rho GTPase signaling complexes with Rac1, a member of the Rho GTPase family, and control GTPase activity during cell migration and invasion.24 DEPDC1B has been found to be a guanine nucleotide exchange factor (GEF) and physically interacts with Rac1 protein, thereby contributing to Rac1 activation and mediating a downstream signaling pathway involved in oral cancer.9 Furthermore, Palak Ahuja and Kailash Singh predicted 3D model of DEPDC1B to identify the interacting amino acid residues of the DEPDC1B protein with Rac1.25 Similarly, our analysis of data from TCGA project showed that DEPDC1B expression was well related to Rac1 expression, and Co-IP results also demonstrated that DEPDC1B may directly interact with the Rac1 protein. DEPDC1B overexpression can contribute to increasing the relative expression of GTP-Rac1, and DEPDC1B suppression can lead to the opposite results in PC cell lines. However, it is notable that exogenous expression of DEPDC1B in HeLa cells inhibited Rac1 activation.26 The manner and mechanism by which DEPDC1B interacts with Rac1 in different cells may be different, which may be the cause of this inconsistency, remains to be elucidated. Another interesting finding is that, although TCGA data showed that the mRNA levels of DEPDC1B and RAC1 well correlated, our results indicated that DEPDC1B regulated the activity of Rac1 without altering the protein level. Our present study could not provide a definitive answer for this discrepancy. Of note, TCGA data included in our analysis were based on mRNA expression information derived from chip data of tissues of clinical PC patients. Protein and mRNA expression in tumor tissue could be more complicated due to complex microenvironment and regulation factors. This phenomenon warrants further exploration.
The Rac/PAK pathway, one of the downstream signaling pathways of Rho GTPases, increases the phosphorylation of Lin-Isl-Mlc three homologous protein kinases (LIMKs) and cofilin to promote actin aggregation, cell migration, invasion and EMT.18,27–29 Rhein, a potential inhibitor of Rac1, can inhibit the migratory and invasive capabilities of SKOV3-PM4 cells by decreasing the phosphorylation of Rac1/PAK1-LIMK1-cofilin1 pathway associated protein.30 DADS (Diallyl disulfide) downregulated the Rac1-Pak1/Rock1-LIMK1 pathway in MGC803 cells, suppressing cell migration and invasion.31 Consistent with the results of these studies, our results revealed that DEPDC1B interference inhibited the phosphorylation of Rac1/PAK1-LIMK1-cofilin1 pathway-associated proteins in PC cell lines. Accordingly, DEPDC1B overexpression enhanced the levels of phosphorylated proteins in PC cells. Furthermore, our data showed that EHop-016, an inhibitor of Rac1 and potentially an anti-metastatic cancer drug,32 could effectively suppress the Rac1/PAK1-LIMK1-cofilin1 pathway activation induced by DEPDC1B. Moreover, we observed that EHop-016 can reverse the cell migration and invasion induced by DEPDC1B in vitro. PC-derived liver metastasis models revealed that DEPDC1B could promote the metastasis of PC in vivo and that EHop-016 could reverse metastasis without apparent toxicity. To the best of our knowledge, our study is the first to use an animal model to investigate the function of DEPDC1B in PC in vivo.
Previous studies have clearly documented that miRNAs could directly decrease the expression of DEPDC1B in human diseases.33 In addition, lncRNAs may function as competing endogenous RNAs (ceRNAs) of miRNAs to antagonize miRNAs’ ability to inhibit downstream protein translation.34,35 Moreover, increasing evidence shows that the interaction mechanism between lncRNAs and miRNAs plays a key role in the oncogenesis and development of PC, and both of types of molecules may be important biomarkers and targets for cancer diagnosis, treatment and prognosis.36 In subsequent studies, we will integrate published multi-level expression data and a computational bioinformatics approach to explore whether lncRNA positively regulates the expression of DEPDC1B by interacting with a miRNA, thus activating the Rac1/PAK1-LIMK1-cofilin1 signaling pathway to promote cancer cell migration and invasion. Then, the migration and invasion of PC cells can be more effectively suppressed by targeting the upstream molecules of DEPDC1B.
In summary, we have demonstrated that DEPDC1B is overexpressed in PC cell lines, which indicates that DEPDC1B may be a biomarker of PC. DEPDC1B can promote migration and invasion in PC cells by activating the Rac1/PAK1-LIMK1-cofilin1 signaling pathway. Our novel finding supports the future consideration of DEPDC1B and the Rac1/PAK1-LIMK1-cofilin1 signaling pathway as therapeutic targets against PC.
Acknowledgments
The research was supported by the National Natural Science Foundation of China (nos. 81602093, 81572393, and 81972888), the Natural Science Foundation of Jiangsu Province (nos. BK20160118 and BK20141324), Key Project supported by the Medical Science and Technology Development Foundation, Nanjing Municipality Health Bureau (nos. ZKX15020 and ZKX17022), the Fundamental Research Funds for the Central Universities (nos. 021414380215, 021414380242, 021414380258, and 14380329/3), the Jiangsu Provincial Key Research and Development Program (BE2018701), and the Foundation of Health and Family planning Commission of Lianyungang (201802).
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Hidalgo M. Pancreatic cancer. N Engl J Med 2010;362(17):1605–1617. doi: 10.1056/NEJMra0901557 [DOI] [PubMed] [Google Scholar]
- 2.Zhou S, Ma X, Wang ZJ, et al. Research on the establishment of a TPM3 monoclonal stable transfected PANC-1 cell line and the experiment of the EMT occurrence in human pancreatic cancer. Onco Targets Ther. 2019;12:5577–5587. doi: 10.2147/OTT.S212689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker AE, Hernandez YG, Frucht H, Lucas AL. Pancreatic ductal adenocarcinoma: risk factors, screening, and early detection. World J Gastroenterol. 2014;20(32):11182–11198. doi: 10.3748/wjg.v20.i32.11182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet (London, England). 2011;378(9791):607–620. doi: 10.1016/S0140-6736(10)62307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Sanagapalli S, Stoita A. Challenges in diagnosis of pancreatic cancer. World J Gastroenterol. 2018;24(19):2047–2060. doi: 10.3748/wjg.v24.i19.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science (New York, N.Y.). 2008;321(5897):1801–1806. doi: 10.1126/science.1164368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou B, Xu JW, Cheng YG, et al. Early detection of pancreatic cancer: where are we now and where are we going? Int J Cancer. 2017;141(2):231–241. doi: 10.1002/ijc.30670 [DOI] [PubMed] [Google Scholar]
- 8.Boudreau HE, Broustas CG, Gokhale PC, et al. Expression of BRCC3, a novel cell cycle regulated molecule, is associated with increased phospho-ERK and cell proliferation. Int J Mol Med. 2007;19(1):29–39. [PubMed] [Google Scholar]
- 9.Su YF, Liang CY, Huang CY, et al. A putative novel protein, DEPDC1B, is overexpressed in oral cancer patients, and enhanced anchorage-independent growth in oral cancer cells that is mediated by Rac1 and ERK. J Biomed Sci. 2014;21:67. doi: 10.1186/s12929-014-0067-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Liu L, Cai J, et al. DEPDC1B enhances migration and invasion of non-small cell lung cancer cells via activating Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2014;450(1):899–905. doi: 10.1016/j.bbrc.2014.06.076 [DOI] [PubMed] [Google Scholar]
- 11.Bai S, Chen T, Du T, et al. High levels of DEPDC1B predict shorter biochemical recurrence-free survival of patients with prostate cancer. Oncol Lett. 2017;14(6):6801–6808. doi: 10.3892/ol.2017.7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollino S, Benassi MS, Pazzaglia L, et al. Prognostic role of XTP1/DEPDC1B and SDP35/DEPDC1A in high grade soft-tissue sarcomas. Histol Histopathol. 2018;33(6):597–608. doi: 10.14670/HH-11-959 [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, Zhou Q. Knockdown of BRCC3 exerts an antitumor effect on cervical cancer in vitro. Mol Med Rep. 2018;18(6):4886–4894. doi: 10.3892/mmr.2018.9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Sun W, Zheng B, et al. DEPDC1B knockdown inhibits the development of malignant melanoma through suppressing cell proliferation and inducing cell apoptosis. Exp Cell Res. 2019;379(1):48–54. doi: 10.1016/j.yexcr.2019.03.021 [DOI] [PubMed] [Google Scholar]
- 15.Peck J, Douglas G, Wu CH, Burbelo PD. Human RhoGAP domain-containing proteins: structure, function and evolutionary relationships. FEBS Lett. 2002;528(1–3):27–34. doi: 10.1016/S0014-5793(02)03331-8 [DOI] [PubMed] [Google Scholar]
- 16.Chen A, Zhang Y, Meng G, et al. Oncolytic measles virus enhances antitumour responses of adoptive CD8(+)NKG2D(+) cells in hepatocellular carcinoma treatment. Sci Rep. 2017;7(1):5170. doi: 10.1038/s41598-017-05500-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu XH, Yang YF, Fang HY, Wang XH, Zhang MF, Wu DC. CEP131 indicates poor prognosis and promotes cell proliferation and migration in hepatocellular carcinoma. Int J Biochem Cell Biol. 2017;90:1–8. doi: 10.1016/j.biocel.2017.07.001 [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Luo J, Wan P, Wu J, Laski F, Chen J. Regulation of cofilin phosphorylation and asymmetry in collective cell migration during morphogenesis. Development. 2011;138(3):455–464. doi: 10.1242/dev.046870 [DOI] [PubMed] [Google Scholar]
- 19.Martemyanov KA, Lishko PV, Calero N, et al. The DEP domain determines subcellular targeting of the GTPase activating protein RGS9 in vivo. J Neurosci. 2003;23(32):10175–10181. doi: 10.1523/JNEUROSCI.23-32-10175.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 2006;126(6):1079–1093. doi: 10.1016/j.cell.2006.07.030 [DOI] [PubMed] [Google Scholar]
- 21.Sokol S. A role for Wnts in morpho-genesis and tissue polarity. Nat Cell Biol. 2000;2(7):E124–E125. doi: 10.1038/35017136 [DOI] [PubMed] [Google Scholar]
- 22.Kyriazis AP, McCombs WB 3rd, Sandberg AA, Kyriazis AA, Sloane NH, Lepera R. Establishment and characterization of human pancreatic adenocarcinoma cell line SW-1990 in tissue culture and the nude mouse. Cancer Res. 1983;43(9):4393–4401. [PubMed] [Google Scholar]
- 23.Phenotype and genotype of pancreatic cancer cell lines: erratum. Pancreas. 2018;47(6):e37. doi: 10.1097/MPA.0000000000001087 [DOI] [PubMed] [Google Scholar]
- 24.Lawson CD, Ridley AJ. Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol. 2018;217(2):447–457. doi: 10.1083/jcb.201612069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahuja P, Singh K. In silico approach for SAR analysis of the predicted model of DEPDC1B: a novel target for oral cancer. Adv Bioinformatics. 2016;2016:3136024. doi: 10.1155/2016/3136024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu D, Zhu X, Jimenez-Cowell K, et al. Identification of the GTPase-activating protein DEP domain containing 1B (DEPDC1B) as a transcriptional target of Pitx2. Exp Cell Res. 2015;333(1):80–92. doi: 10.1016/j.yexcr.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santibanez JF, Kocic J, Fabra A, Cano A, Quintanilla M. Rac1 modulates TGF-beta1-mediated epithelial cell plasticity and MMP9 production in transformed keratinocytes. FEBS Lett. 2010;584(11):2305–2310. doi: 10.1016/j.febslet.2010.03.042 [DOI] [PubMed] [Google Scholar]
- 28.Al-Azayzih A, Gao F, Somanath PR. P21 activated kinase-1 mediates transforming growth factor beta1-induced prostate cancer cell epithelial to mesenchymal transition. Biochim Biophys Acta. 2015;1853(5):1229–1239. doi: 10.1016/j.bbamcr.2015.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park KS, Gumbiner BM. Cadherin-6B stimulates an epithelial mesenchymal transition and the delamination of cells from the neural ectoderm via LIMK/cofilin mediated non-canonical BMP receptor signaling. Dev Biol. 2012;366(2):232–243. doi: 10.1016/j.ydbio.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang M, Hong LI, Zhou GM, Xie YH, Ruan H, Dan-Rong LI. Rhein inhibits the movement and invasion of human ovarian carcinoma cells through Rac1/LIMK1/cofilin signaling pathway. Chin Pharmaco Bull. 2016;32(3):366–372. [Google Scholar]
- 31.Xia L, Lin J, Su J, et al. Diallyl disulfide inhibits colon cancer metastasis by suppressing Rac1-mediated epithelial-mesenchymal transition. Onco Targets Ther. 2019;12:5713–5728. doi: 10.2147/OTT.S208738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castillo-Pichardo L, Humphries-Bickley T, De La Parra C, et al. The rac inhibitor EHop-016 inhibits mammary tumor growth and metastasis in a nude mouse model. Transl Oncol. 2014;7(5):546–555. doi: 10.1016/j.tranon.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grey F, Tirabassi R, Meyers H, et al. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5ʹUTRs. PLoS Pathog. 2010;6(6):e1000967. doi: 10.1371/journal.ppat.1000967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–22525. doi: 10.18632/oncotarget.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao B, Lu Y, Cao X, et al. MiRNA-124 inhibits the proliferation, migration and invasion of cancer cell in hepatocellular carcinoma by downregulating lncRNA-UCA1. Onco Targets Ther. 2019;12:4509–4516. doi: 10.2147/OTT.S205169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng JF, Zhuang YY, Huang FT, Zhang SN. Noncoding RNAs and pancreatic cancer. World J Gastroenterol. 2016;22(2):801–814. doi: 10.3748/wjg.v22.i2.801 [DOI] [PMC free article] [PubMed] [Google Scholar]