Abstract
Introduction
Enterococcus faecalis is one of the most common pathogens in urinary tract infections (UTIs). Fluoroquinolones have been frequently used to treat E. faecalis UTIs, and the emergence of fluoroquinolone-resistant E. faecalis strains has recently been reported in several countries. This study aimed to elucidate the mechanisms involved in fluoroquinolone resistance in clinical E. faecalis isolates by analyzing mutations in quinolone- resistance-determining regions (QRDRs) of gyrA and parC and investigating the role of some efflux pumps.
Methods
In total, 70 clinical E. faecalis isolates collected from UTIs were identified by phenotypic and genotypic methods. Antimicrobial susceptibility testing was performed and multidrug-resistant (including ciprofloxacin resistant) isolates were studied for minimum inhibitory concentrations to ciprofloxacin, levofloxacin, and ofloxacin. In the following, mutations in QRDRs of gyrA and parC and expression of EfrA, EfrB, and EmeA efflux pumps were investigated in 20 high-level ciprofloxacin resistant and two ciprofloxacin susceptible isolates.
Results
High-level resistance to ciprofloxacin was detected in 97.5% of isolates. Sequencing of QRDRs revealed that 65% and 75% of isolates carried mutations in gyrA and parC, respectively. The presence of efflux genes was detected in all studied isolates, but expression of efrA, emeA, and efrB was demonstrated in 50%, 40%, and 30% of resistant isolates, respectively. Neither QRDR mutation nor the expression of efflux genes showed any significant association with MIC.
Conclusion
Co-incidence of mutation and efflux gene expression in more than half of isolates (13/20) suggests that both mechanisms may play a role in fluoroquinolone resistance. The other unknown mechanisms including different efflux pumps and probably other QRDRs mutations may contribute to fluoroquinolone resistance in E. faecalis.
Keywords: E. faecalis, UTI, gyrA, parC, efflux pump
Introduction
Fluoroquinolones, as broad-spectrum drugs with lesser side effects and favorable oral dosage, are of convenient therapeutic agents recommended for the treatment of urinary tract infections (UTIs) caused by both gram-positive and gram-negative organisms.1
Among nosocomial pathogens, enterococci have caused major concerns because of their typical antibiotic resistance. Within various body systems, urinary tract is a common site of enterococcal infections. Reportedly, Enterococcus faecalis is responsible for more than half of enterococcal UTIs.2 Since these drugs can interact with various cellular components, different resistance strategies including intrinsic or acquired mechanisms may be proposed.3
Resistance to fluoroquinolones is mediated through chromosomes and/or plasmid by various mechanisms, including mutations in the structural genes targeted by fluoroquinolones, gyrA and gyrB coding for DNA gyrase, or parC and parE coding for topoisomerase IV, as well as some efflux pumps.4
Mutational changes in quinolone-resistance-determining region (QRDR) of type II topoisomerases, DNA gyrase and topoisomerase IV are among common resistance mechanisms. Both enzymes are critical during bacterial replication. Thus, it is vital to inhibit their enzymatic activities. While interaction of fluoroquinolones to these enzymes differs in various bacteria, association of higher-level fluoroquinolone resistance with mutations in both target enzymes has been demonstrated.5
Decreased accumulation of fluoroquinolones in bacterial cells mediated by efflux pumps is another major resistance mechanism. EmeA and EfrAB belong to Major Facilitator Superfamily (MFS) and ATP-binding cassette (ABC) family transporters, respectively, and are multidrug efflux pumps contributing in the extrusion of fluoroquinolones in enterococci.3,6
Having knowledge about resistance mechanisms will be crucial to find new suitable drug targets that can subvert established bacterial traits. Therefore, this study aimed to analyze QRDR mutations conferring resistance and the expression of some multidrug efflux pumps in a population of MDR E. faecalis isolated from UTIs.
Materials and Methods
Bacterial Population and Antimicrobial Susceptibility Testing
During 2 years, E. faecalis isolates were collected from patients with clinical symptoms of UTIs at two university-affiliated hospitals (Kerman, Iran). Following identification and confirmation,7,8 disk diffusion test was performed to detect susceptibility, in order to find MDR isolates (i.e. resistance to more than three different classes of antibiotics). Next, for defined MDR isolates (n= 40), minimal inhibitory concentrations (MICs) were determined for ciprofloxacin, levofloxacin, and ofloxacin (obtained from Exir pharmaceutical company, Iran) using two-fold agar dilution method. Results were interpreted according to susceptibility breakpoints defined by the Clinical Laboratory Standard Institute (CLSI) guidelines.9 At last, 20 MDR isolates (with high-level resistance to ciprofloxacin; MIC ≥ 64 µg/mL) and two susceptible isolates (non-MDR) were randomly selected for the next phases.
Sequencing the QRDR Coding Regions in gyrA and parC
Bacterial DNA was extracted using DNA extraction kit (Sinaclon, Iran) according to the manufacturer's instructions. Amplification of DNA fragments containing QRDRs of gyrA and parC genes was carried out using specific primers as described previously.10 Amplicons were sequenced using the same primers by Bioneer Company on an applied Biosystems 3730/3730X1 DNA analyzer (Applied Biosystems, Foster City, CA, USA). Sequences were compared with E. faecalis V583 (GenBank Accession No. NC_004668.1) using Vector NTI AdvanceTM 10.
Expression of EmeA, EfrA/B Efflux Pumps
RNA Extraction and cDNA Synthesis
Overnight grown cultures of E. faecalis isolates were diluted (1:50) and inoculated to fresh brain heart infusion broth media (Merck, Darmstadt, Germany) as the following: one containing ciprofloxacin (at concentration of 1/2 MIC) and the other without any antibiotic supplementation. Following incubation with gentle shaking until reaching the late exponential phase (OD600 = 0.8),1 mL of each culture was harvested by centrifugation (10,000 rpm/2 min) at 4°C and the bacterial pellet was immediately used for RNA extraction using RNA extraction Kit (Yekta Tajhiz, Tehran, Iran) following manufacturer’s instruction. Residual chromosomal DNA was removed by treating samples with the DNase I, RNase-free kit (Sinaclon, Iran). RNA quantification and quality assessment were carried out by NanoDrop NDe1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Then, cDNA was synthesized using the cDNA Synthesis Kit (Yekta Tajhiz, Tehran, Iran) according to the manufacturer’s recommended protocol.
Quantitative RT-PCR
Real-time PCR was carried out in an ABI Step One plus Real-time PCR system with Eva Green PCR Master Mix (Applied Biosystems) (90ºC for 15 min, 40 cycles of 95ºC for 20 s, 57ºC (for efrA), 56ºC (for efrB)11 and 60ºC (for both emeA and gyrB). Real-time PCR reactions were carried out in duplicate. The primer pairs used for emeA in real-time PCR were different from those in conventional PCR.10,12 Threshold values were calculated at a constant level of fluorescence. To determine the relative expression level of emeA, efrA, and efrB genes, the method described by Pfaffl13 was employed using gyrB as housekeeping gene.10
Results
Totally, 40 isolates (40/70, 57%) were defined as MDR. As shown in Table 1, all these isolates were included in the resistant category for ciprofloxacin, levofloxacin, and ofloxacin according to CLSI standards.9 In this study, ciprofloxacin MIC ≥64 μg/mL was considered high-level ciprofloxacin resistance. In the following, 20 MDR E. faecalis isolates which showed high-level resistance to ciprofloxacin and two ciprofloxacin susceptible isolates were randomly selected for further investigations.
Table 1.
The Fluoroquinolone MIC Distribution for 40 Clinical MDR E. faecalis Isolated from UTIs
| Fluoroquinolones | MIC (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| 8 | 16 | 32 | 64 | 128 | 256 | >256 | |
| Ciprofloxacin | 1 | 0 | 1 | 10 | 16 | 7 | 5 |
| Levofloxacin | 1 | 5 | 17 | 13 | 4 | 0 | 0 |
| Ofloxacin | 1 | 0 | 4 | 18 | 13 | 4 | 0 |
Abbreviations: MIC, minimum inhibitory concentration; UTI, urinary tract infection.
Sequence Variations in QRDRs of gyrA and parC
Regarding two susceptible isolates, 100% similarity was observed with the reference sequence (E. faecalis V583) in both QRDRs of gyrA and parC.
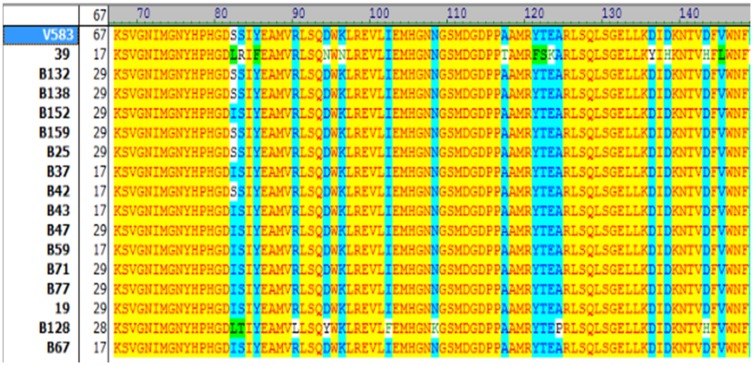
For gyrA, amino acid mutations were seen in 65% of the studied isolates (13/20). Among these isolates, two types of mutations were observed: serine 84 to isoleucine (n=12, 92%) and serine 84 to tyrosine (n=1, 7.6%) (Figure 1). Notably, seven isolates did not possess any mutational changes.
Figure 1.
Amino acid sequences of QRDR domain of gyrA in ciprofloxacin-resistant Enterococcus faecalis isolates with different MIC values. The amino acid substitutions are marked with different colors. Sequences of gyrA were compared with reference sequence (E.feacalis V583) using Vector NTI AdvanceTM 10. Amino acids identical to the corresponding reference sequence are indicated by yellow color. S=Serine (Ser); I=Isoleucine (Ile); L=Leucine (Leu); R=Arginin (Arg); Y=Tyrosine (Tyr); F= Phenylalanine (Phe); D= Aspartic acid (Asp); N= Asparagine (Asn); T= Threonine (Thr); K= Lysine (Lys).
For parC QRDR, totally three types of mutations were found among 75% of resistant isolates (15/20). In 86.6% of isolates (n =13), only one amino acid substitution, serine 82 to isoleucine, was detected. The two other mutation types were seen among two different isolates as follows: isolate 39: amino acid changes at positions serine 82 to leucine, serine 83 to arginine, tyrosine 85 to phenylalanine, aspartic acid 94to asparagine and lysine 96 to asparagine, and isolate B128: amino acid changes at positions serine 82 to leucine, serine 83 to threonine, arginine 90 to leucine, aspartic acid 94 to tyrosine, isoleucine 102 to phenylalanine and asparagine 106 to lysine (Figure 2). Interestingly, the two latter isolates exhibited high MIC to all tested quinolones.
Figure 2.
Amino acid sequences of QRDR domain of parC in ciprofloxacin-resistant Enterococcus faecalis isolates with different MIC values. The amino acid substitutions are marked with different colors. Sequences of parC were compared with reference sequence (E.feacalis V583) using Vector NTI AdvanceTM10. Amino acids identical to the corresponding reference sequence are indicated by yellow color. S=Serine (Ser); I=Isoleucine (Ile); L=Leucine (Leu); R=Arginin (Arg); Y=Tyrosine (Tyr); F= Phenylalanine (Phe); D= Aspartic acid (Asp); N= Asparagine (Asn); T= Threonine (Thr); K= Lysine (Lys).
Herein, 75% of isolates (n= 15), carried mutations in one or both QRDRs of DNA gyrase (GyrA) and topoisomerase IV (ParC). As shown in Table 2, co-incidence of mutations in QRDRs of gyrA and parC was detected in 65% of isolates (n= 13). Only two isolates contained mutation in parC, and for five other isolates, no mutation was found. Statistical analysis did not show any significant association between mutations and MICs level (P > 0.05).
Table 2.
Characteristics of Enterococcus faecalis Isolated from UTIs with Different MICs to Ciprofloxacin: Polymorphisms in the QRDRs of the gyrA and parC Genes and Relative Quantities of the emeA, efrA, and efrB Expression
| Samples | MIC(μg/mL) | Amino Acid Changes ina: | emeAb | efrAb | efrBb | ||||
|---|---|---|---|---|---|---|---|---|---|
| Ciprofloxacin | Levofloxacin | Ofloxacin | gyrA parC | ||||||
| 1 | 39 | 256 | >64 | >128 | S83 I | S82L, S82L, S83R, Y85F, D94N, K96N | 0.976 | 7.015 | 0.199 |
| 2 | 575 | 256 | 32 | 32 | S83 I | S82 I | 1.911 | 0.035 | 0.094 |
| 3 | B128 | >256 | 32 | 128 | S83 I | S82L, S83T, R90L, D94Y, I102F.N108K | 0.598 | 6.276 | 0.871 |
| 4 | B1f | 64 | 32 | 64 | 0.093 | 0.144 | 0.219 | ||
| 5 | B138 | >256 | 32 | 8 | 1.398 | 0.414 | 0.533 | ||
| 6 | B152 | 256 | 32 | 32 | S83 I | S82 I | 0.439 | 1.972 | 1.955 |
| 7 | B153 | >256 | 32 | 64 | S83T | S82 I | 0.599 | 0.702 | 0.751 |
| 8 | B159 | 256 | 32 | 64 | 0.55 | 0.444 | 0.761 | ||
| 9 | B21 | 64 | 16 | 64 | S83 I | S82 I | 0.863 | 1.231 | 2.346 |
| 10 | B25 | 64 | 16 | 64 | 0.185 | 0.112 | 0.393 | ||
| 11 | B37 | 128 | 32 | 64 | S83 I | S82 I | 1.551 | 0.51 | 0.235 |
| 12 | B42 | 64 | 32 | 64 | 0.026 | 0.062 | 0.039 | ||
| 13 | B43 | 128 | 32 | 64 | S83 I | S82 I | 1.119 | 1.283 | 1.035 |
| 14 | B47 | 128 | 32 | 64 | S83 I | S82 I | 1.55 | 1.741 | 1.409 |
| 15 | B59 | 128 | 32 | 64 | S83 I | S82 I | 0.724 | 7.621 | 1.186 |
| 16 | B67 | 128 | 32 | 64 | S83 I | S82 I | 1.048 | 1.239 | 4.32 |
| 17 | B77 | 128 | 64 | 128 | S83 I | S82 I | 0.934 | 2.62 | 0.546 |
| 18 | B80 | 128 | >64 | 128 | S82 I | 2.004 | 0.411 | 0.217 | |
| 19 | B71 | 128 | 64 | 128 | S83I | S82 I | 1.078 | 2.77 | 0.588 |
| 20 | 19 | 256 | 64 | 64 | S82 I | 0.111 | 0.13 | 0.008 | |
| 21 | B123 | 1 | 2 | 1 | 0.135 | 0.027 | 0.014 | ||
| 22 | B132 | 1 | 1 | 1 | 0.115 | 0.017 | 0.011 | ||
Notes: Ciprofloxacin MIC ≥ 64 µg/mL was considered as high-level resistance to ciprofloxacin. B123 and B132 are ciprofloxacin susceptible. aAmino acid changes with respect to the reference sequence of E. faecalis V583 (GenBank Accession No. NC_004668.1). bmRNA expression levels as measured by real-time PCR.
Abbreviations: MIC, minimum inhibitory concentration; S, Serine; Y, Tyrosine; N, Asparagine; I, Isoleucine; K, Lysine; F, Phenylalanine; L, Leucine; D, Aspartic acid.
Expression of emeA, efrA and efrB Genes
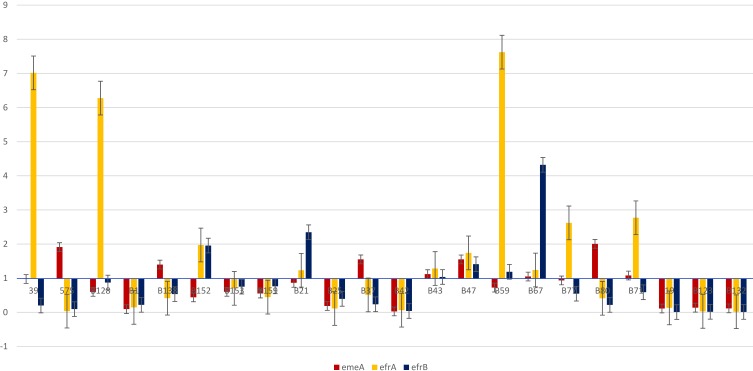
The emeA, efrA and efrB genes were detected in all studied isolates. The ratio of relative expression of emeA, efrA and efrB mRNA among the resistant isolates varied from 0.026 to 2.004 for emeA, 0.062 to 7.621 for efrA, and 0.008 to 4 for efrB (Table 2). The abundance of the emeA, efrA and efrB mRNA expression varied considerably among the isolates with 1.88-fold, 2.08-fold and 2.69-fold difference between the least to the most expressed ones, respectively.
According to our results, the most frequent expressed efflux gene was efrA (10/20, 50%), followed by emeA (8/20, 40%) and efrB (6/20, 30%). Furthermore, co-expression of efflux genes was mostly detected for efrA+ efrB (6/20, 30%), followed by efrB+ emeA (4/20, 20%) and efrA+ emeA (3/20, 15%). Altogether, co-expression of two efflux pumps was seen in 13 isolates. In addition, in three isolates (B43, B47, B67), three efflux pumps were co-expressed simultaneously. All of these isolates contained similar mutations both in gyrA and parC (Figure 3).
Figure 3.
Fold changes expression of three different efflux genes (emeA, efrA, efrB) in 22 clinical MDR E. faecalis isolates.
As shown in Table 2, no expression of efflux genes was found in six high-level ciprofloxacin-resistant isolates (6/20). Interestingly, four of these isolates did not have any mutations in gyrA and parC. Statistical analysis did not show any significant association between efflux gene expression and MICs (P> 0.05).
Discussion
Resistance to fluoroquinolones in bacteria is mainly mediated by spontaneous mutations in the QRDRs of DNA gyrase and topoisomerase IV especially at the highly conserved residues.5 Whether gyrA mutation is more important in fluoroquinolone resistance or mutation in parC, is controversial. Leavis et al14 found parC mutation in all ciprofloxacin resistant isolates while additional mutation in gyrA was detected only in 42% of them. Thus, they concluded that the main target in mutation-mediated ciprofloxacin resistance in enterococci is mutation in parC. On the other hand, López et al15 suggested DNA gyrase as the primary target for resistance to levofloxacin and ciprofloxacin in E. faecalis isolates. In contrast to the mentioned studies and similar to Yasufuku et al,16 our results did not show any significant difference between the frequency of mutations in gyrA and parC (65% versus 75%, respectively).
Different frequencies of mutations have also been described: Jafari-Sales et al17 reported gyrA mutation in 92.5% of ciprofloxacin resistant isolates, while in the study by Piekarska et al,18 19% and 36% of isolates and in a report by Yasufuku et al,16 28% and 18% of isolates harbored mutations in gyrA and parC, respectively.
The type of mutations is also different: In a study conducted by Lopez et al,15 all strains with ciprofloxacin MIC >32 mg/l presented amino acid changes in GyrA protein (S83I, S83Y, S83R or S83I, and E87G) with/without changes in ParC protein (S80I or S80R or S80I). A report by Kanematsu et al19 demonstrated the amino acid mutation of S83I in GyrA in 28.3% and the amino acid mutation of S80I in ParC in 17.9% of clinical strains of E. faecalis. In addition, Kim and Woo20 reported that all high-level ciprofloxacin-resistant enterococcus isolates recovered from chicken meat in Korea showed mutations in both parC and gyrA consisting of S83I-S80I (94.2%), S83F-S80I (2.3%), S83Y-S80I (2.3%), and S83Y-S80F (1.2%). However, in our study, 92% of mutations in QRDR gyrA and 80% of parC mutations were related to S83I and S82I, respectively.
In this study, similar to Kim et al in 2017 and Oyamada et al in 2006, no correlation was found between mutation pattern and the MIC.10,20 This is in contrast to Yasufuku et al16 who found that mutations were significantly correlated with the MICs of levofloxacin.
Correlations between mutations and their effects on drug–target interactions have not been fully explored. Published crystallographic structures suggest that Serine and acidic residues anchor the water–metal ion bridge that coordinates drug binding.21 Accordingly, the number of mutations is a determining factor in reduced affinity between the drug and target enzymes resulting in drug resistance. In this context, Urushibara et al in 2014 found that MIC elevations significantly correlated with the number of QRDR mutations which cannot be generalized to our study with limited number of mutations in QRDRs.5
Resistance through active efflux of quinolones and increased expression of endogenous efflux pumps are alternative quinolone resistance mechanisms reported for E. faecalis.3,6 In our study, the presence of emeA, efrA/B was detected in all ciprofloxacin resistant and susceptible isolates. Wei Jia et al detected emeA gene in 73.8% of ciprofloxacin-resistant (but not susceptible) enterococci and suggested that the distribution of the emeA gene is associated with the resistance to fluoroquinolones in enterococcus species.12 In contrast, Shiadeh et al in 2018 and Valenzuela et al in 2013 detected efrA and efrB in 100% and 96% of isolates, respectively.4,22
In this study, different levels of efflux gene expression were obtained which can be attributable to bacterial isolates as reported by Lerma et al in 2014.11 In addition, no significant relationship was found between the expression of efflux pumps and level of MIC. However, Lubelski et al in 2007 reported that elevated drug resistance was only observed when efrA and efrB genes were co-expressed.23
It is yet unclear which mechanism is more effective in development of resistance because there are resistant isolates with no expression of efflux genes but the presence of QRDR mutations and the presence of one isolate without any mutation with an expression of an efflux gene. Bioinformatic approaches have demonstrated 34 potential multidrug efflux pump genes, nine of which, including emeA and a bmr homologue, belong to the MF superfamily. In addition, 23 ABC transporter homologues have been detected in E. faecalis isolates.24 Thus, it is suggested that another pump of MF superfamily rather than EmeA, or ABC transporter rather than EfrA/B may be involved in fluoroquinolone resistance. Also, other mechanisms such as mutation in parE or gyrB may be proposed. However, it has already been established that alterations in GyrB and ParE are associated with fluoroquinolone resistance in some Gram-positive bacteria such as S. aureus, S. pneumoniae and E. faecium, but not in E. faecalis.25
In conclusion, co-incidence of mutation and efflux gene expression in 65% of isolates suggests that both mechanisms may be important. More investigations are required to detect different strategies involved in this process, which will be critical to tackle antimicrobial resistance problem in clinical care, using novel therapeutic agents such as efflux pumps inhibitors to control multidrug antibiotic resistance.
Acknowledgment
This work was financially supported by the research council of Kerman University of Medical Sciences, Kerman, Iran (Grant Number: 96001119). This study was performed under approval of institutional review board of Kerman University of Medical Sciences (IR.KMU.AH.REC.1397.004).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Adam HJ, Hoban DJ, Gin AS, Zhanel GG. Association between fluoroquinolone usage and a dramatic rise in ciprofloxacin-resistant Streptococcus pneumoniae in Canada, 1997–2006. Int J Antimicrob Agents. 2009;34(1):82–85. doi: 10.1016/j.ijantimicag.2009.02.002 [DOI] [PubMed] [Google Scholar]
- 2.Hidron AI, Edwards JR, Patel J, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006_2007. Infect Control Hosp Epidemiol. 2008;29(11):996–1011. doi: 10.1086/591861 [DOI] [PubMed] [Google Scholar]
- 3.Jonas BM, Murray BE, Weinstock GM. Characterization of emeA, anorA homolog and multidrug resistance efflux pump, in Enterococcus faecalis. Antimicrob Agents Chemother. 2001;45(12):3574–3579. doi: 10.1128/AAC.45.12.3574-3579.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiadeh SMJ, Hashemi A, Fallah F, Lak P, Azimi L, Rashidan M. First detection of efrAB, an ABC multidrug efflux pump in Enterococcus faecalis in Tehran, Iran. Acta Microbiol Immunol Hun. 2018;66(1):57–68. doi: 10.1556/030.65.2018.016 [DOI] [PubMed] [Google Scholar]
- 5.Urushibara N, Suzaki K, Kawaguchiya M, et al. Contribution of type II topoisomerase mutations to fluoroquinolone resistance in Enterococcus faecium from Japanese clinical setting. Microb Drug Resist. 2018;24(1):1–7. doi: 10.1089/mdr.2016.0328 [DOI] [PubMed] [Google Scholar]
- 6.Phillips-Jones MK, Harding SE. Antimicrobial resistance (AMR) nanomachines—mechanisms for fluoroquinolone and glycopeptide recognition, efflux and/or deactivation. Biophys Rev. 2018;10(2):347–362. doi: 10.1007/s12551-018-0404-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/JCM.27.4.731-734.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/JCM.33.1.24-27.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Performance CLSI. Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2019. [Google Scholar]
- 10.Oyamada Y, Ito H, Inoue M, Yamagishi JI. Topoisomerase mutations and efflux are associated with fluoroquinolone resistance in Enterococcus faecalis. J Med Microbiol. 2006;55(10):1395–1401. doi: 10.1099/jmm.0.46636-0 [DOI] [PubMed] [Google Scholar]
- 11.Lerma LL, Benomar N, Valenzuela AS, Muñoz M, Gálvez A, Abriouel H. Role of EfrAB efflux pump in biocide tolerance and antibiotic resistance of Enterococcus faecalis and Enterococcus faecium isolated from traditional fermented foods and the effect of EDTA as EfrAB inhibitor. Food Microbiol. 2014;44:249–257. doi: 10.1016/j.fm.2014.06.009 [DOI] [PubMed] [Google Scholar]
- 12.Jia W, Li G, Wang W. Prevalence and antimicrobial resistance of Enterococcus species: a hospital-based study in China. Int J Environ Res Public Health. 2014;11(3):3424–3442. doi: 10.3390/ijerph110303424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leavis HL, Willems RJ, Top J, Bonten MJ. High-level ciprofloxacin resistance from point mutations in gyrA and parC confined to global hospital-adapted clonal lineage CC17 of Enterococcus faecium. J Clin Microbiol. 2006;44(3):1059–1064. doi: 10.1128/JCM.44.3.1059-1064.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez M, Tenorio C, Del Campo R, Zarazaga M, Torres C. Characterization of the mechanisms of fluoroquinolone resistance in vancomycin-resistant enterococci of different origins. J Chemother. 2011;23(2):87–91. doi: 10.1179/joc.2011.23.2.87 [DOI] [PubMed] [Google Scholar]
- 16.Yasufuku T, Shigemura K, Shirakawa T, et al. Mechanisms of and risk factors for fluoroquinolone resistance in clinical Enterococcus faecalis isolates from patients with urinary tract infections. J Clin Microbiol. 2011;49(11):3912–3916. doi: 10.1128/JCM.05549-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jafari-Sales A, Sayyahi J, Akbari-Layeg F, Mizabi-Asl M, Rasi-Bonab F. Identification of gyrA gene in ciprofloxacin-resistant enterococcus faecalis in strains isolated from clinical specimens in hospitals and clinics of Tabriz and Marand cities. Arch Clin Microbiol. 2017;8(5):63. [Google Scholar]
- 18.Piekarska K, Gierczynski R, Lawrynowicz-Paciorek M, Kochman M, Jagielski M. Novel gyrase mutations and characterization of ciprofloxacin-resistant clinical strains of Enterococcus faecalis isolated in Poland. Pol J Microbiol. 2008;57(2):121–124. [PubMed] [Google Scholar]
- 19.Kanematsu E, Deguchi T, Yasuda M, Kawamura T, Nishino Y, Kawada Y. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV associated with quinolone resistance in Enterococcus faecalis. Antimicrob Agents Chemother. 1998;42(2):433–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MC, Woo GJ. Characterization of antimicrobial resistance and quinolone resistance factors in high‐level ciprofloxacin‐resistant Enterococcus faecalis and Enterococcus faecium isolates obtained from fresh produce and fecal samples of patients. J Sci Food Agric. 2017;97(9):2858–2864. doi: 10.1002/jsfa.8115 [DOI] [PubMed] [Google Scholar]
- 21.Aldred KJ, McPherson SA, Turnbough CL Jr, Kerns RJ, Osheroff N. Topoisomerase IV-quinolone interactions are mediated through a water-metal ion bridge: mechanistic basis of quinolone resistance. Nucleic Acids Res. 2013;41(8):4628–4639. doi: 10.1093/nar/gkt124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez Valenzuela A, Lavilla Lerma L, Benomar N, Gálvez A, Perez Pulido R, Abriouel H. Phenotypic and molecular antibiotic resistance profile of Enterococcus faecalis and Enterococcus faecium isolated from different traditional fermented foods. Foodborne Pathog Dis. 2013;10(2):143–149. doi: 10.1089/fpd.2012.1279 [DOI] [PubMed] [Google Scholar]
- 23.Lubelski J, Konings WN, Driessen AJ. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol Mol Biol Rev. 2007;71(3):463–476. doi: 10.1128/MMBR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pazoles J, Talbot MK, Alder EA, et al. Enterococcus faecalis multi-drug resistance transporters: application for antibiotic discovery. J Mol Microbiol Biotechnol. 2001;3(2):179–184. [PubMed] [Google Scholar]
- 25.Oyamada Y, Ito H, Fujimoto K, et al. Combination of known and unknown mechanisms confers high-level resistance to fluoroquinolones in Enterococcus faecium. J Med Microbiol. 2007;55(6):729–736. doi: 10.1099/jmm.0.46303-0 [DOI] [PubMed] [Google Scholar]