Salmonella enterica serovar Pullorum is the pathogen of pullorum disease, which leads to severe economic losses in many developing countries. In contrast to the strong inflammatory response induced by Salmonella enterica serovar Typhimurium and Salmonella enterica serovar Enteritidis, S. Pullorum causes systemic infection with little inflammation. The effector proteins secreted by Salmonella often play a crucial role in modulating host signal transduction and cellular processes to the pathogen’s advantage.
KEYWORDS: NF-κB pathway, IpaJ protein, anti-inflammatory response, Salmonella enterica serovar Pullorum
ABSTRACT
Salmonella enterica serovar Pullorum is the pathogen of pullorum disease, which leads to severe economic losses in many developing countries. In contrast to the strong inflammatory response induced by Salmonella enterica serovar Typhimurium and Salmonella enterica serovar Enteritidis, S. Pullorum causes systemic infection with little inflammation. The effector proteins secreted by Salmonella often play a crucial role in modulating host signal transduction and cellular processes to the pathogen’s advantage. In the present study, the invasion plasmid antigen J (IpaJ) protein specifically identified in S. Pullorum was found to significantly inhibit activation of the key proinflammatory transcription factor, NF-κB, which was induced by tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), and lipopolysaccharide (LPS). IpaJ inhibited the NF-κB pathway in cells infected with S. Pullorum through the stabilization of IκBα. Deletion of ipaJ in S. Pullorum caused a significantly increased level of ubiquitinated IκBα that was subsequently degraded by the proteasome in HeLa cells. Moreover, IpaJ was efficient in the prevention of NF-κB translocation to the nucleus and ultimately interfered with the secretion of the proinflammatory cytokines IL-1β, IL-6, and IL-8 in infected HeLa cells. Additionally, the transformation of ipaJ into S. Enteritidis decreased the secretion of proinflammatory cytokines in HeLa cells through suppression of the NF-κB pathway. The infection of chicken peripheral blood monocyte-derived macrophages (chMDM) confirmed that ipaJ-deleted S. Pullorum induced a stronger expression of proinflammatory cytokines than the wild-type and complementary strains. In summary, the present study revealed that IpaJ functions as an important anti-inflammatory protein involved in S. Pullorum infection through inhibition of the NF-κB pathway and the subsequent inflammatory response.
INTRODUCTION
Salmonella enterica is a facultative intracellular pathogen with more than 2,600 serotypes causing multistage systemic infection in humans and animals. The host immune response to pathogenic bacteria is often caused by the stimulation of pattern recognition receptors by conserved bacterial products commonly known as pathogen-associated molecular patterns (PAMPs), including flagellin, nucleic acids, or bacterial envelope components, such as lipoproteins, peptidoglycan, and lipopolysaccharide (1). The recognition of these products by transmembrane Toll-like receptors (TLRs), cytoplasmic nucleotide oligomerization domain-like receptors (NLRs), or RIG-I-like (RLR) receptors triggers cascades of cellular signals, which finally lead to the production of proinflammatory cytokines (2–4).
A critical signaling pathway induced by many bacterial pathogens is the nuclear factor-kappa B (NF-κB) pathway (5–7). NF-κB comprises a family of transcription factors that play an important role in transcribing genes that govern innate and adaptive immune responses, apoptosis, proliferation, and cellular differentiation (8, 9). Since its discovery in 1986, when it was initially described as a B lymphocyte-specific immunoglobulin κ-light chain enhancer-binding protein, NF-κB has been shown to be present in and have important biological functions in almost every nucleated cell type examined so far (10).
Numerous virulence proteins are involved in the process of invasion, colonization, and infection during Salmonella host infection (11). Several of these have been proposed to or shown to manipulate the cellular processes and pathways required for the induction of an innate immune response. For instance, the effectors SseK1 and SseK3 are suggested to inhibit Salmonella-induced NF-κB activation (12). Another effector from Salmonella enterica serovar Typhimurium, SseL, was shown to deubiquitinate IκBα, leading to the inhibition of inflammatory responses in the host (13), but the following study showed that SseL failed to play a role in the downregulation of the host immune response and the NF-κB pathway in particular (14). AvrA, the bacterial effector that exists in 80% of Salmonella, was tested to be a deubiquitinase that removes ubiquitin from IκBα and β-catenin, thereby inhibiting the NF-κB signaling pathway (15). The YopJ protein of Yersinia species and OspG kinase expressed in Shigella flexneri are examples of bacterial virulence factors that interfere with NF-κB activation (16–18).
Currently, the pathogenesis of Salmonella enterica serovar Pullorum is poorly understood, and the study on the mechanism of Salmonella infection mostly concentrates on other serotypes, such as S. Typhimurium. S. Pullorum specifically infects poultry, leading to a high incidence and mortality rate in chicks. The infected adult birds show weight loss, decreased egg laying, dysentery, deformity of the reproductive tract, etc. (19, 20). In contrast to the strong inflammatory response induced by Salmonella enterica serovar Enteritidis and S. Typhimurium, S. Pullorum causes systemic infection of chicks without evident inflammation. It has also been reported that S. Pullorum is inclined to induce a Th2-dominated response, while S. Enteritidis induces both Th1 and Th2 responses (21). In the present study, the protein invasion plasmid antigen J (IpaJ), which is present specifically in S. Pullorum, was identified to inhibit the activation of the NF-κB signaling pathway during S. Pullorum infection. The ipaJ gene is located in the pSPI12 plasmid, which is prevalent in 99.08% of S. Pullorum isolates (22, 23). Our previous study suggested that IpaJ contributes to the early stage of S. Pullorum infection, but did not establish a particular role for this protein (22). In the present study, IpaJ was confirmed to be a specific anti-inflammatory protein in S. Pullorum responsible for inhibition of the NF-κB signaling pathway.
RESULTS
IpaJ prevents NF-κB activation induced by TNF-α, IL-1β, and LPS.
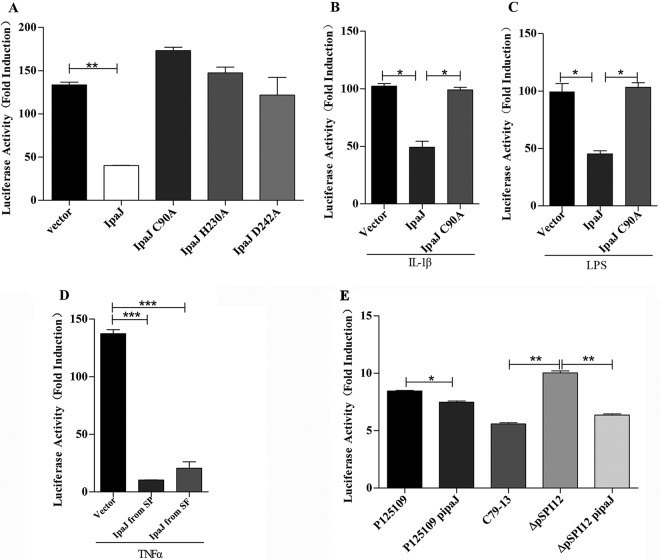
Dual NF-κB reporter gene assays were performed using HeLa cells transfected with plasmids expressing IpaJ, IpaJ (C90A), IpaJ (H230A), or IpaJ (D242A), in which the predicted active sites of cysteine (C), histidine (H), and aspartic acid (D), respectively, were mutated to alanine (A). The cells were cotransfected with a luciferase reporter gene plasmid containing NF-κB binding sites within its promoter and renilla luciferase as an internal control. IpaJ significantly inhibited tumor necrosis factor alpha (TNF-α)-induced NF-κB activation while IpaJ (C90A), IpaJ (H230A), and IpaJ (D242A) could not interfere with the NF-κB signaling pathway (Fig. 1A). NF-κB activation was also inhibited by IpaJ when the stimulants were interleukin-1β (IL-1β) or lipopolysaccharide (LPS) (Fig. 1B and C).
FIG 1.
IpaJ inhibits activation of the NF-κB pathway. (A) HeLa cells were transfected with empty vector (250 ng) or plasmids encoding IpaJ, IpaJ (C90A), IpaJ (H230A), and IpaJ (D242A). NF-κB activity induced by TNF-α (25 ng/ml) was monitored using gene reporter assays (mean ± SD; n = 3). (B and C) NF-κB activity was also induced using IL-1β (25 ng/ml) or LPS (100 mg/ml). (D) HeLa cells were transfected with empty vector (250 ng), plasmids encoding IpaJ in S. Pullorum (SP), or IpaJ expressed by Shigella flexneri (SF). NF-κB activity induced using TNF-α was measured as described for panel A. (E) HeLa cells were cotransfected with plasmids encoding an NF-κB-dependent firefly luciferase gene and renilla luciferase as an internal control. After transfection for 24 h, HeLa cells were infected with S. Enteritidis (P125109), the P125109 strain complemented with pBR322-ipaJ (P125109-pipaJ), the wild-type S. Pullorum strain (C79-13), the pSPI12-deleted strain (ΔpSPI12), or ΔpSPI12-pipaJ for 3 h. Then, cells were washed three times and treated for 30 min with TNF-α and gentamicin. The NF-κB activity induced by different Salmonella strains was depicted as fold increase over negative control (the value was set as 1), which was transfected with NF-κB luciferase reporter vector and Renilla luciferase vector but without TNF-α stimulation (mean ± SD; n = 3). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Although IpaJ from Shigella flexneri showed only 49% homology with IpaJ in S. Pullorum (22), the expressed protein could also inhibit activation of the NF-κB signaling pathway induced by TNF-α in an NF-κB reporter gene assay with plasmids carrying the ipaJ gene from Shigella flexneri (Fig. 1D).
IpaJ inhibits the NF-κB pathway during Salmonella infection.
To investigate the effect of the IpaJ protein on the NF-κB pathway during Salmonella infection, HeLa cells were transfected with a plasmid encoding an NF-κB-dependent firefly luciferase gene and a renilla luciferase. These cells were infected with different Salmonella strains for 3 h followed by TNF-α stimulation for 30 min and gentamicin treatment to kill the remaining bacteria. The cells were washed three times for detection of the luciferase levels. As shown in Fig. 1E, infection with the ΔpSPI12 strain (S. Pullorum C79-13 with the pSPI12 plasmid deleted) activated the reporter by approximately 2-fold compared with that of the wild-type (WT) S. Pullorum strain C79-13 and ΔpSPI12 complemented by the expression of IpaJ from the low-copy-number plasmid pBR322, showing the sensitivity of the NF-κB pathway in the cell line to Salmonella infection. Interestingly, the transformation of pBR322-ipaJ into the S. Enteritidis strain P125109 resulted in decreased NF-κB reporter activity in HeLa cells compared with that of the P125109 group. All of these results showed that IpaJ affected NF-κB activation in HeLa cells during Salmonella infection (Fig. 1E).
IpaJ is required for the inhibition of NF-κB translocation to the nucleus.
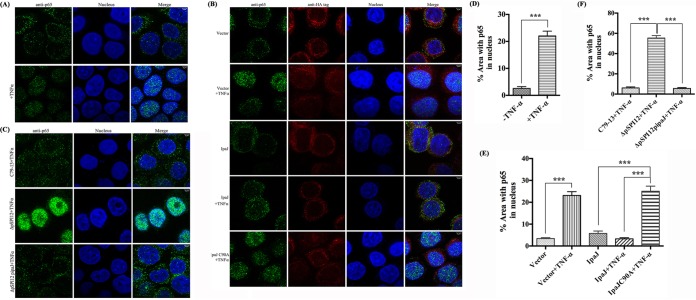
Many Salmonella type III secretion system (T3SS) effectors affect the NF-κB pathway by blocking the nuclear translocation of the canonical NF-κB subunit p65 (24). To determine the potential role of IpaJ in inhibiting the nuclear translocation of p65, confocal microscopy was used to detect the translocation of the p65 subunit to the nucleus of HeLa cells transfected with plasmids expressing IpaJ or IpaJ (C90A) upon TNF-α treatment. The cells were immunolabeled with anti-p65 antibody and anti-hemagglutinin (HA) antibody against HA-IpaJ and stained with 4′,6-diamidino-2-phenylindole (DAPI) for the nucleus. TNF-α induced the translocation of p65 from the cytoplasm to the nucleus (Fig. 2A and D). In contrast, the expression of HA-IpaJ in HeLa cells prevented the nuclear translocation of p65 with the stimulation of TNF-α, while IpaJ (C90A) did not interfere with p65 translocation (Fig. 2B and E). Moreover, nuclear translocation of p65 was detected during the infection of HeLa cells by the ΔpSPI12 strain without the ipaJ gene. The wild-type C79-13 and complementary strain ΔpSPI12-pipaJ, but not the ΔpSPI12 mutant, blocked the translocation of p65 to the nucleus (Fig. 2C and F). These results were in agreement with the transfection analysis, supporting the inference that the presence of IpaJ blocked the translocation of p65 to the nucleus.
FIG 2.
IpaJ is sufficient for blocking NF-κB translocation to the nucleus. (A, D) HeLa cells were treated with TNF-α for 30 min or remained untreated, after which they were fixed and stained with anti-p65 (green) and 4′,6-diamidino-2-phenylindole (DAPI) (blue). (B, E) HeLa cells transfected with plasmids expressing HA tag, HA-IpaJ, or HA-IpaJ (C90A) were treated with TNF-α for 30 min or remained untreated, after which they were fixed and stained with anti-p65 (green), anti-HA (red), and DAPI (blue). (C, F) HeLa cells were infected with wild-type (WT), a pSPI12 mutant, or ΔpSPI12-pipaJ. After 3 h, cells were washed and treated with TNF-α and gentamicin. At 30 min post-TNF-α induction, they were fixed and stained with anti-p65 (green) and DAPI (blue). The measurement of p65 in HeLa cells were performed in duplicate, and the percentage of area in nucleus occupied by p65 was analyzed to demonstrate the translocation ability of p65 into the nucleus (D, E, F). ***, P ≤ 0.001.
IpaJ deficiency increases the IκBα degeneration in TNF-α-stimulated Salmonella-infected cells.
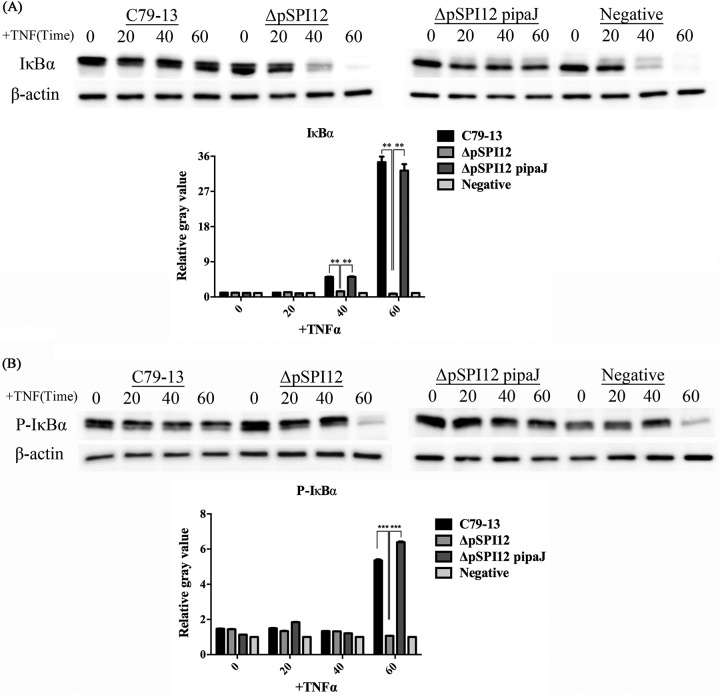
Because IκBα is a key factor in the NF-κB pathway, analysis of the levels of IκBα is commonly considered a measure of the activation of the canonical NF-κB pathway (25). Immunoblotting analysis was used to detect the level of IκBα and phospho-IκBα in HeLa cells infected with different S. Pullorum strains, and the cells were then stimulated by TNF-α to induce IκBα degradation at different time points. In control cells, TNF-α induced a rapid decline in IκBα levels. In cells infected with wild-type S. Pullorum and the complementary strain, IκBα levels remained essentially unchanged throughout the time course of the TNF-α stimulation studied. Compared with that of the wild-type and complementary strains of S. Pullorum, infection with the ΔpSPI12 mutant resulted in strong IκBα degradation in cells at 40 min upon TNF-α treatment, decreasing to barely detectable levels at 60 min similar to the negative control (Fig. 3A). Meanwhile, upon stimulation with TNF-α at 60 min, the phospho-IκBα content remained stable in the wild-type and complementary strain infection groups but decreased in the mutant group, which is similar to the negative control (Fig. 3B).
FIG 3.
IpaJ prevents IκBα degradation upon TNF-α treatment during Salmonella infection. HeLa cells were infected for 3 h with different strains or remained untreated, followed by TNF-α treatment as indicated. Proteins were extracted at 0, 20, 40, and 60 min post-TNF-α treatment. The Western blots were developed with anti-IκBα (A), anti-phospho-IκBα (B), or anti-β-actin antibodies (loading control). **, P ≤ 0.01; ***, P ≤ 0.001.
IpaJ deficiency increases IκBα ubiquitination in Salmonella-infected cells.
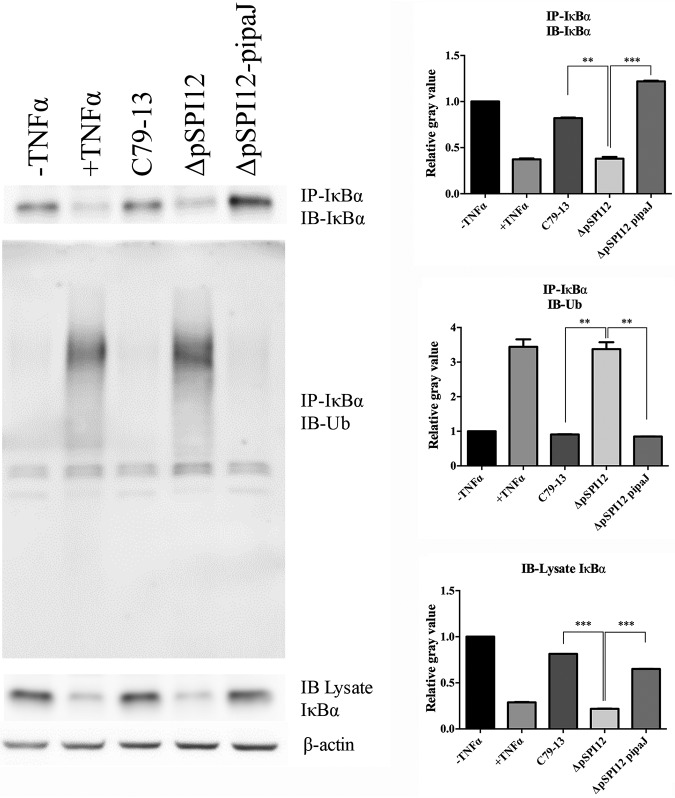
Next, the effects of the C79-13, pSPI12 mutant, and complementary strains were compared with respect to IκBα ubiquitination in infected HeLa cells. Compared with that of C79-13 and ΔpSPI12-pipaJ strains, the ΔpSPI12 mutant infection caused increased IκBα degradation and ubiquitination in the infected cells as shown by IκBα ubiquitination (determined by the immunoprecipitation of IκBα, followed by immunoblot analysis with antiubiquitin antibody) (Fig. 4). This demonstrated that IpaJ is responsible for IκBα stabilization and deubiquitination in the context of Salmonella infection.
FIG 4.
IpaJ prevents IκBα ubiquitination upon TNF-α treatment during Salmonella infection. HeLa cells were infected for 3 h with different strains or remained untreated, followed by TNF-α treatment as indicated. Proteins were extracted at 30 min post-TNF-α treatment. Cell lysates were normalized for protein contents and subjected to IP using anti-IκBα antibody. IκBα expression levels and ubiquitination status in the immunoprecipitates and lysates were analyzed via immunoblotting using anti-IκBα and antiubiquitin antibodies, respectively. Data are representative of three independent experiments (n = 3). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
IpaJ inhibits the secretion of proinflammatory cytokines in infected HeLa cells.
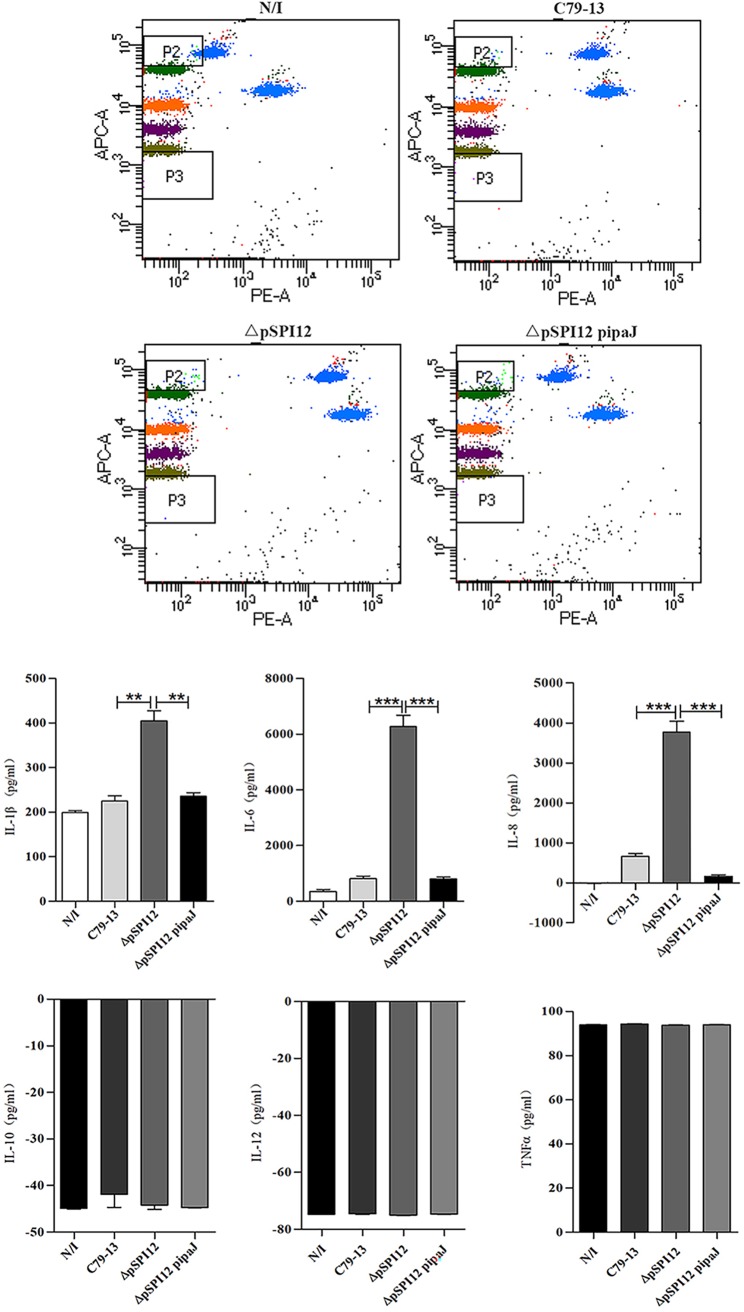
To explore whether the inhibition of NF-κB activation would lead to the secretion of cytokines through S. Pullorum infection, the secretion of proinflammatory cytokines was measured in the supernatant of HeLa cells infected with Salmonella using the BD cytometric bead array (CBA) human inflammatory cytokines kit (BD, USA). Briefly, different S. Pullorum strains were used to infect HeLa cells for 7 h, and the supernatants of the cells were then collected and subjected to measurement of proinflammatory cytokine levels following the manufacturer’s instructions. In comparison with that of the ΔpSPI12 mutant, the wild-type and the complementary strains showed significant inhibition of IL-1β, IL-6, and IL-8 secretion in infected HeLa cells (Fig. 5). There was a 5-fold increase in the production of the NF-κB-dependent cytokine IL-8 in the supernatant of HeLa cells infected with the ΔpSPI12 strain compared with that of the wild-type or complementary strains. However, the secretion of the other cytokines, IL-10, TNF-α, and IL-12, did not show significant changes at this time point. These results thus provide additional confirmation of the inhibitory effect of IpaJ on NF-κB activity in the context of Salmonella infection.
FIG 5.
IpaJ is required to block IL-8, IL-1β, and IL-6 secretion. HeLa cells were infected with wild-type S. Pullorum (C79-13), pSPI12-deleted strain (ΔpSPI12), and ΔpSPI12-pipaJ or remained uninfected (N/I). After 1 h of infection, cells were washed and treated with gentamicin to kill the extracellular bacteria. After an additional 6 h of incubation, the supernatants of the cultures were harvested and the secretion of proinflammatory cytokines was analyzed using the CBA human inflammatory cytokines kit. The BD FACSAria SORP (BD, USA) was used to detect the expression level of cytokines. The experiment was performed three times in duplicates, and typical results are shown. Bars indicate standard errors. **, P ≤ 0.01; ***, P ≤ 0.001.
IpaJ shows an anti-inflammatory response in avian cells infected with Salmonella.
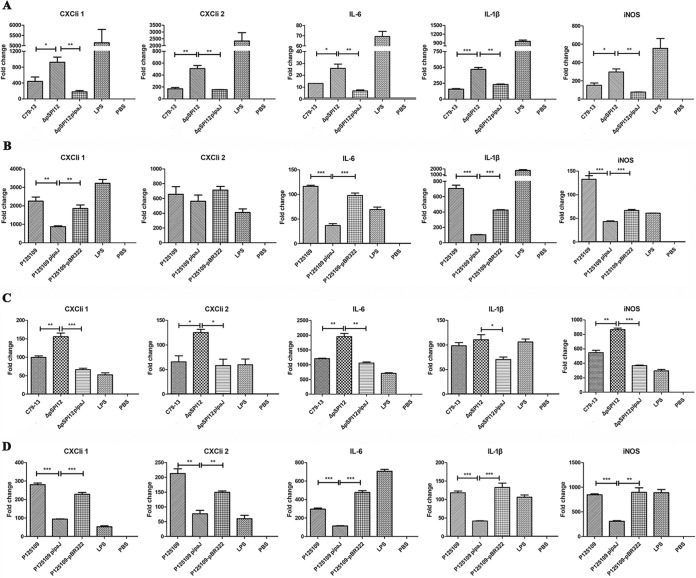
The immune response was also detected in avian cells infected by the pathogen considering that S. Pullorum is a host-restricted serotype of Salmonella. The avian macrophage HD-11 cell line and the primary monocytes chicken peripheral blood monocyte-derived macrophages (chMDM) were infected with S. Pullorum and S. Enteritidis. As shown in Fig. 6A and Fig. S1A in the supplemental material, the TaqMan probe quantitative real-time PCR (qRT-PCR) analysis demonstrated that the expression levels of CXCLi1, CXCLi2, IL-6, IL-1β, and iNOS in the HD-11 cells infected with the ΔpSPI12 mutant were significantly higher than those in cells infected with C79-13 or a complementary strain carrying the ipaJ gene. Additionally, in HD-11 cells infected with S. Enteritidis P125109-pipaJ, the expression levels of CXCLi1, CXCLi2, IL-6, IL-1β, and iNOS were significantly downregulated in comparison to those in HD-11 cells infected with wild-type P125109 or P125109-pBR322 without the ipaJ gene (Fig. 6B; see also Fig. S1B). These results were consistent with those of our previous study (26). However, because the HD-11 cell is a cell line derived from chicken hematopoietic cells (27), the primary chicken blood monocytes (chMDM) were isolated and purified for further analysis. As shown in Fig. 6C and Fig. S1C, similar to the cytokines expressed in HD-11 cells, CXCLi1, CXCLi2, IL-6, IL-1β, and iNOS were also significantly increased in ΔpSPI12-infected chMDM cells compared with those in the other two groups. Further, in chMDM cells infected with S. Enteritidis P125109-ipaJ, the decreased expression levels of the five cytokines compared with those of the groups infected with ipaJ-negative strains were much more evident than those in the HD-11 cells (Fig. 6D; see also Fig. S1D).
FIG 6.
mRNA expression level of proinflammatory cytokines in avian cells at 6 h postinfection with S. Pullorum (C79-13, ΔpSPI12, ΔpSPI12-pipaJ) or S. Enteritidis (P125109, P125109-pipaJ, P125109-pBR322) evaluated using TaqMan qRT-PCR. The expression levels of proinflammatory cytokines were measured in HD-11 cells infected with S. Pullorum (A) and S. Enteritidis (B). The corrected threshold cycle (CT) values are shown as fold change in the mRNA level of cytokines in comparison to those from uninfected controls. Asterisks indicate differences between levels of cytokines induced by different strains, *, P < 0.05; **, P < 0.01; ***, P < 0.001. mRNA expression level of proinflammatory cytokines in chMDM cells infected with S. Pullorum (C) and S. Enteritidis (D). chMDM cells were primary blood monocyte-derived macrophages isolated from chicken blood.
The expression and secretion of IpaJ are not manipulated by T3SS1 or T3SS2.
S. Pullorum SPI1 or SPI2 mutant strains were supplemented with the expression of IpaJ containing an HA tag from pBR322. The IpaJ-HA expressed in bacteria and secreted to the supernatant was measured using immunoblotting analysis. The results revealed that the deletion of SPI1 and/or SPI2 did not affect the expression and secretion of IpaJ (Fig. 7). IpaJ protein was normally expressed under in vitro culture conditions and was secreted to the extracellular environment. This demonstrated that the expression and secretion of IpaJ were not regulated by T3SS.
FIG 7.
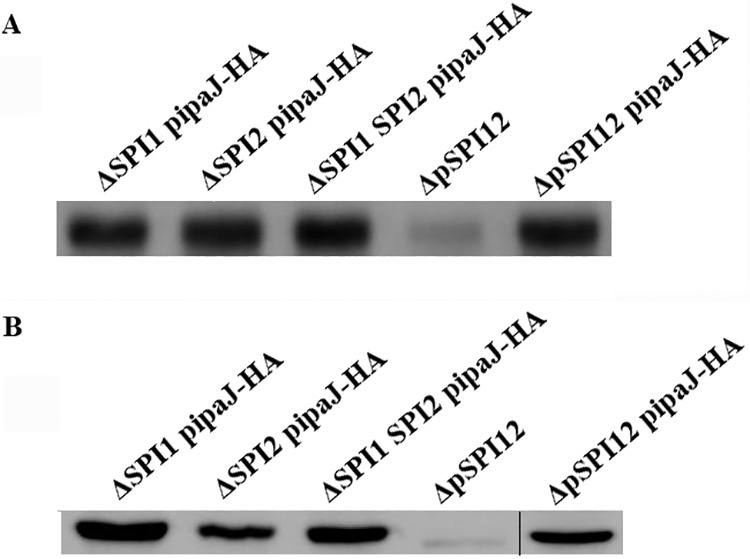
The expression and secretion of IpaJ were not regulated by T3SS. (A) Immunoblotting analysis of bacterial cell lysates to detect IpaJ-Flag using anti-Flag antibody. (B) Immunoblotting analysis of secreted proteins isolated from cultured LB medium supernatant. The plasmid pipaJ carrying the ipaJ gene was transformed into S. Pullorum knock-out strains of SPI1 (ΔSPI1), SPI2 (ΔSPI2), or SPI1 and SPI2 (ΔSPI1 SPI2) to detect whether the secretion of IpaJ is dependent on SPI1 or SPI2. The mutant (ΔpSPI12) and the complementary strain (ΔpSPI12-pipaJ) were used as the negative control and positive control, respectively. The thin black line shows that the left samples and the right sample were performed in two gels and analyzed by Western blotting separately.
DISCUSSION
Effector proteins secreted by Salmonella often play a crucial role in modulating host signal transduction and cellular processes to benefit the survival of bacteria in host cells. In the present study, we demonstrated that the IpaJ protein from S. Pullorum significantly inhibited the activation of the key proinflammatory transcription factor NF-κB, which was induced by the cytokines TNF-α, IL-1β, and LPS. It could also restrain the NF-κB signaling pathway during Salmonella infection. IpaJ from S. Pullorum harbors the catalytic cysteine (C), histidine (H), and aspartate (D) residues required for its function. As predicted by this alignment, alanine substitutions at C90, H230, or D242 abolished the ability of IpaJ to inhibit the activation of the host NF-κB signaling pathway. We suggest that IpaJ interferes with the ubiquitination of the NF-κB inhibitor IκBα upon TNF-α stimulation, thus inhibiting the rapid degradation of IκBα induced by TNF-α. The pSPI12 plasmid-deleted strain was deficient at blocking IκBα degradation compared with that of the wild-type strain. Compared with that of the S. Pullorum wild-type strain, the ΔpSPI12 mutant led to increased ubiquitination and degradation of IκBα in infected cells, but IpaJ did not interfere with the phosphorylation of IκBα induced by TNF-α. Moreover, IpaJ was efficient at inhibiting NF-κB translocation to the nucleus and ultimately interfered with the secretion of proinflammatory cytokines in infected HeLa and avian cells.
The ipaJ gene was first identified as an invasion plasmid gene encoding a novel 27-kDa antigen in Shigella flexneri (28). Further studies showed that the IpaJ expressed in Shigella flexneri functions as a type III secretion system (T3SS) effector and plays a significant role in signal transduction. The Shigella flexneri protease IpaJ was found to cleave myristoylated glycine in eukaryotic proteins and IpaJ substrates comprising proteins with diverse molecular functions, including E3 ubiquitin ligases (29, 30). The IpaJ identified in S. Pullorum contained 279 amino acids that shared just 49% identity with IpaJ from Shigella flexneri, but both harbored the catalytic cysteine (C), aspartic acid (D), and histidine (H) that are closely related to the cysteine peptidase C39-like family of domains with unknown function (31). In our studies, we have shown that the IpaJ protein from Shigella flexneri also has the same ability to inhibit the activation of the NF-κB signaling pathway. IpaJ from S. Pullorum also contributes to the fragmentation of the Golgi apparatus (see Fig. S2 in the supplemental material).
According to a previous study, anti-inflammatory Salmonella serotypes, such as S. Pullorum and proinflammatory species, have the same ability to induce the phosphorylation of IκBα, but S. Pullorum depresses the subsequent ubiquitination of IκBα, thus preventing its degradation. The study suggested the existence of a hypothetical factor that could be translocated from the bacteria into the host cells via T3SS or an alternative specific route (32). In the present study, we found that IpaJ stabilized IκBα but did not affect the phosphorylation of IκBα, consistent with the previous study. Therefore, we propose that IpaJ functions as a proteinase that could increase a specific deubiquitinating activity and finally inhibit the NF-κB pathway and the subsequent inflammatory response (see Fig. S3 in the supplemental material). In addition, the deficiency of SPI1 and/or SPI2 did not interfere with the expression and secretion of IpaJ, demonstrating that the expression and secretion of S. Pullorum IpaJ are not regulated by T3SS, which manipulated expression of Shigella flexneri IpaJ.
We suggest that the IpaJ protein is a novel bacterial protein from S. Pullorum responsible for an anti-inflammatory state. Although the molecular mechanism of this inhibitory action and the IpaJ secretion pathway are not deeply understood, such an ability conferred by nonpathogenic organisms could provide us with new insight for understanding the mechanisms involved in Salmonella infection.
MATERIALS AND METHODS
Bacterial strains, cells, plasmids, and primers.
The bacterial strains, plasmids, and primers used in this study are listed in Tables 1 and Table S1 in the supplemental material. All eukaryotic expression vectors and cloning vectors were constructed using the primers listed in Table S1 following the manufacturer’s instructions for the ClonExpress one step cloning kit (Vazyme, Nanjing, China). The primers eu-ipaJ-F/R and ipaJ-HA-F/R were designed based on the nucleotide sequence of the ipaJ gene harbored in pSPI12 (GenBank accession number GU949535) from S. Pullorum using the Primer Premier 5.0 software (Premier Biosoft, Palo Alto, CA, USA). The primers eu-FipaJ-F/R were designed based on the genomic sequence of Shigella flexneri strain M90T (GenBank accession number NC_002698) using Primer Premier 5.0. C-F/R, H-R/F, and D-F/R used for overlapping PCR converting Cys90, His230, and Asp242 to alanine were designed by Primer X (http://www.bioinformatics.org/primerx/). The rapid one-step site-directed substitution method was described previously (33).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| C79-13 | Wild-type S. Pullorum strain (WT) | Chinese Institute of Veterinary Drug Control |
| ΔpSPI12 | pSPI12 plasmid mutant strain of C79-13 | 26 |
| ΔpSPI12-pipaJ | ΔpSPI12 complemented with expression of IpaJ from pBR322 | 26 |
| P125109 | Wild-type S. Enteritidis strain | Laboratory stock |
| P125109-pipaJ | P125109 complemented by pBR322-ipaJ | 26 |
| P125109-pBR322 | P125109 transformed with the pBR322 plasmid | 26 |
| ΔSPI1 | S. Pullorum strain S06004 with SPI1 deleted | 39 |
| ΔSPI2 | S. Pullorum strain S06004 with SPI2 deleted | 39 |
| ΔSPI1 SPI2 | S. Pullorum strain S06004 with both SPI1 and SPI2 deleted | 39 |
| ΔSPI1 pipaJ-HA | ΔSPI1 complemented with pBR322-ipaJ-HA | This work |
| ΔSPI2 pipaJ-HA | ΔSPI2 complemented with pBR322-ipaJ-HA | This work |
| ΔSPI1 SPI2 pipaJ-HA | ΔSPI1 SPI2 complemented with pBR322-ipaJ-HA | This work |
| ΔpSPI12 pipaJ-HA | ΔpSPI12 complemented with pBR322-ipaJ-HA | This work |
| SNI | S. flexneri strain | Laboratory stock |
| Plasmids | ||
| pCMV-HA | Mammalian expression vector expressing proteins containing an N-terminal hemagglutinin (HA) epitope tag | Laboratory stock |
| pCMV-HA-ipaJ | Vector expressing IpaJ from S. Pullorum | This work |
| pCMV-HA-ipaJ(C90A) | Vector expressing IpaJ replacing cysteine at position 90 with alanine | This work |
| pCMV-HA-ipaJ(H230A) | Vector expressing IpaJ replacing histidine at position 230 with alanine | This work |
| pCMV-HA-ipaJ(D242A) | Vector expressing IpaJ replacing aspartic acid at position 242 with alanine | This work |
| pCMV-HA-FipaJ | Vector expressing IpaJ from S. flexneri | This work |
| pBR322 | Cloning vector | TaKaRa |
| pBR322-ipaJ | Cloning vector expressing IpaJ | 26 |
| pBR322-ipaJ-HA | Cloning vector expressing IpaJ with HA tag | This work |
Nonagitated microaerophilic bacterial cultures were grown overnight in Luria-Bertani (LB) medium with or without ampicillin (50 μg/ml). Then, the bacteria were washed three times in sterile phosphate-buffered saline (PBS) before in vivo and in vitro infection experiments. HeLa cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin and cultured at 37°C in 5% CO2 air. Chicken peripheral blood monocyte-derived macrophages (chMDM) were prepared and cultured as previously described (34, 35). Chicken macrophage-like cells (HD11) were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin and cultured at 41°C in 5% CO2 air.
Luciferase gene reporter assays.
HeLa cells were seeded into 24-well plates and transfected the next day using the Lipofectamine 3000 kit (Invitrogen, CA, USA), keeping the total DNA content constant, using the pCMV-HA vector per the manufacturer’s protocol. At 24 h posttransfection, the cells were left untreated or treated with TNF-α (25 ng/ml), LPS (100 ng/ml), or IL-1β (100 ng/ml) for 5 h, and then the transcriptional activity was assessed. The experiment was performed in triplicate, and the data were expressed as fold induction relative to basal activity (mean ± standard deviation [SD]; n = 3) in relative light units.
Measurement of p65 translocation to nuclei.
Nuclear p65 translocation was monitored in HeLa cells grown in 24-well plates and transfected with 250 ng of DNA. At 24 h posttransfection, cells were cultured with or without TNF-α (25 ng/ml) for 30 min. Next, the cells were fixed (with 4% paraformaldehyde in PBS for 20 min and washed three times with PBS), perforated (with 0.25% Triton X-100 in PBS for 20 min and washed three times with PBS), and blocked (5% bovine serum albumin [BSA] in phosphate-buffered saline with Tween 20 [PBST]) at 37°C for 1 h. The cells were then stained using the anti-NF-κB p65 antibody (1 μg/ml in PBST) (ab16502; Abcam) overnight and further stained with goat anti-rabbit IgG H&L (Alexa Fluor 488) (1:1,000 in PBST) (ab150077; Abcam) for 1 h. Then, the cells were incubated with anti-HA tag antibody (1:1,000) (ab18181; Abcam) and goat anti-mouse IgG2b cross-adsorbed secondary antibody, Alexa Fluor 546 (1:100) (A-21143; Thermo Fisher Scientific). Finally, 4′,6-diamidino-2-phenylindole (DAPI) (C1002; Beyotime) was added at a final concentration of 0.5 μg/ml to label the double-stranded DNA (dsDNA) in the cell nucleus at 37°C for 15 min. The coverslips were mounted on glass slides, and stained cells were observed using laser confocal microscopy (Leica TCS SP8; Leica, Germany). Simultaneously, HeLa cells were infected with Salmonella strains at a multiplicity of infection (MOI) of 100:1 for 3 h and subsequently activated by 25 ng/ml TNF-α for 30 min. Next, the cells were treated as described above.
Analysis of IκBα stability and IκBα phosphorylation.
HeLa cells cultured in 4-cm plates were infected with bacteria (MOI, 100:1) at 5% CO2 and 37°C. Following 3 h of infection, the medium was replaced with fresh DMEM either with or without 25 ng/ml TNF-α for 0, 20, 40, or 60 min. To terminate the infection, cells were washed three times with cold PBS. Next, the pellets were resuspended in 40 ml RIPA lysis and extraction buffer (Thermo Fisher Scientific) and centrifuged (14,000 × g for 10 min at 4°C). The supernatants were boiled in 5× sample buffer and analyzed using immunoblotting with anti-IκBα antibody (1:5,000) (ab32518; Abcam), β-actin antibody as a loading verification control (1:1,000) (sc-517582; Santa Cruz), or anti-IκBα (phospho S32) antibody (1:5,000) (ab92700; Abcam), followed by horseradish peroxidase-conjugated goat anti-rabbit IgG or goat anti-mouse IgG and SuperSignal Western Pico PLUS chemiluminescent substrate (Thermo Scientific, USA). The density of bands was analyzed using ImageJ software (NIH, USA) and normalized by the arbitrary units of β-actin as indicated. The results are presented as the mean ± SD from three independent experiments.
Immunoprecipitation.
For immunoprecipitation, infected HeLa cells were lysed in RIPA lysis and extraction buffer. Cell lysates (100 μl) were incubated with 20 μl of agarose-conjugated antibodies against IκBα and allowed to bind overnight at 4°C. Samples were then washed five times, boiled in 5× sample buffer, and analyzed using immunoblotting with antiubiquitin antibody (1:1,000) (ab19247; Abcam) as described above.
Determination of proinflammatory cytokine levels in culture supernatant.
HeLa cells in six-well plates were inoculated with a 1:100 dilution in the DMEM of bacteria statically grown overnight at 37°C (MOI, 100:1) and incubated for 3 h (5% CO2, 37°C). To terminate the infection and induce cytokine expression, the medium was replaced with fresh DMEM supplemented with 2% fetal calf serum (FCS), 100 μg/μl gentamicin, and with or without 25 ng/ml TNF-α and incubated for an additional 7 h. Next, the cytokine levels were measured using the cytometric bead array (CBA) human inflammatory cytokines kit (BD Biosciences, NJ, USA) following the manufacturer’s protocol. Each sample was measured in duplicate, with results expressed as mean ± SD.
Quantitative real-time PCR analysis.
The infected cell samples were stored in Cryovials containing RNAlater (76106; Qiagen) to protect RNA with immediate RNase inactivation. The total RNA was prepared from infected cells using the RNeasy plus minikit (74136; Qiagen) following the manufacturer’s instructions. cDNA synthesis was performed with the PrimeScript RT reagent kit with genomic DNA (gDNA) eraser (RR047A; TaKaRa) using 1 μg RNA per sample in each reaction mixture following the manufacturer’s guidelines.
The 7500 real-time PCR system (4351105; Applied Biosystems) was used for qRT-PCR to measure the gene expression of selected cytokines and chemokines. The primer and probe sequences used have been described previously (36–38). RT-PCR was performed using Premix Ex Taq (RR390A; TaKaRa). The qPCR mixture for each sample tested consisted of 10 μl of 2× Premix Ex Taq, 0.4 μM forward and reverse primers, 0.2 μM TaqMan probe, 0.2 μl of ROX reference dye II, 2 μl of cDNA, and 5.8 μl of distilled water (dH2O). qPCR was performed following the thermal profile of one cycle at 95°C for 30 s, 40 cycles at 95°C for 5 s, and 60°C for 34 s. The assay was performed in triplicate and repeated three times.
Secretion assays.
Overnight cultures of different S. Pullorum strains were centrifuged and the supernatant passed through a 0.22-μm Millipore filter. Trichloroacetic acid was added to a final concentration of 10% (vol/vol), and the samples were incubated at 4°C overnight. The precipitated proteins were centrifuged at 14,000 × g for 30 min at 4°C and then washed in ice-cold acetone. The samples were then washed four times, boiled in 5× sample buffer, and analyzed using immunoblotting with anti-HA tag antibody (1:1,000) (ab18181; Abcam) and goat anti-mouse IgG H&L (horseradish peroxidase [HRP]) (1:2,000) (ab6708; Abcam).
Statistical analysis.
All of the data are expressed as the mean ± SD. One-way analysis of variance (ANOVA) was performed to analyze the differences. P values of <0.05 were considered significantly different between two groups.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31730094), the National Key Research and Development Program of China (2017YFD0500700, 2017YFD0500100), Jiangsu Province Agricultural Science and Technology Independent Innovation Funds—CX(16)1028, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
We declare that we have no conflicts of interest regarding this work.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Medzhitov R. 2007. Recognition of microorganisms and activation of the immune response. Nature 449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 2.Creagh EM, O'Neill LAJ. 2006. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol 27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Barton GM, Medzhitov R. 2003. Toll-like receptor signaling pathways. Science 300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 4.Doyle SL, O'Neill LAJ. 2006. Toll-like receptors: from the discovery of NF-κB to new insights into transcriptional regulations in innate immunity. Biochem Pharmacol 72:1102–1113. doi: 10.1016/j.bcp.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Johannessen M, Askarian F, Sangvik M, Sollid JE. 2013. Bacterial interference with canonical NF-κB signalling. Microbiology 159:2001–2013. doi: 10.1099/mic.0.069369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhavsar AP, Guttman JA, Finlay BB. 2007. Manipulation of host-cell pathways by bacterial pathogens. Nature 449:827–834. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- 7.Rahman MM, McFadden G. 2011. Modulation of NF-κB signalling by microbial pathogens. Nat Rev Microbiol 9:291–306. doi: 10.1038/nrmicro2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenner D, Blaser H, Mak TW. 2015. Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol 15:362–374. doi: 10.1038/nri3834. [DOI] [PubMed] [Google Scholar]
- 9.Lawrence T. 2009. The nuclear factor NF-κB pathway in inflammation. Cold Spring Harb Perspect Biol 1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayden MS, Ghosh S. 2011. NF-κB in immunobiology. Cell Res 21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibarra JA, Steele-Mortimer O. 2009. Salmonella–the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol 11:1579–1586. doi: 10.1111/j.1462-5822.2009.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Günster RA, Matthews SA, Holden DW, Thurston T. 2017. SseK1 and SseK3 type III secretion system effectors inhibit NF-κB signaling and necroptotic cell death in Salmonella-infected macrophages. Infect Immun 85:e00010-17. doi: 10.1128/IAI.00010-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Negrate G, Faustin B, Welsh K, Loeffler M, Krajewska M, Hasegawa P, Mukherjee S, Orth K, Krajewski S, Godzik A, Guiney DG, Reed JC. 2008. Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-κB, suppresses IκBα ubiquitination and modulates innate immune responses. J Immunol 180:5045–5056. doi: 10.4049/jimmunol.180.7.5045. [DOI] [PubMed] [Google Scholar]
- 14.Mesquita FS, Holden DW, Rolhion N. 2013. Lack of effect of the Salmonella deubiquitinase SseL on the NF-κB pathway. PLoS One 8:e53064. doi: 10.1371/journal.pone.0053064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. 2007. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol 171:882–892. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Staskawicz B, Dixon JE. 2000. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H, Monack DM, Kayagaki N, Wertz I, Yin J, Wolf B, Dixit VM. 2005. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-κB activation. J Exp Med 202:1327–1332. doi: 10.1084/jem.20051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. 2005. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A 102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrow PA, Neto O. 2011. Pullorum disease and fowl typhoid—new thoughts on old diseases: a review. Avian Pathol 40:1–13. doi: 10.1080/03079457.2010.542575. [DOI] [PubMed] [Google Scholar]
- 20.Shivaprasad HL. 2000. Fowl typhoid and pullorum disease. Rev Sci Tech 19:405–424. doi: 10.20506/rst.19.2.1222. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y. 2016. Immune modulation of Salmonella enterica serotype Pullorum in the chicken. PhD dissertation University of Nottingham, Nottingham, UK. [Google Scholar]
- 22.Li Q, Hu Y, Xu Y, Chen J, Fang L, Liu Z, Jiao X. 2014. A gene knock-in method used to purify plasmid pSPI12 from Salmonella enterica serovar Pullorum and characterization of IpaJ. J Microbiol Methods 98:128–133. doi: 10.1016/j.mimet.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Xu L, Liu Z, Li Y, Yin C, Hu Y, Xie X, Li Q, Jiao X. 2018. A rapid method to identify Salmonella enterica serovar Gallinarum biovar Pullorum using a specific target gene ipaJ. Avian Pathol 47:238–244. doi: 10.1080/03079457.2017.1412084. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell S, Vargas J, Hoffmann A. 2016. Signaling via the NF-κB system. Wiley Interdiscip Rev Syst Biol Med 8:227–241. doi: 10.1002/wsbm.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verstrepen L, Bekaert T, Chau TL, Tavernier J, Chariot A, Beyaert R. 2008. TLR-4, IL-1R and TNF-R signaling to NF-κB: variations on a common theme. Cell Mol Life Sci 65:2964–2978. doi: 10.1007/s00018-008-8064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin C, Xu L, Li Y, Liu Z, Gu D, Li Q, Jiao X. 2018. Construction of pSPI12-cured Salmonella enterica serovar Pullorum and identification of IpaJ as an immune response modulator. Avian Pathol 47:410–417. doi: 10.1080/03079457.2018.1471195. [DOI] [PubMed] [Google Scholar]
- 27.Beug H, von Kirchbach A, Döderlein G, Conscience JF, Graf T. 1979. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 18:375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- 28.Buysse JM, Dunyak DS, Hartman AB, Venkatesan MM. 1997. Identification and molecular characterization of a 27 kDa Shigella flexneri invasion plasmid antigen, IpaJ. Microb Pathog 23:357–369. doi: 10.1006/mpat.1997.0164. [DOI] [PubMed] [Google Scholar]
- 29.Burnaevskiy N, Peng T, Reddick LE, Hang HC, Alto NM. 2015. Myristoylome profiling reveals a concerted mechanism of ARF GTPase deacylation by the bacterial protease IpaJ. Mol Cell 58:110–122. doi: 10.1016/j.molcel.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnaevskiy N, Fox TG, Plymire DA, Ertelt JM, Weigele BA, Selyunin AS, Way SS, Patrie SM, Alto NM. 2013. Proteolytic elimination of N-myristoyl modifications by the Shigella virulence factor IpaJ. Nature 496:106–109. doi: 10.1038/nature12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michiels J, Dirix G, Vanderleyden J, Xi C. 2001. Processing and export of peptide pheromones and bacteriocins in Gram-negative bacteria. Trends Microbiol 9:164–168. doi: 10.1016/s0966-842x(01)01979-5. [DOI] [PubMed] [Google Scholar]
- 32.Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, Karmali V, Rao AS, Madara JL. 2000. Prokaryotic regulation of epithelial responses by inhibition of IκB-α ubiquitination. Science 289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 33.Wu D, Guo X, Lu J, Sun X, Li F, Chen Y, Xiao D. 2013. A rapid and efficient one-step site-directed deletion, insertion, and substitution mutagenesis protocol. Anal Biochem 434:254–258. doi: 10.1016/j.ab.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 34.Wigley P, Hulme SD, Bumstead N, Barrow PA. 2002. In vivo and in vitro studies of genetic resistance to systemic salmonellosis in the chicken encoded by the SAL1 locus. Microbes Infect 4:1111–1120. doi: 10.1016/s1286-4579(02)01635-0. [DOI] [PubMed] [Google Scholar]
- 35.Menck K, Behme D, Pantke M, Reiling N, Binder C, Pukrop T, Klemm F. 2014. Isolation of human monocytes by double gradient centrifugation and their differentiation to macrophages in Teflon-coated cell culture bags. JoVE 18:e51554. doi: 10.3791/51554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaiser P, Stäheli P. 2014. Avian cytokines and chemokines, p 189–204. In Schat KA, Kaspers B, Kaiser P (ed), Avian immunology. Elsevier, Philadelphia, PA. [Google Scholar]
- 37.Weining KC, Sick C, Kaspers B, Staeheli P. 1998. A chicken homolog of mammalian interleukin-1 beta: cDNA cloning and purification of active recombinant protein. Eur J Biochem 258:994–1000. doi: 10.1046/j.1432-1327.1998.2580994.x. [DOI] [PubMed] [Google Scholar]
- 38.Setta A, Barrow PA, Kaiser P, Jones MA. 2012. Immune dynamics following infection of avian macrophages and epithelial cells with typhoidal and non-typhoidal Salmonella enterica serovars; bacterial invasion and persistence, nitric oxide and oxygen production, differential host gene expression, NF-κB signalling and cell cytotoxicity. Vet Immunol Immunopathol 146:212–224. doi: 10.1016/j.vetimm.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Yin J, Cheng Z, Wang X, Xu L, Li Q, Geng S, Jiao X. 2015. Evaluation of the Salmonella enterica serovar Pullorum pathogenicity island 2 mutant as a candidate live attenuated oral vaccine. Clin Vaccine Immunol 22:706–710. doi: 10.1128/CVI.00130-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.