Coxiella burnetii is an obligate intracellular bacterial pathogen that replicates inside the lysosome-derived Coxiella-containing vacuole (CCV). To establish this unique niche, C. burnetii requires the Dot/Icm type IV secretion system (T4SS) to translocate a cohort of effector proteins into the host cell, which modulate multiple cellular processes.
KEYWORDS: Coxiella burnetii, SILAC, TFE3, TFEB, autophagy, host-pathogen interactions, lysosome biogenesis
ABSTRACT
Coxiella burnetii is an obligate intracellular bacterial pathogen that replicates inside the lysosome-derived Coxiella-containing vacuole (CCV). To establish this unique niche, C. burnetii requires the Dot/Icm type IV secretion system (T4SS) to translocate a cohort of effector proteins into the host cell, which modulate multiple cellular processes. To characterize the host-pathogen interactions that occur during C. burnetii infection, stable-isotope labeling by amino acids in cell culture (SILAC)-based proteomics was used to identify changes in the host proteome during infection of a human-derived macrophage cell line. These data revealed that the abundances of many proteins involved in host cell autophagy and lysosome biogenesis were increased in infected cells. Thus, the role of the host transcription factors TFEB and TFE3, which regulate the expression of a network of genes involved in autophagy and lysosomal biogenesis, were examined in the context of C. burnetii infection. During infection with C. burnetii, both TFEB and TFE3 were activated, as demonstrated by the transport of these proteins from the cytoplasm into the nucleus. The nuclear translocation of these transcription factors was shown to be dependent on the T4SS, as a Dot/Icm mutant showed reduced nuclear translocation of TFEB and TFE3. This was supported by the observation that blocking bacterial translation with chloramphenicol resulted in the movement of TFEB and TFE3 back into the cytoplasm. Silencing of the TFEB and TFE3 genes, alone or in combination, significantly reduced the size of the CCV, which indicates that these host transcription factors facilitate the expansion and maintenance of the organelle that supports C. burnetii intracellular replication.
INTRODUCTION
Coxiella burnetii is a Gram-negative obligate intracellular pathogenic bacterium and the causative agent of Q (query) fever, a life-threatening zoonosis (1, 2). The clinical presentation of infection is quite varied, ranging from acute influenza-like symptoms to debilitating chronic illness, such as hepatitis or endocarditis, with significant mortality (2).
Human alveolar macrophages are the primary targets of C. burnetii (3). Following internalization, C. burnetii transits through the endolysosomal pathway. Once the pathogen reaches the acidic lysosomal environment, C. burnetii becomes metabolically active and begins remodeling the vacuolar environment to support its replication. This bacterially modified spacious and unique replicative niche is termed the Coxiella-containing vacuole (CCV) (4). The Dot/Icm type IVB secretion system (T4SS) is essential for intracellular replication and biogenesis of the CCV (5, 6). This secretion system translocates over 130 bacterial effector proteins into the host cytoplasm, where they modulate various cellular processes (7). To date, the functional roles of very few of these effectors have been defined.
The CCV is highly fusogenic and merges with endocytic vesicles, other CCVs, and autophagosomes to create a large organelle that will rapidly occupy much of the host cell’s cytoplasmic space. The CCV is positive for lysosomal markers, including lysosome-associated membrane glycoproteins, CD63, the vacuolar ATPase, and cathepsin D (8–10). More recent findings have demonstrated that the CCV remains in an autolysosomal state of maturation, with autophagosome-CCV fusion contributing to CCV biogenesis (11, 12). In agreement with this, the internal pH of the mature CCV was recently shown to be slightly elevated from the normal lysosomal pH (13).
Degradative membrane transport pathways, both endocytic and autophagic, are highly dynamic systems that play important roles in eukaryotic cell adaptation to nutrient availability and defense against microbes (14). The transcription factors TFEB and TFE3 are bona fide master transcriptional regulators of lysosomal biogenesis, facilitating the expression of many genes implicated in lysosome-related processes, including phagocytosis and autophagy (15, 16). When present in the nucleus, TFEB and TFE3 form homo- or heterodimers that specifically bind to CLEAR (coordinated lysosomal expression and regulation) elements to induce the transcription of genes critical for autophagosome and lysosome biogenesis (17, 18).
The activation of TFEB and/or TFE3 is controlled by a multifaceted lysosome-to-nucleus signaling pathway. Under nutrient-rich conditions, TFEB/TFE3 is phosphorylated by active mammalian/mechanistic target of rapamycin complex 1 (mTORC1) at the lysosomal membrane (19–21). The phosphorylation of TFEB/TFE3 promotes interactions with 14-3-3 proteins, leading to their retention in the cytoplasm (20, 21). Under nutrient-limited conditions, mTORC1 dissociates from the lysosome and is inactivated. Calcium is released from the lysosome into the cytoplasm via the lysosomal calcium channel mucolipin-1 (MCOLN1) (also known as TRPML1), leading to the localized activation of the phosphatase calcineurin (22). Activated calcineurin dephosphorylates TFEB and TFE3, facilitating their nuclear translocation and subsequent influence on the cellular transcriptional profile (22).
Because C. burnetii creates a large lysosome-derived vacuole that subverts the host autophagy pathway, we hypothesized that TFEB and TFE3 activation may be an important element of infection. In support of this idea, an unbiased analysis of the host proteome during infection demonstrated a significant increase in the abundance of many proteins within the CLEAR network and that the activation of the transcription factors TFEB and TFE3 during C. burnetii infection is important for CCV biogenesis.
RESULTS
C. burnetii infection leads to a host cell proteome shift that supports increased lysosome biogenesis.
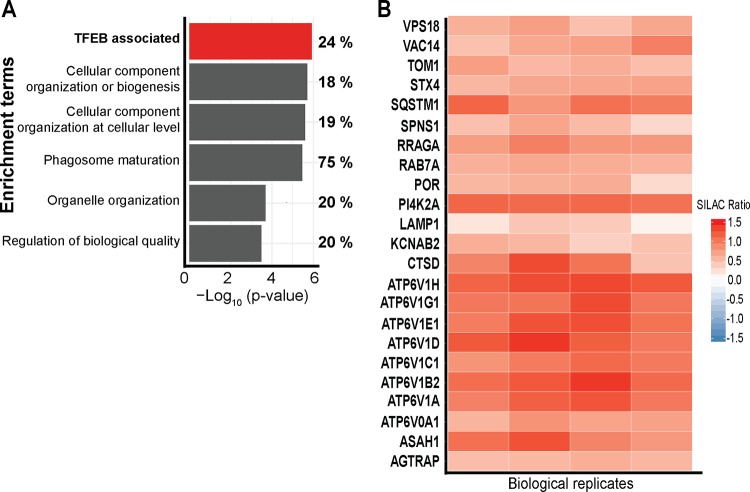
To gain insight into the human host cell response to C. burnetii infection, quantitative proteomics using stable-isotope labeling by amino acids in cell culture (SILAC) was employed to examine changes to the host proteome of human macrophage-like THP-1 cells during infection. Labeled and differentiated THP-1 cells were infected with C. burnetii expressing mCherry for 3 days, allowing CCV establishment and intracellular replication. To ensure that all cells were infected, cells were sorted for mCherry fluorescence prior to mixing with mock-sorted uninfected cells. Proteins were digested and analyzed by mass spectrometry, identifying a total of 5,112 proteins at a 1% false discovery rate (FDR), with 83% (4,266) of these proteins being derived from the host proteome and 846 being derived from C. burnetii (see Table S1 in the supplemental material). Of the 4,266 host proteins, 851 were altered in abundance when comparing infected to uninfected cells, as determined by a one-sample t test with a Benjamini-Hochberg multiple-hypothesis correction of 5%, leading to a significance cutoff of a >1.85 −log10(P value) (47). Analysis of these altered proteins using Fisher’s exact enrichment analysis of protein-associated gene ontology (GO) terms identified significant enrichment of proteins involved in “phagosome maturation” (15 out of 20 proteins identified; 75%), “cellular component organization or biogenesis” (228 out of 1,257 proteins identified; 18%), and “organelle organization” (128 out of 653 proteins identified; 20%) (Fig. 1A and Table S2). Importantly, 24% (60 of the 252) of the proteins regulated by TFEB (23) were significantly altered upon infection with C. burnetii (Fig. 1A). The SILAC ratios revealed significant increases in proteins within the TFEB/TFE3 regulatory network, which were consistent across all four biological replicates (Fig. 1B), including many components of the vacuolar ATPase and the autophagy receptor Sequestosome 1 (SQSTM1), which has previously been shown to be increased in abundance during C. burnetii infection (12, 24). This proteomics-based analysis demonstrated that 3 days after infection with C. burnetii, the abundances of proteins associated with lysosomal biogenesis were increased.
FIG 1.
C. burnetii infection leads to a host cell proteome shift that supports increased lysosome biogenesis. (A) GO-based enrichment of proteins involved in various biological processes analyzed using Fisher’s exact enrichment analysis. Percentages represent the proportions of proteins enriched in each category compared to the observable proteome. (B) Heat map of proteins with TFEB-associated gene expression. Color represents the SILAC ratio for each of four biological replicates, where red represents an increase in C. burnetii-infected THP-1 cells.
Nuclear translocation of TFEB-GFP during C. burnetii infection of HeLa TFEB-GFP cells.
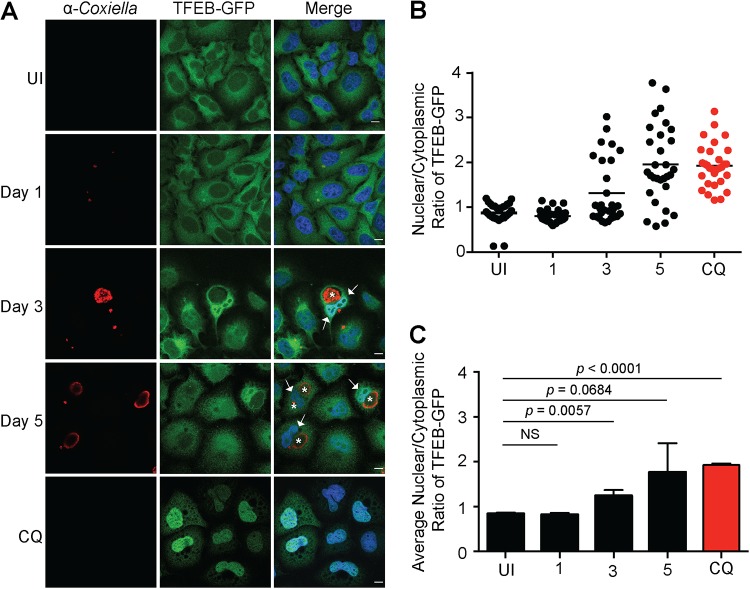
Given the increased abundance of proteins involved in lysosome biogenesis and autophagy observed during C. burnetii infection, the nuclear translocation of TFEB was examined. The subcellular localization of TFEB-green fluorescent protein (GFP) was monitored in uninfected HeLa cells and compared to that in cells infected with C. burnetii for 1, 3, or 5 days. In uninfected cells, the majority of the TFEB-GFP was retained within the cytoplasm (Fig. 2A). Cells infected with C. burnetii for 1 day (24 h) showed a similar cytoplasmic TFEB-GFP distribution. In contrast, at 3 and 5 days postinfection, when C. burnetii was replicating within the CCV, TFEB-GFP nuclear translocation was observed (Fig. 2). The nuclear translocation of TFEB-GFP was quantified by calculating the nuclear-to-cytoplasmic ratio of the TFEB-GFP ratio. This ratio was quantified for at least 30 cells under each experimental condition for each of three independent biological replicates. Regardless of individual cell variability (Fig. 2B), a consistent increase in the nuclear/cytoplasmic TFEB-GFP signal was observed at days 3 and 5 of C. burnetii infection (Fig. 2C). By day 5 postinfection, this increase was equivalent to that in uninfected cells treated with chloroquine (CQ), a lysosomotropic agent that induces TFEB nuclear translocation by causing lysosomal stress (20) (Fig. 2).
FIG 2.
Nuclear translocation of TFEB-GFP during C. burnetii infection of HeLa TFEB-GFP cells. (A) Representative confocal images of HeLa TFEB-GFP cells that were uninfected (UI); infected for 1, 3, or 5 days with C. burnetii NMII at an MOI of 100; or treated for 24 h with 50 μg/ml chloroquine (CQ). Following infection and/or treatment, cells were fixed and immunolabeled with anti-C. burnetii (red), and nuclei were stained with DAPI (blue). White arrows and asterisks denote TFEB nuclear translocation and CCVs, respectively. Bars, 10 μm. (B) Representative nuclear/cytoplasmic ratios of TFEB-GFP from 30 uninfected, infected, or treated cells at the time points indicated. (C) Average nuclear/cytoplasmic ratios of TFEB-GFP from three biological replicates. The means and SD for each time point are presented. An unpaired two-tailed t test was used to determine the statistical significance between samples. NS, not significant.
C. burnetii infection induces nuclear translocation of endogenous TFEB.
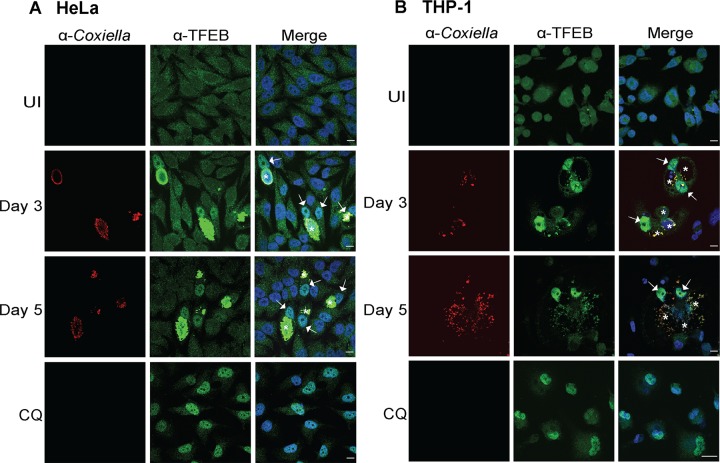
To discount the potential impact of TFEB overexpression on its nuclear translocation during C. burnetii infection, the localization of endogenous TFEB was examined. HeLa cells (Fig. 3A) and differentiated THP-1 cells (Fig. 3B) were fixed and stained with anti-TFEB and anti-C. burnetii antibodies under the following conditions: uninfected, treated with CQ for 24 h, or infected with C. burnetii for 3 or 5 days. Consistent with our previous findings, TFEB was observed to be enriched within the nucleus 3 and 5 days after infection with C. burnetii in both cell lines (Fig. 3). The anti-TFEB antibody demonstrated significant cross-reactivity with C. burnetii, hampering the possibility of quantifying the nuclear/cytoplasmic intensity of TFEB during infection. Visual observation confirms that TFEB is recruited into the host cell nucleus during C. burnetii infection, which may account for the increased production of host proteins that contribute to lysosome biogenesis and autophagy observed in our SILAC studies.
FIG 3.
C. burnetii infection induces nuclear translocation of endogenous TFEB. (A) Representative immunofluorescence images of HeLa CCL2 cells that were uninfected, infected with C. burnetii NMII at an MOI of 100 for 3 or 5 days, or treated using 50 μg/ml chloroquine (CQ) for 24 h. (B) Differentiated THP-1 macrophage cells were uninfected, infected with C. burnetii NMII at an MOI of 100 for 3 or 5 days, or treated using 50 μg/ml CQ for 24 h. Cells after infection and upon treatment were fixed for immunolabeling with antibodies against C. burnetii (red) and TFEB (green), and nuclei were stained with DAPI (blue). Bars, 10 μm. White arrows and asterisks denote endogenous TFEB nuclear translocation and CCVs, respectively.
Endogenous nuclear translocation of TFE3 is induced during C. burnetii infection.
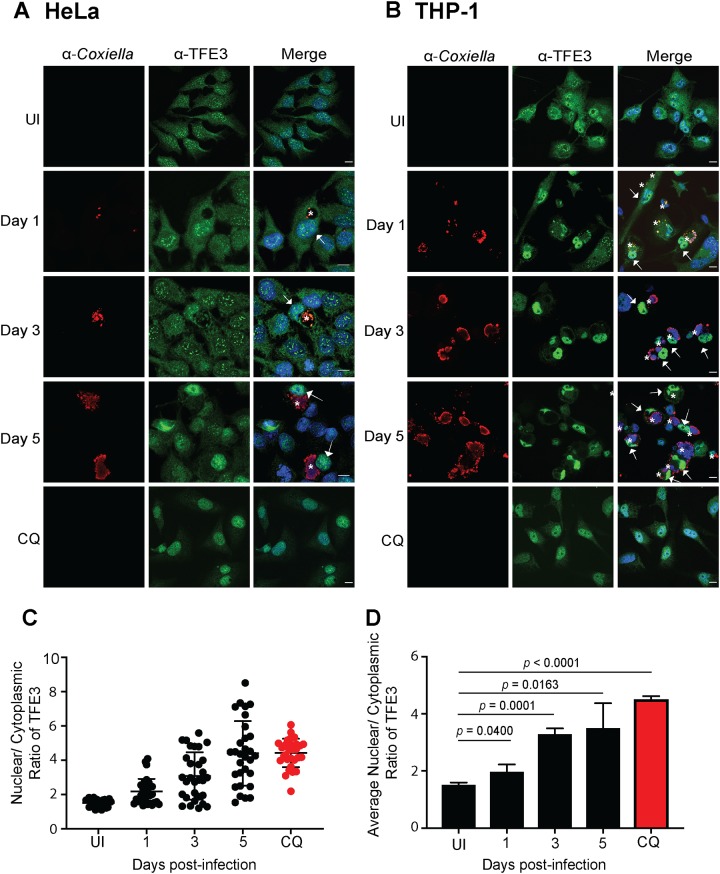
Given that both TFEB and TFE3 upregulate the expression of genes involved in lysosomal biogenesis and autophagy, the subcellular localization of TFE3 during C. burnetii infection was also examined. HeLa cells (Fig. 4A) and differentiated THP-1 cells (Fig. 4B) were infected with C. burnetii for 1, 3, and 5 days before being fixed and stained with anti-TFE3 and anti-C. burnetii. These samples were compared to uninfected cells that remained untreated or were treated with CQ for 24 h. As expected, in uninfected cells, the majority of TFE3 was present in the cytoplasm. TFE3 was observed to translocate into the host cell nucleus throughout the course of infection. The nuclear/cytoplasmic intensity of TFE3 was calculated for at least 30 HeLa cells under each experimental condition (Fig. 4C) for each of three independent experiments (Fig. 4D). A statistically significant increase in nuclear TFE3 was observed from day 1 postinfection, compared to uninfected cells, and this ratio increased at day 3 and day 5 postinfection, approaching the TFE3 nuclear translocation observed with CQ treatment (Fig. 4D). These results demonstrate that TFE3, in addition to TFEB, is actively recruited into the host nucleus during C. burnetii infection. Interestingly, TFE3 nuclear localization was observed at an earlier stage of infection than TFEB.
FIG 4.
C. burnetii infection induces nuclear translocation of endogenous TFE3. (A and B) Representative confocal images of HeLa CCL2 cells (A) or THP-1 cells (B) that were uninfected or infected with C. burnetii NMII at an MOI of 100 for 1, 3, or 5 days. After infection or following chloroquine (CQ) treatment, cells were fixed and immunolabeled with antibodies against C. burnetii (red) and TFE3 (green), and nuclei were stained with DAPI (blue). White arrows and asterisks denote endogenous TFE3 nuclear translocation and CCVs, respectively. Bars, 10 μm. (C) Representative nuclear/cytoplasmic ratios of endogenous TFE3 from 30 uninfected, infected, or treated HeLa CCL2 cells at the time points indicated. (D) Average nuclear/cytoplasmic ratios of endogenous TFE3 in HeLa CCL2 cells from three independent experiments. The means and SD for each time point are presented. An unpaired two-tailed t test was used to determine the statistical significance between samples.
Nuclear localization of TFEB-GFP and TFE3 during C. burnetii infection is Dot/Icm dependent and reversed upon inhibition of bacterial protein translation.
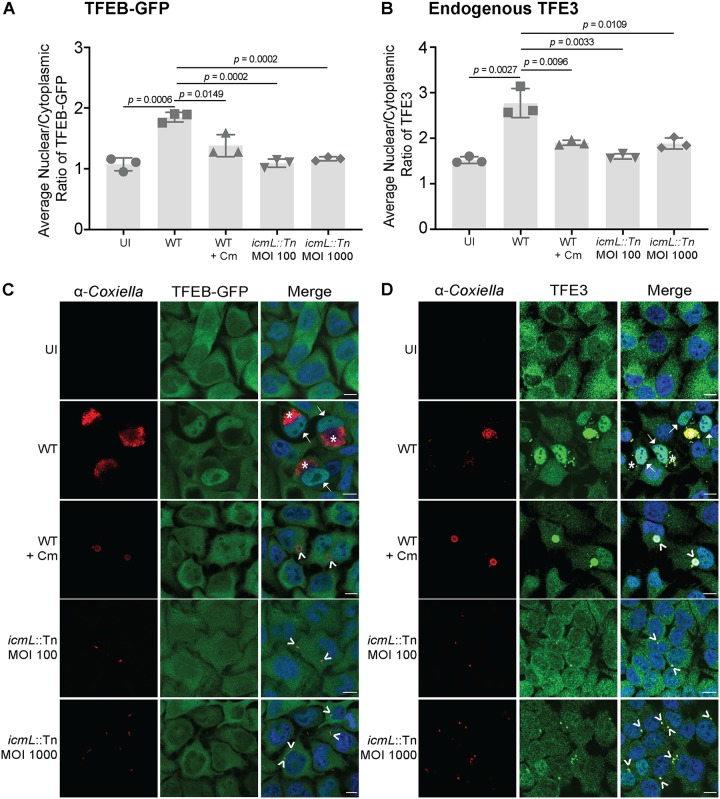
To determine whether the nuclear translocation of TFEB and TFE3 is driven by C. burnetii, the localization of these host proteins was visualized in HeLa cells infected for 3 days with Dot/Icm-deficient C. burnetii. C. burnetii with a disrupted T4SS did not induce the nuclear translocation of TFEB-GFP or TFE3 3 days after infection of HeLa cells (Fig. 5). In order to account for the absence of replication of this C. burnetii mutant, a 10-fold-increased multiplicity of infection (MOI) of C. burnetii icmL::Tn (6) was included, but there was still no observable increase in the nuclear-to-cytoplasmic ratio of TFEB-GFP (Fig. 5A and C) or TFE3 (Fig. 5B and D).
FIG 5.
C. burnetii drives nuclear translocation of TFEB-GFP and TFE3. (A) HeLa cells constitutively expressing TFEB-GFP were infected with either wild-type (WT) C. burnetii or a Dot/Icm-deficient mutant (icmL::Tn) and treated as indicated before being fixed at 3 days postinfection and stained for C. burnetii. “Cm” refers to addition of chloramphenicol 8 h before fixation. The average nuclear/cytoplasmic ratio of TFEB-GFP was calculated from three independent experiments. (B) HeLa cells were infected as described above and stained for endogenous TFE3 and C. burnetii at 3 days postinfection. The average nuclear/cytoplasmic ratio of TFE3 was calculated from three independent experiments. In both scenarios, the signal ratio from a minimum of 30 individual cells was calculated under each condition tested in each independent experiment. The mean ratio for each independent experiment is shown, and the gray bar represents the average mean from the independent experiments. Error bars represent standard deviations. An unpaired two-tailed t test was used to determine statistical significance between WT infection and other samples, and P values of <0.05 are displayed. (C and D) Representative images of TFEB-GFP (C) and TFE3 (D). WT CCVs are labeled in the merged images with asterisks, and nucleus-localized TFEB-GFP or TFE3 is highlighted with an arrow. Collapsing CCVs and nonreplicating C. burnetii cells are indicated with arrowheads. Bars, 10 μm.
In order to examine whether the sustained nuclear localization of TFEB-GFP and TFE3 required C. burnetii activity, HeLa cells infected with C. burnetii for 3 days, to induce the nuclear translocation of TFEB-GFP and TFE3, were treated with 10 μg/ml of chloramphenicol for 8 h (Fig. 5). Chloramphenicol blocks bacterial protein synthesis and has previously been used to demonstrate the importance of C. burnetii protein synthesis during infection (6, 25). Similar to the results seen with Dot/Icm-deficient C. burnetii, blocking of bacterial protein translation with chloramphenicol led to a loss of nuclear TFEB-GFP and TFE3. Quantification of the ratio of nuclear to cytoplasmic signals was confirmed by immunofluorescence microscopy (Fig. 5C and D). Together, these data indicate that the sustained nuclear localization of TFEB-GFP and TFE3 during C. burnetii infection depends on bacterial protein synthesis and the activity of the Dot/Icm secretion system.
TFEB and TFE3 are important for vacuole biogenesis during C. burnetii infection.
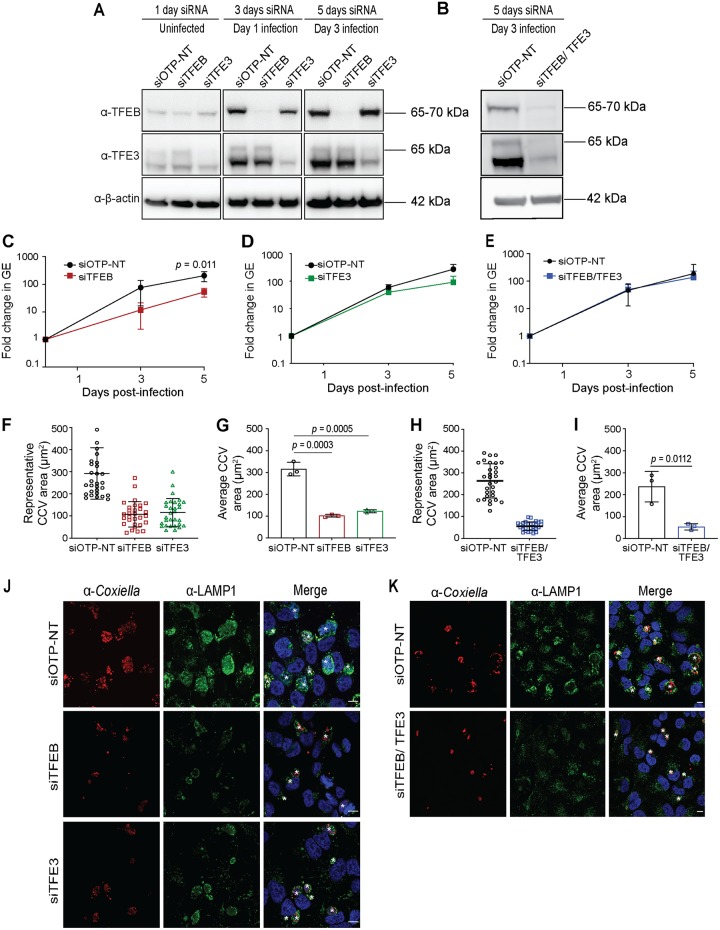
The nuclear translocation of TFEB and TFE3 and the increase in the abundances of many proteins controlled by TFEB and TFE3 during C. burnetii infection call for a direct assessment of these transcription factors toward a productive C. burnetii infection. To examine the effects of these transcription factors on CCV biogenesis and the replication of C. burnetii, small interfering RNA (siRNA) was used to silence the expression of TFEB or TFE3. HeLa cells were transfected with siRNA targeting TFEB (siTFEB) or TFE3 (siTFE3) or the OnTarget Plus nontargeting (OTP-NT) control (siOTP-NT). Reductions in TFEB and TFE3 were confirmed by immunoblotting lysates collected at 1, 3, and 5 days posttransfection (Fig. 6A). Two days after siRNA transfection, HeLa cells were infected with C. burnetii, and bacterial replication was measured by collecting cell lysates at 4 h (day 0), 3 days, and 5 days postinfection (corresponding to 2, 5, and 7 days posttransfection). Across 4 independent experiments, siTFEB treatment led to a small but significant (P = 0.011) reduction in C. burnetii replication (Fig. 6C). HeLa cells treated with siOTP-NT supported a fold change in C. burnetii genome equivalents of 205.00 ± 81.60 over 5 days, compared to 52.81 ± 18.81 for siTFEB treatment (P = 0.011). Gene silencing of TFE3 induced a similar trend toward less C. burnetii replication, with siOTP-NT control-treated cells supporting a fold change in C. burnetii genome equivalents of 272.24 ± 131.84 across 5 days, compared to 91.84 ± 58.12 for TFE3-silenced cells (P = 0.055) (Fig. 6D). At 3 days postinfection, samples were fixed and stained with anti-Coxiella and anti-lysosome-associated membrane protein 1 (LAMP1) to monitor CCV size upon the depletion of TFEB and TFE3 (Fig. 6F, G, and J). The microscopy analysis independently validated the functional impact of TFEB and TFE3 gene silencing, as significantly less LAMP1 was observed in gene-silenced cells (Fig. 6J and K). The sizes of the CCVs formed during gene silencing of TFEB or TFE3 were significantly reduced compared to those of vacuoles that formed with siOTP-NT treatment (Fig. 6F, G, and J). Figure 6F demonstrates the range of CCV sizes in individual host cells in one experiment, and Fig. 6G shows the average CCV sizes and standard deviations from three independent experiments. With siOTP-NT treatment, CCVs were 315.40 ± 30.95 μm2, compared to 101.43 ± 6.04 μm2 in the absence of TFEB (P = 0.0003) and 122.03 ± 7.21 μm2 in the absence of TFE3 (P = 0.0005).
FIG 6.
Functional importance of TFEB and TFE3 during C. burnetii infection. HeLa cells were transfected with nontargeting siRNA (siOTP-NT) or siRNA specifically targeting TFEB (siTFEB), TFE3 (siTFE3), or both siTFEB and siTFE3 (siTFEB/TFE3) and harvested at days 0, 3, and 5 days posttransfection. (A) Immunoblot analysis of whole-cell lysate showing the absence of TFEB and TFE3 with siTFEB and siTFE3 treatment. (B) Immunoblot analysis of whole-cell lysates showing the loss of both TFEB and TFE3 5 days after siTFEB/TFE3 treatment. α-β-Actin was used as a loading control in both panels A and B. (C to E) siRNA-transfected HeLa cells were infected with C. burnetii at an MOI of 100, and intracellular replication was quantified by enumerating genome equivalents (GE) by qPCR at 0, 3, and 5 days postinfection. Nontargeting siRNA (siOTP-NT) treatment was compared to siTFEB (C), siTFE3 (D), and siTFEB/TFE3 (E) treatments. (F) CCV area measurement from 30 individual infected cells from one experiment showing variation in CCV size following siRNA treatments. (G) Quantification of the average CCV area from three biological replicates comparing siOTP-NT treatment with silencing of the expression of either TFEB or TFE3. (H) CCV area measurement from 30 individual infected cells from one experiment showing variation in CCV size following treatment with either siOTP-NT or siTFEB/TFE3. (I) Quantification of the average CCV area from three biological replicates comparing siOTP-NT and siTFEB/TFE3 treatments. Error bars in panels F to I represent the SD. An unpaired two-tailed t test was used to determine statistical significance between siOTP-NT and the other siRNAs tested, and all P values of <0.05 are displayed. (J and K) Immunofluorescence images of HeLa cells depicting CCVs at 3 days postinfection. Cells were fixed and immunolabeled with antibodies against LAMP1 (green) and C. burnetii (red), and DNA was stained with DAPI (blue). White asterisks indicate CCVs. Bars, 10 μm.
To determine whether TFEB and TFE3 act independently or have overlapping roles during C. burnetii infection of HeLa cells, double-knockdown experiments were performed. Here, the expression of both TFEB and TFE3 was silenced (Fig. 6B), and C. burnetii infection was compared to that in siOTP-NT control-treated cells. C. burnetii intracellular replication in the absence of both TFEB and TFE3 was comparable to that with siOTP-NT treatment (Fig. 6E). Even at 5 days postinfection, there was no observable difference in bacterial numbers (n = 5; P = 0.6467). However, in agreement with the silencing of either TFEB or TFE3, the CCV area was significantly impacted by the absence of both TFEB and TFE3 (Fig. 6H, I, and K). Consistent with previous measurements, the average CCV size 3 days after infection of siOTP-NT-treated HeLa cells was 236.41 ± 69.44 μm2, compared to 53.80 ± 14.63 μm2 for cells treated with both siTFEB and siTFE3 (P = 0.0112). These data demonstrate that TFEB and TFE3 contribute to the biogenesis of the normally spacious CCV.
DISCUSSION
Intracellular bacterial pathogens mediate complex interactions with eukaryotic host cells that similarly have evolved many strategies to recognize and respond to infection. Deciphering this arms race can inform our understanding of bacterial pathogenesis and reveal a novel understanding of eukaryotic cell function and dysfunction. Here, a SILAC-based proteomics approach was employed to capture a global snapshot of the host proteomic changes that occur during C. burnetii infection. This revealed a significant increase in many lysosomal proteins within the TFEB and TFE3 CLEAR regulatory network.
In support of previous findings, the SILAC experiments performed here demonstrated an increase in the abundance of the autophagy receptor SQSTM1 during C. burnetii infection (12, 24). A previous study showed the noncanonical recruitment of SQSTM1 to the CCV during infection, where it is proposed to act as a signaling molecule activating the Nrf2-Keap1 cytoprotective pathway (24). Rab7, a marker of late endosomes, showed a marked increase in abundance during infection with C. burnetii. As a key regulator of endocytic maturation and autophagosome-lysosome fusion, Rab7 is required for the biogenesis of the CCV and the activation of the C. burnetii T4SS (26, 27). Additionally, the abundances of eight subunits of the vacuolar ATPase, the proton pump responsible for the acidification of the endocytic network, were increased during C. burnetii infection.
Examination of TFEB and TFE3 localization throughout infection demonstrated significant nuclear translocation of these host transcription factors. Interestingly, significant nuclear localization of TFE3 was detected at 1 day postinfection, and TFEB nuclear translocation was notable from 3 days postinfection, corresponding to CCV expansion and C. burnetii replication. This nuclear translocation of TFEB and TFE3 likely accounts for the increased abundance of CLEAR network proteins observed during C. burnetii infection.
In HeLa cells, the nuclear translocation of TFEB and TFE3 appears to be controlled by C. burnetii, as blocking bacterial translation with chloramphenicol leads to TFEB/TFE3 movement back into the cytoplasm. In support of this hypothesis, a Dot/Icm-deficient C. burnetii mutant was unable to induce the nuclear recruitment of TFEB and TFE3. Intriguingly, these data suggest that C. burnetii may encode Dot/Icm effectors that promote the activation of this signaling pathway to facilitate increased lysosomal biogenesis and autophagy.
To determine the impact of TFEB or TFE3 on C. burnetii infection, we used siRNA gene silencing in HeLa cells and measured the consequences for C. burnetii intracellular success. The loss of TFEB or TFE3 had a significant impact on CCV expansion but little impact on the intracellular replication of C. burnetii. The absence of both TFEB and TFE3 recapitulated a nonredundant role for these proteins in CCV expansion. Despite these host transcription factors not influencing bacterial replication, C. burnetii replication within a noncanonical, tightly bound CCV may have important downstream consequences for C. burnetii fitness that can be observed only in multicellular models of infection.
The findings reported here should be considered in the context of similar research published during the preparation of this article (28). In agreement with our findings, that study also demonstrated TFE3 nuclear translocation during C. burnetii infection of HeLa cells and reported that this may be linked to the observation that mTORC1 activity is inhibited during infection in a Dot/Icm-dependent manner (28). Knockout of both TFEB and TFE3 in RAW 264.6 macrophage-like cells also led to the production of smaller CCVs but, surprisingly, more C. burnetii replication across 3 days. This finding supports our observation that TFEB and TFE3 are important for the formation of the hallmark spacious CCV but are not necessary for bacterial replication. Interestingly, Larson et al. demonstrated that infection with Dot/Icm-deficient C. burnetii can also induce the nuclear translocation of TFE3 in HeLa cells (28). This TFE3 activation appears to be independent of mTORC1 inhibition and may represent a host response to bacterial infection independent of the pathogen-driven activation of this pathway that we observed here. Given the opposing findings presented here and by Larson et al., it will be interesting to explore whether any Dot/Icm effector proteins can, either directly or indirectly, influence the nuclear localization of TFEB and TFE3 and the activity of mTORC1.
Research into the relationship between other bacterial pathogens and TFEB has demonstrated that the activation of this pathway can have divergent consequences for different infections. Acinetobacter baumannii, a Gram-negative pathogen predominantly associated with hospital-acquired infections, induces the activation of TFEB in alveolar epithelial cells, and this promotes the internalization and intracellular success of this pathogen (29). In contrast, TFEB activation during both Staphylococcus aureus and Salmonella enterica serovar Typhimurium infections is considered an important host response to infection (30–32). Interestingly, Mycobacterium tuberculosis, the causative agent of tuberculosis, has been shown to inhibit TFEB activation by inducing the increased expression of the host microRNA miR-33 (33).
To date, the factors that influence the host cell transcriptome during C. burnetii infection are not clearly understood. The proteomics data presented here reflect the complex interplay between host and bacterial signaling influences, both transcriptional and posttranscriptional. Not all TFEB/TFE3-regulated proteins demonstrate increased abundances during C. burnetii infection. Despite the observed nuclear translocation of TFEB and TFE3, there are other host and bacterial factors that contribute to the host transcriptome during infection. For example, the Nrf2-Keap1 antioxidant pathway, regulated by SQSTM1, was recently shown to be exploited by C. burnetii with the transcription factor Nrf2 present in the nucleus of infected cells (24). C. burnetii can influence host transcription through Dot/Icm effector proteins. C. burnetii has many effectors with the capacity to enter the host nucleus, including CaeA and AnkG, both of which possess antiapoptotic activity, and CBU1314, which has been shown to interact with chromatin and induce transcriptional changes when ectopically expressed in eukaryotic cells (34–36). Both host and bacterial factors may be acting in potentially cooperative and antagonistic manners to impact the host proteome profile defined here.
This research demonstrates that C. burnetii actively manipulates host cell transcription to promote autophagy and lysosomal biogenesis. The mechanism by which the Q fever pathogen controls this pathway is important for developing our understanding of the virulence strategies possessed by this emerging pathogen. Cumulative evidence suggests that the regulation of lysosomal biogenesis and autophagy by TFEB can influence important human diseases such as lysosomal storage disorders and neurodegenerative diseases (37). Thus, understanding how C. burnetii controls the lysosome-nucleus signaling axis may provide novel insights into understanding mammalian cell function and the mechanisms contributing to several important human disease states.
MATERIALS AND METHODS
Mammalian cell culture and C. burnetii.
HeLa CCL2 and HeLa TFEB-GFP stable cells (a kind gift from Shawn Ferguson, Yale University) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with GlutaMAX (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37°C with 5% CO2. THP-1 cells were maintained in RPMI 1640 supplemented with 10% FBS at 37°C with 5% CO2. C. burnetii Nine Mile phase II (NMII) strain RSA439, clone 4, and C. burnetii icmL::Tn (6) were axenically cultured in liquid ACCM-2 (38) with chloramphenicol added at 3 μg/ml, when required, at 37°C with 5% CO2 and 2.5% O2 for 7 days.
C. burnetii infections.
HeLa cell lines were plated at a density of 5 × 104 cells/well into 24-well tissue culture plates 1 day prior to infection, with or without 10-mm glass coverslips. THP-1 cells were seeded into 24-well tissue culture plates at a density of 5 × 105 cells/well and treated with 10 nM phorbol 12-myristate 13-acetate (PMA) for 72 h to induce differentiation into adherent, macrophage-like cells. Axenically grown C. burnetii cells were quantified using the Quant-iT PicoGreen double-stranded DNA (dsDNA) assay kit (Thermo Fischer Scientific) according to the manufacturer’s instructions. Sample values were compared to DNA standards of known concentrations, and quantification of C. burnetii genome equivalents was performed using the mass of the Coxiella genome (2.2 × 10−9 μg) to enumerate the multiplicity of infection (MOI). In all experiments, HeLa cells were infected with C. burnetii at an MOI of 100, and THP-1 cells were infected at an MOI of 25. Cells were incubated for 4 h at 37°C with 5% CO2, washed once in phosphate-buffered saline (PBS), and further incubated in fresh DMEM with 5% FBS (HeLa cells) or RPMI 1640 with 10% FBS (THP-1 cells). At defined times postinfection, samples were either fixed with 4% paraformaldehyde (PFA) for microscopy analysis or lysed with H2O and collected for C. burnetii genomic DNA quantification.
Western blot analysis.
Samples for Western blotting were collected at the indicated time points throughout infection by resuspension in 2× SDS-PAGE sample buffer. Proteins were resolved using Bolt 4 to 12% Bis-Tris Plus gels (Thermo Fischer Scientific, Invitrogen) and transferred to nitrocellulose membranes using the iBlot 2 transfer system (Life Technologies, Thermo Fischer Scientific). Membranes were blocked using 5% skim milk in Tris-buffered saline containing 0.1% Tween 20 (TBST). The following antibodies were diluted in 5% bovine serum albumin (BSA) or skim milk in TBST and incubated with a nitrocellulose membrane at 4°C overnight: polyclonal anti-TFEB (1:1,000; Cell Signaling Technology), polyclonal anti-TFE3 (1:1,000; Sigma), monoclonal anti-β-actin AC-74 (1:5,000; Sigma), anti-mouse horseradish peroxidase (HRP) (1:3,000; Perkin Elmer), and anti-rabbit HRP (1:3,000; Perkin Elmer). Blots were developed using Clarity Western ECL reagent (Bio-Rad) and exposed on an MF-ChemiBIS system, version 3.2 (DNR Bio-Imaging Systems, Ltd.). Immunoblots were analyzed using GelCapture software, version 7.0.18 (DNR Bio-Imaging Systems, Ltd.).
SILAC labeling and infection of the THP-1 cell line.
SILAC labeling of undifferentiated THP-1 cells was performed as previously described (39, 40). Briefly, RPMI 1640 (Cambridge Isotope Laboratories, Inc.) lacking arginine and lysine was supplemented with 10% dialyzed FBS (Sigma). Arginine and lysine were added in either a light (l-arginine and l-lysine [Sigma] [Arg0; Lys0]) or heavy (l-[13C6, 15N4]arginine and l-[13C6, 15N2]lysine [Cambridge Isotope Laboratories] [Arg6; Lys4]) combination to final concentrations of 1.15 mM for arginine and 0.274 mM for lysine. THP-1 cells were split and passaged independently 5 times to allow for the incorporation of labeled amino acids into the cellular proteome. THP-1 cells were PMA differentiated 72 h prior to infection with C. burnetii expressing mCherry (5) at an MOI of 100. Cells were washed with PBS at 24 h postinfection to remove any extracellular bacteria and maintain infections within a 24-h window. Infected cells were incubated for a further 2 days prior to isolation and sorting for mCherry fluorescence using fluorescence-activated cell sorting (FACS). An equal number of uninfected cells were similarly counted via FACS and combined 1:1 with the infected cells. Cells were snap-frozen in liquid nitrogen prior to preparation for proteomic analysis.
Proteomics of whole-cell SILAC samples.
Whole-cell SILAC samples were lysed in guanidine hydrochloride (GdHCl) lysis buffer [6 M GdHCl, 100 mM Tris (pH 8.0), 10 mM tris(2-carboxyethyl)phosphine, 40 mM 2-chloroacetamide (CAA)] at 95°C at 1,400 rpm (41). Lysates were cooled, and the protein concentration was determined using a bicinchoninic acid (BCA) kit (Pierce). A total of 50 to 100 μg of protein was acetone precipitated (4 volumes of acetone, 1 volume of water, and 1 volume of the sample) overnight at −20°C. Precipitated material was then pelleted at 16,000 × g for 10 min at 4°C and dried, to remove the remaining acetone, at 65°C. Pellets were resuspended in buffer containing 6 M urea, 2 M thiourea, and 10 mM dithiothreitol (DTT) and incubated at room temperature (RT) in the dark for 1 h to reduce disulfide bonds. Samples were then alkylated by the addition of 40 mM CAA and incubated for a further hour. Alkylation was halted by the addition of 20 mM DTT, and the samples were incubated at RT for 15 min. Samples were digested with endoproteinase Lys-C (1/200 [wt/wt]; Wako Lab Chemicals) for 4 h at RT before dilution with 20 mM ammonium bicarbonate and digestion with trypsin (1/50 [wt/wt]; Sigma) overnight at 25°C. Digested peptides were then acidified by the addition of 2% formic acid and desalted using homemade C18 (Empore C18; Sigma) stage tips (42) before being dried down for analysis by liquid chromatography-mass spectrometry (LC-MS). Prior to loading, samples were reconstituted in MS running buffer (2% acetonitrile, 0.1% trifluoroacetic acid) to a concentration of 0.5 μg/μl, of which 2 μg was loaded for analysis.
Samples were analyzed by LC-MS on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) coupled to a Dionex Ultimate 3000 ultraperformance liquid chromatography system (Thermo Fisher Scientific) using a two-column chromatography setup composed of a PepMap100 C18 20-mm by 75-μm trap column and a PepMap C18 500-mm by 75-μm analytical column (Thermo Fisher Scientific). Samples were concentrated at 5 μl/min onto the trap column for 5 min with buffer A (2% acetonitrile and 0.1% formic acid) prior to the initiation of analytical separation, which was performed at 300 nl/min. Liquid chromatography separation was undertaken by altering the composition of buffer B from 3% to 30% over 180 min and then from 30% to 40% over 7 min and 40% to 90% over 5 min, with holding at 90% for 5 min and then a drop to 3% buffer B (80% ACN and 0.1% formic acid) over 3 min, with the column then equilibrated by holding at 3% buffer B for 5 min. Data from experiments were acquired using data-dependent acquisition with a 120,000-resolution MS1 scan of a mass-to-charge (m/z) range of 400 to 1,600 (automatic gain control [AGC] set to 5 × 105 or a maximum injection time of 50 ms) acquired every 3 s, followed by MS2 scans. MS2 scans were acquired using high-energy collision dissociation fragmentation with a normalized collision energy of 35, a resolution of 15,000, and an AGC of 2 × 105 or a maximum injection time of 40 ms.
All raw files were analyzed using the MaxQuant platform (version 1.6.3.4), searching against the UniProt human and C. burnetii databases (UniProt accession numbers UP000005640 and UP000215556, respectively; downloaded July 2017) containing reviewed, canonical, and isoform variants and a database containing common contaminants generated by the Andromeda search engine (43). A multiplicity of 2 was used, denoting either the canonical amino acids (Arg0 and Lys0) or the SILAC amino acids (Arg6 and Lys4). Searches were performed with cysteine carbamidomethylation as a fixed modification and methionine oxidation and N-terminal acetylation as variable modifications. A false discovery rate (FDR) of 1% was applied as defined within the default parameters. “Re-quantify” and “match between runs” functions were enabled, with a match time window of 2 min. Unique and razor peptides were used for identification, with a minimum ratio count of 2. The “protein groups” output file was then imported into the Perseus platform (version 1.6.2.3) for further analysis (44). Identifications labeled by MaxQuant as “only identified by site,” “potential contaminant,” and “reverse hit” were removed, and the SILAC ratios were log2 transformed. Identifications were matched to human and C. burnetii annotation files (UniProt) using gene name identifiers. Mean log2-transformed heavy/light ratios and P values for each group were calculated using a single-sample two-sided t test. Proteins determined to be significantly altered between groups were assessed using Fisher’s exact enrichment analysis to identify overrepresented GO terms. Both the determination of altered proteins and Fisher’s exact enrichment analysis were subjected to multiple-hypothesis correction using Benjamini-Hochberg correction at an FDR of 5%. Data were exported into the R framework for visualization.
C. burnetii intracellular replication.
To enumerate C. burnetii genome equivalents by quantitative PCR (qPCR), infected cells were lysed in sterile distilled H2O and collected. Lysed samples were pelleted to obtain the C. burnetii and mammalian cell debris before being resuspended in 100 μl of sterile H2O. C. burnetii genomic DNA was extracted using the Quick-DNA miniprep kit (Zymo Research), and bacterial genome equivalents were enumerated by qPCR using ompA-specific primers and comparison to reference samples (45).
Confocal microscopy.
Immunofluorescence labeling was performed in TFEB-GFP stable cells to monitor TFEB nuclear localization. At the desired times postinfection, the cells were washed once with PBS and fixed with 4% (wt/vol) paraformaldehyde for 20 min at room temperature. The fixed cells were blocked and permeabilized using 0.05% saponin and 2% BSA in PBS, followed by staining using the following primary and secondary antibodies: rabbit polyclonal anti-C. burnetii (1:10,000; in-house [6]) and secondary antibody conjugated to Alexa Fluor 568 (1:2,000; Life Technologies). To monitor the endogenous nuclear translocation of TFEB and TFE3, cells were washed once with PBS, fixed with 4% PFA, and permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. This was followed by primary antibody staining in 10% FBS and 0.1% saponin for 1 h at RT with the following antibodies: rabbit polyclonal anti-TFEB (1:100; Cell Signaling) or rabbit polyclonal anti-TFE3 (1:200; Sigma), mouse monoclonal anti-LAMP1 (1:200; Developmental Studies Hybridoma Bank), and mouse anti-C. burnetii (1:1,000; in-house). Secondary antibodies conjugated to rabbit Alexa Fluor 488 (1:2,000; Invitrogen) and mouse Alexa Fluor 568 (1:2,000; Invitrogen) were used in the same blocking buffer. Antibody incubations were followed by PBS washes and incubation with 4′,6-diamidino-2-phenylindole (DAPI) (diluted 1:10,000 in PBS) to stain DNA. Coverslips were mounted using ProLong gold antifade mountant (Invitrogen). Images were acquired with a Zeiss LSM700 confocal laser scanning microscope (Zeiss, Germany) and processed using Fiji software (46).
siRNA gene silencing of TFEB and TFE3.
For siRNA-mediated gene silencing of TFEB and TFE3 in HeLa cells, cells were reverse transfected with small interfering RNA (siRNA) siGenome SMARTpools (Dharmacon, GE Life Sciences) against human TFEB or TFE3 or OnTarget Plus nontargeting (OTP-NT) siRNA using DharmaFECT-1 (Dharmacon, GE Life Sciences). Cells were seeded at a density of 2.5 × 105 cells in 24-well plates with a final concentration of 40 nM siRNA. Cells treated with both siTFEB and siTFE3 were transfected with 40 nM each siRNA. At 24 h posttransfection, the medium was changed, and cells were incubated for a further 2 days. Transfected cells were then infected with C. burnetii at an MOI of 100 for 4 h and washed once with PBS, and the medium was replaced with DMEM containing 5% FBS. This time point was designated day 0. Samples for immunoblotting were collected by resuspending them in 2× SDS-PAGE sample buffer at 1, 3, and 5 days posttransfection. Cells were processed for C. burnetii genome quantification at days 0, 3, and 5 postinfection as described above. At 3 days postinfection, cells were fixed and processed for immunofluorescence as described above.
Quantification of the nuclear-to-cytoplasmic ratio.
The nuclear-to-cytoplasmic ratios of TFEB and TFE3 were measured using Fiji (46). The image analysis workflow was semiautomated with a custom ImageJ macro where the microscope images were imported and the fluorescence intensity on the segmented nucleus and cytoplasm was measured automatically. The cytoplasm, nucleus, and infected area were segmented from TFEB/TFE3, DAPI, and infection channels, respectively. The segmentation threshold was determined from the background of each channel or by using the negative-control image and applied identically to every image. In noninfected cells, the applied threshold yielded no positive segmentation. The applied threshold was further checked by visual inspection and minimally adjusted, if necessary, prior to use for segmentation. Following segmentation, cells that were touching the boundary of the image were excluded. All complete cells were sorted based on infection status. For each cell, the mean intensity of cytoplasm and nucleus areas were measured, and the nucleus-to-cytoplasm ratio was calculated as nuclear mean intensity/cytoplasmic mean intensity.
Statistical analysis and quantification.
Data graphs were generated using GraphPad Prism, version 7.03. All graphs display means ± standard deviations (SD). Unpaired two-tailed Student’s t test was used for comparisons between two groups.
Supplementary Material
ACKNOWLEDGMENTS
This research was financially supported by NHMRC grant 1120344 awarded to H.J.N.; ARC grant DP180101298 awarded to H.J.N., D.S., and N.E.S.; and NIH grant R01AI114760 awarded to C.R.R. A.H. is supported by the H2020-MSCA-Global Fellowship, grant number 657766. B.P. is supported by a Melbourne International Research Scholarship and a Melbourne International Fee Remission Scholarship, The University of Melbourne.
Confocal imaging was performed at the Biological Optical Microscopy Platform, The University of Melbourne (www.microscopy.unimelb.edu.au). We thank Shawn M. Ferguson, Yale University, for the TFEB-GFP stable HeLa cell line.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Angelakis E, Raoult D. 2010. Q fever. Vet Microbiol 140:297–309. doi: 10.1016/j.vetmic.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 2.Schneeberger PM, Wintenberger C, van der Hoek W, Stahl JP. 2014. Q fever in the Netherlands—2007-2010: what we learned from the largest outbreak ever. Med Mal Infect 44:339–353. doi: 10.1016/j.medmal.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Graham JG, MacDonald LJ, Hussain SK, Sharma UM, Kurten RC, Voth DE. 2013. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell Microbiol 15:1012–1025. doi: 10.1111/cmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newton HJ, Roy CR. 2011. The Coxiella burnetii Dot/Icm system creates a comfortable home through lysosomal renovation. mBio 2:e00226-11. doi: 10.1128/mBio.00226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2:e00175-11. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey KL, Newton HJ, Luhrmann A, Roy CR. 2011. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luhrmann A, Newton HJ, Bonazzi M. 2017. Beginning to understand the role of the type IV secretion system effector proteins in Coxiella burnetii pathogenesis. Curr Top Microbiol Immunol 413:243–268. doi: 10.1007/978-3-319-75241-9_10. [DOI] [PubMed] [Google Scholar]
- 8.Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. 1996. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun 64:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe D, Mallavia LP. 2000. Coxiella burnetii exhibits morphological change and delays phagolysosomal fusion after internalization by J774A.1 cells. Infect Immun 68:3815–3821. doi: 10.1128/iai.68.7.3815-3821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howe D, Shannon JG, Winfree S, Dorward DW, Heinzen RA. 2010. Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect Immun 78:3465–3474. doi: 10.1128/IAI.00406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohler LJ, Reed SR, Sarraf SA, Arteaga DD, Newton HJ, Roy CR. 2016. Effector protein Cig2 decreases host tolerance of infection by directing constitutive fusion of autophagosomes with the Coxiella-containing vacuole. mBio 7:e01127-16. doi: 10.1128/mBio.01127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latomanski EA, Newton HJ. 2018. Interaction between autophagic vesicles and the Coxiella-containing vacuole requires CLTC (clathrin heavy chain). Autophagy 14:1710–1725. doi: 10.1080/15548627.2018.1483806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulye M, Samanta D, Winfree S, Heinzen RA, Gilk SD. 2017. Elevated cholesterol in the Coxiella burnetii intracellular niche is bacteriolytic. mBio 8:e02313-16. doi: 10.1128/mBio.02313-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onyenwoke RU, Brenman JE. 2015. Lysosomal storage diseases—regulating neurodegeneration. J Exp Neurosci 9:81–91. doi: 10.4137/JEN.S25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Settembre C, Fraldi A, Medina DL, Ballabio A. 2013. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raben N, Puertollano R. 2016. TFEB and TFE3: linking lysosomes to cellular adaptation to stress. Annu Rev Cell Dev Biol 32:255–278. doi: 10.1146/annurev-cellbio-111315-125407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. 2009. A gene network regulating lysosomal biogenesis and function. Science 325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 18.Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, Puertollano R. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martina JA, Chen Y, Gucek M, Puertollano R. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, Settembre C, Wang WY, Gao Q, Xu HX, Sandri M, Rizzuto R, De Matteis MA, Ballabio A. 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol 17:288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmieri M, Impey S, Kang H, di Ronza A, Pelz C, Sardiello M, Ballabio A. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet 20:3852–3866. doi: 10.1093/hmg/ddr306. [DOI] [PubMed] [Google Scholar]
- 24.Winchell CG, Dragan AL, Brann KR, Onyilagha FI, Kurten RC, Voth DE. 2018. Coxiella burnetii subverts p62/Sequestosome 1 and activates Nrf2 signaling in human macrophages. Infect Immun 86:e00608-17. doi: 10.1128/IAI.00608-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howe D, Melnicakova J, Barak I, Heinzen RA. 2003. Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell Microbiol 5:469–480. doi: 10.1046/j.1462-5822.2003.00293.x. [DOI] [PubMed] [Google Scholar]
- 26.Romano PS, Gutierrez MG, Beron W, Rabinovitch M, Colombo MI. 2007. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol 9:891–909. doi: 10.1111/j.1462-5822.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 27.Newton HJ, McDonough JA, Roy CR. 2013. Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One 8:e54566. doi: 10.1371/journal.pone.0054566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larson CL, Sandoz KM, Cockrell DC, Heinzen RA. 2019. Noncanonical inhibition of mTORC1 by Coxiella burnetii promotes replication within a phagolysosome-like vacuole. mBio 10:e02816-18. doi: 10.1128/mBio.02816-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parra-Millan R, Guerrero-Gomez D, Ayerbe-Algaba R, Pachon-Ibanez ME, Miranda-Vizuete A, Pachon J, Smani Y. 2018. Intracellular trafficking and persistence of Acinetobacter baumannii requires transcription factor EB. mSphere 3:e00106-18. doi: 10.1128/mSphere.00106-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AF, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE. 2014. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurer K, Reyes-Robles T, Alonzo F III, Durbin J, Torres VJ, Cadwell K. 2015. Autophagy mediates tolerance to Staphylococcus aureus alpha-toxin. Cell Host Microbe 17:429–440. doi: 10.1016/j.chom.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Najibi M, Labed SA, Visvikis O, Irazoqui JE. 2016. An evolutionarily conserved PLC-PKD-TFEB pathway for host defense. Cell Rep 15:1728–1742. doi: 10.1016/j.celrep.2016.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouimet M, Koster S, Sakowski E, Ramkhelawon B, van Solingen C, Oldebeken S, Karunakaran D, Portal-Celhay C, Sheedy FJ, Ray TD, Cecchini K, Zamore PD, Rayner KJ, Marcel YL, Philips JA, Moore KJ. 2016. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat Immunol 17:677–686. doi: 10.1038/ni.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bisle S, Klingenbeck L, Borges V, Sobotta K, Schulze-Luehrmann J, Menge C, Heydel C, Gomes JP, Lührmann A. 2016. The inhibition of the apoptosis pathway by the Coxiella burnetii effector protein CaeA requires the EK repetition motif, but is independent of survivin. Virulence 7:400–412. doi: 10.1080/21505594.2016.1139280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schafer W, Eckart RA, Schmid B, Cagkoylu H, Hof K, Muller YA, Amin B, Luhrmann A. 2017. Nuclear trafficking of the anti-apoptotic Coxiella burnetii effector protein AnkG requires binding to p32 and importin-alpha1. Cell Microbiol 19:e12634. doi: 10.1111/cmi.12634. [DOI] [PubMed] [Google Scholar]
- 36.Weber MM, Faris R, McLachlan J, Tellez A, Wright WU, Galvan G, Luo ZQ, Samuel JE. 2016. Modulation of the host transcriptome by Coxiella burnetii nuclear effector Cbu1314. Microbes Infect 18:336–345. doi: 10.1016/j.micinf.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Bajaj L, Lotfi P, Pal R, Ronza AD, Sharma J, Sardiello M. 2019. Lysosome biogenesis in health and disease. J Neurochem 148:573–589. doi: 10.1111/jnc.14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, Samuel JE, Heinzen RA. 2011. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol 77:3720–3725. doi: 10.1128/AEM.02826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoehr G, Schaab C, Graumann J, Mann M. 2013. A SILAC-based approach identifies substrates of caspase-dependent cleavage upon TRAIL-induced apoptosis. Mol Cell Proteomics 12:1436–1450. doi: 10.1074/mcp.M112.024679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott NE, Rogers LD, Prudova A, Brown NF, Fortelny N, Overall CM, Foster LJ. 2017. Interactome disassembly during apoptosis occurs independent of caspase cleavage. Mol Syst Biol 13:906. doi: 10.15252/msb.20167067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humphrey SJ, Azimifar SB, Mann M. 2015. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat Biotechnol 33:990–995. doi: 10.1038/nbt.3327. [DOI] [PubMed] [Google Scholar]
- 42.Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 43.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 44.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 45.Jaton K, Peter O, Raoult D, Tissot JD, Greub G. 2013. Development of a high throughput PCR to detect Coxiella burnetii and its application in a diagnostic laboratory over a 7-year period. New Microbes New Infect 1:6–12. doi: 10.1002/2052-2975.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JR Statist Soc B 57:289–300. https://www.jstor.org/stable/2346101. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.