Staphylococcus aureus is a significant human pathogen due to its capacity to cause a multitude of diseases. As such, S. aureus efficiently pillages vital nutrients from the host; however, the molecular mechanisms that support sulfur acquisition during infection have not been established. One of the most abundant extracellular sulfur-containing metabolites within the host is cysteine, which acts as the major redox buffer in the blood by transitioning between reduced and oxidized (cystine) forms.
KEYWORDS: Staphylococcus aureus, sulfur, cystine, cysteine, homocystine, TcyP, TcyABC, glutathione, N-acetyl cysteine
ABSTRACT
Staphylococcus aureus is a significant human pathogen due to its capacity to cause a multitude of diseases. As such, S. aureus efficiently pillages vital nutrients from the host; however, the molecular mechanisms that support sulfur acquisition during infection have not been established. One of the most abundant extracellular sulfur-containing metabolites within the host is cysteine, which acts as the major redox buffer in the blood by transitioning between reduced and oxidized (cystine) forms. We therefore hypothesized that S. aureus acquires host-derived cysteine and cystine as sources of nutrient sulfur during systemic infection. To test this hypothesis, we used the toxic cystine analogue selenocystine to initially characterize S. aureus homologues of the Bacillus subtilis cystine transporters TcyABC and TcyP. We found that genetic inactivation of both TcyA and TcyP induced selenocystine resistance. The double mutant also failed to proliferate in medium supplemented with cystine, cysteine, or N-acetyl cysteine as the sole sulfur source. However, only TcyABC was necessary for proliferation in defined medium containing homocystine as the sulfur source. Using a murine model of systemic infection, we observed tcyP-dependent competitive defects in the liver and heart, indicating that this sulfur acquisition strategy supports proliferation of S. aureus in these organs. Phylogenetic analyses identified TcyP homologues in many pathogenic species, implying that this sulfur procurement strategy is conserved. In total, this study is the first to experimentally validate sulfur acquisition systems in S. aureus and establish their importance during pathogenesis.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is endemic in hospital and community settings in the United States and is the leading cause of morbidity and mortality resulting from infections with antibiotic-resistant bacteria (1–3). This stems from the fact that S. aureus infection can lead to wide-ranging disease manifestations, including skin and soft tissue infections, infective endocarditis, sepsis, osteomyelitis, and pneumonia (4, 5). In addition to invasive disease, S. aureus innocuously colonizes the skin and nasal passages of 30% of the population (6–9). Together, these facts highlight the dynamic tissue tropism of this organism.
During infection, pathogens must procure nutrients from host environments. The mechanisms S. aureus employs to acquire carbon sources and trace metals during infection are becoming increasingly clear (10–17). However, a complete understanding of the physiology that contributes to the persistence and invasiveness of S. aureus is imperative to control infection. Therefore, impairing nutrient acquisition offers potential for therapeutic intervention. S. aureus employs a sophisticated metabolism that allows it to both persist during nasal carriage and proliferate to cause systemic disease (18–24). One clear deficiency in our knowledge of S. aureus nutrient acquisition is the strategy by which the pathogen scavenges sulfur during infection.
Sulfur is an essential building block for life because it transitions between numerous oxidation states, allowing for broad functionality (25). Once sulfur is assimilated to a functional oxidation state, most cells flux sulfur to cysteine (Cys) and methionine to generate other sulfur-containing compounds (25–27). The sulfur demand for a cell is sizable due to the many sulfur-containing compounds required to synthesize proteins and power central metabolism. Consequently, Escherichia coli imports 2.2 mM min−1 of sulfur to support a growth rate of 0.0173 min−1 (28). To fulfill the sulfur requirement some bacteria utilize two broad categories of sulfur-containing substrates: inorganic sulfur sources that include sulfate and thiosulfate or organosulfur sources such as methionine, Cys, and others (29). S. aureus lacks enzymes necessary to assimilate sulfate and consequently it cannot convert sulfate to sulfite (30–32). Accordingly, S. aureus relies on either organosulfur sources or select inorganic sulfur sources, such as thiosulfate, to satisfy its sulfur requirement. Previous studies have shown S. aureus acquires cystine (CSSC), Cys, glutathione (GSH), sulfide, and thiosulfate as sulfur sources in vitro (30), but how S. aureus imports and catabolizes these metabolites is not known. Consistent with this, sulfur transporters have yet to be experimentally validated in S. aureus, despite the fact that the pathogen likely encounters many potential sulfur sources during dissemination from the bloodstream to various internal organs (33). For example, human serum contains CSSC, Cys, homocysteine, cysteinyl-glycine, GSH, sulfate, sulfide, and methionine (34, 35), but whether these potential sources of sulfur satisfy the sulfur requirement in vivo is not known.
To understand how S. aureus fulfills its sulfur requirement, we evaluated two putative CSSC transporters encoded by the closely related bacterium Bacillus subtilis, TcyABC and TcyP (36–38). B. subtilis encodes three CSSC transporter systems: TcyP, TcyJKLMN, and TcyABC. TcyABC is a low-affinity ATP-binding cassette (ABC) transporter whereby the lipid-anchored substrate-binding protein TcyA delivers substrate to the permease TcyB, which is then translocated, aided by the hydrolysis of ATP by TcyC into the cytoplasm (37). B. subtilis TcyP is a high-affinity CSSC and sodium symporter (37). Whether S. aureus TcyABC and TcyP act in similar ways to the B. subtilis homologues is unknown; however, studies have reported that tcyABC and tcyP are transcriptionally controlled by the master Cys metabolism regulator CymR in both organisms (31, 39–41).
We hypothesized that TcyABC and TcyP facilitate S. aureus CSSC utilization as a sulfur source in vivo. To test this hypothesis, we utilized a genetic approach to characterize tcyA and tcyP mutants in two different strain backgrounds, the laboratory-derived methicillin-resistant strain endemic in the United States, community-acquired USA300 (JE2), and the methicillin-sensitive strain Newman (NWMN) (42, 43). We report the first experimental evidence that S. aureus tcyABC and tcyP encode CSSC acquisition systems. Additionally, we demonstrate that TcyABC and TcyP are required for growth in media supplemented with Cys or N-acetyl cysteine (NAC) as the sole sulfur source. Moreover, TcyABC supports proliferation in medium supplemented with homocystine (hCSSC). Expression of both systems is induced upon sulfur starvation, but tcyP expression is stimulated to a greater extent. Notably, our results implicate TcyABC and TcyP as nutrient sulfur acquisition systems that are important during S. aureus pathogenesis. This study represents the first time sulfur source transporters have been experimentally validated and demonstrated to affect S. aureus virulence.
RESULTS
Genetic inactivation of tcyA increases S. aureus resistance to the toxic CSSC analogue selenocystine.
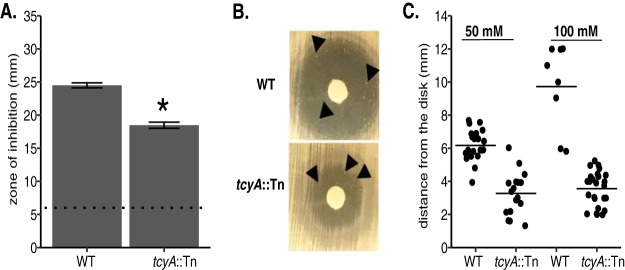
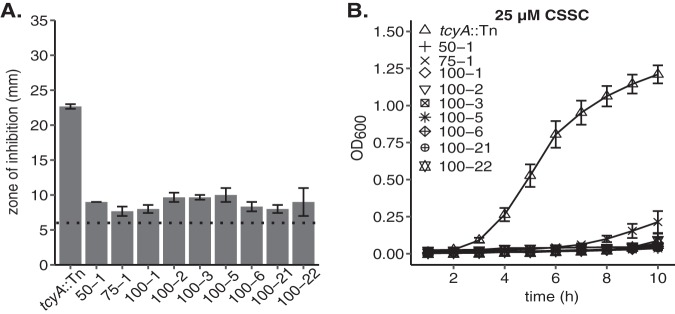
S. aureus encodes homologues of two of the three CSSC transporter systems present in B. subtilis, TcyABC and TcyP (37). Functional characterization of the CSSC transporters in B. subtilis was determined using the toxic CSSC analogue selenocystine (37). In B. subtilis, enhanced resistance to selenocystine correlated with reduced capacity to transport CSSC. We hypothesized that the S. aureus tcyABC homologue encodes a CSSC transporter, and that genetic inactivation would lead to decreased sensitivity to selenocystine. To test this hypothesis, we compared selenocystine sensitivity of a wild-type (WT) USA300 methicillin-resistant strain (strain JE2) to that of an isogenic tcyA::Tn mutant using a Kirby Bauer disk diffusion assay. The tcyA::Tn strain demonstrates a reduced zone of inhibition compared to WT, indicating TcyA has a potential role in selenocystine acquisition (Fig. 1A). However, the fact that the tcyA::Tn strain displays a zone of inhibited growth suggests that TcyABC is not the exclusive putative transporter in S. aureus. To test this idea, we incubated the selenocystine plates for an additional amount of time and examined them for the presence of resistant colonies within the zone of inhibited growth. We reasoned colonies able to grow in the zone of inhibition harbor mutations in additional transporters, thereby allowing them to grow in the presence of the toxic metabolite. After 4 days of further incubation, resistant colonies grew within the WT and tcyA::Tn zones of inhibited growth (Fig. 1B and C). Colonies generated in the tcyA::Tn mutant background grew closer in proximity to the selenocystine-containing disk than colonies of the otherwise WT background (Fig. 1C), indicating that the tcyA::Tn colonies are more resistant than the WT colonies. Similar selenocystine resistance results were obtained using the methicillin-sensitive strain Newman (NWMN) and an isogenic tcyA transposon (tcyA::Tn) mutant (Fig. S1 in the supplemental material), demonstrating functional conservation between S. aureus strains. The JE2 tcyA::Tn selenocystine-resistant colonies were isolated and tested for selenocystine resistance. All the isolates demonstrated nearly complete resistance to selenocystine, indicating that the strains likely contain inactivating mutations in at least one other transporter (Fig. 2A). Next, we tested the effect the secondary mutations had on the ability of the isolates to grow in defined medium (PN) containing CSSC. Selenocystine-resistant tcyA::Tn mutants displayed reduced growth in medium containing CSSC relative to the parental tcyA::Tn mutant (Fig. 2B). Together these data suggest that TcyABC is a major contributor to selenocystine sensitivity in S. aureus but that another transporter system (or systems) that is not required for growth in rich medium is also active.
FIG 1.
S. aureus tcyA mutants demonstrate enhanced resistance to the toxic cystine analogue selenocystine. (A) WT and tcyA::Tn strains were plated as a lawn on tryptic soy agar (TSA), and a paper disk supplemented with 100 mM selenocystine was added to the plate. The mean zone of inhibition of at least three independent trials is presented. The dotted line represents the disk diameter (6 mm). (B) Colonies develop within the zone of inhibition after 96 h of growth when the WT or tcyA::Tn strain is plated in the presence of 50 mM selenocystine. Arrows point to resistant colonies. (C) Selenocystine-resistant WT or tcyA::Tn colonies grew the indicated distance from a sterile Whatman paper disk containing 50 mM or 100 mM selenocystine. The bar represents the mean distance from the disk. Error bars represent ± 1 standard error of the mean. * indicates P < 0.05 determined from Student's t test.
FIG 2.
Characterization of selenocystine-resistant tcyA::Tn mutant isolates. (A) Representative tcyA::Tn selenocystine-resistant colonies were isolated and tested for their resistance in the presence of 100 mM selenocystine. The first number for each bar indicates the concentration of selenocystine (50 mM, 75 mM, or 100 mM) added via the Whatman disk to the plate the mutants first appeared, and the second number represents the isolate number from that concentration of selenocystine. The dotted line represents the disk diameter (6 mm). (B) Growth of tcyA::Tn selenocystine-resistant mutants in 25 μM cystine. Data represent the mean of at least three independent trials. Error bars represent ± 1 standard error of the mean.
Disruption of both tcyA and tcyP leads to complete selenocystine resistance and reduced growth in a defined medium supplemented with CSSC as the sole sulfur source.
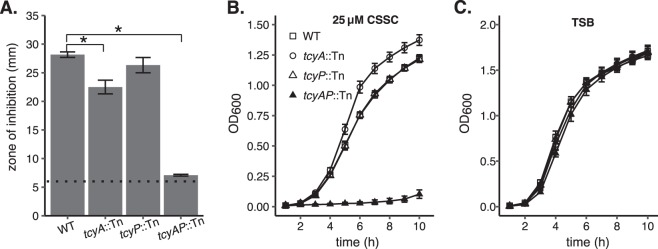
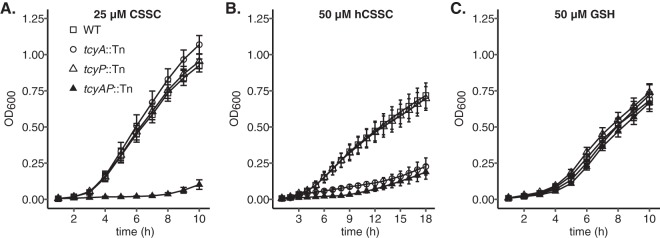
We hypothesized that the selenocystine-resistant tcyA::Tn mutants harbor mutations in the tcyP open reading frame or promoter sequence. To test this, we sequenced the tcyP locus in nine of these mutants. All nine harbored WT tcyP promoter sequence (not shown); however, four isolates contained mutations within the tcyP open reading frame. The mutation identified in each isolate was unique and included a nonsynonymous point mutation, two nonframeshift deletions, and a frameshift deletion (Table 1). These mutations likely inactivate TcyP, as they promote selenocystine resistance and disrupt the ability of the tcyA::Tn mutant to proliferate in medium supplemented with CSSC (Fig. 1 and Fig. 2). To further define the contribution of both transporters toward S. aureus selenocystine resistance and CSSC acquisition, we generated a tcyA::Tn tcyP::Tn (tcyAP::Tn) double mutant. Compared to WT, the tcyP::Tn single mutant exhibits similar selenocystine sensitivity, but the tcyAP::Tn double mutant demonstrates considerably enhanced resistance (Fig. 3A). Comparable selenocystine-resistance results were obtained using a tcyAP::Tn mutation generated in strain NMWN (Fig. S2). These results indicate that tcyABC and tcyP are the dominant selenocystine acquisition systems in S. aureus. To directly test the hypothesis that tcyABC and tcyP are CSSC acquisition systems, we cultured the strains in PN supplemented with 25 μM CSSC as the sole sulfur source. WT, tcyA::Tn, and tcyP::Tn strains displayed similar growth patterns in this medium; however, the tcyAP::Tn double mutant demonstrated significantly impaired proliferation relative to WT and the single mutants (Fig. 3B). Notably, in rich medium the four strains grew similarly, indicating the proliferation defect of the tcyAP::Tn strain is specific to the inability to utilize CSSC as a sulfur source (Fig. 3C). To ensure the selenocystine resistance of the tcyAP::Tn strain is due in part to tcyP or tcyABC inactivation, we constructed complementation vectors carrying tcyP or tcyABC under the control of their native promoters (pKK22 PtcyP::tcyP or pKK22 PtcyABC::tcyABC, respectively). The tcyAP::Tn strain harboring either pKK22 PtcyP::tcyP or pKK22 PtcyABC::tcyABC demonstrates increased sensitivity to selenocystine (Fig. S3A). Additionally, ectopic expression of tcyP or tcyABC restores growth of the tcyAP mutant in PN medium supplemented with 25 μM CSSC to WT levels (Fig. S3B and C). These data validate the roles of TcyP and TcyABC in selenocystine resistance and CSSC utilization.
TABLE 1.
Mutations identified in the tcyP opening reading frame of tcyA::Tn selenocystine-resistant mutants
| Isolate | Mutation | Protein effect |
|---|---|---|
| 50-1 | Deletion 580–591 (12 bp) | Nonframeshift |
| 75-1 | G593A | G198E |
| 100-2 | Deletion 715–777 (63 bp) | Nonframeshift |
| 100-3 | Deletion 799–802 (4 bp) | Frameshift |
FIG 3.
TcyABC and TcyP are required for sensitivity to selenocystine and utilization of cystine as a nutrient sulfur source. (A) WT and tcyA::Tn strains were plated as a lawn on TSA, and a disk supplemented with 100 mM selenocystine was added to the plate. The mean zone of inhibition of at least three independent trials is presented. The dotted line represents the disk diameter (6 mm). (B) WT, tcyA::Tn, tcyP::Tn, and tcyA tcyP (tcyAP::Tn) double mutant strains were cultured in PN medium supplemented with 25 μM cystine as the sole sulfur source. (C) WT, tcyA::Tn, tcyP::Tn, and tcyAP::Tn strains were cultured in TSB. Measurements represent at least three independent trials. Error bars represent ± 1 standard error of the mean. * indicates P < 0.05 determined from Student's t test.
S. aureus TcyABC and TcyP support proliferation in medium containing Cys or N-acetyl cysteine as the sole source of sulfur.
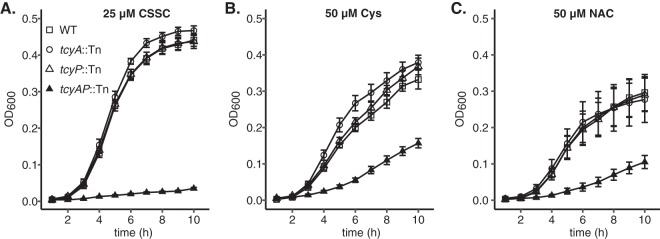
Some bacterial ABC-transporters are capable of transporting both dimeric and monomeric substrates (36, 37). For example, in Streptococcus mutans TcyABC-mediated import of CSSC is inhibited by Cys, indicating that TcyABC is capable of binding and transporting both substrates (36). To test whether TcyABC and TcyP are important for S. aureus utilization of monomeric substrates such as Cys, we quantified growth in media supplemented with Cys as the sole source of sulfur. Because Cys is prone to oxidation in environments containing O2, quantification of growth was carried out in anaerobic conditions in medium containing the alternative electron acceptor nitrate to stimulate anaerobic respiration. Consistent with the results obtained in aerobic conditions, the tcyAP::Tn strain was the only strain that displayed a growth defect relative to WT when CSSC was supplied as the sole sulfur source (Fig. 4A). Similarly, the tcyAP::Tn strain exhibits reduced anaerobic growth relative to WT and the two single mutants in PN supplemented with Cys (Fig. 4B). This observation indicates that both TcyABC and TcyP support growth on Cys and are functioning redundantly. The capacity of TcyABC and TcyP to support growth on CSSC and Cys implies an extended substrate-binding potential. To test this further, we examined whether both systems support proliferation in medium supplemented with N-acetyl cysteine (NAC), a Cys-containing metabolite present in human blood (44). We cultured the S. aureus WT strain and tcyA::Tn, tcyP::Tn, or tcyAP::Tn mutants in PN supplemented with 50 μM N-acetyl cysteine. In keeping with the previous CSSC and Cys results, only proliferation of tcyAP::Tn was reduced relative to WT (Fig. 4C). Notably, WT S. aureus achieves a greater maximum optical density at 600 nm (OD600) in PN medium containing CSSC than in PN containing Cys or NAC, despite the addition of stoichiometrically equivalent levels of sulfur. These results imply that CSSC is the preferred sulfur source for S. aureus in these growth conditions.
FIG 4.
TcyABC and TcyP support proliferation in medium supplemented with cysteine (Cys) or N-acetyl-cysteine (NAC) as the sole sulfur source. WT, tcyA::Tn, tcyP::Tn, and tcyAP::Tn strains were cultured anaerobically in PN medium supplemented with 100 mM sodium nitrate to induce anaerobic respiration and 25 μM cystine (A), 50 μM cysteine (B), or 50 μM N-acetyl-cysteine (C) as the sole sulfur source. The mean of at least three independent trials is presented. Error bars represent ± 1 standard error of the mean.
TcyABC supports growth on homocystine but not glutathione.
To further define potential TcyABC and TcyP substrates, we monitored growth of WT, tcyA::Tn, tcyP::Tn, or tcyAP::Tn strains in PN medium supplemented with homocystine (hCSSC), the oxidized derivative of homocysteine, a four-carbon compound that is an intermediate in the methionine synthesis pathway. Homocysteine directly enters S. aureus sulfur metabolism and can be fluxed to Cys or methionine (31, 32, 45). Based on the structural similarity between hCSSC and CSSC, we hypothesized that TcyABC would support growth on hCSSC. To test this, 50 μM hCSSC was supplemented in PN medium as the sole sulfur source. In the hCSSC-supplemented medium, tcyA::Tn and tcyAP::Tn strains failed to proliferate (Fig. 5B), indicating that TcyABC but not TcyP is involved in hCSSC acquisition.
FIG 5.
TcyABC supports growth of S. aureus in medium containing homocystine but not GSH as a sulfur source. WT, tcyA::Tn, tcyP::Tn, and tcyAP::Tn strains were cultured in PN supplemented with 25 μM cystine (CSSC) (A), 50 μM homocystine (hCSSC) (B), or 50 μM reduced glutathione (GSH) (C) as the sole source of sulfur. Error bars represent ± 1 standard error of the mean. The average of at least three independent trials is presented.
In Streptococcus mutans, import of the Cys-containing tripeptide glutathione (GSH) is mediated by the TcyBC permease and ATP hydrolysis subunits (46). We tested whether S. aureus TcyABC could also be involved in the acquisition of GSH as a sulfur source by culturing WT, tcyA::Tn, tcyP::Tn, or tcyAP::Tn strains in medium containing 50 μM GSH. Compared to the differential growth patterns in the other sulfur sources, no difference was observed between WT and mutant growth in PN with GSH (Fig. 5C), indicating that neither transporter is required for GSH utilization as a source of sulfur.
tcyP and tcyA are induced in sulfur starvation conditions.
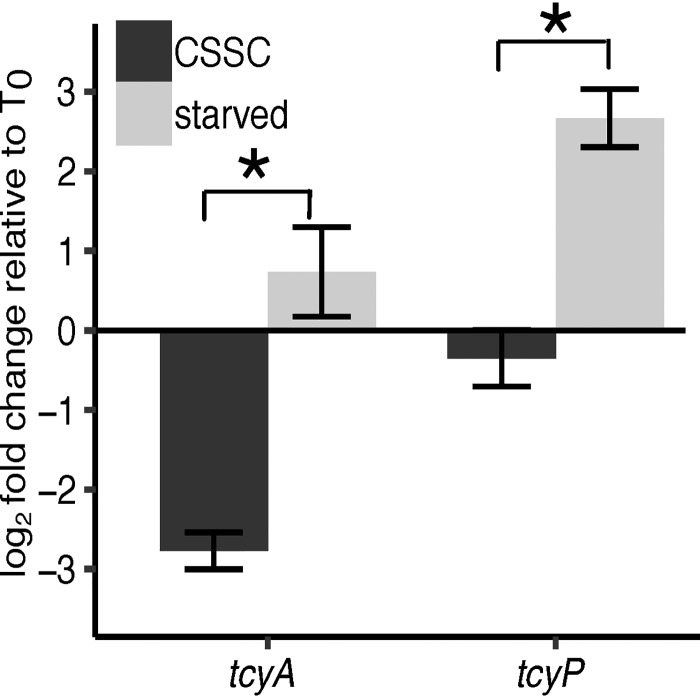
The transcription of tcyP and tcyA is controlled by the sulfur regulator CymR (31, 39). A limitation of the previous CymR studies is that they monitored expression in sulfur-replete conditions using a cymR mutant. To understand how tcyP and tcyA expression changes in response to sulfur starvation, we subcultured cells from a sulfur-replete medium to a sulfur-depleted medium. Specifically, S. aureus was cultured in PN supplemented with 25 μM CSSC (T0) and then transferred to PN medium supplemented with 25 μM CSSC (sulfur replete) or a medium devoid of a sulfur source. We found that tcyP and, to a lesser extent, tcyA are significantly upregulated in response to sulfur starvation relative to sulfur replete-medium as determined by a greater log2 fold change after 2 h of incubation (Fig. 6). Interestingly, tcyA was repressed when S. aureus was transferred from CSSC-supplemented medium into fresh medium containing CSSC (Fig. 6). These data demonstrate that tcyP and tcyA expression is responsive to the abundance of sulfur in the environment.
FIG 6.
Expression of tcyP and tcyA is induced in sulfur starvation conditions. The comparative ΔΔCT methodology was used to calculate fold change and rho was used as a reference gene. Log2 fold change of tcyA and tcyP transcripts relative to T0 was determined. S. aureus was cultured in PN supplemented with 25 μM CSSC and then transferred to medium containing 25 μM CSSC or no sulfur source (starved). Bars represent the mean of at least three independent trials. The error bars represent ± 1 standard error of the mean. * indicates P < 0.05 determined from Student's t test.
TcyP is required for maximal fitness in the murine liver while TcyABC and TcyP are required for maximal fitness in the murine heart.
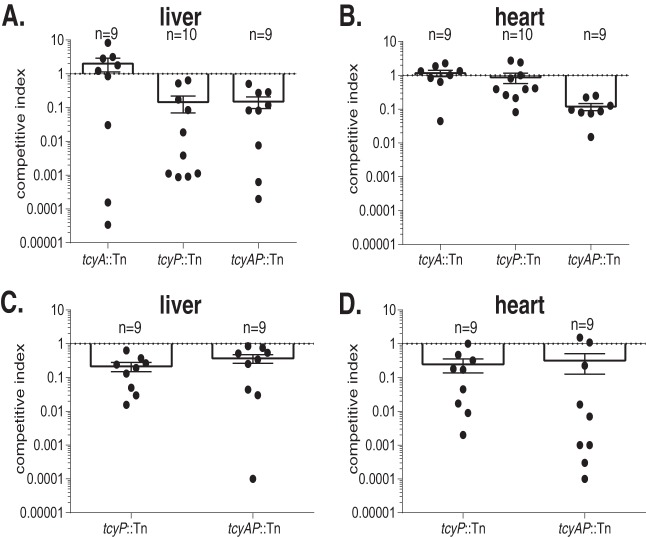
The role of sulfur source acquisition systems during S. aureus pathogenesis has not been elucidated. Additionally, how TcyP and TcyABC support the proliferation of a bacterial pathogen in any infection model has not been described. We hypothesized that TcyP and TcyABC function cooperatively to satisfy the S. aureus sulfur requirement in vivo. To test this, we performed competition infections between NWMN WT and isogenic tcyA::Tn, tcyP::Tn, or tcyAP::Tn mutant strains in a murine model of systemic infection (47). The competitive index in the liver demonstrates that the tcyP::Tn strain was outcompeted by WT (Fig. 7A and B). The tcyA::Tn strain did not display a competitive defect in the heart, liver, or kidneys. Neither single mutant exhibited reduced fitness in the heart; however, the tcyAP double mutant displayed a competitive defect in this organ. Collectively, these data indicate that S. aureus NWMN sulfur acquisition is mediated predominantly by TcyP in the liver and that TcyABC is capable of compensating for the loss of TcyP in the heart.
FIG 7.
TcyP is required for maximal fitness in the murine heart and liver. BALB/c mice were systemically infected with a 1:1 ratio WT and the indicated mutant strain in Newman (A and B) or JE2 (C and D) strains of S. aureus. Competitive indices were calculated as the output ratio of mutant to WT CFU ml−1 over the input mutant to WT CFU ml−1. The mean competitive index for liver (A and C) and heart (B and D) is presented. Error bars represent ± 1 standard error of the mean.
To determine if the involvement of TcyP and TcyABC during infection is similar between S. aureus strains, we repeated the competition infections using the USA300 derivative JE2 (42). Due to the lack of an observed competitive defect for the NWMN tcyA::Tn mutant, we excluded this mutant from the JE2 competition studies. Consistent with the results using NWMN, the competitive index in the liver demonstrated that the tcyP::Tn strain is outcompeted by WT (Fig. 7C). Additionally, the tcyP::Tn strain demonstrated a competitive defect in the murine heart (Fig. 7D). These data show that S. aureus sulfur acquisition in JE2 is mediated by TcyP in both the liver and heart, indicating that sulfur acquisition strategies differ between S. aureus strains in vivo.
TcyP is conserved across many bacterial phyla.
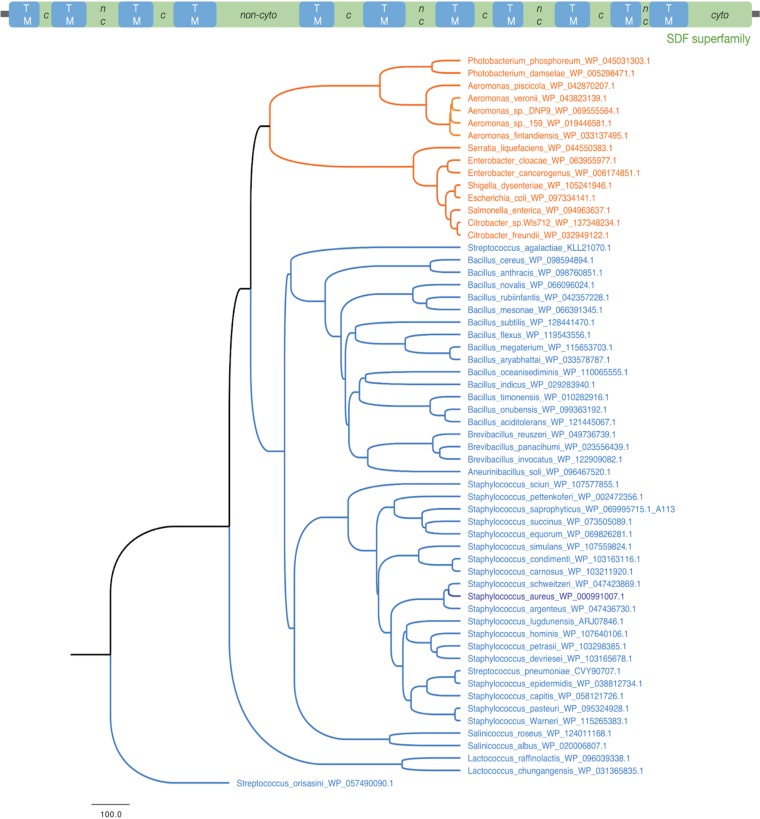
TcyP is a member of the sodium:dicarboxylate symporter superfamily (IPR036458 specifically, PF00375; IPR001991) with several characteristic transmembrane regions and a C-terminal cytoplasmic tail (Fig. 8, top). Previous functional characterization of TcyP has only been performed in E. coli and B. subtilis (28, 36). To determine the conservation of TcyP throughout the bacterial kingdom, we used iterative BLAST (see Materials and Methods). We identified Staphylococcus TcyP homologues in numerous pathogen-containing bacterial phyla and orders, including Enterobacteriaceae within the Proteobacteria (Fig. 8, orange nodes), and Bacillales within the Firmicutes (Fig. 8, blue nodes). While many homologues are present across bacteria, TcyP sequences form a distinct cluster with the Staphylococcus, Salinococcus, and a few members of the Streptococcus, all of which are firmicutes. Similarly, the proteobacterial homologues in Citrobacter, Salmonella, Enterobacter, and Aeromonas cluster together (Fig. 8). These data demonstrate that while many bacterial phyla carry homologues of TcyP, the variations are lineage-specific. We find that there are homologous transporter proteins in other bacterial clades, such as the Actinobacteria, Fusobacteria, Deinococcus, Thermotogae, and Spirochaetes, which belong to the TcyP or the broader sodium:dicarboxylate symporter family that includes the aerobic C4-dicarboxylate transporter protein DctA (Fig. S4 and Data Set S1 in the supplemental material). Further experimentation is needed to establish the functional roles of these homologues in CSSC or Cys acquisition and pathogenesis.
FIG 8.
TcyP is broadly conserved in bacteria. (Top) Domain architecture and cellular localization of the S. aureus TcyP protein (462aa; WP_000991007.1), as characterized by Interproscan, Phobius (see Materials and Methods). The predominant domain, sodium:dicarboxylate symporter superfamily, is indicated in green with the transmembrane (TM) regions shown in blue. The predicted cytoplasmic (c) and noncytoplasmic (nc) regions are labeled for the loop residues. (Bottom) Homologues of TcyP in Firmicutes (blue) and Proteobacteria (orange) using the S. aureus TcyP as the query are shown. Staphylococcus aureus is highlighted in dark blue.
DISCUSSION
Acquisition of nutrient sulfur is essential for all organisms; however, we lack a basal understanding of the sulfur sources used by S. aureus and the proteins necessary to facilitate their acquisition. To synthesize critical metabolites required for cell proliferation, organisms have evolved assimilatory processes that exploit both inorganic and organic sulfur sources (26, 32, 48, 49). S. aureus sulfur catabolism is limited due to the inability to convert sulfate, a fully oxidized and highly abundant plasma sulfur source, into a reduced and utilizable form (30–32, 50). In the current study, we examined CSSC acquisition by S. aureus through the characterization of TcyABC and TcyP. Human serum levels of CSSC have been reported to be as high as 62.9 μM (34). While the ability of S. aureus to use CSSC as a sulfur source in vitro was previously described (30), the transporters involved in this process have not been experimentally validated.
Our results demonstrate that TcyP and TcyABC support growth on a wide range of Cys analogues. We exploited this fundamental aspect to determine whether the respective mutant strains demonstrated enhanced resistance to selenocystine, which differs from CSSC by the replacement of the sulfur atoms with selenium atoms. The ability to transport Cys and CSSC by the same transporters has been previously speculated in other organisms (36, 37). Evidence presented here demonstrates that disruption of both tcyA and tcyP impairs growth in medium supplemented with Cys relative to WT S. aureus. These data suggest that TcyP and TcyABC are capable of importing both Cys and CSSC, consistent with previous investigations that provide precedent for the observed substrate promiscuity of TcyP and TcyABC (36, 37).
Broadly, ABC-transporters can be categorized into three types (51, 52). Type I encompasses transporters that translocate metabolites a cell needs in bulk quantities, including amino acids and sugars (51–53). Type I ABC transporters and their corresponding substrate-binding proteins contain a substrate binding and recognition pocket that is surrounded by flexible loops, whereas type II substrate-binding proteins are confined by a nonflexible helix (52). Consequently, type II ABC transporters have a higher affinity for substrates than type I (52). TcyABC likely belongs to the type I group of ABC transporters due to the speculated low-affinity for CSSC and the fact that its cognate metabolite is required in large quantities (37). These facts could explain why TcyABC might be able to accommodate both CSSC and Cys in its substrate-binding site. Further studies will provide insight into the flexible substrate-binding site of TcyABC, but TcyA has been crystalized and the structure has been determined with either CSSC or Cys bound to the binding site, revealing that it has affinity for both derivatives (54).
Additional studies in B. subtilis have also shown that TcyP transport of CSSC is inhibited by Cys, indicating that Cys competes with CSSC for access to the channel of the transporter (37). Similar findings have been reported in Streptococcus mutans TcyABC (36). Our data agree with these previous studies showing that TcyP and TcyABC display substrate promiscuity in transporting CSSC and Cys analogues. We show that while TcyABC and TcyP support growth on Cys and derivatives, WT S. aureus does not grow to the same levels when these sulfur sources are present at equivalent sulfur levels to CSSC. Calculations in E. coli reveal that a cell contains as much as 230 μmol sulfur atoms and this amount of sulfur can be provided by the transport of 115 μmol of CSSC (28). Assuming that S. aureus has a similar sulfur demand to E. coli, theoretically equimolar concentrations of CSSC, Cys, or NAC would lead to equivalent levels of growth; however, this is not observed. In fact, S. aureus growth in CSSC-containing medium exceeds growth when either NAC or Cys is present, indicating that CSSC must be more efficiently acquired by S. aureus than NAC or Cys. Both the Streptococcus mutans and B. subtilis studies show a moderate decrease in CSSC transport in the presence of Cys, but a limitation of these studies is that Cys transport was measured through the ability to inhibit radiolabeled CSSC import (36, 37). Based upon prior results and data presented here, both transporters appear to have greater affinity for CSSC but can likely transport Cys with some efficiency (31). Therefore, as a consequence of decreased efficiency, the same sulfur molar concentration of NAC or Cys does not lead to the same intracellular concentration of sulfur and ultimately a greater environmental sulfur concentration is required to support the same level of growth. Our study provides further resolution due to our examination of TcyABC- and TcyP-mediated growth on Cys and derivatives.
We next wanted to broaden our studies to investigate transport of structurally similar intermediates in the sulfur assimilation pathway and show for the first time that S. aureus utilizes hCSSC as a sulfur source. Prior studies established reduced homocysteine as a viable sulfur source for S. aureus but did not examine growth on the oxidized form (31). hCSSC is the disulfide, oxidized form of homocysteine, a four-carbon compound that is an intermediate in the pathway that produces methionine using Cys. Cys is a substrate for a condensation reaction with O-acetyl-homoserine that generates cystathionine (32, 45). Cystathionine is then cleaved to generate homocysteine (32, 38). We hypothesize that homocysteine directly enters S. aureus sulfur metabolism and can be fluxed to Cys or methionine (31, 32, 45). We demonstrate that genetic inactivation of tcyABC leads to a failure to grow in medium supplemented with hCSSC as the sole sulfur source. These data further imply that the TcyA substrate-binding domain is promiscuous, as it must act as a receptor for multiple sulfur-containing metabolites, including CSSC, Cys, NAC, hCSSC, and selenocystine.
While it is known that CymR represses TcyP and TcyP, we wanted to determine the responsiveness of tcyA and tcyP expression to the abundance of sulfur in the environment. Our results demonstrate that sulfur-depleted environments induce tcyP expression and, to a lesser extent, tcyA expression. Previous studies showed tcyP and tcyA are transcriptionally repressed by CymR, the master Cys metabolism regulator in Firmicutes (31, 39). In cymR mutants, tcyP transcript levels increase 25-fold relative to WT (31, 39). Furthermore, gel-shift assays demonstrated that CymR directly binds the tcyP promoter (31). Enhanced transcriptional control of tcyP likely has several physiological implications. First, B. subtilis TcyP is predicted to have a greater affinity for CSSC than TcyABC (37). Induction of TcyP by S. aureus in sulfur-depleted environments is an adept strategy to accrue CSSC because of the greater affinity relative to TcyABC. Second, upon expression, the increased affinity of TcyP for CSSC likely ensures the sulfur requirement is satisfied by CSSC in sulfur-depleted environments. Third, increased CSSC import leads to a greater intracellular Cys pool, which potentiates Fenton-chemistry and oxidative damage in E. coli (28, 55). Additionally, studies have shown that growth of a cymR mutant in B. subtilis in the presence of CSSC is severely impaired, further suggesting the necessity to control CSSC levels (40). Therefore, strict transcriptional control of S. aureus tcyP likely reduces a potentially toxic accumulation of intracellular Cys. An interesting observation from our transcriptional analysis of tcyA and tcyP is the apparent tcyA repression that occurs when the cells are cultured in CSSC and then subcultured into the same medium. This finding seems counterintuitive as it appears the cell is repressing transporters necessary to obtain the only viable sulfur source in the medium; however, given the potential toxicity associated with Cys accumulation (28, 55), repression of TcyABC could be a tactic to modulate CSSC import to thereby limit toxicity.
This work and others suggest that there are at least two different strategies for bacterial Cys and CSSC import. Some organisms encode mechanisms supported by high-affinity CSSC transporters that are also capable of transporting Cys at a reduced efficiency, while others have evolved strategies in which dedicated Cys and CSSC importers are specific for their cognate metabolite (36, 37, 56). One model predicts that the strategy employed is dependent on the oxygenic environments in which the organism resides (57). For instance, some anaerobes and facultative anaerobes have homologues of a distinct Cys transporter present in Campylobacter jejuni (56, 57). In an anaerobic environment, Cys will likely be reduced, while CSSC is the dominant form in aerobic environments. E. coli has evolved a strategy in which CSSC is imported and converted to Cys intracellularly; however, excess Cys is exported and subsequently rapidly oxidized in the external environment (28). This strategy suggests that E. coli predominantly encounters CSSC in its environment and has evolved to target CSSC. S. aureus does not encode a predicted Cys-specific transporter. Based on data presented here, S. aureus employs a strategy that utilizes dedicated CSSC transporters that also transport Cys with lower affinity and efficiency. The preference for CSSC over Cys is also interesting due to the fact that the cell must invest energy in reducing the CSSC disulfide bond to liberate Cys. Cys, on the other hand, can be directly incorporated into the translational or cofactor synthesis pathways. Evolutionarily the preference for CSSC might provide some insights into the environments in which S. aureus primarily resides and the state of Cys in these environments. An integral aspect of S. aureus pathogenesis is the upregulation of secreted proteases (58, 59). Protease-null strains demonstrate pronounced growth reductions in peptide-rich environments, including serum (60). Protease secretion likely increases Cys abundance at the host-pathogen interface. Additionally, the reactive Cys thiol is sensitive to effects of the host oxidative burst, increasing CSSC, or other Cys-containing thiol adducts that result from the formation of mixed disulfides (61). These conditions could enhance the importance of Cys and CSSC metabolism to S. aureus pathogenesis. Future work will further characterize TcyABC and TcyP by examining their role in the transport of Cys-containing mixed disulfides.
A previous study has noted a potential link between Cys acquisition and pathogenesis. The Listeria monocytogenes protein CtaP is required for in vitro proliferation in low Cys conditions and ctaP mutants demonstrate reduced colonization in the livers and spleens of mice following intravenous challenge. A direct correlation between Cys acquisition and L. monocytogenes pathogenesis is difficult due to the fact that CtaP was also shown to be involved in host cell adhesion (62). Using a murine model of systemic infection, we show that in the S. aureus strain JE2, TcyP is sufficient for proliferation of S. aureus in the heart and liver. However, in strain NWMN, TcyP is required for maximal fitness in the liver but both TcyP and TcyABC are important for proliferation in the heart. While a prior study demonstrated that a mutant strain of S. aureus inactivated for thiosulfate assimilation is as virulent as the corresponding WT strain in a murine skin lesion infection model (30), these data show for the first time that sulfur-source acquisition systems support S. aureus infection. The fact that TcyABC and TcyP are required for growth in medium containing different sulfur sources makes it challenging to define the exact sulfur-containing metabolite that is acquired in vivo. Due to the competitive defect observed for JE2 tcyP::Tn in murine liver and heart, we can narrow the acquired sulfur source to CSSC, Cys, or NAC. Similarly, we can conclude that NWMN could be catabolizing CSSC, Cys, or NAC during liver colonization. In contrast, due to the observation that inactivation of both tcyA and tcyP in S. aureus strain NWMN is necessary for a colonization defect in murine heart, we can only conclude that the catabolism of sulfur sources occurring could include CSSC, Cys, NAC, and/or hCSSC. The different in vivo phenotypes observed between the two strains could be due to regulatory factors present in JE2 but not in NWMN. JE2 harbors the staphylococcal cassette chromosome (SCCmec) and the arginine catabolism mobile element (ACME) (5, 63), and previous work has shown that ACME and SCCmec mutants differentially regulate a subset of genes which includes some annotated but uncharacterized transporters (63). The presence of ACME and SCCmec in JE2 may cause a differential sulfur requirement during proliferation in the heart compared to NWMN, due to transcriptional changes. This might explain why the JE2 tcyP mutant demonstrates a heart colonization defect phenotype while inactivation of both tcyP and tcyABC is required to observe a virulence defect in strain NWMN.
An interesting observation from the infection experiments is that loss of TcyABC and TcyP does not completely ablate the capacity of the strains to compete with WT, as tcyAP double mutants are still recovered from the organs. This result suggests that other sulfur sources are likely present that support proliferation in the absence of cognate TcyP- and TcyABC-dependent metabolites. Our data indicate that TcyABC and TcyP do not support growth on GSH as a sulfur source. Therefore, GSH present in the heart and liver could allow the tcyAP mutants to establish infection in these organs. Further investigation into the sulfur catabolism of S. aureus will be necessary to determine the identity and abundance of TcyP- and TcyABC-dependent and independent sulfur sources present within host organs. Additionally, the identification of GSH acquisition systems will support direct investigation into the importance of GSH acquisition during infection.
We found that many bacteria encode TcyP homologues and that the homologues predominantly cluster into groups based on their lineages. Whether these transport systems are unique to CSSC and whether they are functional and relevant to pathogenesis in the other phyla remain to be explored. Based on the homology to S. aureus TcyP and the data presented here showing that TcyP is involved in CSSC, Cys, and NAC transport, these results provide a platform to characterize TcyP-dependent sulfur transport in other pathogenic bacteria.
MATERIALS AND METHODS
Bacterial strains.
Strains used in this study are described in Table S1 in the supplemental material. JE2 is derived from the community-acquired methicillin-resistant (CA-MRSA) S. aureus clinical isolate USA300 LAC (42). Strain Newman is a methicillin-sensitive strain (43). Mutants were created by transducing the transposon (Tn) inactivated gene insertion provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources into the desired strain using previously described transduction methodology (42, 64). For the tcyA tcyP double mutant, the erythromycin resistance cassette in the tcyP::Tn strain was replaced with a tetracycline resistance cassette using a previously described allelic replacement strategy (65). Tn insertion was confirmed by PCR.
Generation of complementation vectors.
The plasmid pKK22 was used for complementation studies (66). Integration of inserts was achieved using Gibson assembly methodology and employed the HiFi assembly master mix (New England Biolabs, Ipswich, MA). The tcyP or tcyABC open reading frame sequence, including the native promoter, was amplified from JE2 genomic DNA and pKK22 was amplified from plasmid DNA using primers listed in Table S2. HiFi assembly was used to construct pKK22 PtcyP::tcyP or PtcyABC::tcyABC, and the constructs were transformed into a pir+ Escherichia coli strain. The PtcyP::tcyP and PtcyABC::tcyABC sequence was verified by Sanger sequencing. Plasmids were isolated from E. coli and transformed into RN4220, and the RN4220-passaged plasmids were transduced into the final recipient strains.
Growth analysis.
Strains were cultured in trypic soy broth (TSB) overnight, washed with phosphate-buffered saline (PBS), and normalized to an optical density at 600 nm (OD600) of 1. Cells were then added to 96-well round-bottom plates at a starting OD600 of 0.01. Growth was monitored by measuring OD600 at hourly time points using a BioTek plate reader set at 37°C with continuous, linear shaking. Growth analysis was carried out with PN medium supplemented with 5 mg ml−1 glucose and indicated sulfur sources. PN medium was prepared as previously described and contained seventeen amino acids, excluding Cys, asparagine, and glutamine (67). Cys was omitted to test the ability of S. aureus to grow on various sulfur sources. Blank measurements were subtracted from measurements and experiments were performed in triplicate, which were subsequently averaged. To fully ensure Cys and N-acetyl cysteine were maintained in the reduced state, chemicals were weighed aerobically and immediately transferred to a Coy anaerobic chamber with a 95:5 N2:H2 atmosphere (Grass Lake, MI) where it was dissolved in a degassed solution of 1N HCl. Anaerobic growth analysis was carried out in the presence of 100 mM sodium nitrate as an alternative electron acceptor.
Selenocystine disk diffusion assays and isolation of resistant mutants.
Overnight cultures grown in TSB at 37°C were swabbed onto 20 ml trypic soy agar (TSA) plates to generate a lawn of bacterial growth. A volume of 10 μl of 50 mM, 75 mM, or 100 mM selenocystine dissolved in 1N HCl was added to a sterile Whatman paper disk and was transferred to the TSA plate. Plates were incubated at 37°C for 24 h. The zone of inhibition was measured in millimeters and was defined as the diameter between bacterial lawn growth. Colonies arising in the zone of inhibition after 96 h were restreaked onto TSA plates, subsequently cultured in TSB, and archived at –80°C.
Genomic DNA isolation and gene sequencing.
Selenocystine-resistant mutants were cultured in TSB. Cells were centrifuged and resuspended in buffer containing 60 μg ml−1 lysostaphin from ABMI (Lawrence, NY) and were incubated for 1 h at 37°C to remove the cell wall. Genomic DNA was isolated using a Promega Wizard genomic DNA purification kit (Madison, WI) following the manufacturer’s directions. The tcyP ORF and promoter were amplified using GoTaq polymerase (Promega, Madison, WI). The amplicon was purified using the Wizard PCR cleanup kit (Promega, Madison, WI) according to the manufacturer’s instructions. Sanger sequencing was performed by the Michigan State University Research Technology Support Facility (East Lansing, MI).
qRT-PCR analysis.
Overnight cultures of JE2 were grown in TSB at 37°C and subcultured 1:100 into 50 ml of PN supplemented with 5 mg ml−1 glucose and 25 μM CSSC in three 250-ml Erlenmeyer flasks for 3 h at 37°C. A volume of 50 ml was centrifuged for 10 min at 4,700 rpm at room temperature, washed once in PBS, and centrifuged a second time. One sample was processed as the T0 and was lysed. Upon resuspension in the indicated growth medium, PN supplemented with 5 mg ml−1 glucose and 25 μM CSSC or PN supplemented with 5 mg ml−1 glucose lacking a sulfur source, cells were incubated for 2 h at 37°C in 250-ml Erlenmeyer flasks. A volume of 50 ml was centrifuged for 10 min at 4,700 rpm at room temperature, and the cell pellet was resuspended in 750 μl LETS buffer containing 0.1 M LiCl, 10 mM EDTA, 10 mM Tris-HCl (pH 7.4) and 1% SDS. Cells were transferred to a 2 ml bead-beating tube with 500 μl volume of 0.1 mm zirconia/silica beads (BioSpec, Bartlesville, OK). The lysates were incubated at 55°C for 5 min and centrifuged for 10 min at 13,700 rpm at 4°C. Supernatant was transferred to 1 ml TRIzol. RNA was precipitated using chloroform phase separation and isopropanol and then purified using an RNAeasy kit following the manufacturer’s instructions. Contaminating DNA was removed via treatment with an on-column DNase treatment. RNA was treated with Turbo DNase and cDNA was synthesized using SuperScript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions and random hexamer methodology (Invitrogen, Carlsbad, CA). Quantitative real-time PCR (qRT-PCR) was set up using SYBR green master mix with 20 μl reaction mixtures containing 10 μl SYBR green master mix, 2 μl of 10 μM forward primer, 2 μl of 10 μM reverse primer, and 50 ng cDNA. Primers used for each gene are presented in Table S2. qRT-PCR was performed on a QuantStudio 3 Real-Time PCR thermocycler (Thermo Fisher Scientific, Waltham, MA) and was performed in technical triplicate with minus reverse transcriptase controls to determine genomic DNA contamination. Comparative ΔΔCT methodology was used to compare transcript levels using rho as a reference target (68–70).
Competition in a murine model of systemic infection.
WT and mutant strains were grown in TSB overnight at 37°C, subcultured 1:100, and grown in TSB for 3 h at 37°C and 225 rpm. Strains were washed in PBS and normalized to an OD600 equal to 0.4. The competitions were prepared by mixing equal volumes of WT and mutant strains. To quantify the input ratio, the mixture was serially diluted and plated onto TSA and TSA supplemented with 10 μg ml−1 erythromycin (erm10) to discern between WT and mutant, which were erythromycin resistant. The mutant strains utilized in these experiments harbored at least one erm resistance cassette. WT CFU were calculated by subtracting CFU generated on TSA-erm10 from CFU present on TSA. Ten female 8-week-old BALB/c mice were retro-orbitally infected with 100 μl of 107 CFU of the WT and mutant mixtures. After 96 h, the heart and liver were collected and homogenized in 1 ml PBS. Homogenates were serially diluted and plated onto TSA or TSA-erm10. Competitive indices were calculated as dividing the mutant:WT output CFU ratio by the mutant:WT input CFU ratio.
Ethics approval.
Infections were performed at Michigan State University under the principles and guidelines described in the Guide for the Care and Use of Laboratory Animals (77). Animal work was followed as approved by Michigan State University Institutional Animal Care and Use Committee (IACUC) approved protocol number 12/16-205-00.
Phylogenetic analyses of TcyP.
The TcyP amino acid sequence (WP_000991007.1 from Staphylococcus aureus USA300_FRP3757, assembly GCF_000013465.1) was used as the query sequence for a position-specific iterative BLAST (PSI-BLAST) for two iterations against the NR database. The protein sequences of select representative homologues from pathogen-containing clades Proteobacteria and Firmicutes were aligned to the S. aureus TcyP sequence using ClustalO (71, 72). A phylogenetic tree was generated from the alignment using FigTree (73). The protein domain and localization predictions were done using CDD (74), Interproscan (75), and Phobius (76). For the supplemental analyses, S. aureus TcyP (WP_00991007.1) was used as the query for one iteration of PSI-BLAST across all bacteria using the RefSeq protein database (to limit redundancy). The results from the top 3,000 hits in the similarity search were used to identify TcyP-like proteins in other bacterial clades (Data Set S1 in the supplemental material). To include representative members across diverse genera, matches with closely related S. aureus strains and multispecies were filtered out followed by selection of the top hit (based on percent identity) per genus. The resulting data were used to construct the phylogenetic tree, as described above.
Supplementary Material
ACKNOWLEDGMENTS
The following reagents were provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources, NIAID, NIH: Staphylococcus aureus subsp. aureus strain JE2, tcyA transposon mutant (NE1592), and tcyP transposon mutant (NE625).
We thank Jeffery Bose’s laboratory at the University of Kansas Medical Center for supplying the pKK22 complementation vector.
J.P.D. is supported by an American Society for Microbiology undergraduate research fellowship. This work is funded by the American Heart Association 16SDG30170026, start-up funds provided by Michigan State University, and the National Institutes of Health R01 AI139074.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core surveillance (ABCs) MRSA Investigators. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Cabell CH, Jollis JG, Peterson GE, Corey GR, Anderson DJ, Sexton DJ, Woods CW, Reller LB, Ryan T, Fowler VG. 2002. Changing patient characteristics and the effect on mortality in endocarditis. Arch Intern Med 162:90–94. doi: 10.1001/archinte.162.1.90. [DOI] [PubMed] [Google Scholar]
- 3.Kallen AJ, Mu Y, Bulens S, Reingold A, Petit S, Gershman K, Ray SM, Harrison LH, Lynfield R, Dumyati G, Townes JM, Schaffner W, Patel PR, Fridkin SK, Active Bacterial Core surveillance (ABCs) MRSA Investigators of the Emerging Infections Program. 2010. Health care-associated invasive MRSA infections, 2005–2008. JAMA 304:641–648. doi: 10.1001/jama.2010.1115. [DOI] [PubMed] [Google Scholar]
- 4.Salgado-Pabón W, Schlievert PM. 2014. Models matter: the search for an effective Staphylococcus aureus vaccine. Nat Rev Microbiol 12:585–591. doi: 10.1038/nrmicro3308. [DOI] [PubMed] [Google Scholar]
- 5.Gordon RJ, Lowy FD. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46(Suppl 5):S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kluytmans J, van Belkum A, Verbrugh H. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 10:505–520. doi: 10.1128/CMR.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miles AA. 1941. Some problems of wound infection. Lancet 238:507–510. doi: 10.1016/S0140-6736(00)76850-4. [DOI] [Google Scholar]
- 8.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 9.Gould JC, McKillop EJ. 1954. The carriage of Staphylococcus pyogenes var. aureus in the human nose. J Hyg (Lond) 52:304–310. doi: 10.1017/s0022172400027509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitko NP, Grosser MR, Khatri D, Lance TR, Richardson AR. 2016. Expanded glucose import capability affords Staphylococcus aureus optimized glycolytic flux during infection. mBio 7:e00296-16. doi: 10.1128/mBio.00296-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halsey CR, Lei S, Wax JK, Lehman MK, Nuxoll AS, Steinke L, Sadykov M, Powers R, Fey PD. 2017. Amino acid catabolism in Staphylococcus aureus and the function of carbon catabolite repression. mBio 8:e01434-16. doi: 10.1128/mBio.01434-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson AR, Libby SJ, Fang FC. 2008. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319:1672–1676. doi: 10.1126/science.1155207. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Sun F, Cho H, Yelavarthi V, Sohn C, He C, Schneewind O, Bae T. 2010. CcpA mediates proline auxotrophy and is required for Staphylococcus aureus pathogenesis. J Bacteriol 192:3883–3892. doi: 10.1128/JB.00237-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. 2004. Iron-source preference of Staphylococcus aureus infections. Science 305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 15.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 16.Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, Skaar EP. 2013. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun 81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dale SE, Doherty-Kirby A, Lajoie G, Heinrichs DE. 2004. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect Immun 72:29–37. doi: 10.1128/iai.72.1.29-37.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107. doi: 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan L, Cheng A, Dunman PM, Missiakas D, He C. 2010. Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J Bacteriol 192:3068–3077. doi: 10.1128/JB.00928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Indriati Hood M, Skaar EP. 2013. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio 4:e00241-13. doi: 10.1128/mBio.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammer ND, Schurig-Briccio LA, Gerdes SY, Gennis RB, Skaar EP. 2016. CtaM is required for menaquinol oxidase aa3 function in Staphylococcus aureus. mBio 7:e00823-16. doi: 10.1128/mBio.00823-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dastgheyb SS, Otto M. 2015. Staphylococcal adaptation to diverse physiologic niches: an overview of transcriptomic and phenotypic changes in different biological environments. Future Microbiol 10:1981–1995. doi: 10.2217/fmb.15.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krismer B, Liebeke M, Janek D, Nega M, Rautenberg M, Hornig G, Unger C, Weidenmaier C, Lalk M, Peschel A. 2014. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog 10:e1003862. doi: 10.1371/journal.ppat.1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weidenmaier C, Goerke C, Wolz C. 2012. Staphylococcus aureus determinants for nasal colonization. Trends Microbiol 20:243–250. doi: 10.1016/j.tim.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Beinert H. 2000. A tribute to sulfur. Eur J Biochem 267:5657–5664. doi: 10.1046/j.1432-1327.2000.01637.x. [DOI] [PubMed] [Google Scholar]
- 26.Lill R, Mühlenhoff U. 2006. Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu Rev Cell Dev Biol 22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- 27.Meister A, Anderson ME. 1983. Glutathione. Annu Rev Biochem 52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 28.Chonoles Imlay KR, Korshunov S, Imlay JA. 2015. Physiological roles and adverse effects of the two cystine importers of Escherichia coli. J Bacteriol 197:3629–3644. doi: 10.1128/JB.00277-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lensmire JM, Hammer ND. 2019. Nutrient sulfur acquisition strategies employed by bacterial pathogens. Curr Opin Microbiol 47:52–58. doi: 10.1016/j.mib.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Lithgow JK, Hayhurst EJ, Cohen G, Aharonowitz Y, Foster SJ. 2004. Role of a cysteine synthase in Staphylococcus aureus. J Bacteriol 186:1579–1590. doi: 10.1128/jb.186.6.1579-1590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soutourina O, Poupel O, Coppée J-Y, Danchin A, Msadek T, Martin-Verstraete I. 2009. CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol Microbiol 73:194–211. doi: 10.1111/j.1365-2958.2009.06760.x. [DOI] [PubMed] [Google Scholar]
- 32.Guédon E, Martin-Verstraete I. 2007. Cysteine metabolism and its regulation in bacteria, p 195–218. In Wendisch VF. (ed), Amino acid bioynthesis—pathways, regulation and metabolic engineering. Springer, Berlin, Germany. [Google Scholar]
- 33.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. 2009. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J 23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, Bouatra S, Sinelnikov I, Krishnamurthy R, Eisner R, Gautam B, Young N, Xia J, Knox C, Dong E, Huang P, Hollander Z, Pedersen TL, Smith SR, Bamforth F, Greiner R, McManus B, Newman JW, Goodfriend T, Wishart DS. 2011. The human serum metabolome. PLoS One 6:e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ubbink JB, Hayward Vermaak WJ, Bissbort S. 1991. Rapid high-performance liquid chromatographic assay for total homocysteine levels in human serum. J Chromatogr 565:441–446. doi: 10.1016/0378-4347(91)80407-4. [DOI] [PubMed] [Google Scholar]
- 36.Kim J, Senadheera DB, Lévesque CM, Cvitkovitch DG. 2012. TcyR regulates L-cystine uptake via the TcyABC transporter in Streptococcus mutans. FEMS Microbiol Lett 328:114–121. doi: 10.1111/j.1574-6968.2011.02492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burguière P, Auger S, Hullo M-F, Danchin A, Martin-Verstraete I. 2004. Three different systems participate in L-cystine uptake in Bacillus subtilis. J Bacteriol 186:4875–4884. doi: 10.1128/JB.186.15.4875-4884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baptist EW, Kredich NM. 1977. Regulation of L-cystine transport in Salmonella typhimurium. J Bacteriol 131:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soutourina O, Dubrac S, Poupel O, Msadek T, Martin-Verstraete I. 2010. The pleiotropic CymR regulator of Staphylococcus aureus plays an important role in virulence and stress response. PLoS Pathog 6:e1000894. doi: 10.1371/journal.ppat.1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hullo M-F, Martin-Verstraete I, Soutourina O. 2010. Complex phenotypes of a mutant inactivated for CymR, the global regulator of cysteine metabolism in Bacillus subtilis. FEMS Microbiol Lett 309:201–207. doi: 10.1111/j.1574-6968.2010.02043.x. [DOI] [PubMed] [Google Scholar]
- 41.Even S, Burguière P, Auger S, Soutourina O, Danchin A, Martin-Verstraete I. 2006. Global control of cysteine metabolism by CymR in Bacillus subtilis. J Bacteriol 188:2184–2197. doi: 10.1128/JB.188.6.2184-2197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duthie ES. 1952. Variation in the antigenic composition of staphylococcal coagulase. J Gen Microbiol 7:320–326. doi: 10.1099/00221287-7-3-4-320. [DOI] [PubMed] [Google Scholar]
- 44.Longo A, Di Toro M, Galimberti C, Carenzi A. 1991. Determination of N-acetylcysteine in human plasma by gas chromatography-mass spectrometry. J Chromatogr 562:639–645. doi: 10.1016/0378-4347(91)80614-i. [DOI] [PubMed] [Google Scholar]
- 45.Kredich NM. 1996. Biosynthesis of cysteine, p 514–527. In Neidhardt FC. (ed), Escherichia coli and Salmonella, cellular and molecular biology, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 46.Vergauwen B, Verstraete K, Senadheera DB, Dansercoer A, Cvitkovitch DG, Guédon E, Savvides SN. 2013. Molecular and structural basis of glutathione import in Gram-positive bacteria via GshT and the cystine ABC importer TcyBC of Streptococcus mutans. Mol Microbiol 89:288–303. doi: 10.1111/mmi.12274. [DOI] [PubMed] [Google Scholar]
- 47.Kim HK, Missiakas D, Schneewind O. 2014. Mouse models for infectious diseases caused by Staphylococcus aureus. J Immunol Methods 410:88–99. doi: 10.1016/j.jim.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sekowska A, Kung HF, Danchin A. 2000. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J Mol Microbiol Biotechnol 2:145–177. [PubMed] [Google Scholar]
- 49.Kredich NM. 2008. Biosynthesis of cysteine. Ecosal Plus 2008 doi: 10.1128/ecosalplus.3.6.1.11. [DOI] [PubMed] [Google Scholar]
- 50.Hoppe B, Kemper MJ, Hvizd MG, Sailer DE, Langman CB. 1998. Simultaneous determination of oxalate, citrate and sulfate in children’s plasma with ion chromatography. Kidney Int 53:1348–1352. doi: 10.1046/j.1523-1755.1998.00891.x. [DOI] [PubMed] [Google Scholar]
- 51.ter Beek J, Guskov A, Slotboom DJ. 2014. Structural diversity of ABC transporters. J Gen Physiol 143:419–435. doi: 10.1085/jgp.201411164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewinson O, Livnat-Levanon N. 2017. Mechanism of action of ABC importers: conservation, divergence, and physiological adaptations. J Mol Biol 429:606–619. doi: 10.1016/j.jmb.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 53.Rice AJ, Park A, Pinkett HW. 2014. Diversity in ABC transporters: type I, II and III importers. Crit Rev Biochem Mol Biol 49:426–437. doi: 10.3109/10409238.2014.953626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar P, Kesari P, Kokane S, Ghosh DK, Kumar P, Sharma AK. 2019. Crystal structures of a putative periplasmic cystine-binding protein from Candidatus Liberibacter asiaticus: insights into an adapted mechanism of ligand binding. FEBS J 286:3450. doi: 10.1111/febs.14921. [DOI] [PubMed] [Google Scholar]
- 55.Park S, Imlay JA. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J Bacteriol 185:1942–1950. doi: 10.1128/jb.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bulut H, Moniot S, Licht A, Scheffel F, Gathmann S, Saenger W, Schneider E. 2012. Crystal structures of two solute receptors for L-cystine and L-cysteine, respectively, of the human pathogen Neisseria gonorrhoeae. J Mol Biol 415:560–572. doi: 10.1016/j.jmb.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 57.Müller A, Thomas GH, Horler R, Brannigan JA, Blagova E, Levdikov VM, Fogg MJ, Wilson KS, Wilkinson AJ. 2005. An ATP-binding cassette-type cysteine transporter in Campylobacter jejuni inferred from the structure of an extracytoplasmic solute receptor protein. Mol Microbiol 57:143–155. doi: 10.1111/j.1365-2958.2005.04691.x. [DOI] [PubMed] [Google Scholar]
- 58.Cassat JE, Hammer ND, Campbell JP, Benson MA, Perrien DS, Mrak LN, Smeltzer MS, Torres VJ, Skaar EP. 2013. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe 13:759–772. doi: 10.1016/j.chom.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burlak C, Hammer CH, Robinson M-A, Whitney AR, McGavin MJ, Kreiswirth BN, Deleo FR. 2007. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol 9:1172–1190. doi: 10.1111/j.1462-5822.2006.00858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kolar SL, Ibarra JA, Rivera FE, Mootz JM, Davenport JE, Stevens SM, Horswill AR, Shaw LN. 2013. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2:18–34. doi: 10.1002/mbo3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wymann MP, von Tscharner V, Deranleau DA, Baggiolini M. 1987. The onset of the respiratory burst in human neutrophils. Real-time studies of H2O2 formation reveal a rapid agonist-induced transduction process. J Biol Chem 262:12048–12053. [PubMed] [Google Scholar]
- 62.Xayarath B, Marquis H, Port GC, Freitag NE. 2009. Listeria monocytogenes CtaP is a multifunctional cysteine transport-associated protein required for bacterial pathogenesis. Mol Microbiol 74:956–973. doi: 10.1111/j.1365-2958.2009.06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, Des Etages S-A, Jones A, Palazzolo-Ballance AM, Perdreau-Remington F, Sensabaugh GF, DeLeo FR, Chambers HF. 2008. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis 197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 64.Schneewind O, Missiakas D. 2014. Genetic manipulation of Staphylococcus aureus. Curr Protoc Microbiol 32:Unit 9C.3. doi: 10.1002/9780471729259.mc09c03s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krute CN, Krausz KL, Markiewicz MA, Joyner JA, Pokhrel S, Hall PR, Bose JL. 2016. Generation of a stable plasmid for in vitro and in vivo studies of staphylococcus species. Appl Environ Microbiol 82:6859–6869. doi: 10.1128/AEM.02370-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pattee PA, Neveln DS. 1975. Transformation analysis of three linkage groups in Staphylococcus aureus. J Bacteriol 124:201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Theis T, Skurray RA, Brown MH. 2007. Identification of suitable internal controls to study expression of a Staphylococcus aureus multidrug resistance system by quantitative real-time PCR. J Microbiol Methods 70:355–362. doi: 10.1016/j.mimet.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 69.Opperman TJ, Williams JD, Houseweart C, Panchal RG, Bavari S, Peet NP, Moir DT, Bowlin TL. 2010. Efflux-mediated bis-indole resistance in Staphylococcus aureus reveals differential substrate specificities for MepA and MepR. Bioorg Med Chem 18:2123–2130. doi: 10.1016/j.bmc.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 70.Sihto H-M, Tasara T, Stephan R, Johler S. 2014. Validation of reference genes for normalization of qPCR mRNA expression levels in Staphylococcus aureus exposed to osmotic and lactic acid stress conditions encountered during food production and preservation. FEMS Microbiol Lett 356:134–140. doi: 10.1111/1574-6968.12491. [DOI] [PubMed] [Google Scholar]
- 71.Sievers F, Higgins DG. 2018. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci 27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rambaut A. 2009. FigTree. http://tree.bio.ed.ac.uk/software/figtree/.
- 74.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. 2017. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Käll L, Krogh A, Sonnhammer E. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 77.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.