The neglected tropical disease Buruli ulcer (BU) is an infection of subcutaneous tissue with Mycobacterium ulcerans. There is no effective vaccine. Here, we assessed an experimental prime-boost vaccine in a low-dose murine tail infection model. We used the enoyl reductase (ER) domain of the M. ulcerans mycolactone polyketide synthases electrostatically coupled with a previously described Toll-like receptor 2 (TLR-2) agonist-based lipopeptide adjuvant, R4Pam2Cys.
KEYWORDS: Buruli ulcer, Mycobacterium ulcerans, machine learning, mycolactone, polyketide synthase, vaccine
ABSTRACT
The neglected tropical disease Buruli ulcer (BU) is an infection of subcutaneous tissue with Mycobacterium ulcerans. There is no effective vaccine. Here, we assessed an experimental prime-boost vaccine in a low-dose murine tail infection model. We used the enoyl reductase (ER) domain of the M. ulcerans mycolactone polyketide synthases electrostatically coupled with a previously described Toll-like receptor 2 (TLR-2) agonist-based lipopeptide adjuvant, R4Pam2Cys. Mice were vaccinated and then challenged via tail inoculation with 14 to 20 CFU of a bioluminescent strain of M. ulcerans. Mice receiving either the experimental ER vaccine or Mycobacterium bovis bacillus Calmette-Guérin (BCG) were equally protected, with both groups faring significantly better than nonvaccinated animals (P < 0.05). To explore potential correlates of protection, a suite of 29 immune parameters were assessed in the mice at the end of the experimental period. Multivariate statistical approaches were used to interrogate the immune response data to develop disease-prognostic models. High levels of interleukin 2 (IL-2) and low gamma interferon (IFN-γ) produced in the spleen best predicted control of infection across all vaccine groups. Univariate logistic regression revealed vaccine-specific profiles of protection. High titers of ER-specific IgG serum antibodies together with IL-2 and IL-4 in the draining lymph node (DLN) were associated with protection induced by the ER vaccine. In contrast, high titers of IL-6, tumor necrosis factor alpha (TNF-α), IFN-γ, and IL-10 in the DLN and low IFN-γ titers in the spleen were associated with protection following BCG vaccination. This study suggests that an effective BU vaccine must induce localized, tissue-specific immune profiles with controlled inflammatory responses at the site of infection.
INTRODUCTION
Buruli ulcer (BU) is a disease primarily of the subcutaneous tissue caused by infection with Mycobacterium ulcerans. BU initially presents as redness of the skin that is often accompanied by edema and swelling. As the disease progresses, edema may increase or an open ulcer might develop (1, 2); the latter is typically characterized by deep undermined edges with a necrotic core composed of bacteria, dead skin cells, and immune cells (3, 4). Ulcers are predominately found on the extremities of the body, such as the upper limbs (27% of cases) and lower limbs (70% of cases) (note that these percentages are for cases from southeastern Australia) (5). The disease is rarely fatal, but if left untreated, extensive destruction of subcutaneous tissue can leave victims with significant deformities and lifelong disabilities (3, 6–9).
BU is likely caused when M. ulcerans is introduced beneath the skin. This can occur if a region of contaminated skin surface is punctured or by insertion of an object contaminated with the bacteria into subcutaneous tissue (e.g., via an insect bite) (10–12). Areas of endemicity for BU include certain regions of West and Sub-Saharan Africa and southeastern Australia (13, 14).
M. ulcerans is a slow-growing bacterium, with a doubling time of greater than 48 h (15, 16), making it difficult for early disease diagnosis, as symptoms can take between 4 and 5 months to appear after primary infection (17, 18). If diagnosed early, however, BU can be treated effectively by combination antibiotic therapy (19–21). Unfortunately, in many cases, the disease can initially be misdiagnosed as other, more common skin infections. Additionally, a large proportion of BU cases in African countries occur in rural villages and poorer areas with limited or no access to health care, with patients facing disfigurement and permanent disability. Given that diagnoses are delayed and usually occur after a lesion has become relatively advanced and ulceration is extensive (22), the development of an effective BU vaccine to protect those in areas of high endemicity is of paramount importance.
Currently, the only licensed vaccine against mycobacterial infections approved for human use is the M. bovis-derived bacillus Calmette-Guérin (BCG) vaccine for the prevention of tuberculosis. This vaccine is cross-protective against M. ulcerans but only delays the onset of disease (23–25). Several experimental vaccines have been tested against M. ulcerans infection, as summarized in Table 1. Although different animal models have been utilized to study M. ulcerans infections, including guinea pigs, primates, pigs, and armadillos (10, 23, 26–31), most studies assessing vaccine efficacy have used mice. Vaccines tested in these murine challenge models have included DNA-based, virus-based, protein subunit, and whole-cell vaccines (25, 32–35) (Table 1). Among the various vaccines, BCG expressing M. ulcerans antigens appears to offer the best protective effect against challenge. Hart et al. (36, 37) showed enhanced protection against BU using a recombinant BCG vaccine that expressed M. ulcerans Ag85A or recombinant Ag85B-EsxH fusion protein in a mouse footpad challenge model. While improving the immunogenicity of BCG may be a promising route, there are also some drawbacks; exposure to environmental mycobacteria is believed to decrease the efficacy of the BCG vaccine, and administration in areas where people have been exposed to BCG may be problematic (38).
TABLE 1.
Summary of putative M. ulcerans vaccines tested in a murine model of BU infectiona
| Vaccine type | Description of components | M. ulcerans challenge dose (strain)b | Challenge model | Efficacy compared to BCG | Reference(s) |
|---|---|---|---|---|---|
| DNA based | pCDNA3 vector encoding Hsp65 | 104 AFB (1615 ATCC 35840) | Tail | Less protective | 52 |
| DNA based | V1Jns.tPA vector encoding Ag85A | 3 × 104 AFB (5150) or 105 AFB (04-855) | Footpad | Less protective | 32, 33 |
| DNA based | Primary vaccination with V1Jns.tPA plasmid encoding mycolactone polyketide domains and boosted with recombinant domain proteins emulsified in Gerbu adjuvant | 105 AFB (1615) | Footpad | Less protective | 49 |
| Viral | Vesicular stomatitis virus replicon particles expressing M. ulcerans codon optimized antigens MUL_2232 and MUL_3720 | 30 μl of 2.8 × 105 CFU/ml stock (8.4 × 103 CFU/dose) (S1013) | Footpad | Less protective | 108 |
| Subunit | MUL2232 and MUL3720 adjuvanted with GLA-SE (EM408) | 1.5 × 106 or 1.5 × 105 CFU (S1013) | Footpad | Less protective | 34 |
| Live cell | M. ulcerans | 106.3 or 104.3 viable bacteria | Footpad | Less protective | 109 |
| Live cell | M. marinum | 105 bacteria (1615) | Footpad | More protectivec | 35 |
| Live cell recombinant | M. marinum expressing Ag85A (on vector) | 105 bacteria (1615) | Footpad | More protectivec | 35 |
| Live cell recombinant | M. bovis BCG expressing Ag85A (on vector pMV261) | 105 bacteria (1615) | Footpad | More protective | 36 |
| Live cell recombinant | M. bovis BCG expressing Ag85B-EsxH fusion protein Ag85A (on vector pMV261) | 105 bacteria (1615) | Footpad | More protective | 37 |
| Inactivated whole cell | Mycolactone-negative M. ulcerans (strain 5114) | 4 log10 or 3 log10 CFU (98-912) | Footpad | Less protective | 25 |
| Inactivated whole cell | Mycolactone-deficient attenuated M. ulcerans (strain ATCC 19423) | 106 bacteria (TMC1615) | Footpad | Not compared | 54 |
| Inactivated whole cell | Formalin-treated M. ulcerans (strain TMC1615) | 106 bacteria (TMC1615) | Footpad | Not compared | 54 |
| Inactivated whole cell | Dewaxed M. ulcerans (strain TMC1615) | 106 bacteria (TMC1615) | Footpad | Not compared | 54 |
| Phage | Mycobacteriophage D29 (therapeutic vaccine) | 5.5 log10 AFB (1615) | Footpad | Not comparable | 53 |
No test of whether the bacterial culture used for challenge was assessed for mycolactone production before infection was mentioned with any of the vaccines.
AFB, acid-fast bacilli.
Vaccine was more protective than the BCG vaccine; however, all mice eventually developed footpad swelling.
M. ulcerans causes disease primarily through the production of a lipid toxin called mycolactone (39). Mycolactone modulates cell function, in particular, the secretion of critical cytokines by specifically inhibiting the Sec61 translocon, enabling M. ulcerans to escape host immune defenses (40–46). The toxin is formed from simple acetate and propionate precursors by three polyketide synthases (PKS) encoded by genes on the plasmid pMUM001 (47, 48). Within each PKS are enzymatic domains that form the mycolactone molecule. Some of these domains have been found to be immunogenic, and, in particular, immune responses against the enoyl reductase (ER) domain have previously been shown to significantly reduce M. ulcerans bacterial load in the footpad of a murine prime-boost vaccination study (49). Based on the immunogenic and protective qualities of ER, we have utilized it as a target antigen for a BU subunit vaccine.
Protein antigens generally require an adjuvant to boost immunogenicity and shape immune responses. A known Toll-like receptor 2 (TLR-2) ligand, R4Pam2Cys, has been previously shown to induce robust antibody responses, as well as augmented CD4+ and CD8+ T cell responses, possibly through promoting dendritic cell antigen uptake and trafficking to lymph nodes (50, 51). Given that BU is a disease where the bacteria can be both extracellular and intracellular, the ability of R4Pam2Cys to robustly engage multiple arms of the adaptive immune system may be beneficial for a BU subunit vaccine.
Previous research has shown that as few as three CFU of bacteria are required to initiate infection (10); however, most animal models challenge with >104 CFU (see Table 1) (25, 32–37, 49, 52–54). This dose is likely to be far higher than the dose of bacteria that leads to natural infection in humans and other animals (10). Such an unrealistic high dose may overwhelm immune responses and underestimate the true efficacy of potential M. ulcerans vaccines. For these reasons, we have used a low dose of M. ulcerans in a tail infection challenge model to evaluate an experimental prime-boost subunit vaccine against BU. The experimental subunit vaccine developed here comprised the M. ulcerans mycolactone ER domain protein formulated with the adjuvant R4Pam2Cys. Our murine challenge model with physiologically relevant dosing enabled us to more accurately measure vaccine-induced protection and to explore immune correlates of protection against BU.
RESULTS
Formulation of the ER-R4Pam2Cys subunit vaccine candidate.
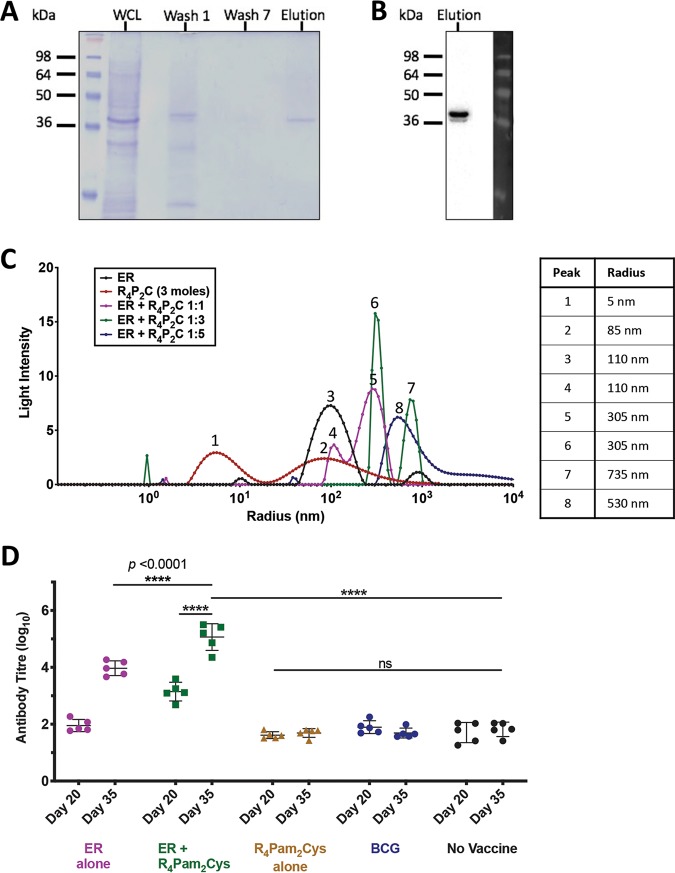
Mycolactone is the key virulence factor produced by M. ulcerans and an attractive vaccine target, but the molecule is poorly immunogenic (55). However, the PKS enzymes used by the bacterium to synthesize mycolactones are highly conserved and immunogenic (49, 56–58). Therefore, we hypothesized that targeting the conserved enzymatic domains of the mycolactone PKS could be an effective vaccine strategy. One domain in particular, the enoyl reductase (ER) protein domain, elicits serum antibodies in BU patients and healthy controls in regions endemic for BU (57). The ER protein expressed as an antigen in a DNA-protein prime-boost vaccine has also been shown to reduce bacterial burden in a mouse footpad M. ulcerans challenge model (49). Here, we utilized the ER protein to create a novel BU vaccine candidate by electrostatically associating it with the TLR-2 agonist-based lipopeptide R4Pam2Cys. To formulate this vaccine, recombinant 6×His-tagged ER protein (37 kDa) was first produced and confirmed by SDS-PAGE (Fig. 1A) and Western blotting (Fig. 1B). This protein antigen was formulated with R4Pam2Cys at various ratios to first optimize the formation of protein-lipopeptide complexes. A ratio of 1:1 was sufficient to produce complexes that were larger than each constituent on its own (Fig. 1C). While the majority of these complexes were ∼300 nm in radius (peak 5), the presence of smaller particles of ∼100 nm (peak 4) suggests that not all antigen was incorporated within a complex, as this size range corresponds to the size of ER protein (peak 3) or R4Pam2Cys alone (peak 2). Although a 1:3 ratio appears to be more effective for the association of these constituents, the size distribution of form complexes was not monodispersed and appeared as two distinct populations (peaks 6 and 7). A 1:5 ratio, however, produced a uniform population of complexes that were ∼500 nm in radius (peak 8), and this formulation was subsequently used to vaccinate animals.
FIG 1.
Analysis of purified recombinant ER protein antigen characteristics and formulation with R4Pam2Cys. (A) The presence of recombinant ER protein (∼37 kDa) was monitored by SDS-PAGE at each stage of the purification process. Lane 1, whole-cell lysate (WCL); lane 2, wash 1; lane 3, wash 7; lane 4, ER protein elution (containing 10 μg protein). (B) Western blot using an anti-6×His tag antibody to detect the presence of a single band corresponding to the correct molecular weight of the ER protein in the final eluate. (C) To analyze the formation of antigen-lipopeptide complexes, a constant amount of antigen was mixed with lipopeptide at different protein/lipopeptide molecular ratios in 50 μl of PBS. The size distribution of particles was then analyzed by DLS, with each profile depicting the hydrodynamic radius (in nanometers) of the complexes in each solution. The average radius of each formulation is highlighted in the accompanying table. (D) BALB/c mice (n = 5/group) were vaccinated on days 0 and 21 with R4Pam2Cys alone, ER antigen alone, or antigen formulated with R4Pam2Cys or vaccinated with BCG on day 0 only. Total serum (IgG) antibody against recombinant ER protein was measured by ELISA after the primary dose (day 20) and 2 weeks after the secondary dose (day 35). Statistical tests were conducted at the 5% significance level. The null hypothesis was rejected if there was a significant difference in mean antibody responses between treatment groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant. The error bars represent the standard deviation (n = 5).
Evaluation of ER-specific antibody responses in vaccinated mice.
To evaluate the ability of the vaccine to induce ER-specific antibody responses, mice were vaccinated and serum samples obtained after priming and boosting with the subunit vaccine. Our results showed that primary vaccination with ER plus R4Pam2Cys resulted in significantly higher levels of ER-specific antibody than vaccination with unadjuvanted ER antigen (P < 0.0001) (Fig. 1D). In fact, there was no significant difference in responses between unvaccinated mice and those vaccinated with a single dose of ER alone, R4Pam2Cys only or BCG. Although a second dose of ER alone was able to increase these responses, the titers were still ∼100-fold less than levels achieved by boosting mice with ER plus R4Pam2Cys (P < 0.0001). These results not only indicate that ER-specific antibodies can be generated in mice and that the use of R4Pam2Cys can significantly enhance these responses, they also indicate that BCG does not induce cross-reactive antibodies to the ER protein.
Characterization of a low-dose M. ulcerans murine tail infection model.
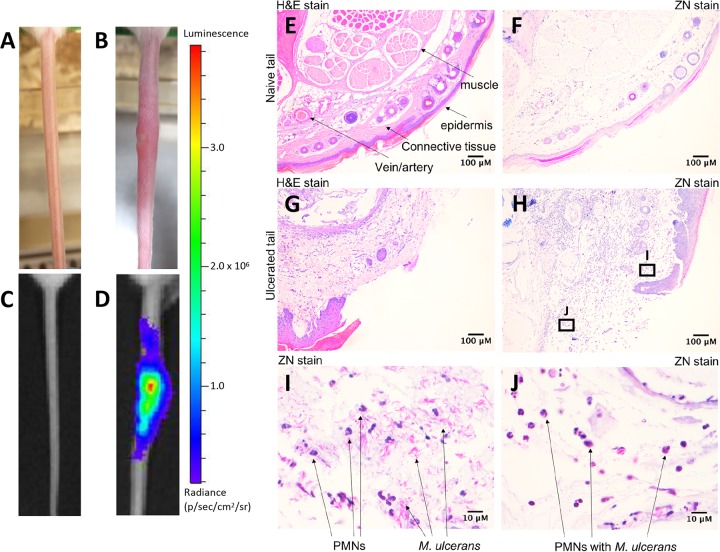
We have previously described the use of a low-dose tail infection model for studying insect-mediated transmission of M. ulcerans (10). We reasoned that because BU patients were likely to be initially infected with a low bacterial inoculum (10–12), we could use this model to test the protective efficacy of responses induced by the ER plus R4Pam2Cys vaccine. This model features the use of a bioluminescent strain of M. ulcerans (10, 59), and its infectious characteristics are summarized in Fig. 2. Compared to an uninfected tail (Fig. 2A and C), subcutaneous infection of a tail results in the appearance of a visible ulcer (Fig. 2B) exhibiting the highest levels of bioluminescence concentrated around the center of the lesion (Fig. 2D), i.e., where swelling appears to be the greatest, and beginning to diminish around the periphery, reflecting a positive correlation between bacterial burden and light emission (60). Histological cross-sections revealed that while the tissue integrity of an uninfected tail appears as defined and intact (Fig. 2E and F), dramatic differences are observed in the infected tail, typified by loss of muscle, vasculature, and epidermis structure and disruptions to surrounding connective tissue (Fig. 2G and H). Further examination of this tissue showed the presence of acid-fast bacilli as well as an infiltration of polymorphonuclear cells (PMNs) (Fig. 2I). Despite evidence of bacterial engulfment by these cells (Fig. 2J), it would appear that this response was insufficient for controlling the infection and preventing disease progression.
FIG 2.
Characterization of infection using a low-dose bioluminescent M. ulcerans strain. (A and B) Representative light camera images of tails from an uninfected BALB/c mouse (A) or at the point of ulceration (16 weeks) following intradermal inoculation with 20 CFU of bioluminescent M. ulcerans (B). (C and D) The same tails were visualized under an IVIS camera to detect and quantify bioluminescence intensity (as photons [p]/s). (E to H) Histological cross-section of an uninfected (E and F) or infected tail tissue (G and H) following hematoxylin & eosin (H&E) and Ziehl-Neelsen (ZN) staining. (I and J) Zoomed images of the regions indicated within the denoted boxes of panel H depict the presence of polymorphonuclear cells (PMNs) and acid-fast bacilli (ZN staining) within tissue.
Monitoring vaccine efficacy using bioluminescent M. ulcerans and in vivo imaging system.
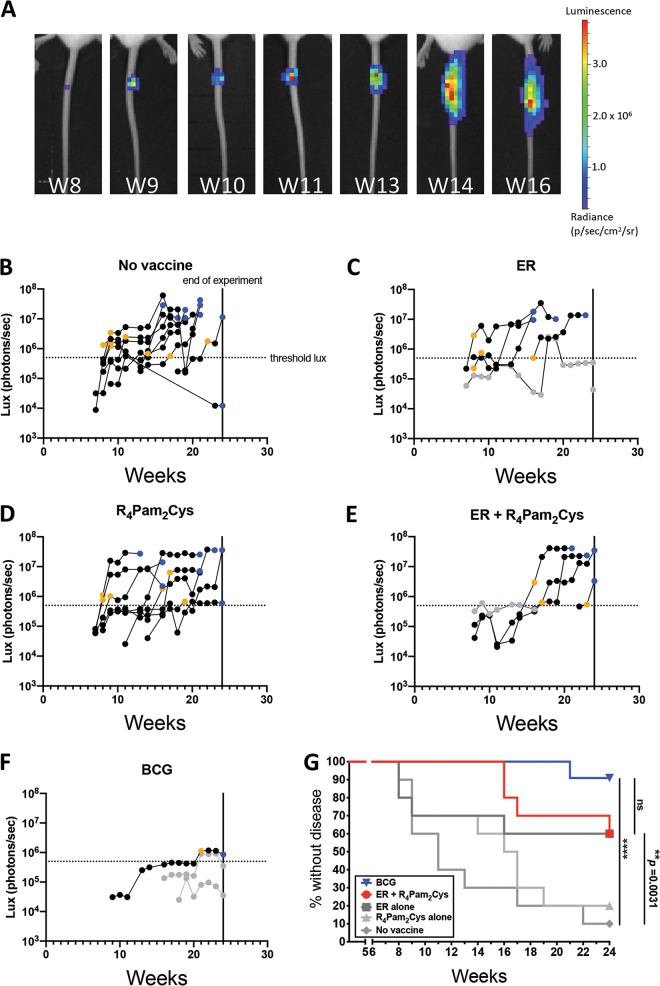
We have previously established a correlation between M. ulcerans bioluminescence and bacterial burden (60). Here, we set a threshold of ≥5 × 105 photons/s (equivalent to 2.8 × 105 CFU) as a measure of ineffective disease control, i.e., appearance of disease. Mice were vaccinated subcutaneously and then challenged with an estimated 17 CFU (range, 14 to 20 CFU) of bioluminescent M. ulcerans via intradermal tail inoculation. Animals were monitored weekly for changes in bioluminescence using IVIS for up to 24 weeks. Figure 3A shows an example of the progression of bioluminescence (and therefore disease) in an unvaccinated mouse, up to week 16, whereupon the clinical endpoint of the experiment was reached. Bioluminescence for all mice was recorded across the experimental period. Plots for the different treatment groups show the progression in bioluminescence over time (Fig. 3B to F). Mice from the ER-vaccinated, R4Pam2Cys-vaccinated, and unvaccinated treatment groups displayed the first detectable bioluminescence at week 7. There also appeared to be a threshold in bioluminescence, whereby animals expressing ≥5 × 105 photons/s from tail lesions became less able to control the infection and progressed to the clinical endpoint (Fig. 3B). The immune response data for all mice are provided in Table S1 in the supplemental material. Using these data, failure to protect was defined as tail bioluminescence equal to or greater than 5 × 105 photons/s at or before week 24 (end of the experiment). Therefore, mice were defined as “protected” if bioluminescence was less than 5 × 105 photons/s at week 24.
FIG 3.
Development of BU over time after vaccination. (A) Tails of mice were intradermally infected with 20 CFU of bioluminescent M. ulcerans and imaged weekly by IVIS. Representative panels depict the weekly progression of bioluminescent M. ulcerans burden in the tail of an unvaccinated mouse over the course of 16 weeks expressed as photons/s. (B to F) BALB/c mice (n = 10/group) were left unvaccinated (B) or vaccinated on days 0 and 21 with ER antigen alone (C), R4Pam2Cys alone (D), ER antigen formulated with R4Pam2Cys (E), or on day 0 with BCG (F), followed by challenge on week 5 with bioluminescent M. ulcerans. Threshold bioluminescence (threshold lux) for disease was defined as ≥5 × 105 photons/s, as mice that reached this level typically progressed to the clinical (ethical) endpoint. Mice were classified as diseased if they reached this endpoint within the 24 weeks (vertical lines) following challenge or if their bioluminescence value at week 24 was ≥5 × 105 p/s. Mice were classified as protected if they did not reach this clinical endpoint and their bioluminescence value was <5 × 105 photons/s. The data point depicting when an infected mouse first exhibited bioluminescence at ≥5 × 105 lb/in2 is represented with a yellow symbol. The data point denoting when a mouse reached clinical endpoint is represented with a blue symbol. Protected mice with detectable bioluminescence are depicted as gray symbols. (G) Time to bioluminescence measured by IVIS. A survival curve was utilized to analyze the time (in weeks) taken for each BU diseased mouse to first reach threshold bioluminescence of ≥5 × 105 photons/s. The BCG group (upside-down triangle) is labeled in blue, ER plus R4Pam2Cys (circle) is red, and ER alone (square), R4Pam2Cys alone (triangle), and no-vaccine (diamond) groups are depicted in gray. Statistical tests were conducted at the 5% significance level. The null hypothesis was rejected if there was a significant difference in survival between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
Vaccination with ER plus R4Pam2Cys offers similar protection to M. bovis BCG vaccine.
Survival analysis was conducted to assess vaccine efficacy by measuring the time from infection until tail bioluminescence at the threshold of 5 × 105 photons/s was reached. Mice that reached this threshold were defined as “not protected.” Significantly fewer mice vaccinated with ER plus R4Pam2Cys (4/10 animals) developed disease than did unvaccinated mice (9/10 animals), indicating that ER plus R4Pam2Cys provided some level of protection against disease progression compared to no vaccination (Fig. 3B, E, and G) (P < 0.01). Mice vaccinated with BCG were best protected, with only 1 animal exceeding the bioluminescence threshold (Fig. 3F and G). Although this number of mice was reduced compared to the animals vaccinated with ER plus R4Pam2Cys, the difference was not significant (Fig. 3G). However, the bacterial burden (as indicated by mean photon counts/s at the clinical endpoint) in BCG-vaccinated mice was lower than that in animals that received the ER plus R4Pam2Cys vaccine (mean, 6 × 105 [n = 2; range, 3.5 × 105 to 8.6 × 105] versus 3.3 × 107 [n = 3; range, 2.4 × 107 to 4.1 × 107] photons/s, respectively), consistent with the protective superiority of BCG. There were also no significant differences between the protective efficacy of vaccination with ER alone and that with ER plus R4Pam2Cys, although mice vaccinated with ER plus R4Pam2Cys exhibited delayed disease progression (onset at weeks 16 to 24) compared to that with ER-vaccinated mice (onset at weeks 8 to 16), indicating that formulation of the antigen with the R4Pam2Cys adjuvant improved immunity (Fig. 3D and E).
Measuring immune parameters following vaccination and challenge.
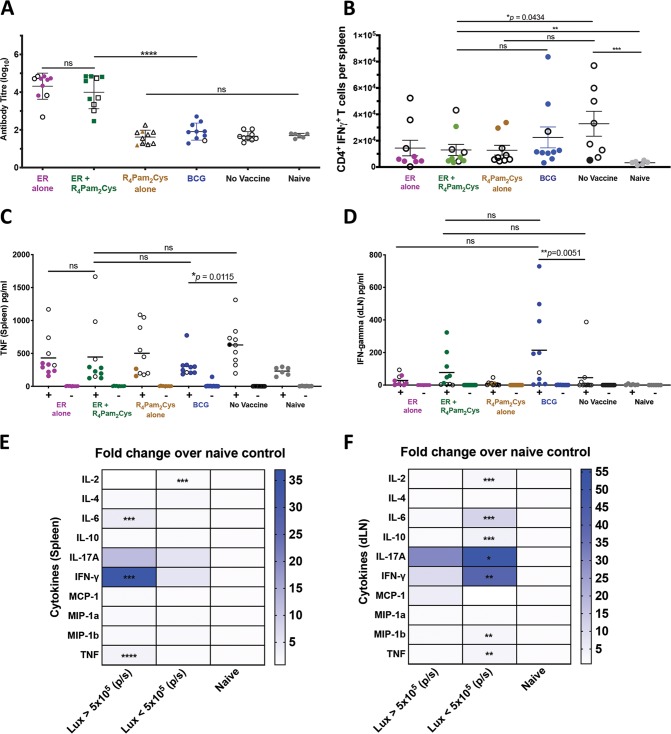
At the experimental endpoint, serum samples, spleens, and draining lymph nodes (DLN) from all animals were collected, and several parameters were further analyzed, as follows: ER-specific antibodies, CD4+ and CD8+ T cells, and a panel of 10 murine Th1, Th2, and Th17 cytokines. After M. ulcerans challenge, mice vaccinated with ER alone or ER plus R4Pam2Cys were found to exhibit significantly more ER-specific antibodies than did the other treatment groups (Fig. 4A) despite not being fully protected. This indicates that even though the ER protein is highly immunogenic, anti-ER antibodies do not appear to play a major role in controlling infection.
FIG 4.
Immune responses after M. ulcerans infection. BALB/c mice (n = 10/group) were left unvaccinated or vaccinated on day 0 and day 21 with ER antigen alone, ER antigen formulated with R4Pam2Cys, R4Pam2Cys alone, or on day 0 with BCG followed by challenge on day 36 with bioluminescent M. ulcerans. (A) Total serum (IgG) antibody against recombinant ER protein was measured by ELISA after the experimental endpoint. All data points for diseased mice (bioluminescence ≥5 × 105 p/s) are depicted with white symbols. Statistical tests were conducted at the 5% significance level. The null hypothesis was rejected if there was a significant difference in mean antibody responses between treatment groups. The error bars represent the standard deviation. (B) After experimental endpoint was reached, CD4+ IFN-γ+ T cells were enumerated from the spleen of mice in response to ER protein. The null hypothesis was rejected if there was a significant difference in mean CD4+ IFN-γ+ T cells between treatment groups. Once the experimental endpoint was reached, cytokines from draining lymph nodes and spleens of M. ulcerans challenged mice were also measured in response to in vitro cell stimulation with recombinant ER protein (Table S1). (C and D) Shown here are TNF produced from immune cells in the spleen (C) and cytokine titers of IFN-γ produced from immune cells in the draining lymph nodes (D). The null hypothesis was rejected if there was no difference in mean cytokine titers between treatment groups. The black bars represent the mean. (E and F) The fold changes of mean cytokine titers from protected mice (bioluminescence, <5 × 105 p/s) and diseased mice (bioluminescence, ≥5 × 105 p/s) over naive mice were compared in the spleen (E) and draining lymph nodes (F). The null hypothesis was rejected if there was a significant difference in mean cytokine titers between treatment groups. All statistical tests were conducted at the 5% significance level. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
We next investigated if there were any differences in the ability of T cells harvested from the spleens of vaccinated mice to produce cytokines following stimulation with recombinant ER protein (Table S1). Our results showed that the numbers of gamma interferon (IFN-γ)-producing CD4+ T cells across all vaccine groups did not differ and in some cases were higher in unvaccinated mice than in those vaccinated with ER plus R4Pam2Cys (Fig. 4B), indicating that there was no clear correlation between the frequencies of these cells and protection. Similarly, there also did not appear to be any correlation between tumor necrosis factor alpha (TNF-α)-positive CD4+ T cells, IFN-γ+ CD8+ T cells, or TNF-α+ CD8+ T cells and protection (Table S1).
A comparison of levels of cytokine production between vaccine groups also did not clearly identify any cytokines that correlated with protection (see Table S1). For example, TNF-α was present in the spleens of unvaccinated mice at higher levels than in BCG-vaccinated mice (Fig. 4C), and even though BCG vaccination resulted in significantly more IFN-γ in draining lymph nodes than that in unvaccinated mice, this was not observed for mice vaccinated with ER plus R4Pam2Cys (Fig. 4D).
We therefore based our analysis on comparisons between diseased or protected mice irrespective of the vaccines they received (Fig. 4E and F). Herein, we identified significant increases in interleukin 2 (IL-2), IL-6, IL-10 IL-17A, IFN-γ, macrophage inflammatory protein 1b (MIP-1b), and TNF-α in the lymph nodes of protected mice (Fig. 4F) and significant increases of IFN-γ, IL-6, and TNF-α in the spleens of diseased mice (Fig. 4E). While these data implicate these cytokines as correlates of protection and disease, they did not rank the importance of each cytokine toward either outcome.
Identifying immune responses associated with vaccination outcome.
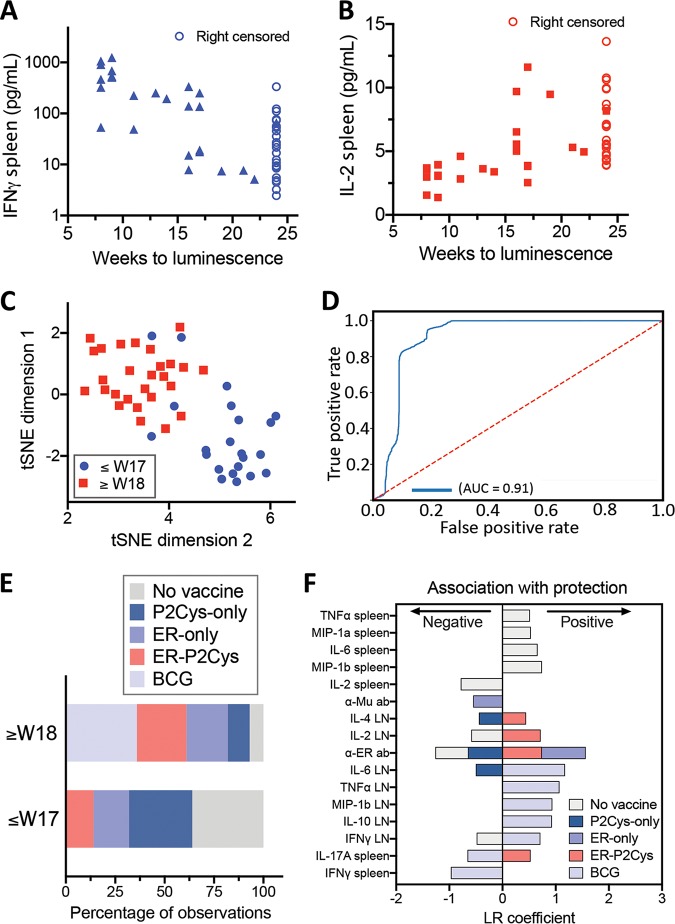
To identify the immune parameters (features) that associate with the response variable “vaccine protection” (here measured as the time to reach our bioluminescence detection threshold) independent of the vaccine used, we conducted univariate survival analyses via the Cox proportional hazards model. We used this model, as it accounts for the fact that a subset of mice (observations) were right censored, as vaccination outcomes were not measured after 24 weeks postchallenge. For each of the 28 immunological features, their association with the response variable (time to bioluminescence measured in weeks) was ranked using concordance index (CI) scores. The CI is analogous to the area under the receiver operating characteristic (ROC) curve, with a CI value of 0.5 indicating a random correlation and 1 indicating a perfect, positive correlation (61). The CI for each univariate regression analysis was used to rank the strength of association for each of the 28 features against the response variable. Using a CI cutoff of 0.70, the top six features were identified, as well as the direction of their association with the prevention of development of bioluminescence (Table 2). Low levels of IFN-γ and high levels of IL-2 produced in mouse splenocytes were the top two immune parameter features influencing this model, as reflected in the individual correlations between their respective titers and time to bioluminescence (Fig. 5A and B).
TABLE 2.
High-scoring immune features associated with delayed bioluminescence
| Feature no. | Feature | Tissue site | Association with delayed bioluminescence | Concordance index |
|---|---|---|---|---|
| 1 | IFN-γ | Spleen | Negative | 0.786 |
| 2 | IL-2 | Spleen | Positive | 0.769 |
| 3 | IL-6 | Spleen | Negative | 0.759 |
| 4 | TNF-α | Spleen | Negative | 0.745 |
| 5 | IL-2 | Lymph node | Positive | 0.715 |
| 6 | IFN-γ CD4+ T-cell | Spleen | Negative | 0.707 |
FIG 5.
Statistical modeling to identify immune parameters (features) associated with vaccine protection. Univariate Cox proportional hazards models were specified for each of the 28 immunological features to test their association with the response variable (time to bioluminescence measured in weeks). The resulting concordance index (CI) scores were obtained, and the six features with a CI of >0.7 were retained. (A and B) The inverse associations of the top two features IFN-γ (A) and IL-2 (B) produced in murine splenocytes at the experimental endpoint are shown. (C) Plot depicting a two-dimensional representation of the top six features that associate with time to bioluminescence from the unsupervised t-SNE. The shapes/colors indicate the two groups identified through K-means clustering of bioluminescence by 8 to 17 weeks or at 18 weeks and beyond (up to 24 weeks). (D) Receiver operating characteristic curve (and corresponding area under the curve) displaying the trade-off between sensitivity and specificity across all thresholds for 1,000 random train-test splits of a logistic regression classifier (90% of observations used for training). The red dotted line depicts the expectation of a random classifier, and the blue line depicts the model performance. (E) Proportion of observations (mice) across treatment groups for each of the classes, both 8 to 17 weeks and 18 to 24 weeks, and those with no detection. (F) Group-specific univariate logistic regression analyses for each of the five treatment groups. Model coefficients were used to determine both the strength and direction of association of each feature with that of each treatment group. Depicted are those features with a corresponding P value of <0.05 (Table S2).
Next, using these top six features, we performed unsupervised machine learning to reveal any structure without the influence of labels, such as arbitrarily imposed bioluminescence thresholds. Here, the data separated into two main clusters. K-means clustering was then applied to assign mice (observations) to the two cluster groups. Inspection of the resulting cluster membership with respect to time to bioluminescence showed a separation of the mice either side of 17 weeks post-M. ulcerans challenge (Fig. 5C).
Given that the clusters identified through unsupervised learning closely resembled a temporal breakpoint at 17 weeks, we further investigated this binary divide in the data. We used multivariate logistic regression and developed a low-error classifier that could generalize to unseen data, using the underlying structure apparent in the immunological data. We then tested the classifier through extensive cross-validation. Observations (mice) with bioluminescence above threshold between weeks 8 to 17 were assigned a “0,” and for those with bioluminescence detected in weeks 18 to 24 or no detection throughout the experiment period, a “1” was assigned. The model included the top six features (Table 2) and was validated through 1,000 random train test splits, with 90% of observations comprising the training groups at each split. The resulting classifier probabilities were used to calculate the area under the ROC curve (AUC), which was 0.91 (number of true negatives, 1,774; number of true positives, 2,662; number of false negatives, 120; number of false positives, 444). The low error and generalizable nature of this classifier demonstrate the existence of a robust structure in the data, in the form of two clusters separated around week 17 (Fig. 5D), and highlight the strong association of the six identified immune parameters with outcome. Most notably, here, it appears that tissue specific immune responses are important, with a correlation between the appearance of a tail ulcer and evidence of a systemic (spleen) response and protection correlating with both local (draining lymph nodes) and systemic (spleen) responses (Table 2). This association of BU and the production of inflammatory cytokines (IL-6, IFN-γ, and TNF-α) as possible markers of infection indicated that we could also identify correlates of immune protection against BU.
Assessment of vaccine-specific immune responses.
To dissect the immune responses associated with ER plus R4Pam2Cys vaccination versus BCG vaccination and the different controls, we noted the differences in the percentage of observations (mice) belonging to specific treatment groups between the two clusters described above (Fig. 5C) separated by week 17. This summary was reflective of the survival analysis and showed that unvaccinated mice and those that received the adjuvant or ER alone were predominantly in the cluster of weeks 8 to 17, while ER plus R4Pam2Cys- and BCG-vaccinated mice were predominantly in the cluster of weeks 18 to 24 (Fig. 5E). In order to obtain the individual immune profiles of the different treatment groups, group-specific univariate logistic regression analyses were undertaken. Here, the response variable was coded as a “1” for membership in a particular group and “0” for membership in all other groups. Analyses were conducted for each of the five treatment groups, and the model coefficient weights were used to determine both the strength and direction of association of each feature with that of each treatment group. The resulting P values from the model coefficients were used to assess significance of the associations (Fig. 5F and Table S2). The combination of the ER antigen and R4Pam2Cys was important for inducing protective responses that associated with local production of IL-4, IL-2, and IL-17A in the DLN, in addition to ER-specific antibody responses. Neither ER antigen nor R4Pam2Cys alone induced this profile, and protection using the former was only linked to ER- but not M. ulcerans-specific antibody responses (postchallenge time points). In comparison, BCG vaccination-mediated protection was associated with a greater breadth of localized cytokine responses than vaccination with ER plus R4Pam2Cys, with higher IL-6, TNF-α, and MIP-1β levels in the DLN. Of note, evidence of systemic inflammation, such as IL-17A and IFN-γ in the spleen, was associated with poorer BCG performance.
DISCUSSION
In this study, we investigated various immune parameters induced by the use of BCG and an experimental subunit vaccine against BU and sought to identify immune correlates associated with protection in a mouse tail infection model. Studies have shown that the tail is a suitable location for infection, as BU predominantly affects extremities (11, 29, 52) and in combination with the use of bioluminescent M. ulcerans offers several advantages over footpad, hock, or ear infections used in most BU vaccine studies (as shown in Table 1 and reference 30). Tail infections are less likely to affect mouse mobility or cause rapid tissue loss, and they may also prevent added trauma, inflammation, or secondary infections at the challenge site (62). A key feature of this model also allows the use of a significantly lower bacterial challenge dose than that in other studies (approximately 14 to 20 CFU compared to 104 to 106 CFU) (25, 32–37, 49, 52–54) (Table 1) and enables the measurement of bacterial growth in the same animal over time. This lower dose is likely to be more physiologically relevant in terms of reflecting the bacterial inoculum that occurs during M. ulcerans transmission to humans (10–12). Sporadic healing of BU disease was also seen in this model, an observation that has been noted in humans and other animals (29, 63–65).
Vaccination with the ER plus R4Pam2Cys formulation resulted in protection of 60% of mice from BU challenge. The fact that vaccination with ER plus R4Pam2Cys was significantly more protective than was vaccination with R4Pam2Cys alone indicates that any non-antigen-specific triggering of innate immune responses by this adjuvant (66–68) was not sufficient for conferring long-lasting protection and that the inclusion of the ER protein was necessary to achieve any protective effects. This was further evidenced by vaccinating with ER alone. Despite not seeing any clear significant differences in the clinical outcomes at the end of the trial compared to ER plus R4Pam2Cys vaccination, the inclusion of R4Pam2Cys delayed disease onset by ∼8 weeks and correlated with the induction of significantly more ER-specific antibodies after a primary and booster vaccination (Fig. 3).
ER-specific antibodies were nonetheless insufficient to provide total protection against M. ulcerans challenge. This is perhaps not unexpected given that other studies have also reported little correlation between strong BU antibodies and protection in mice (34) or sera of BU patients (57, 69). To identify other immune correlates associated with the protective effects observed in our study, a Cox proportional hazards regression model was utilized. Here, the univariate analyses allowed us to identify the top six features that most strongly associated with differences in time to detection of bioluminescence. These models assume normalized data, and although we cannot infer how much an increase or decrease in units could affect the clinical outcome, we are able to rank each factor based on its contribution to either disease or protection. This model also considers the effect of each factor in delaying the onset of disease rather than just modeling through a binary “protected” or “diseased” outcome. Similar regression modeling has been described to predict outcomes for tuberculosis patients after treatment, effects of hospital-acquired Clostridium difficile on hospital stay, and survival of Staphylococcus aureus in milk (70–72). The model assumes that eventually all mice will succumb to disease, and due to the constraints in our data, it cannot determine the threshold levels of cytokines that will predict disease outcomes.
The cytokines most associated with protection in BCG mice were different from those identified in mice vaccinated with ER plus R4Pam2Cys, which is not surprising given that BCG is a multiantigen live-attenuated vaccine and thus likely to utilize protective responses and mechanisms both common to and distinct from those induced by our vaccine candidate. Through multivariate logistic regression modeling, we identified the presence of IL-2 in the spleen and lymph nodes as markers that were most strongly associated with protection. Although there are no studies that directly link IL-2 with protection against M. ulcerans, it plays a key role in the differentiation, proliferation, and maintenance of T cell responses (73). Therefore, it is perhaps not surprising that its role in this murine model is likely to be important for the induction of protective adaptive immunity against M. ulcerans.
M. ulcerans-specific CD4+ T cells migrate to the site of infection from draining lymph nodes early in infection but are depleted as the infection persists (74), an effect attributed to the ability of the M. ulcerans exotoxin mycolactone to impair T cell and macrophage function (41, 44, 75). However, we observed no correlation between levels of cytokine-producing CD4+ and CD8+ T cells and protection in vaccinated groups.
We did identify systemic IL-6, TNF-α, and IFN-γ (in the spleen) strongly associated with disease. IL-6 is a proinflammatory cytokine produced by many cell types in response to pathogens and is linked to the production of TNF-α, both of which can be detected in BU lesions and serum of BU patients (75–77). TNF-α in particular plays a key role in inflammatory cell recruitment, and in conjunction with IFN-γ, it increases the phagocytic ability of macrophages to enhance killing of mycobacteria (78, 79). However, many studies have shown that mycolactone suppresses TNF-α production by T cells and especially macrophages (45, 80), decreasing their ability to control BU infection (81).
IFN-γ is important for controlling M. ulcerans infection (82) and is also detected at high levels in patients with both developed ulcers and early lesions (83) and healed ulcers (84), where it is believed to mediate macrophage function (46) and drive inducible nitric oxide synthase (iNOS) expression to facilitate bacterial killing (85). The fact that these cytokines are elevated at a systemic level in the diseased animals in our study but not in lymph nodes draining from the tail suggests that their activity is being dampened at the site of infection. These effects do not appear to be present in protected animals where increased levels of cytokines are detected in the draining lymph node and not the spleen. The localized, but not systemic, presence of these and other cytokines, including IL-17A, MIP-1b, and IL-10, is strongly associated with protection.
Although most of the immunosuppression during BU infection can be attributed to mycolactone, chronic inflammation can also be key driver, as noted by the increased splenic cytokine levels. Many cell types have been implicated as the cause of immunosuppression in cancers, chronic viral infections (such as hepatitis C virus [HCV], human immunodeficiency virus [HIV], and hepatitis B virus [HBV]), and even Mycobacterium tuberculosis infection. These include myeloid-derived suppressor cells (MDSCs) (86), regulatory T (Treg) cells (87), and T helper 17 (Th17) cells (88), which can suppress effector T cell function and inhibit natural killer (NK) and dendritic cell activity through direct cell-to-cell interactions or the production of immunosuppressive cytokines. MDSCs in particular can be recruited by IFN-γ (89) and IL-6 (90), both of which are found in higher levels in our unprotected mice. On the other hand, while Th17 cells are crucial for the control of infection, especially extracellular bacterial and fungal infections, elevated frequencies can lead to tissue inflammation alongside matrix destruction, autoimmunity, and vascular activation (91). The observation that higher systemic IL-17A correlates with the lack of protection suggests that these cells play a role in determining BU disease outcomes.
Tissue changes due to chronic infection could also have a compounding effect on the severity of BU disease outcomes. Our histological analysis of BU-infected tail tissue showed a loss of muscle and epidermis, changes in connective tissue, and loss of vasculature, which may explain why lymphocytes and other immune cells are unable to access the sites of greatest infection and tissue damage. BU tissue necrosis can also extend some distance from the site of bacterial colonization, an observation that led to the identification of mycolactone as the cause of coagulative necrosis (39, 92, 93). Mycolactone has been well described as causing cell death to skin-resident cells such as fibroblasts, adipocytes, keratinocytes, and endothelial cells (39, 94, 95). Primary human dermal microvascular endothelial cells are especially sensitive to mycolactone and after exposure lose their ability to activate a key anticoagulant protein (protein C), causing a reduction in intravascular fluidity and preventing immune cell infiltration to the infection site (94). Thus, the combination of immunosuppressive immune host responses and tissue destruction, in conjunction with mycolactone at the site of infection, may increase the risk for poorer disease outcomes for those chronically infected with BU.
Although we have identified several factors associated with disease and protection, our results provide impetus to further expand these profiles and establish their importance. For example, changes in cytokine levels before challenge and throughout the infection phase could be monitored and integrated into models, the frequencies of various other innate- and adaptive-immune cell populations could be analyzed, and populations that produce cytokines of interest could be identified. In evaluating and demonstrating that a subunit vaccine can protect against BU in our mouse challenge model, albeit not as efficacious as BCG, our results showed that protection can be mediated through different immune mechanisms. Disease progression was also commonly linked to the presence of proinflammatory cytokines in the spleen and not the lymph node. These profiles suggest that localized and not systemic responses are more important for conferring protection and also provide a template that could guide the design and development of novel vaccination strategies against BU.
Finally, we conclude that the mycolactone biosynthesis pathway constitutes a viable vaccine target to protect against M. ulcerans. As M. ulcerans is slow growing and requires its highly conserved mycolactone PKS for virulence, the development of resistance is unlikely. As such, approaches based on the use of multiple PKS enzymatic domains may prove even more efficacious. Moreover, studies that have introduced M. ulcerans- and Mycobacterium marinum-specific proteins into BCG have been shown to increase its protective effect, while BCG alone has no long-term efficacy (24, 32, 36, 37). Collectively, this demonstrates the additive power of using a broader suite of antigens and the potential for a viable vaccine against BU.
MATERIALS AND METHODS
Strains and culture conditions.
Escherichia coli ClearColi BL21(DE3) containing the plasmid pJexpress-ER (strain TPS847) was grown at 37°C in Luria-Bertani (LB) broth (Difco; Becton, Dickinson, MD, USA) supplemented with 100 μg/ml ampicillin (Sigma-Aldrich, USA) to express the enoyl reductase (ER) protein (57). Log-phase bioluminescent Mycobacterium ulcerans (strain JKD8049 containing integrated plasmid pMV306 hsp::luxG13) (10, 59) was grown at 30°C in 7H9 broth or 7H10 agar (Middlebrook; Becton, Dickinson) supplemented with oleic acid-albumin-dextrose-catalase (OADC) growth supplement (Middlebrook; Becton, Dickinson), 0.5% glycerol (vol/vol), and 25 μg/ml kanamycin (Sigma-Aldrich). M. bovis BCG (strain Danish 1331) used for vaccinations was grown at 37°C in 7H9 broth or 7H10 agar supplemented with OADC. Mycobacterial colony counts from cultures or tissue specimens were performed using spot plating, as previously described, with plates read at 10 weeks and then rechecked at 18 weeks (10). All culture extracts were screened by liquid chromatography-mass spectrometry (LC-MS) for the presence of mycolactones, as previously described, to ensure that the bacteria used in the challenge experiments remained fully virulent (96).
Recombinant protein expression.
Overnight culture of E. coli TPS847 was diluted to an optical density at 600 nm (OD600) of 0.05 in LB broth. The culture was incubated at 37°C with shaking at 200 rpm until an OD600 of 0.6 to 0.7 was reached, followed by the addition of 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) to induce protein expression for a further 4 h. Cells were then resuspended in wash buffer (8 M urea, 150 mM sodium chloride, 10% glycerol) and sonicated at amplitude 60 (QSonica ultrasonic liquid processor S-4000; Misonix) until the solution turned clear. The lysate was filtered with a 0.22-μm filter (Millipore) to remove cellular debris, and protein was column purified using anti-histidine resin (Clontech). The resin was washed with wash buffer, which was gradually replaced with Tris buffer (20 mM Tris-HCl, 150 mM sodium chloride, 10% glycerol) over 10 washes, followed by two washes with Tris buffer containing 20 mM imidazole. Protein was eluted in Tris buffer containing 200 mM imidazole and dialyzed in phosphate-buffered saline (PBS) before concentration using a Microcon column (Millipore). Proteins were tested for endotoxin contamination using the Pierce limulus amebocyte lysate assay (Thermo Scientific), and relative size was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Samples were denatured in an equal volume of 2× sample loading buffer (40% [vol/vol] 0.5 M Tris-HCl [pH 6.8], 10% glycerol, 1.7% [wt/vol] SDS, 10% 2-β-mercaptoethanol, 0.13% [wt/vol] bromophenol blue in distilled water) at 100°C for 5 min. Ten microliters of each sample and SeeBlue Plus2 prestained protein standard (Invitrogen) was loaded onto a 0.5-mm 12% polyacrylamide gel under reducing conditions, as previously described (97). The gel was run in buffer containing 0.3% (wt/vol) Tris, 1.44% (wt/vol) glycine, and 0.1% (wt/vol) SDS in distilled water for 1 h at 150 V (Mini-Protean vertical electrophoresis cell; Bio-Rad), stained in Coomassie stain (45% methanol, 10% acetic acid, 0.25% [wt/vol] Coomassie brilliant blue in distilled water) for 1 h, and destained in Coomassie destain (33% methanol, 10% acetic acid, 60% distilled water) until protein bands were visualized.
Western blotting.
Protein separated on a 12% polyacrylamide gel was transferred to a nitrocellulose membrane in a Tris-glycine transfer buffer (1.5 mM Tris, 12 mM glycine, 15% methanol [vol/vol] in distilled water) for 1 h at 100 V (Mini Trans-Blot cell; Bio-Rad) and incubated in blocking buffer (5% [wt/vol] skim milk powder and 0.1% Tween 20 in PBS) overnight at 4°C. The membrane was then incubated with anti-6×His-horseradish peroxidase (anti-6×His-HRP) antibody (Roche Applied Science, USA) at a 1:500 dilution for 2 h and washed in PBS containing 0.1% Tween 20 prior to exposure to developing solution (Western Lighting chemiluminescence kit; PerkinElmer, USA), according to the manufacturer’s guidelines. Chemiluminescence was detected using an MF ChemiBIS gel imaging system (DNR Bio-Imaging Systems, Israel).
Particle size analysis of protein antigen and lipopeptide formulations by DLS.
The association between protein and R4Pam2Cys was measured using dynamic light scattering (DLS) by mixing 5 μg of protein with increasing amounts of lipopeptide in 50 μl PBS. The size distribution of particles in solution (presented as hydrodynamic radius) was measured in 4 μl of cyclin olefin copolymer cuvettes using a DynaPro NanoStar DLS instrument (Wyatt Technology, CA, USA) equipped with a 658-nm laser with a scattering angle of 90°. Measurements were acquired in triplicate, with each measurement consisting of 30 readings at 5-s intervals at 25°C. Data were analyzed using the Dynamics software (v7.1.7.16).
Vaccination of animals.
The synthesis and purification of the branched cationic lipopeptide R4Pam2Cys were performed as previously described (98, 99). Each vaccine dose contained 25 μg protein formulated in PBS with R4Pam2Cys at a 1:5 molar ratio of protein to lipopeptide in a final volume of 100 μl. Live-attenuated M. bovis BCG strain Danish 1331 was grown to log phase and stored at –80°C in 20% glycerol until used. Bacteria were washed with PBS and resuspended in 200 μl before administration at 4.7 × 105 bacteria per dose. All vaccines and control formulations were sonicated for 5 min in a water bath sonicator before being administered.
Female 6-week-old BALB/c mice were sourced from ARC (Canning Vale, Australia) and housed in individual ventilated cages. Food and water were given ad libitum. Experiments were approved by the University of Melbourne Animal Ethics Committee (approval identification number 1613870). For vaccination using R4Pam2Cys, animals were inoculated subcutaneously at the base of the tail (100 μl per dose at 50 μl per flank) and boosted 21 days later with the same formulations. Mice vaccinated with M. bovis BCG were given one dose subcutaneously in a similar manner (200 μl per dose at 100 μl per flank). There were 10 mice in each vaccination group.
M. ulcerans challenge.
Mice were challenged with bioluminescent M. ulcerans on day 35, as described previously (10). Briefly, tails of isoflurane-anesthetized mice were dipped in 7H9 culture containing log-phase bioluminescent M. ulcerans bacteria (concentration, 1.27 × 106 CFU/ml [range, 1.07 × 106 to 1.46 × 106 CFU/ml]). Contaminated tails were then pierced once subcutaneously with a sterile 25-G needle. The infectious dose was calculated to be 17 CFU (range, 14 to 20 CFU), using methods previously described (10). Mice were allowed to recover, were monitored for up to 24 weeks after infection, and were sacrificed when tail ulceration was observed, wherein spleen, lymph node, and serum samples were harvested for immunological analysis.
IVIS imaging.
Infected mice were imaged weekly from 6 weeks postinfection to detect the emission of bioluminescence. Images were captured using the Lumina XRS series III in vitro imaging system (IVIS; PerkinElmer, MA, USA) and Living Image Software v3.2 with the following settings: field of view, 24; relative aperture, f’1.2; medium binning; and 60-s exposure. Bioluminescence was calculated using the Living Image Software v3.2.
Serum antibody titer measurements.
Serum antibody titers were measured with an enzyme-linked immunosorbent assay (ELISA) (100) using plates (Nunc; Thermo Scientific) that were previously coated with antigen overnight, either purified recombinant ER protein or heat-killed whole-cell M. ulcerans lysate. The presence of bound antibodies was detected by incubating serum-exposed wells with horseradish peroxidase-conjugated rabbit anti-mouse IgG (Dako, Glostrup, Denmark) for 2 h, followed by the addition of the enzyme substrate [0.2 mM 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] (ABTS) in 50 mM citric acid containing 0.004% hydrogen peroxide and left to develop for 10 to 15 min before the addition of 50 nM sodium fluoride to stop the reaction. Plates were read at dual wavelengths of 505 and 595 nm on a plate reader (LabSystems Multiskan Multisoft microplate reader) and antibody titers expressed as the reciprocal of the highest dilution of serum required to achieve an optical density of 0.2.
Intracellular cytokine staining.
Single-cell suspensions were derived from the spleen and draining lymph nodes and resuspended in RP10 medium, comprising RPMI 1640 (Sigma) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Waltham, MA, USA), 2 mM l-glutamine, 1 mM sodium pyruvate, 55 μM 2-mercaptoethanol, 12 μg gentamicin, 100 U/ml penicillin, and 100 μg/ml streptomycin. Spleen- and lymph node-derived cells were cultured in 96-well plates (Costar, Corning, USA) at 1 × 107 cells/per well and 1 × 105 cells/well, respectively, with 200 μl of RP10 containing 10 U/ml IL-2 (Roche, Mannheim, Germany), 1 μg/ml plate-bound anti-CD28 (clone 37.51; BD Pharmingen, Becton, Dickinson), and 20 μg/ml ER protein for 12 h at 37°C in 5% CO2. GolgiPlug (1 μg/ml; Becton, Dickinson) was added for the last 4 h of incubation. Cells were then stained with 7AAD-LIVE/DEAD stain dye (BioLegend, CA, USA), BV510-anti-B220 (clone RA3-6B2; BD Horizon, Becton, Dickinson), BV605-antiCD4 (clone RM4-5; BioLegend), allophycocyanin (APC)-Cy7-anti-TCRb (clone H57-597; BD Pharmingen), and phycoerythrin (PE)-Cy7-anti-CD8 (clone 53-6.7; BD Pharmingen) anti-mouse monoclonal antibodies at 4°C in the dark. Intracellular staining was performed by fixing cells with Cytofix/Cytoperm solution (Becton, Dickinson, USA), followed by permeabilization and intracellular staining with Perm/Wash buffer (Becton, Dickinson) and BV786-IFN-γ (clone XM G1.2; BD Horizon), AF647-IL-17A (clone TCII-18H10; BD Pharmingen, Becton, Dickinson), and PE-TNF-α (clone MP6-XT22; BD Biosciences) antibodies for 30 min at 4°C before analysis on an LSRFortessa flow cytometer (BD Biosciences). Data analyses were performed using FlowJo (Tree Star, OR, USA).
Cytokine bead array.
Spleen- and lymph node-derived cells were incubated in 500 μl RP10 supplemented with 25 μg/ml ER protein for 72 h at 37°C in 5% CO2. The supernatant was collected, and a cytokine bead array was performed using a mouse flex set (BD Biosciences, USA) to detect IL-2, IL-4, IL-6, IL-10, IL-17, IFN-γ, TNF-α, MCP-1, MIP-1α, and MIP-1β, as per the manufacturer’s instructions. Samples were acquired using a FACSCanto II flow cytometer (BD Biosciences) and cytokine quantities calculated using the FCAP Array software v3.0.
Histology and microscopy.
Tail tissues from the site of infection were fixed in PBS containing 10% nonbuffered formalin and then embedded in paraffin and sliced into 10-μm-thick segments. The sliced segments were Ziehl-Neelsen or hematoxylin and eosin (H&E) stained prior to microscope imaging. Images of tail segments were captured using a light microscope (BX53 light microscope; Olympus Life Science).
Statistical analysis.
The Prism v7 software (GraphPad, CA, USA) was used to perform statistical analyses on the antibody titer, time to luminescence, T cell numbers, and cytokine titer data. Antibody titers were analyzed using one-way analysis of variance (ANOVA) with Tukey’s correction for multiple comparisons. The time to bioluminescence data were displayed as a Kaplan-Meier plot and differences determined using a log rank (Mantel-Cox) test. Mann-Whitney tests were performed to compare cytokine titers between protected and diseased mice and for comparisons between vaccination groups. All tests were conducted at the 5% significance level.
Statistical modeling.
Twenty-eight data features (i.e., the immune parameters measured in each mouse; refer to Table S1) were transformed using the R package bestNormalize (101). Transformed features were then normalized (between 0 and 1 for each feature) using the MinMaxScaler function of Scikit-learn (102). As many of the vaccination outcome observations (time to bioluminescence) were right censored, we employed the Cox proportional hazards regression analysis using the Scikit-survival module of Scikit-learn in Python (103). Here, univariate analyses were run for each of the 28 features using the continuous response variable of time to bioluminescence. The standard metric for assessing the predictive performance of a survival model is the concordance index (CI) (61, 104, 105). A CI of >0.7 was used to identify the top six features of this model. Unsupervised learning and data dimensionality reduction represent an ideal way to identify structure in continuous data without the influence of labels. The method of t-distributed stochastic neighbor embedding (t-SNE) is a dimensionality reduction technique that retains both the global structure and local layout of the high-dimensional data through exchanging the Euclidean distances between all pairs of data points into heavy-tailed conditional probabilities (106). This method is advantageous over conventional principal-component analysis (PCA), as it does not rely on a linear assumption and can capture nonlinear relationships (106). We explored the data, independent of labels, by reducing the top six features obtained from the Cox proportional hazards regression analysis to a two-dimensional space using the t-SNE package in Scikit-learn (102). The two clusters detected through visual inspection were objectively defined, with observations assigned to two groups using K-means clustering, as implemented in Scikit-learn (102). A multivariate logistic regression classifier was then built using the top six features, with the two clusters identified by t-SNE as the response variables. To reduce the possibility of overfitting, the model was validated through 1,000 random train-test splits, in which 90% of the observations made up each training set. These models were built using the logistic regression classifier as implemented in Scikit-learn (102) and receiver operating characteristic curves were used to evaluate model performance (107). In order to assess the immune features that were associated with different vaccination groups, a univariate logistic regression analysis was then conducted for each group using R (107). The estimated model coefficients were used to assess the direction and strength of the association, and the corresponding P values were used to determine statistical significance at the 5% significance level.
Supplementary Material
ACKNOWLEDGMENTS
We thank Laura Leone for expert assistance with histology.
This research was supported by the National Health and Medical Research Council, Australia (grant GNT1008549).
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Guarner J, Bartlett J, Whitney EA, Raghunathan PL, Stienstra Y, Asamoa K, Etuaful S, Klutse E, Quarshie E, van der Werf TS, van der Graaf WT, King CH, Ashford DA. 2003. Histopathologic features of Mycobacterium ulcerans infection. Emerg Infect Dis 9:651–656. doi: 10.3201/eid0906.020485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent QB, Ardant M-F, Adeye A, Goundote A, Saint-André J-P, Cottin J, Kempf M, Agossadou D, Johnson C, Abel L, Marsollier L, Chauty A, Alcaïs A. 2014. Clinical epidemiology of laboratory-confirmed Buruli ulcer in Benin: a cohort study. Lancet Glob Health 2:e422–e430. doi: 10.1016/S2214-109X(14)70223-2. [DOI] [PubMed] [Google Scholar]
- 3.Hayman J, McQueen A. 1985. The pathology of Mycobacterium ulcerans infection. Pathology 17:594–600. doi: 10.3109/00313028509084759. [DOI] [PubMed] [Google Scholar]
- 4.Oliveira MS, Fraga AG, Torrado E, Castro AG, Pereira JP, Filho AL, Milanezi F, Schmitt FC, Meyers WM, Portaels F, Silva MT, Pedrosa J. 2005. Infection with Mycobacterium ulcerans induces persistent inflammatory responses in mice. Infect Immun 73:6299–6310. doi: 10.1128/IAI.73.10.6299-6310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yerramilli A, Tay EL, Stewardson AJ, Kelley PG, Bishop E, Jenkin GA, Starr M, Trevillyan J, Hughes A, Friedman ND, O'Brien DP, Johnson PDR. 2017. The location of Australian Buruli ulcer lesions–implications for unravelling disease transmission. PLoS Negl Trop Dis 11:e0005800. doi: 10.1371/journal.pntd.0005800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Werf TS, van der Graaf WT, Tappero JW, Asiedu K. 1999. Mycobacterium ulcerans infection. Lancet 354:1013–1018. doi: 10.1016/S0140-6736(99)01156-3. [DOI] [PubMed] [Google Scholar]
- 7.Pszolla N, Sarkar MR, Strecker W, Kern P, Kinzl L, Meyers WM, Portaels F. 2003. Buruli ulcer: a systemic disease. Clin Infect Dis 37:e78–e82. doi: 10.1086/377170. [DOI] [PubMed] [Google Scholar]
- 8.Loftus MJ, Tay EL, Globan M, Lavender CJ, Crouch SR, Johnson PDR, Fyfe J. 2018. Epidemiology of Buruli ulcer infections, Victoria, Australia, 2011–2016. Emerg Infect Dis 24:1988–1997. doi: 10.3201/eid2411.171593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toll A, Gallardo F, Ferran M, Gilaberte M, Iglesias M, Gimeno JL, Rondini S, Pujol RM. 2005. Aggressive multifocal Buruli ulcer with associated osteomyelitis in an HIV-positive patient. Clin Exp Dermatol 30:649–651. doi: 10.1111/j.1365-2230.2005.01892.x. [DOI] [PubMed] [Google Scholar]
- 10.Wallace JR, Mangas KM, Porter JL, Marcsisin R, Pidot SJ, Howden B, Omansen TF, Zeng W, Axford JK, Johnson PDR, Stinear TP. 2017. Mycobacterium ulcerans low infectious dose and mechanical transmission support insect bites and puncturing injuries in the spread of Buruli ulcer. PLoS Negl Trop Dis 11:e0005553. doi: 10.1371/journal.pntd.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsollier L, Robert R, Aubry J, Saint Andre JP, Kouakou H, Legras P, Manceau AL, Mahaza C, Carbonnelle B. 2002. Aquatic insects as a vector for Mycobacterium ulcerans. Appl Environ Microbiol 68:4623–4628. doi: 10.1128/aem.68.9.4623-4628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyers WM, Shelly WM, Connor DH, Meyers EK. 1974. Human Mycobacterium ulcerans infections developing at sites of trauma to skin. Am J Trop Med Hyg 23:919–923. doi: 10.4269/ajtmh.1974.23.919. [DOI] [PubMed] [Google Scholar]
- 13.Buultjens AH, Vandelannoote K, Meehan CJ, Eddyani M, de Jong BC, Fyfe JAM, Globan M, Tobias NJ, Porter JL, Tomita T, Tay EL, Seemann T, Howden BP, Johnson PDR, Stinear TP, Buultjens AH, Vandelannoote K, Meehan CJ, Eddyani M, de Jong BC, Fyfe JAM, Globan M, Tobias NJ, Porter JL, Tomita T, Tay EL, Seemann T, Howden BP, Johnson PDR, Stinear TP. 2018. Comparative genomics shows that Mycobacterium ulcerans migration and expansion preceded the rise of Buruli ulcer in southeastern Australia. Appl Environ Microbiol 84:e02612-17. doi: 10.1128/AEM.02612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simpson H, Deribe K, Tabah EN, Peters A, Maman I, Frimpong M, Ampadu E, Phillips R, Saunderson P, Pullan RL, Cano J. 2019. Mapping the global distribution of Buruli ulcer: a systematic review with evidence consensus. Lancet Glob Health 7:e912–e922. doi: 10.1016/S2214-109X(19)30171-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stinear TP, Seemann T, Pidot S, Frigui W, Reysset G, Garnier T, Meurice G, Simon D, Bouchier C, Ma L, Tichit M, Porter JL, Ryan J, Johnson PD, Davies JK, Jenkin GA, Small PL, Jones LM, Tekaia F, Laval F, Daffe M, Parkhill J, Cole ST. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res 17:192–200. doi: 10.1101/gr.5942807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsollier L, Stinear T, Aubry J, Saint Andre JP, Robert R, Legras P, Manceau AL, Audrain C, Bourdon S, Kouakou H, Carbonnelle B. 2004. Aquatic plants stimulate the growth of and biofilm formation by Mycobacterium ulcerans in axenic culture and harbor these bacteria in the environment. Appl Environ Microbiol 70:1097–1103. doi: 10.1128/aem.70.2.1097-1103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loftus MJ, Trubiano JA, Tay EL, Lavender CJ, Globan M, Fyfe J, Johnson P. 2018. The incubation period of Buruli ulcer (Mycobacterium ulcerans infection) in Victoria, Australia - Remains similar despite changing geographic distribution of disease. PLoS Negl Trop Dis 12:e0006323. doi: 10.1371/journal.pntd.0006323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trubiano JA, Lavender CJ, Fyfe JA, Bittmann S, Johnson PD. 2013. The incubation period of Buruli ulcer (Mycobacterium ulcerans infection). PLoS Negl Trop Dis 7:e2463. doi: 10.1371/journal.pntd.0002463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarfo FS, Phillips R, Asiedu K, Ampadu E, Bobi N, Adentwe E, Lartey A, Tetteh I, Wansbrough-Jones M. 2010. Clinical efficacy of combination of rifampin and streptomycin for treatment of Mycobacterium ulcerans disease. Antimicrob Agents Chemother 54:3678–3685. doi: 10.1128/AAC.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nienhuis WA, Stienstra Y, Thompson WA, Awuah PC, Abass KM, Tuah W, Awua-Boateng NY, Ampadu EO, Siegmund V, Schouten JP, Adjei O, Bretzel G, van der Werf TS. 2010. Antimicrobial treatment for early, limited Mycobacterium ulcerans infection: a randomised controlled trial. Lancet 375:664–672. doi: 10.1016/S0140-6736(09)61962-0. [DOI] [PubMed] [Google Scholar]
- 21.Wadagni AC, Barogui YT, Johnson RC, Sopoh GE, Affolabi D, van der Werf TS, de Zeeuw J, Kleinnijenhuis J, Stienstra Y. 2018. Delayed versus standard assessment for excision surgery in patients with Buruli ulcer in Benin: a randomised controlled trial. Lancet Infect Dis 18:650–656. doi: 10.1016/S1473-3099(18)30160-9. [DOI] [PubMed] [Google Scholar]
- 22.Zingue D, Bouam A, Tian RBD, Drancourt M. 2018. Buruli ulcer, a prototype for ecosystem-related infection, caused by Mycobacterium ulcerans. Clin Microbiol Rev 31:e00045-17. doi: 10.1128/CMR.00045-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanghe A, Adnet PY, Gartner T, Huygen K. 2007. A booster vaccination with Mycobacterium bovis BCG does not increase the protective effect of the vaccine against experimental Mycobacterium ulcerans infection in mice. Infect Immun 75:2642–2644. doi: 10.1128/IAI.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Converse PJ, Almeida DV, Nuermberger EL, Grosset JH. 2011. BCG-mediated protection against Mycobacterium ulcerans infection in the mouse. PLoS Negl Trop Dis 5:e985. doi: 10.1371/journal.pntd.0000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraga AG, Martins TG, Torrado E, Huygen K, Portaels F, Silva MT, Castro AG, Pedrosa J. 2012. Cellular immunity confers transient protection in experimental Buruli ulcer following BCG or mycolactone-negative Mycobacterium ulcerans vaccination. PLoS One 7:e33406. doi: 10.1371/journal.pone.0033406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh DS, Dela Cruz EC, Abalos RM, Tan EV, Walsh GP, Portaels F, Meyers WM. 2007. Clinical and histologic features of skin lesions in a cynomolgus monkey experimentally infected with Mycobacterium ulcerans (Buruli ulcer) by intradermal inoculation. Am J Trop Med Hyg 76:132–134. doi: 10.4269/ajtmh.2007.76.132. [DOI] [PubMed] [Google Scholar]
- 27.Ortiz RH, Leon DA, Estevez HO, Martin A, Herrera JL, Romo LF, Portaels F, Pando RH. 2009. Differences in virulence and immune response induced in a murine model by isolates of Mycobacterium ulcerans from different geographic areas. Clin Exp Immunol 157:271–281. doi: 10.1111/j.1365-2249.2009.03941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolz M, Ruggli N, Ruf MT, Ricklin ME, Zimmer G, Pluschke G. 2014. Experimental infection of the pig with Mycobacterium ulcerans: a novel model for studying the pathogenesis of Buruli ulcer disease. PLoS Negl Trop Dis 8:e2968. doi: 10.1371/journal.pntd.0002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marion E, Jarry U-O, Cano C, Savary C-O, Beauvillain C-O, Robbe-Saule M, Preisser L, Altare F, Delneste Y, Jeannin P, Marsollier L. 2016. FVB/N mice spontaneously heal ulcerative lesions induced by Mycobacterium ulcerans and switch M. ulcerans into a low mycolactone producer. J Immunol 196:2690–2698. doi: 10.4049/jimmunol.1502194. [DOI] [PubMed] [Google Scholar]
- 30.Bénard A, Sala C, Pluschke G. 2016. Mycobacterium ulcerans mouse model refinement for pre-clinical profiling of vaccine candidates. PLoS One 8:e0167059. doi: 10.1371/journal.pone.0167059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh DS, Meyers WM, Krieg RE, Walsh GP. 1999. Transmission of Mycobacterium ulcerans to the nine-banded armadillo. Am J Trop Med Hyg 61:694–697. doi: 10.4269/ajtmh.1999.61.694. [DOI] [PubMed] [Google Scholar]
- 32.Tanghe A, Content J, Van Vooren JP, Portaels F, Huygen K. 2001. Protective efficacy of a DNA vaccine encoding antigen 85A from Mycobacterium bovis BCG against Buruli ulcer. Infect Immun 69:5403–5411. doi: 10.1128/iai.69.9.5403-5411.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanghe A, Dangy JP, Pluschke G, Huygen K. 2008. Improved protective efficacy of a species-specific DNA vaccine encoding mycolyl-transferase Ag85A from Mycobacterium ulcerans by homologous protein boosting. PLoS Negl Trop Dis 2:e199. doi: 10.1371/journal.pntd.0000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolz M, Benard A, Dreyer AM, Kerber S, Vettiger A, Oehlmann W, Singh M, Duthie MS, Pluschke G. 2016. Vaccination with the surface proteins MUL_2232 and MUL_3720 of Mycobacterium ulcerans induces antibodies but fails to provide protection against Buruli ulcer. PLoS Negl Trop Dis 10:e0004431. doi: 10.1371/journal.pntd.0004431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart BE, Hale LP, Lee S. 2016. Immunogenicity and protection conferred by a recombinant Mycobacterium marinum vaccine against Buruli ulcer. Trials Vaccinol 5:88–91. doi: 10.1016/j.trivac.2016.04.001. [DOI] [Google Scholar]
- 36.Hart BE, Hale LP, Lee S. 2015. Recombinant BCG expressing Mycobacterium ulcerans Ag85A imparts enhanced protection against experimental Buruli ulcer. PLoS Negl Trop Dis 9:e0004046. doi: 10.1371/journal.pntd.0004046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart BE, Lee S. 2016. Overexpression of a Mycobacterium ulcerans Ag85B-EsxH fusion protein in recombinant BCG improves experimental Buruli ulcer vaccine efficacy. PLoS Negl Trop Dis 10:e0005229. doi: 10.1371/journal.pntd.0005229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gowthaman U, Mushtaq K, Tan AC, Rai PK, Jackson DC, Agrewala JN. 2015. Challenges and solutions for a rational vaccine design for TB-endemic regions. Crit Rev Microbiol 41:389–398. doi: 10.3109/1040841X.2013.859125. [DOI] [PubMed] [Google Scholar]
- 39.George KM, Chatterjee D, Gunawardana G, Welty D, Hayman J, Lee R, Small PL. 1999. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science 283:854–857. doi: 10.1126/science.283.5403.854. [DOI] [PubMed] [Google Scholar]
- 40.Ogbechi J, Hall BS, Sbarrato T, Taunton J, Willis AE, Wek RC, Simmonds RE. 2018. Inhibition of Sec61-dependent translocation by mycolactone uncouples the integrated stress response from ER stress, driving cytotoxicity via translational activation of ATF4. Cell Death Dis 9:397. doi: 10.1038/s41419-018-0427-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baron L, Paatero AO, Morel JD, Impens F, Guenin-Mace L, Saint-Auret S, Blanchard N, Dillmann R, Niang F, Pellegrini S, Taunton J, Paavilainen VO, Demangel C. 2016. Mycolactone subverts immunity by selectively blocking the Sec61 translocon. J Exp Med 213:2885–2896. doi: 10.1084/jem.20160662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coutanceau E, Decalf J, Martino A, Babon A, Winter N, Cole ST, Albert ML, Demangel C. 2007. Selective suppression of dendritic cell functions by Mycobacterium ulcerans toxin mycolactone. J Exp Med 204:1395–1403. doi: 10.1084/jem.20070234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meier JL, Burkart MD. 2011. Proteomic analysis of polyketide and nonribosomal peptide biosynthesis. Curr Opin Chem Biol 15:48–56. doi: 10.1016/j.cbpa.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boulkroun S, Guenin-Mace L, Thoulouze MI, Monot M, Merckx A, Langsley G, Bismuth G, Di Bartolo V, Demangel C. 2010. Mycolactone suppresses T cell responsiveness by altering both early signaling and posttranslational events. J Immunol 184:1436–1444. doi: 10.4049/jimmunol.0902854. [DOI] [PubMed] [Google Scholar]
- 45.Pahlevan AA, Wright DJ, Andrews C, George KM, Small PL, Foxwell BM. 1999. The inhibitory action of Mycobacterium ulcerans soluble factor on monocyte/T cell cytokine production and NF-kappa B function. J Immunol 163:3928–3935. [PubMed] [Google Scholar]
- 46.Torrado E, Fraga AG, Logarinho E, Martins TG, Carmona JA, Gama JB, Carvalho MA, Proença F, Castro AG, Pedrosa J. 2010. IFN-γ–dependent activation of macrophages during experimental infections by Mycobacterium ulcerans is impaired by the toxin mycolactone. J Immunol 184:947. doi: 10.4049/jimmunol.0902717. [DOI] [PubMed] [Google Scholar]
- 47.Stinear TP, Mve-Obiang A, Small PL, Frigui W, Pryor MJ, Brosch R, Jenkin GA, Johnson PD, Davies JK, Lee RE, Adusumilli S, Garnier T, Haydock SF, Leadlay PF, Cole ST. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A 101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stinear TP, Pryor MJ, Porter JL, Cole ST. 2005. Functional analysis and annotation of the virulence plasmid pMUM001 from Mycobacterium ulcerans. Microbiology 151:683–692. doi: 10.1099/mic.0.27674-0. [DOI] [PubMed] [Google Scholar]
- 49.Roupie V, Pidot SJ, Einarsdottir T, Van Den Poel C, Jurion F, Stinear TP, Huygen K. 2014. Analysis of the vaccine potential of plasmid DNA encoding nine mycolactone polyketide synthase domains in Mycobacterium ulcerans infected mice. PLoS Negl Trop Dis 8:e2604. doi: 10.1371/journal.pntd.0002604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chua BY, Zeng W, Jackson DC. 2008. Synthesis of Toll-like receptor-2 targeting lipopeptides as self-adjuvanting vaccines. Methods Mol Biol 494:247–261. doi: 10.1007/978-1-59745-419-3_14. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z, Kedzierski L, Nuessing S, Chua BY, Quinones-Parra SM, Huber VC, Jackson DC, Thomas PG, Kedzierska K. 2016. Establishment of memory CD8+ T cells with live attenuated influenza virus across different vaccination doses. J Gen Virol 97:3205–3214. doi: 10.1099/jgv.0.000651. [DOI] [PubMed] [Google Scholar]
- 52.Coutanceau E, Legras P, Marsollier L, Reysset G, Cole ST, Demangel C. 2006. Immunogenicity of Mycobacterium ulcerans Hsp65 and protective efficacy of a Mycobacterium leprae Hsp65-based DNA vaccine against Buruli ulcer. Microbes Infect 8:2075–2081. doi: 10.1016/j.micinf.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 53.Trigo G, Martins TG, Fraga AG, Longatto-Filho A, Castro AG, Azeredo J, Pedrosa J. 2013. Phage therapy is effective against infection by Mycobacterium ulcerans in a murine footpad model. PLoS Negl Trop Dis 7:e2183. doi: 10.1371/journal.pntd.0002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe M, Nakamura H, Nabekura R, Shinoda N, Suzuki E, Saito H. 2015. Protective effect of a dewaxed whole-cell vaccine against Mycobacterium ulcerans infection in mice. Vaccine 33:2232–2239. doi: 10.1016/j.vaccine.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 55.Dangy JP, Scherr N, Gersbach P, Hug MN, Bieri R, Bomio C, Li J, Huber S, Altmann KH, Pluschke G. 2016. Antibody-mediated neutralization of the exotoxin mycolactone, the main virulence factor produced by Mycobacterium ulcerans. PLoS Negl Trop Dis 10:e0004808. doi: 10.1371/journal.pntd.0004808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Porter JL, Tobias NJ, Pidot SJ, Falgner S, Tuck KL, Vettiger A, Hong H, Leadlay PF, Stinear TP. 2013. The cell wall-associated mycolactone polyketide synthases are necessary but not sufficient for mycolactone biosynthesis. PLoS One 8:e70520. doi: 10.1371/journal.pone.0070520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pidot SJ, Porter JL, Marsollier L, Chauty A, Migot-Nabias F, Badaut C, Benard A, Ruf MT, Seemann T, Johnson PD, Davies JK, Jenkin GA, Pluschke G, Stinear TP. 2010. Serological evaluation of Mycobacterium ulcerans antigens identified by comparative genomics. PLoS Negl Trop Dis 4:e872. doi: 10.1371/journal.pntd.0000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pidot SJ, Hong H, Seemann T, Porter JL, Yip MJ, Men A, Johnson M, Wilson P, Davies JK, Leadlay PF, Stinear TP. 2008. Deciphering the genetic basis for polyketide variation among mycobacteria producing mycolactones. BMC Genomics 9:462. doi: 10.1186/1471-2164-9-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Omansen TF, Porter JL, Johnson PDR, van der Werf TS, Stienstra Y, Stinear TP. 2015. In-vitro activity of avermectins against Mycobacterium ulcerans. PLoS Negl Trop Dis 9:e0003549. doi: 10.1371/journal.pntd.0003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Omansen TF, Marcsisin RA, Chua BY, Zeng W, Jackson DC, Porter JL, Stienstra Y, van der Werf TS, Stinear TP. 2019. In vivo imaging of bioluminescent Mycobacterium ulcerans: a tool to refine the murine Buruli ulcer tail model. Am J Trop Med Hyg 101:1312–1321. doi: 10.4269/ajtmh.18-0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Belle V, Pelckmans K, Van Huffel S, Suykens JA. 2011. Support vector methods for survival analysis: a comparison between ranking and regression approaches. Artif Intell Med 53:107–118. doi: 10.1016/j.artmed.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Kamala T. 2007. Hock immunization: a humane alternative to mouse footpad injections. J Immunol Methods 328:204–214. doi: 10.1016/j.jim.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Brien DP, Murrie A, Meggyesy P, Priestley J, Rajcoomar A, Athan E. 2019. Spontaneous healing of Mycobacterium ulcerans disease in Australian patients. PLoS Negl Trop Dis 13:e0007178. doi: 10.1371/journal.pntd.0007178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marion E, Chauty A, Kempf M, Le Corre Y, Delneste Y, Croue A, Marsollier L. 2016. Clinical features of spontaneous partial healing during Mycobacterium ulcerans infection. Open Forum Infect Dis 3:ofw013. doi: 10.1093/ofid/ofw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silva-Gomes R, Marcq E, Trigo G, Gonçalves CM, Longatto-Filho A, Castro AG, Pedrosa J, Fraga AG. 2015. Spontaneous healing of Mycobacterium ulcerans lesions in the guinea pig model. PLoS Negl Trop Dis 9:e0004265. doi: 10.1371/journal.pntd.0004265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan AC, Mifsud EJ, Zeng W, Edenborough K, McVernon J, Brown LE, Jackson DC. 2012. Intranasal administration of the TLR2 agonist Pam2Cys provides rapid protection against influenza in mice. Mol Pharm 9:2710–2718. doi: 10.1021/mp300257x. [DOI] [PubMed] [Google Scholar]
- 67.Mifsud EJ, Tan AC, Reading PC, Jackson DC. 2016. Mapping the pulmonary environment of animals protected from virulent H1N1 influenza infection using the TLR-2 agonist Pam2Cys. Immunol Cell Biol 94:169–176. doi: 10.1038/icb.2015.81. [DOI] [PubMed] [Google Scholar]
- 68.Mifsud EJ, Tan AC, Short KR, Brown LE, Chua BY, Jackson DC. 2016. Reducing the impact of influenza-associated secondary pneumococcal infections. Immunol Cell Biol 94:101–108. doi: 10.1038/icb.2015.71. [DOI] [PubMed] [Google Scholar]
- 69.Gooding TM, Johnson PD, Campbell DE, Hayman JA, Hartland EL, Kemp AS, Robins-Browne RM. 2001. Immune response to infection with Mycobacterium ulcerans. Infect Immun 69:1704–1707. doi: 10.1128/IAI.69.3.1704-1707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tolosie K, Sharma MK. 2014. Application of Cox proportional hazards model in case of tuberculosis patients in selected Addis Ababa health centres, Ethiopia. Tuberc Res Treat 2014:536976. doi: 10.1155/2014/536976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cole SR, Hudgens MG. 2010. Survival analysis in infectious disease research: describing events in time. AIDS 24:2423–2431. doi: 10.1097/QAD.0b013e32833dd0ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forster AJ, Taljaard M, Oake N, Wilson K, Roth V, van Walraven C. 2012. The effect of hospital-acquired infection with Clostridium difficile on length of stay in hospital. CMAJ 184:37–42. doi: 10.1503/cmaj.110543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalia V, Sarkar S. 2018. Regulation of effector and memory CD8 T cell differentiation by IL-2–a balancing act. Front Immunol 9:2987. doi: 10.3389/fimmu.2018.02987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fraga AG, Cruz A, Martins TG, Torrado E, Saraiva M, Pereira DR, Meyers WM, Portaels F, Silva MT, Castro AG, Pedrosa J. 2011. Mycobacterium ulcerans triggers T-cell immunity followed by local and regional but not systemic immunosuppression. Infect Immun 79:421–430. doi: 10.1128/IAI.00820-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Phillips R, Sarfo FS, Guenin-Mace L, Decalf J, Wansbrough-Jones M, Albert ML, Demangel C. 2009. Immunosuppressive signature of cutaneous Mycobacterium ulcerans infection in the peripheral blood of patients with Buruli ulcer disease. J Infect Dis 200:1675–1684. doi: 10.1086/646615. [DOI] [PubMed] [Google Scholar]
- 76.Kiszewski AE, Becerril E, Aguilar LD, Kader IT, Myers W, Portaels F, Hernandez Pando R. 2006. The local immune response in ulcerative lesions of Buruli disease. Clin Exp Immunol 143:445–451. doi: 10.1111/j.1365-2249.2006.03020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peduzzi E, Groeper C, Schutte D, Zajac P, Rondini S, Mensah-Quainoo E, Spagnoli GC, Pluschke G, Daubenberger CA. 2007. Local activation of the innate immune system in Buruli ulcer lesions. J Invest Dermatol 127:638–645. doi: 10.1038/sj.jid.5700593. [DOI] [PubMed] [Google Scholar]
- 78.Ray JCJ, Wang J, Chan J, Kirschner DE. 2008. The timing of TNF and IFN-gamma signaling affects macrophage activation strategies during Mycobacterium tuberculosis infection. J Theor Biol 252:24–38. doi: 10.1016/j.jtbi.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cavalcanti YV, Brelaz MC, Neves JK, Ferraz JC, Pereira VR. 2012. Role of TNF-alpha, IFN-gamma, and IL-10 in the development of pulmonary tuberculosis. Pulm Med 2012:745483. doi: 10.1155/2012/745483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hall BS, Hill K, McKenna M, Ogbechi J, High S, Willis AE, Simmonds RE. 2014. The pathogenic mechanism of the Mycobacterium ulcerans virulence factor, mycolactone, depends on blockade of protein translocation into the ER. PLoS Pathog 10:e1004061. doi: 10.1371/journal.ppat.1004061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Torrado E, Adusumilli S, Fraga AG, Small PLC, Castro AG, Pedrosa J. 2007. Mycolactone-mediated inhibition of tumor necrosis factor production by macrophages infected with Mycobacterium ulcerans has implications for the control of infection. Infect Immun 75:3979–3988. doi: 10.1128/IAI.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bieri R, Bolz M, Ruf M-T, Pluschke G. 2016. Interferon-γ is a crucial activator of early host immune defense against Mycobacterium ulcerans infection in mice. PLoS Negl Trop Dis 10:e0004450. doi: 10.1371/journal.pntd.0004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Phillips R, Horsfield C, Mangan J, Laing K, Etuaful S, Awuah P, Nyarko K, Osei-Sarpong F, Butcher P, Lucas S, Wansbrough-Jones M. 2006. Cytokine mRNA expression in Mycobacterium ulcerans-infected human skin and correlation with local inflammatory response. Infect Immun 74:2917–2924. doi: 10.1128/IAI.74.5.2917-2924.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Westenbrink BD, Stienstra Y, Huitema MG, Thompson WA, Klutse EO, Ampadu EO, Boezen HM, Limburg PC, van der Werf TS. 2005. Cytokine responses to stimulation of whole blood from patients with Buruli ulcer disease in Ghana. Clin Diagn Lab Immunol 12:125–129. doi: 10.1128/CDLI.12.1.125-129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Flynn JL, Chan J. 2001. Immunology of tuberculosis. Annu Rev Immunol 19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 86.Magcwebeba T, Dorhoi A, Du Plessis N. 2019. The emerging role of myeloid-derived suppressor cells in tuberculosis. Front Immunol 10:917. doi: 10.3389/fimmu.2019.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boer MC, Joosten SA, Ottenhoff TH. 2015. Regulatory T-cells at the interface between human host and pathogens in infectious diseases and vaccination. Front Immunol 6:217. doi: 10.3389/fimmu.2015.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Isailovic N, Daigo K, Mantovani A, Selmi C. 2015. Interleukin-17 and innate immunity in infections and chronic inflammation. J Autoimmun 60:1–11. doi: 10.1016/j.jaut.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 89.Greifenberg V, Ribechini E, Rossner S, Lutz MB. 2009. Myeloid-derived suppressor cell activation by combined LPS and IFN-gamma treatment impairs DC development. Eur J Immunol 39:2865–2876. doi: 10.1002/eji.200939486. [DOI] [PubMed] [Google Scholar]
- 90.Jiang M, Chen J, Zhang W, Zhang R, Ye Y, Liu P, Yu W, Wei F, Ren X, Yu J. 2017. Interleukin-6 trans-signaling pathway promotes immunosuppressive myeloid-derived suppressor cells via suppression of suppressor of cytokine signaling 3 in breast cancer. Front Immunol 8:1840. doi: 10.3389/fimmu.2017.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miossec P, Kolls JK. 2012. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov 11:763–776. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 92.Forbes BR, Wannan JS, Kirkland WB. 1954. Indolent cutaneous ulceration due to infection with Mycobacterium ulcerans. Med J Aust 41:475–479. [DOI] [PubMed] [Google Scholar]
- 93.Adusumilli S, Mve-Obiang A, Sparer T, Meyers W, Hayman J, Small PL. 2005. Mycobacterium ulcerans toxic macrolide, mycolactone modulates the host immune response and cellular location of M. ulcerans in vitro and in vivo. Cell Microbiol 7:1295–1304. doi: 10.1111/j.1462-5822.2005.00557.x. [DOI] [PubMed] [Google Scholar]
- 94.Ogbechi J, Ruf MT, Hall BS, Bodman-Smith K, Vogel M, Wu HL, Stainer A, Esmon CT, Ahnstrom J, Pluschke G, Simmonds RE. 2015. Mycolactone-dependent depletion of endothelial cell thrombomodulin is strongly associated with fibrin deposition in Buruli ulcer lesions. PLoS Pathog 11:e1005011. doi: 10.1371/journal.ppat.1005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.George KM, Pascopella L, Welty DM, Small PL. 2000. A Mycobacterium ulcerans toxin, mycolactone, causes apoptosis in guinea pig ulcers and tissue culture cells. Infect Immun 68:877–883. doi: 10.1128/iai.68.2.877-883.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hong H, Gates PJ, Staunton J, Stinear T, Cole ST, Leadlay PF, Spencer JB. 2003. Identification using LC-MSn of co-metabolites in the biosynthesis of the polyketide toxin mycolactone by a clinical isolate of Mycobacterium ulcerans. Chem Commun (Camb) 22:2822–2823. doi: 10.1039/b308163j. [DOI] [PubMed] [Google Scholar]
- 97.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 98.Sekiya T, Yamagishi J, Gray JHV, Whitney PG, Martinelli A, Zeng W, Wong CY, Sugimoto C, Jackson DC, Chua BY. 2017. PEGylation of a TLR2-agonist-based vaccine delivery system improves antigen trafficking and the magnitude of ensuing antibody and CD8+ T cell responses. Biomaterials 137:61–72. doi: 10.1016/j.biomaterials.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 99.Wijayadikusumah AR, Zeng W, McQuilten HA, Wong CY, Jackson DC, Chua BY. 2019. Geometry of a TLR2-agonist-based adjuvant can affect the resulting antigen-specific immune response. Mol Pharm 16:2037–2047. doi: 10.1021/acs.molpharmaceut.9b00026. [DOI] [PubMed] [Google Scholar]
- 100.Chua BY, Pejoski D, Turner SJ, Zeng W, Jackson DC. 2011. Soluble proteins induce strong CD8+ T cell and antibody responses through electrostatic association with simple cationic or anionic lipopeptides that target TLR2. J Immunol 187:1692–1701. doi: 10.4049/jimmunol.1100486. [DOI] [PubMed] [Google Scholar]
- 101.Beasley TM, Erickson S, Allison DB. 2009. Rank-based inverse normal transformations are increasingly used, but are they merited? Behav Genet 39:580–595. doi: 10.1007/s10519-009-9281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12:2825–2830. [Google Scholar]
- 103.Pölsterl S, Gupta P, Wang L, Conjeti S, Katouzian A, Navab N. 2016. Heterogeneous ensembles for predicting survival of metastatic, castrate-resistant prostate cancer patients. F1000Res 5:2676. doi: 10.12688/f1000research.8231.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Evers L, Messow CM. 2008. Sparse kernel methods for high-dimensional survival data. Bioinformatics 24:1632–1638. doi: 10.1093/bioinformatics/btn253. [DOI] [PubMed] [Google Scholar]
- 105.Shivaswamy PK, Chu W, Jansche M. 2007. A support vector approach to censored targets. ICDM 2007: Proceedings of the Seventh IEEE International Conference on Data Mining, 28 to 31 October 2007, Washington, DC. [Google Scholar]
- 106.van der Maaten L, Hinton G. 2008. Visualizing data using t-SNE. J Mach Learn Res 9:2579–2605. [Google Scholar]
- 107.R Core Team. 2014. R: a language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria: https://www.r-project.org/. [Google Scholar]
- 108.Bolz M, Kerber S, Zimmer G, Pluschke G. 2015. Use of recombinant virus replicon particles for vaccination against Mycobacterium ulcerans disease. PLoS Negl Trop Dis 9:e0004011. doi: 10.1371/journal.pntd.0004011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fenner F. 1957. Homologous and heterologous immunity in infections of mice with Mycobacterium ulcerans and Mycobacterium balnei. Am Rev Tuberc 76:76–89. doi: 10.1164/artpd.1957.76.1.76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.