Abstract
DNA damage repair is implemented by proteins that are coordinated by specialised molecular signals. One such signal in the Fanconi Anemia (FA) DNA-interstrand crosslink repair pathway is the site-specific monoubiquitination of FANCD2 and FANCI. The signal is mediated by a multi-protein FA core complex (FA-CC) however, the mechanics for precise ubiquitination remain elusive. We show that FANCL, the RING-bearing module in FA-CC, allosterically activates its cognate E2 Ube2T to drive site-specific FANCD2 ubiquitination. Unlike typical RING E3 ligases, FANCL catalyses ubiquitination by rewiring Ube2T’s intra-residue network to influence the active site. Consequently, a basic triad unique to Ube2T engages a structured acidic patch near the target lysine on FANCD2. This three-dimensional complementarity, between the E2 active site and substrate surface, induced by FANCL is central to site-specific monoubiquitination in the FA pathway. Furthermore, the allosteric network of Ube2T can be engineered to enhance FANCL catalysed FANCD2-FANCI di-monoubiquitination without compromising site-specificity.
Keywords: DNA repair, E2, RING E3, Ubiquitination, allostery
Introduction
Ubiquitination is an essential and reversible post-translational signal that enables reprogramming of eukaryotic cellular pathways. The modification is mediated by an enzyme cascade where ubiquitin’s C-terminus is activated by an E1, transferred onto the catalytic cysteine of an E2 (E2~Ub) and E3 ligases catalyse an isopeptide bond between ubiquitin and a substrate lysine1. The Really Interesting New Gene (RING) family, which share a zinc-coordinating cross-braced RING domain, represent the largest family of ubiquitin E3 ligases. Mechanistically, non-RING elements specify the substrate, while RING domains bind E2~Ub thioesters and induce substrate ubiquitination by stabilizing a productive E2~Ub conformation2. Further, ubiquitin can be targeted to build polyubiquitin chains facilitating diverse signals. Typically, RING-E2 interactions are transient, allowing the E3 to switch E2 partners and assemble polyubiquitin signals3. Approximately 35 E2s are found in mammals, several specializing in polyubiquitination. Mechanisms of linkage-specific polyubiquitination have been extensively studied, however, it is not clear how E3-E2 enzymes install ubiquitin at specific sites of non-ubiquitin substrates.
Site-specific monoubiquitin signals feature in several fundamental DNA damage response pathways4. In higher eukaryotes, repair of DNA interstrand cross-links (ICLs) is mediated by the Fanconi Anemia (FA) pathway, defects in which give rise to FA, a genome instability disorder typified by bone marrow failure and high predisposition to cancers5. A key signal in the FA pathway is monoubiquitination of a specific lysine on two structurally homologous proteins, FANCD2 and FANCI (K561 and K523, respectively, in humans) which likely recruits DNA repair factors6–8. FANCD2 exists mainly in complex with FANCI9 and the mouse FANCD2-FANCI structure reveals an extensive interface that buries the monoubiquitination sites10. Genetic and cell-based studies indicate that a multi-protein FA Core-Complex (FA-CC) E3 ligase activates Ube2T for site-specific monoubiquitination of FANCD2 and FANCI11,12. Mutations in Ube2T/FANCT, an E2 with a C-terminal extension, are also linked to a FA phenotype13. Within the FA-CC resides FANCL, a multi-domain RING protein that facilitates direct FANCD2/FANCI interaction and preferentially binds Ube2T over other E2s14–17. In vitro, the isolated FANCL and Ube2T enzymes catalyse FANCD2 monoubiquitination17,18 while addition of DNA enhances ubiquitination of a FANCD2-FANCI complex, possibly by heterodimer reconfiguration that facilitates lysine access19,20. Further enhancement is observed when FANCL interacts with other FA-CC proteins (FANCB-FANCL-FAAP100 or FANCB-FANCL-FAAP100-FANCC-FANCE-FANCF sub-complexes)21,22. However, mechanistic roles of FA-CC members in promoting E3 activity or directing site-specific ubiquitination are not understood. Furthermore, several lysines on human FANCD2 (32 sites) and FANCI (47 sites) are ubiquitinated in vivo23 thus the mechanism of lysine prioritisation in ICL repair is unresolved. As the repair of DNA-ICLs is crucial for cellular homeostasis, understanding how specific sites on FANCD2/FANCI are strictly monoubiquitinated would provide insights into this pathway and how specific ubiquitin signals are assembled.
Here we show that FANCL activates Ube2T for ubiquitination through an allosteric mechanism distinct from classical RING E3s. We find FANCL’s interaction with Ube2T perturbs its resting state and rewires the E2-fold. These subtle reconfigurations propagate to the E2 active site where a basic triad unique to Ube2T specifies a conserved acidic patch near FANCD2’s target lysine. This three-dimensional interface between Ube2T and FANCD2 is instrumental for FANCL-induced site-specific ubiquitination. Furthermore, by examining E2 residues that link FANCL binding to Ube2T’s active site, we find the allosteric network is intrinsically regulated. Using this insight, we could engineer Ube2T variants that enhance FANCL-mediated FANCD2-FANCI di-monoubiquitination without compromising site-specificity. Finally, we identify similar allosteric networks in other E2s that are appropriated by RING E3s to drive specific ubiquitination.
Results
FANCL drives site-specificity by an atypical mechanism
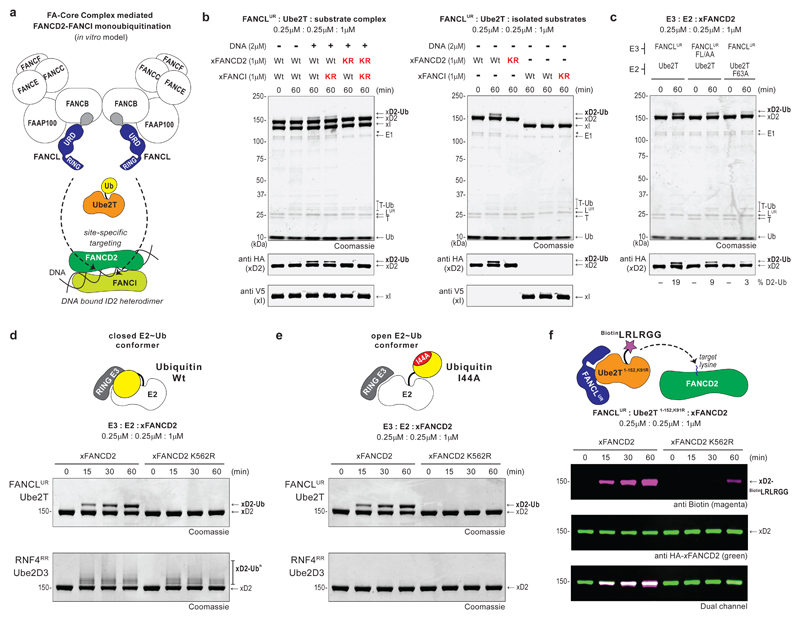
Recent in vitro studies show that a partially reconstituted FA-CC, comprising the dimeric FANCB-FANCL-FAAP100-FANCC-FANCE-FANCF sub-complex, induces efficient di-monoubiquitination of a Xenopus FANCD2-FANCI-DNA complex22,24 (Fig. 1a). The RING-bearing FANCL is embedded in this complex, however, catalytic functions of other FA-CC members are poorly defined. Importantly, FANCL autonomously directs substrate interactions and, in non-vertebrates lacking a FA-CC, mediates site-specific FANCD2 monoubiquitination16,25,26. Thus, to understand mechanisms of site-specific E3 ligase activity we chose to focus on FANCL. Full-length human FANCL is prone to aggregation and has low solubility18,22. Instead, we designed a FANCL URD-RING fragment (FANCLUR) that is stable, monomeric and comprises both substrate (UBC-RWD domain) and E2 (RING domain) binding regions16,17 (Supplementary Fig. 1a). In in vitro assays, sub-stoichiometric levels of FANCLUR and Ube2T catalyse monoubiquitination of Xenopus laevis FANCD2 (xFANCD2) in the xFANCD2-xFANCI-DNA complex or in isolation (Fig. 1b). Modification of xFANCI is undetectable, however, higher FANCLUR-Ube2T concentrations stimulates weak xFANCI monoubiquitination, indicating that xFANCD2 is favoured as a substrate (Supplementary Fig. 1b). While mutations in the substrate-binding patch of FANCLUR (F252A+L254A, FL/AA) and the E3-binding surface of Ube2T (F63A) individually reduce xFANCD2 monoubiquitination, mutation of the physiological site on xFANCD2 (K562R) eliminates modification, confirming site-specific activity of FANCLUR-Ube2T (Fig. 1b,c). In contrast, the well-characterised E3-E2 pair RNF4RR-Ube2D327 ubiquitinates both wildtype and mutant substrate revealing that despite other available ubiquitination sites, FANCLUR-Ube2T targets a particular xFANCD2 lysine (Fig. 1d and Supplementary Fig. 1b). Previous studies show that substrate adaptors and/or additional FA-CC components enhance FANCD2 monoubiquitination19–21. In agreement, we observe FANCLUR-driven xFANCD2 monoubiquitination improves with excess DNA, only with xFANCI, and by swapping FANCLUR with the FANCB-FANCL-FAAP100 complex or SUMO-tagged full-length FANCL (Supplementary Fig. 1d,e). Since FANCLUR-Ube2T alone catalyses xFANCD2 monoubiquitination, we use the minimal components to uncover the underlying mechanism of site-specific E3 activity.
Fig 1. FANCL driven site-specific FANCD2 monoubiquitination does not require the core Ubiquitin fold.
a, Current in vitro model for FANCD2-FANCI monoubiquitination. A partially reconstituted Fanconi Anemia - Core Complex (FA-CC) comprising the dimeric FANCB-FANCL-FAAP100-FANCC-FANCE-FANCF sub-complex activates Ube2T for efficient and site-specific di-monoubiquitination of a Xenopus FANCD2-FANCI heterodimer bound to DNA. The FANCL URD-RING (FANCLUR) fragment is the core E3 ligase module in the FA-CC. b, In in vitro reactions, the FANCLUR-Ube2T pair catalyses site-specific monoubiquitination of Xenopus FANCD2 (xFANCD2/xD2) present in the xFANCD2-xFANCI-DNA substrate complex as well as in isolation. Reactions were carried out in the absence or presence of dsDNA (81 bp) as indicated. ‘Wt’ denotes wildtype substrate, ‘KR’ denotes K562R mutation for xFANCD2 and K524R for xFANCI. A breakdown in wildtype xFANCD2 is denoted by ‘*’. c, In vitro xFANCD2 ubiquitination using a FANCLUR mutant (F252A+L254A, FL/AA) that weakens substrate binding or a Ube2T mutant (F63A) that weakens E3 interaction both result in a decrease in xFANCD2 monoubiquitination. Average percentage of monoubiquitinated FANCD2 from three independent experiments is reported. d, Site-specific monoubiquitination of xFANCD2 is catalysed by the FANCLUR-Ube2T pair but not the RNF4RR-Ube2D3 pair of E3-E2 enzymes. e, Site-specific xFANCD2 monoubiquitination by FANCLUR-Ube2T does not require the I44 Ubiquitin based closed conformation of the E2~Ub thioester. f, The FANCLUR-Ube2T1-152, K91R pair drives site-specific modification of xFANCD2 using a biotinylated Ubiquitin tail peptide (BiotinLRLRGG). Raw images in Supplementary Fig. 11.
Mechanistic studies of RING E3s show RING domains induce a ‘closed’ conformer of the E2~Ub thioester to catalyse ubiquitination (Fig. 1d)2. In this conformation, the I44-centered hydrophobic patch of ubiquitin packs against the E2, while an arginine ‘linchpin’ RING residue braces E2~Ub, priming the thioester for lysine attack. The analogous linchpin position in FANCL is Ser363 which is unlikely to stabilize a closed E2~Ub conformer (Supplementary Fig. 1f). Therefore, we asked if FANCL requires a closed E2~Ub conformation for ubiquitination. Interestingly, the I44A ubiquitin mutant, which renders an open E2~Ub conformer, has no observable effect on site-specific activity of FANCLUR-Ube2T (Fig. 1e, Supplementary Fig. 1b). By contrast, I44A ubiquitin eliminates RNF4RR-Ube2D3 mediated ubiquitination27. As FANCLUR could stabilize other ubiquitin surfaces for E2~Ub activation, we wondered if substrate modification occurs without the globular Ub-fold. To test this, we generated a biotinylated peptide mimicking the ubiquitin tail (BiotinLRLRGG) and is activated by E128. Upon E1-E2 transthioesterification however, the peptide modifies Ube2T and impairs E2 activity. To circumvent this, we generated Ube2T1-152,K91R that lacks the auto-ubiquitination sites11. Remarkably, FANCLUR-Ube2T1-152,K91R readily targets K562 xFANCD2 with BiotinLRLRGG, indicating the Ub-fold is dispensable for site-specific E3 activity (Fig. 1f). Thus, FANCLUR drives site-specific FANCD2 monoubiquitination via a mechanism distinct from generic RING E3s.
Allosteric activation of Ube2T by FANCL
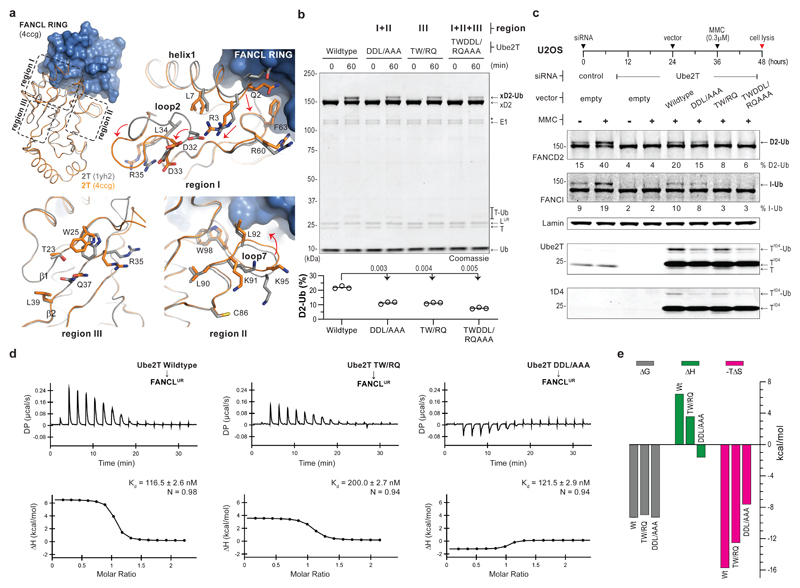
As FANCL directs Ube2T for substrate ubiquitination, we wondered if the mechanism of site-specificity could be revealed by clarifying how FANCL activates Ube2T. To investigate this, we compared structures of Ube2T29 with the FANCL RING (FANCLR) bound Ube2T17. The latter contains two E2 copies in the asymmetric unit, both superpose well with the unbound state and show negligible global differences in the E2-fold (Fig. 2a). However, FANCLR induces local changes in Ube2T’s helix1-loop2 region (R3, D32 and D33, region I) and in loop7 (K91-K95, region II) and loop8 flanking the active site. Further, residues in beta-strands 1-2 (T23, W25, R35 and Q37, region III) are reordered in the FANCLR-Ube2T structure. This region, classically known as ‘backside’ E2, can be influenced by non-RING elements to regulate ubiquitination2. Notably, Ube2T residues in each region are largely conserved (Supplementary Fig. 2a) and not in the E2-FANCLR interface. We therefore wondered if these subtle changes in regions I-III feature in FANCL’s E3 mechanism. To test this, we assayed Ube2T mutants of each region and observe defects in FANCLUR mediated xFANCD2 monoubiquitination (Supplementary Fig. 3a). We extended the analysis by making hybrid mutants spanning regions I+II (D32A+D33A+L92A or DDL/AAA), region III (T23R+W25Q or TW/RQ) and all three regions (T23R+W25Q+D32A+D33A+L92A or TWDDL/RQAAA). In in vitro assays, each hybrid Ube2T mutant confers significant losses in xFANCD2 monoubiquitination (Fig. 2b). These defects could be directly linked to how FANCL activates the Ube2T~Ub thioester, or arise indirectly due to compromised E2~Ub charging and/or lysine conjugation. To discriminate this, we tested E2 charging/auto-ubiquitination and find the hybrid mutants to be comparable to wildtype Ube2T (Supplementary Fig. 2b,c). In contrast, single-turnover assays reveal the mutants impair FANCLUR-E2~Ub productivity and delay xFANCD2 monoubiquitination (Supplementary Fig. 3b). Thus, Ube2T mutations mitigate FANCL’s E3 activity without influencing intrinsic E2 activity.
Fig 2. FANCL induced changes in Ube2T contribute to FANCD2 monoubiquitination.
a, Superpose of FANCL RING (FANCLR, blue) bound Ube2T (orange, PDB ID 4ccg) on unbound Ube2T (grey, PDB ID 1yh2) shows minimal global changes in the E2-fold (residues 1-152, Cα RMSD ~ 0.8Å) upon FANCLR binding. Dashed boxes and corresponding close-ups of Ube2T regions I, II and III reveal local changes in the E2-fold. b, FANCLUR mediated in vitro xFANCD2 ubiquitination is compromised by Ube2T mutations in the indicated regions. A breakdown in xFANCD2 is denoted by ‘*’. DDL/AAA, TW/RQ and TWDDL/RQAAA denote Ube2T mutations D32A+D33A+L92A, T23R+W25Q and T23R+R25Q+D32A+D33A+L92A, respectively. Graphs represent percentage xFANCD2 monoubiquitination (n=3) with a line at mean value. Significant differences were assessed by one-way ANOVA with Dunnett’s multiple-comparison test. Adjusted P values are shown. c, U2OS cells transfected with the indicated siRNA and expression vector (empty/Ube2T-1D4) were treated with DNA crosslinking agent mitomycin-C (MMC, 0.3 µM) as indicated. Experiment schematic is shown above and cell-lysates were analysed by immunoblotting. Mutations in Ube2T reduce monoubiquitination of FANCD2 and FANCI. Average percentage from two biological replicates is reported. Ube2T mutations as in panel b. Raw images in Supplementary Fig. 11. d, Raw and experimental fits of isothermal calorimetric titration of Ube2T to FANCLUR and (e) the associated thermodynamic components (ΔG, ΔH and -TΔS) as bar graphs (values in Supplementary Table 1). Despite comparable binding energies (ΔG), a reduction in binding entropy (-TΔS) suggest smaller conformational changes in the mutant E2-E3 complex. Ube2T mutants as in panel b.
We also wondered if the hybrid Ube2T mutants influenced E3 activity of the FA-CC. In human cell-lines, mitomycin-C (MMC) induced DNA-ICLs trigger FANCD2-FANCI di-monoubiquitination mediated by FA-CC/Ube2T11,13,30,31. Knockdown of Ube2T by siRNA abolishes FANCD2/FANCI modification, however, this can be rescued by transient expression of siRNA-resistant Ube2T-1D4 (Ube2T with C-terminal 1D4-tag, Fig. 2c). Interestingly, Ube2TDDL/AAA-1D4 partially rescues MMC-induced FANCD2/FANCI monoubiquitination while Ube2TTW/RQ-1D4 and Ube2TTWDDL/RQAAA-1D4 impair substrate modification. The contrast between in vitro and cell-based FANCD2 monoubiquitination with Ube2TDDL/AAA (Fig. 2b,c) suggests E2 activation is improved by the FA-CC. These data collectively reveal that Ube2T residues in regions I-III are play a role in efficient E3 activity of FANCL/FA-CC.
We then analysed E3-E2 interactions to understand the basis of restricted activity. Using isothermal titration calorimetry, we observe similar affinities of FANCLUR for wildtype and Ube2TDDL/AAA (average Kd ~ 119 nM) while Ube2TTW/RQ displays a modest binding defect (Kd = 200 nM) (Fig. 2d). The affinity of FANCLR-Ube2T is unaltered by the mutations (average Kd ~ 253 nM) (Supplementary Fig. 3c), but interestingly, is 2-fold weaker than FANCLUR-Ube2T. Thus, a non-RING element in FANCLUR, likely the URD, supports Ube2T interactions via the E2’s backside. The smaller FANCLR fragment, which lacks substrate-binding URD16, is expectedly weaker than FANCLUR in site-specific xFANCD2 monoubiquitination (Supplementary Fig. 3d). However, comparing relative activities of FANCLR and FANCLUR with each hybrid Ube2T mutant reveals that Ube2TDDL/AAA reduces K562 xFANCD2 monoubiquitination regardless of the E3 employed. In contrast, Ube2TTW/RQ impairs only FANCLUR-driven reactions indicating that the longer E3 fragment binds distinct E2 regions to stimulate activity. We also uncover thermodynamic variations that reflect a bimodal influence of FANCLUR for Ube2T activation (Fig. 2e and Supplementary Table 1). Despite comparable binding energies (ΔG), the large binding entropy (-TΔS) observed with FANCLUR-Ube2T interaction (-15.75 kcal/mol) is diminished by TW/RQ and DDL/AAA mutations in Ube2T and by URD deletion in FANCL (-12.55, -7.64 and -12.70 kcal/mol, respectively). The reduced entropy is likely associated with smaller conformational changes in Ube2T upon E3-binding and accounts for weakened substrate ubiquitination. Taken together, the analyses suggest FANCL-binding influences the E2-fold to allosterically activate Ube2T for FANCD2 monoubiquitination.
E2 active site and FANCD2 devise site-specificity
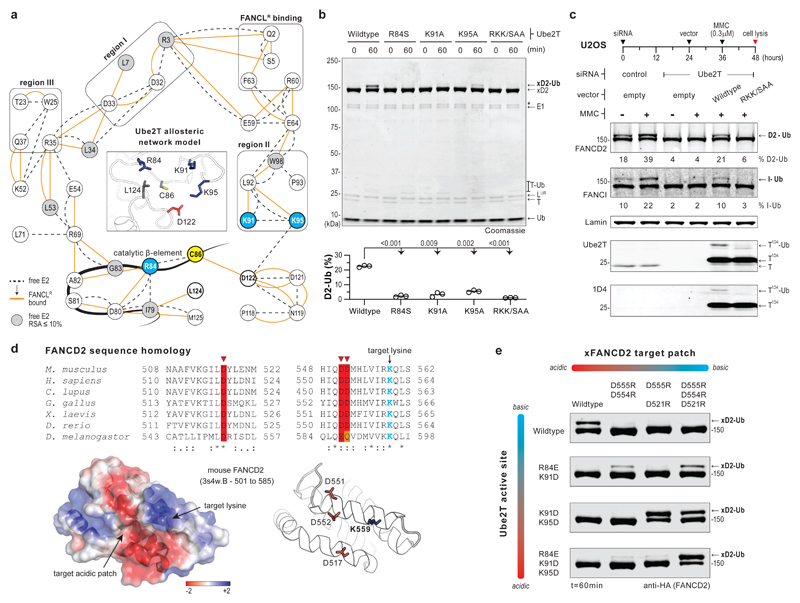
FANCL’s E3 mechanism likely involves subtle changes within Ube2T, therefore, we generated residue interaction networks (RINs) to uncover the mechanics of allosteric activation. Such networks plot E2 residues as nodes while edges represent physicochemical interactions in the tertiary structure. Comparison of unbound and FANCLR-bound Ube2T RINs reveals edges/interactions unique to each network, together representing a ‘dynamic’ RIN (Supplementary Fig. 4a and Supplementary Dataset 1). Here we chose E2 nodes/residues that support FANCLR-binding as initial search nodes to trace connected neighbours, which then serve as successive search nodes. By iteration, we trace possible paths between the FANCLR-binding cluster and E2 active site, focusing on conserved nodes and regions I-III, while filtering out diverging paths. The final allosteric network model represents how FANCLR-binding rewires the intra-molecular connections of Ube2T and likely activates substrate ubiquitination (Fig. 3a). Notably, the network termini in the catalytic beta-element (R84), loop7 (K91 and K95), loop8 and its C-terminal hinge (D122 and L124, respectively) lie within 10Å of Ube2T’s catalytic cysteine (C86, Fig. 3a inset) and could regulate FANCL-induced FANCD2 ubiquitination. Conversely, given their proximity to C86 they could also regulate E2 activity. In several E2s, a hydrophobic residue (L124 in Ube2T) is crucial for E1~Ub-E2~Ub transthioesterification32 while a loop8 residue (D122 in Ube2T) positions and/or deprotonates the target lysine27 (Supplementary Fig. 4b). The remaining Ube2T network termini (R84, K91 and K95) vary among ubiquitin E2s and mutational analysis (R84S, K91A, K95A and R84S+K91A+K95A or RKK/SAA) reveals no impact on E3 interaction or E2~Ub charging with a minor E2 auto-ubiquitination defect11 (Supplementary Fig. 2b,2c,4c and Supplementary Table 1). In contrast, each single mutant impedes xFANCD2 monoubiquitination, while Ube2TRKK/SAA eliminates both in vitro and cell-based substrate monoubiquitination (Fig. 3b,c). Thus, the Ube2T basic triad (R84, K91 and K95) are required exclusively for FANCL/FA-CC driven substrate monoubiquitination but are dispensable for fundamental E2 activity.
Fig 3. Basic triad in Ube2T active site and target acidic patch on FANCD2 licence site-specific ubiquitination.
a, Allosteric network model (38 nodes, 74 edges) shows rewiring of Ube2T intra-residue network upon FANCLR interaction. Dashed and orange lines depict connections in free and FANCLR-bound E2, respectively. Black boxes indicate nodes involved in FANCLR binding and in regions I-III. The catalytic cysteine (C86, yellow) and the proximal network termini (Sγ-Cβ distances ≤ 10Å in unbound Ube2T (inset), PDB ID 1yh2) have a thick outline, with the basic triad (R84, K91 and K95) coloured in blue. The catalytic beta-element includes nodes 82-86. Grey nodes have relative solvent accessibility of <10% in free Ube2T. b, Mutations of Ube2T’s basic triad impair FANCLUR mediated xFANCD2 ubiquitination. RKK/SAA denotes Ube2T mutant R84S+K91A+K95A. Graphs represent percentage xFANCD2 monoubiquitination (n=3). Significant differences were assessed by one-way ANOVA with Dunnett’s multiple-comparison test. Adjusted P values are shown. c, U2OS cells transfected with the indicated siRNA and expression vector (empty/Ube2T-1D4) were treated with mitomycin-C (MMC) as indicated. Experiment schematic is shown above and cell-lysates were analysed by immunoblotting. Average percentage of FANCD2 and FANCI monoubiquitination from two biological replicates is reported. d, FANCD2 sequence alignment showing conservation of acidic residues (red/orange) proximal to target lysine (blue). Structure of mouse FANCD2 (PDB ID 3s4w) depicts acidic residue configuration in relation to target lysine (Cβ-Cβ distances ≤ 13Å) Surface electrostatic potential is colour scaled from −2 kT/e (red) to +2 kT/e (blue). e, Functional significance of Ube2T’s basic triad and xFANCD2’s target acidic patch is assessed by in vitro ubiquitination assays using the indicated charge-reversal mutations. The FANCLUR-Ube2TR84E+K91D and FANCLUR-Ube2TK91D+K95D enzyme pair’s monoubiquitinate xFANCD2D555R+D554R and xFANCD2D555R+D521R, respectively, in a mutually exclusive manner. Consequently, FANCLUR-Ube2TR84E+K91D+K95D exclusively monoubiquitinates xFANCD2D555R+D554R+D521R. Raw images in Supplementary Fig. 11.
We wondered if this catalytic paradox underlies FANCL’s mechanism for site-specific FANCD2 monoubiquitination. Intriguingly, through analysis of the mouse FANCD2 structure10 and sequence homology, we notice a conserved acidic patch proximal to the target lysine (Fig. 3d). In theory, the Ube2T basic triad could mediate electrostatic interactions with FANCD2’s acidic patch and stabilize the E2-substrate intermediate for ubiquitination. To test this, we generated a series of charge-reversal mutations in xFANCD2’s acidic patch (D555R+D554R, D555R+D521R and D555R+D554R+D521R) and Ube2T’s basic triad (R84E+K91D, K91D+K95D and R84E+K91D+K95D). In in vitro assays, none of the acidic patch xFANCD2 mutants are ubiquitinated by FANCLUR-Ube2T (Fig. 3e). Remarkably, FANCLUR catalyses xFANCD2D555R+D554R and xFANCD2D555R+D521R monoubiquitination using Ube2TR84E+K91D and Ube2TK91D+K95D, respectively. Moreover, monoubiquitination of xFANCD2 dyad mutants is mutually exclusive, implying that productive E2-substrate interactions occur in a specific configuration. Finally, FANCLUR-Ube2TR84E+K91D+K95D efficiently monoubiquitinates xFANCD2D555R+D554R+D521R, however, does not modify xFANCD2 or the patch mutant lacking the target lysine (xFANCD2D555R+D554R+D521R+K562A, Fig. 3e and Supplementary Fig. 4d). These results reveal that the basic triad in Ube2T and a target acidic patch on FANCD2 are critical for site-specificity. Furthermore, unlike typical RING E3s3, FANCLUR does not enhance reactivity of the E2~Ub thioester towards free nucleophiles (Supplementary Fig. 4e). Thus, the catalytic outcome of FANCL-Ube2T is fine-tuned for a specific, three-dimensionally structured substrate surface. In summary, FANCL facilitates productive interactions between the basic triad in Ube2T’s active site and a structured target acidic patch on FANCD2, thus enforcing site-specific ubiquitination.
E2 allosteric conduit regulates site-specific E3 activity
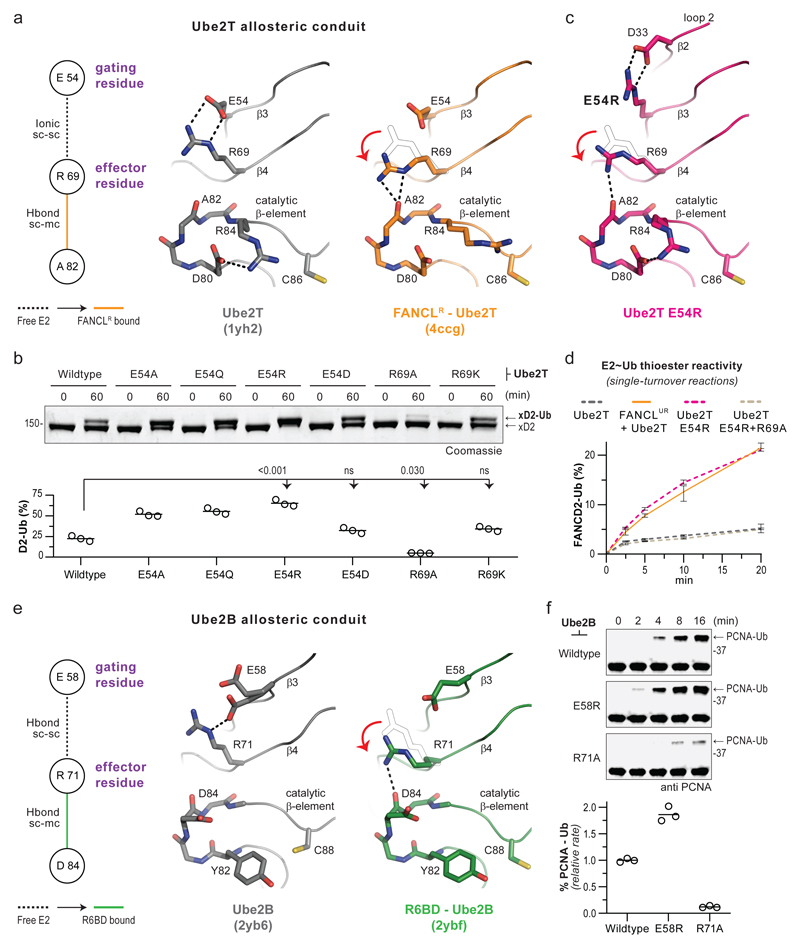
Site-specific FANCD2 monoubiquitination involves modulation of Ube2T’s active site, therefore, the allosteric network model likely includes residue connections that orchestrate FANCL’s activation of Ube2T (Fig. 3a). A long-range network (D33-R35-E54-R69-A82) propagates through Ube2T regions I and III, culminating at the catalytic beta-element where R84 resides. In particular, the R69 side-chain is secured by E54 in unbound Ube2T, however, this link is altered upon FANCL binding (Fig. 4a). Consequently, R69 is free to stabilize the catalytic beta-element, which could facilitate interactions between Ube2T R84 and FANCD2’s target acidic patch. In other words, an effector role for R69 could be gated by E54 in Ube2T’s resting state and allosterically relieved by FANCL to trigger substrate ubiquitination. We tested this hypothesis using Ube2T E54A/Q/R mutants, which would disrupt ionic E54-R69 interactions, and observe enhanced FANCLUR-driven K562 xFANCD2 monoubiquitination, while a conservative Ube2TE54D mutant retains Ube2T-like activity (Fig. 4b, Supplementary Fig. 5a). Meanwhile, Ube2TR69A, which would prevent hydrogen bonding between R69 and catalytic beta-element, reduces FANCLUR-driven K562 xFANCD2 monoubiquitination, while a conservative Ube2TR69K mutant rescues this defect. Notably, E54 and R69 mutants do not alter intrinsic Ube2T activity (Supplementary Fig. 2b,c). To further investigate the gating-effector conduit, we determined the structure of Ube2TE54R to 2.0Å resolution (Supplementary Table 2). The E2-fold of Ube2TE54R is overall similar to unbound Ube2T, however, certain local changes are observed (Supplementary Fig. 5b-d). Loop2 (region I) is reordered while the E54R mutation forms a salt-bridge with D33. Consequently, the R69 side-chain is repositioned within hydrogen bonding distance of the catalytic beta-element (Fig. 4c). These subtle changes in the gating-effector conduit of Ube2TE54R are reminiscent of FANCLR-bound Ube2T and could represent an ‘activated’ E2 state. Remarkably, in single-turnover assays, the Ube2TE54R~Ub thioester catalyses E3-independent xFANCD2 monoubiquitination to a similar extent as the FANCLUR-Ube2T~Ub pair (Fig. 4d, Supplementary Fig. 5e). Therefore, E54 operates as a molecular gate that restricts Ube2T for FANCD2 ubiquitination, however, is rendered permissive by FANCL-binding or targeted mutagenesis.
Fig 4. Specialised residue pair within the E2-fold regulates RING E3 driven substrate ubiquitination.
a, Comparison of a residue network with structures of unbound (grey, PDB ID 1yh2) and FANCLR-bound (orange, PDB ID 1yh2) Ube2T depicting the proposed allosteric conduit. The E54 functions as a gating residue based on its influence on R69, which in turn acts as an effector residue for stabilising the catalytic beta-element. b, FANCLUR mediated in vitro xFANCD2 ubiquitination using Ube2T mutants of the gating and effector residues. Graphs represent percentage xFANCD2 monoubiquitination (n=3) with a line at mean value. Significant differences were assessed by one-way ANOVA with Dunnett’s multiple-comparison test. Adjusted P values are shown. c, Structure of Ube2T gating mutant E54R (magenta) reveals a bonding network in the allosteric conduit similar to the FANCLR-bound Ube2T structure. d, Monoubiquitination of xFANCD2 (1 µM) under single-turnover conditions shows reactivity of the indicated E2~Ub thioester (1 µM) in the absence or presence of FANCLUR (1 µM). Percentage of xFANCD2 monoubiquitination (mean ± range, n=3) is plotted over time. To prevent E2 auto-ubiquitination, Ube2T1-152, K91R is used and indicated mutations incorporated in this background. e, Comparison of a residue network with structures of unbound Ube2B (grey, PDB ID 2yb6) and Rad6 Binding Domain (R6BD) bound Ube2B (green, PDB ID 2ybf) depicting the proposed gating and effector roles for Ube2B residues E58 and R71, respectively. f, Immunoblots of in vitro ubiquitination assays shows Ube2B with a permissive gate (E54R) is more responsive to Rad18 in PCNA monoubiquitination, while the Ube2B effector mutant (R71A) slows the reaction. Percentage of PCNA-Ub (n=3) were quantified, linear rates normalized to Rad18-Ube2B wildtype reaction and plotted with a line at mean value. Raw images in Supplementary Fig. 11.
Curiously, the basic triad configuration in Ube2TE54R resembles unbound more than FANCLR-bound Ube2T (Supplementary Fig. 6a). To investigate active site dynamics, we performed all-atom molecular dynamics simulations with Ube2TE54R and Ube2T structures. We observe no significant differences in the E2 backbone, however, differences in side-chain configurations of the basic triad were discernible (Supplementary Fig. 6b). Notably, the R84/K91 side-chain(s) are more likely to adopt distinct conformation(s), with respect to C86, mainly in Ube2TE54R trajectories. In absence of an E2-substrate intermediate structure, we cannot unambiguously define a ‘catalytic conformation’ for Ube2T’s basic triad. Nevertheless, we speculate that the steadied conformation(s) for R84/K91 in Ube2TE54R may be responsible for substrate ubiquitination and could stem from R69 (effector) influencing the catalytic beta-element. Concurrently, the R69A mutation in Ube2T, that impedes FANCLUR-activation (Fig. 4b), also reverts xFANCD2 modification induced by Ube2TE54R (Fig. 4d, Supplementary Fig. 5e). Thus, FANCL optimises Ube2T’s active site for site-specific FANCD2 monoubiquitination via dynamics of the gating-effector conduit.
We wondered if such regulatory conduits exists in other E2s involved in specific ubiquitin signals. For example, repair pathways that support bypass of stalled DNA replication are initiated upon site-specific (K164) monoubiquitination of Proliferating Cell Nuclear Antigen (PCNA) by the E3-E2 pair Rad18-Ube2B(Rad6B)33. The Rad18-Ube2B interaction is bimodal, with the canonical RING-E2 interface supported by Rad18’s Rad6 binding domain (R6BD) interacting with backside E234. Comparing networks/structures of unbound and R6BD-bound E2s, we notice a potential gating-effector (E58-R71) conduit in Ube2B (Fig. 4e, Supplementary Dataset 2), which is distal from the bimodal Rad18-Ube2B interface. In in vitro assays, Ube2BE58R improves while Ube2BR71A weakens Rad18 mediated PCNA monoubiquitination, however, neither mutant alters intrinsic E2 activity, thus revealing their regulatory roles in substrate modification (Fig. 4f, Supplementary Fig. 7a,b). It is currently unknown if Ube2B active site residues interact with PCNA. We speculate the gating-effector conduit modulates substrate ubiquitination by stabilizing the catalytic beta-element in Ube2B. Taken together, these results suggest that regulatory conduits may operate across the E2 family and could be leveraged by E3s for specific ubiquitin signals.
Ube2T engineering enhances substrate ubiquitination
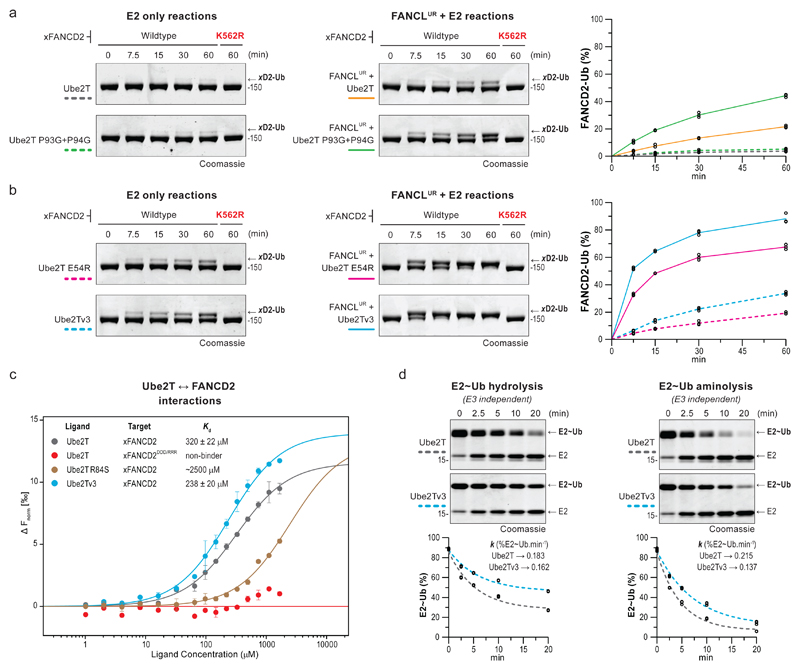
Based on Ube2T’s allosteric network, we successfully engineered a permissive gate (E54R) that enhances FANCD2 monoubiquitination. We wondered if other Ube2T regions could be similarly exploited. The K91/K95 pair in region II are critical for target acidic patch interactions, suggesting the dynamics of loop7 (K91LPPK95) might influence substrate ubiquitination. While FANCLR-binding draws loop7 away from the active site and rewires a short-range network leading into region II (Fig. 2a,3a), the presence of two conserved prolines (P93 and P94, Supplementary Fig. 2a) rigidifies this loop. We reasoned a flexible loop7 could emulate the dynamics observed with FANCL-binding and/or increase the likelihood of K91/K95 interactions with FANCD2’s target acidic patch. Therefore, we engineered a flexible loop7 (P93G+P94G) and observe an improvement in K562 xFANCD2 monoubiquitination by FANCLUR-Ube2TP93G+P94G, however, not with isolated Ube2TP93G+P94G (Fig. 5a, Supplementary Fig. 8a). A plausible explanation for this contrast is the restrictive gate (E54) in isolated Ube2TP93G+P94G. Consequently, we generated another variant, Ube2Tv3, comprising both a permissive gate (E54R) and flexible loop7 (P93G+P94G). The isolated Ube2Tv3 can stimulate efficient K562 xFANCD2 monoubiquitination (Fig. 5b) furthermore, is superior to Ube2TE54R and Ube2TP93G+P94G, revealing a synergistic effect of the mutations. Remarkably, Ube2Tv3 in combination with FANCLUR catalyses xFANCD2 monoubiquitination to near completion without compromising site-specificity. Furthermore, sub-stoichiometric levels of FANCLUR-Ube2Tv3 can also catalyse K524 xFANCI monoubiquitination (Supplementary Fig. 8b), albeit more weakly than xFANCD2. The FANCI surface10 has a spatially constricted acidic patch near the target lysine (Supplementary Fig. 8c), which correlates with lower ubiquitination. Importantly, it reveals how the three-dimensional configuration of the target acidic patch regulates the efficiency of substrate monoubiquitination.
Fig 5. Ube2Tv3 enhances E3-dependent and E3-independent FANCD2 monoubiquitination.
a, b, Monoubiquitination of xFANCD2 (n=3) is improved by a Ube2T variant with a flexible loop7 (Ube2TP93G+P94G) only in the presence of FANCLUR. In contrast, xFANCD2 monoubiquitination is enhanced by Ube2Tv3, which includes a permissive gate (E54R) and a flexible loop7 (P93G+P94G), both in the presence and absence of FANCLUR. Reactions were also performed with xFANCD2 K562R mutant to assess site-specificity. c, MST dose-response-curves for interactions between Ube2T (Ligand) against xFANCD2 (Target). Curves were fitted to a one-site binding model for Kd determination and plotted against baseline corrected normalized fluorescence (Δ Fnorm [‰]). Mutation of the target acidic patch on xFANCD2 (D555R+D554R+D521R or xFANCD2DDD/RRR) or a basic residue in Ube2T’s active site (R84S) weakens or abolishes E2-substrate interactions. A minor improvement in xFANCD2 interaction is observed with Ube2Tv3. To allow for high ligand concentrations, Ube2T1-152 is used and indicated mutations incorporated in this background. All measurements were done in triplicates, error bars indicate standard deviation. d, Single-turnover assays to assess hydrolysis and aminolysis (20 mM lysine) of the E2~Ub thioester (3 µM). To prevent E2 auto-ubiquitination, Ube2T1-152, K91R is used and mutations incorporated in this background. Percentage of E2~Ub (n=3) were plotted over time and fitted with one-phase decay model (see Supplementary Fig. 7d for best-fit values and the goodness of fit). k denotes the rate constant (%E2~Ub.min-1). The Ube2Tv3~Ub thioester is relatively more stable than Ube2T~Ub in both reactions. Raw images in Supplementary Fig. 11.
The engineered permissive gate (Ube2TE54R) influences R84/K91 conformations while loop7 flexibility (Ube2TP93G+P94G) likely permits more conformational freedom for K91/K95. Therefore, enhanced activity of Ube2Tv3 could arise from optimised interactions between the basic triad and the target acidic patch. Alternatively, increased dynamics/flexibility of the engineered E2-fold may improve E2~Ub thioester reactivity. To clarify this, we analysed E2-substrate interactions using microscale thermophoresis. Interestingly, a weak Ube2T-xFANCD2 interaction (Kd = 320±22 μM) is detectable, dependent on both the target acidic patch and the basic triad (Fig. 5c). With Ube2Tv3, we observe a minor improvement in xFANCD2 interaction (Kd = 238±20 μM) that may contribute to enhanced substrate ubiquitination. We also examined E2~Ub thioester stability and its nucleophile reactivity. Ube2Tv3~Ub is marginally more stable than Ube2T~Ub while, unexpectedly, aminolysis is slowed (Fig. 5d, Supplementary Fig. 8d). These data suggest Ube2Tv3 is more adept at targeting the structured substrate lysine with an organised acidic patch than a free lysine. In summary, the underlying enhancement in Ube2Tv3-driven substrate ubiquitination is multifactorial and includes subtle reordering of the basic triad, minor improvements in substrate interaction, increased E2~Ub thioester stability and an optimal configuration of the target acidic patch.
FANCL-Ube2Tv3 catalyse FANCD2-FANCI ubiquitination
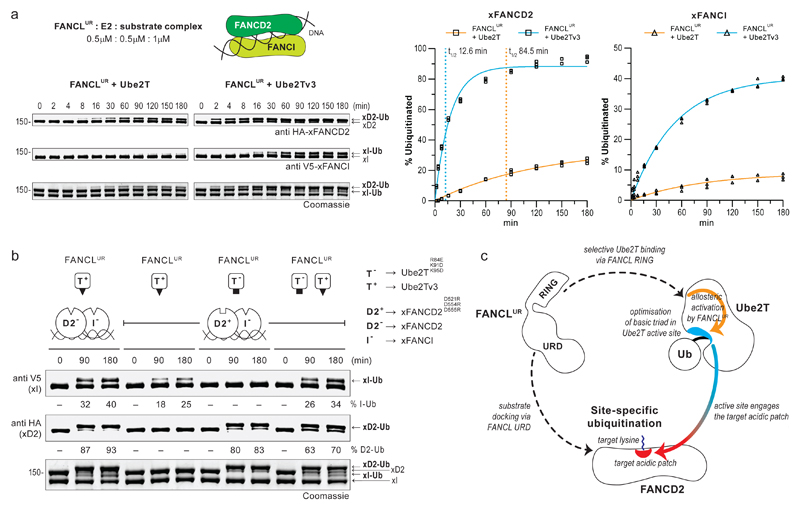
As the engineered Ube2Tv3 efficiently catalyses monoubiquitination of isolated FANCD2/FANCI, we assessed ubiquitination of the xFANCD2-xFANCI-DNA complex and find FANCLUR-Ube2Tv3 improves both the rate and level of xFANCD2 monoubiquitination (7- and 4-fold, respectively) relative to FANCLUR-Ube2T (Fig. 6a, Supplementary Fig. 9a). Further, we observe a remarkable improvement in xFANCI monoubiquitination with ~40% of the substrate modified. Overall, di-monoubiquitination of the DNA-bound heterodimer by the minimal FANCLUR-Ube2Tv3 module is comparable to the reconstituted FA-CC/Ube2T setup22. Moreover, FANCLUR-Ube2Tv3 does not compromise site-specificity and partially overcomes the requirement of DNA for efficient xFANCD2-xFANCI di-monoubiquitination (Supplementary Fig. 9b). The contrasting monoubiquitination profiles for xFANCI, isolated versus the heterodimer, indicates that xFANCD2 influences the efficiency of xFANCI modification (Fig. 6a, Supplementary Fig. 8b). In vivo, phosphorylation of S/TQ motifs in FANCI is reported to regulate substrate ubiquitination35,36 while FANCD2-binding reconfigures FANCI’s S/TQ motifs10 placing them proximal to the target lysine (Supplementary Fig. 8c). Thus, phosphorylation and the presence of FANCD2 could together optimise FANCI’s target acidic patch and promote its monoubiquitination.
Fig 6. Monoubiquitination of FANCD2 regulates efficiency of FANCI monoubiquitination.
a, A time-course ubiquitination assay of the xFANCD2-xFANCI-DNA complex using FANCLUR-Ube2T (left) or FANCLUR-Ube2Tv3 enzyme pairs (right). Percentage of substrate monoubiquitination (n=3) was determined from anti HA (xFANCD2) and anti V5 (xFANCI) immunoblots. Values were plotted over time and fitted with one-phase accumulation model (see Supplementary Fig. 8a for best-fit values and the goodness of fit). t1/2 denotes reaction half-time in minutes. b, FANCLUR catalysed ubiquitination assays of the DNA bound substrate heterodimer using orthogonal E2-substrate pairs. T− denotes Ube2T with an acidic active site (black square, Ube2TR84E+K91D+K95D) that can modify xFANCD2 with a basic target patch (open white square, D2+, xFANCD2D521R+D554R+D555R). T+ denotes the enhanced Ube2Tv3 (Ube2TE54R+P93G+P94G) with a basic active site (black triangle) that can modify substrates with a target acidic patch (open white triangles, xFANCD2 (D2−) or xFANCI (I−)). In the I−-D2+-DNA complex, the FANCLUR-T+ and FANCLUR-T− enzyme pair’s monoubiquitinate I− and D2+, respectively, in a mutually exclusive manner. However, levels of I− monoubiquitination increase both T− and T+ are present indicating that monoubiquitinated D2+ improves efficiency of I− modification. Raw images in Supplementary Fig. 11. c, Model for FANCL catalysed site-specific FANCD2 ubiquitination. FANCL mediates substrate docking via its central UBC-RWD domain (URD) and selectively recruits Ube2T primarily via the RING domain. FANCLUR binding of Ube2T allosterically influences a basic triad in the E2 active site. The optimised active site promotes productive encounters with target acidic patch on FANCD2 and thereby catalyses site-specific ubiquitination.
Curiously, levels of xFANCI monoubiquitination in a xFANCD2K562R-xFANCI-DNA complex regress to those observed with isolated xFANCI. (Supplementary Fig. 8b,9b). In the former, a K562R mutation prevents xFANCD2 modification, thus implying efficient xFANCI modification depends both on xFANCD2 and its monoubiquitinated state. To test this, we devised an orthogonal E2-substrate assay using Ube2TR84E+K91D+K95D (T−), which specifies a basic patch on xFANCD2D555R+D554R+D521R (D2+), and Ube2Tv3 (T+), which requires the acidic patch on xFANCI (I–). In ubiquitination assays of a DNA-bound D2+-I– complex, FANCLUR-T+ exclusively monoubiquitinates I–, while conversely, FANCLUR-T– selectively monoubiquitinates D2+, validating the orthogonal setup (Fig. 6b, Supplementary Fig. 9c). Surprisingly, I– monoubiquitination is improved when both T– and T+ are present in the reaction, despite the former E2 being nonreactive on I–. Therefore, the D2+-Ub product, catalysed by FANCLUR-T–, stimulates I– monoubiquitination by FANCLUR-T+, confirming our hypothesis. Overall, these results indicate that precise ubiquitination is mediated by FANCL-Ube2T acting on the target acidic patch, however, di-monoubiquitination of FANCD2-FANCI is inherently sequential and is regulated by an interplay between the DNA-bound heterodimer.
Discussion
Monoubiquitination of FANCD2 and FANCI is a critical signal in the FA-ICL repair pathway37. Previous studies on FANCL, the catalytic FA-CC subunit, revealed substrate binding features of the UBC-RWD16 and an extended RING-E2 interface that underlies selective E2 recruitment17. In this study, we uncover the molecular mechanism by which FANCL-Ube2T catalyses site-specific FANCD2 monoubiquitination. Unlike most RING E3s, FANCL lacks the classical ‘linchpin’ that primes a productive E2~Ub thioester conformation2. Rather than stabilizing the ubiquitin-fold, FANCL instead stimulates the E2-fold of Ube2T through the RING-E2 interface and backside E2 interactions, likely via the UBC-RWD. This bimodal and high-affinity FANCL-Ube2T interaction rewires the E2’s intra-residue network thereby allosterically influencing its active site. Notably, we find a basic triad unique to Ube2T’s active site and a conserved acidic patch near FANCD2’s target lysine license FANCL’s site-specific E3 activity. Reports on site-specific modification by ubiquitin/ubiquitin-like proteins reveal how E3-substrate interactions guide the E2-E3 complex to a lysine targeting zone38–41. Meanwhile, studies on linkage-specific polyubiquitination show how E2 interactions with ubiquitin residues near the target lysine regulate site-specificity42–46. Mechanistically, FANCL-Ube2T mirrors these strategies in that substrate docking by FANCL’s UBC-RWD restricts the lysine-targeting area of the RING-bound E2, while electrostatic complementarity between Ube2T’s basic triad and FANCD2’s target acidic patch fine-tunes the lysine-targeting zone. However, unique to this E3-E2 pair is the allosteric activation of Ube2T by FANCL that optimises the E2’s active site for productive interactions with FANCD2’s target acidic patch, thereby driving site-specific ubiquitination (Fig. 6c). In fact, FANCL’s allosteric activation of Ube2T is uniquely adapted for a three-dimensionally structured surface thus precluding any spurious ubiquitination. As we do not observe processive modification, either on the installed ubiquitin or a different FANCD2 site, neither surface is optimally recognized by FANCL-Ube2T, restricting ubiquitination to a single event.
Our structure-guided analysis also reveals the allosteric trigger, i.e. the gating-effector couple in Ube2T, which underlies FANCL’s activation mechanism. In unbound Ube2T, a gating residue (E54) restricts an effector residue (R69) from influencing the active site. However, FANCL binding frees this restraint, which likely restructures the active site to promote site-specific substrate ubiquitination. Interestingly, mutations of the gating residue yield deregulated E2s that improve both FANCL-dependent and -independent FANCD2 monoubiquitination, without compromising site-specificity. We find analogous pairs in several other E2s that could potentially function as gating-effector residues (Supplementary Fig. 10) and validate this feature for Ube2B/Rad6B, an E2 responsible for site-specific PCNA monoubiquitination. Recent efforts in designing E2 inhibitors have uncovered promising fragments that bind near this regulatory pair and allosterically alter E2 activities47,48. These observations collectively suggest that allosteric networks operate across the E2 family and could prove instrumental for understanding specificity in ubiquitin signalling.
By elucidating FANCL’s atypical E3 activity and the underlying mechanics of site-specificity, we successfully engineered a catalytically-enhanced E2 variant, Ube2Tv3, which substantially improves FANCD2-FANCI di-monoubiquitination. Thus, substrate-binding and site-specificity are essentially inherent to the FANCL-Ube2T module, consistent with studies in non-vertebrates lacking a FA-CC25,26. Importantly, we show that Ube2T activation and optimization of the target acidic patch on FANCD2/FANCI are two critical factors that influence the efficiency of site-specific monoubiquitination. We propose that substrate adaptors and/or FA-CC members serve these functions. Indeed, several studies have demonstrated how FANCI/DNA improve FANCD2 monoubiquitination18–20. Given their intimate relationship with FANCD2, FANCI/DNA are ideal candidates for reconfiguring FANCD2’s target acidic patch. The FANCC-FANCE-FANCF complex has also been suggested to modulate the FANCI-FANCD2-DNA complex to promote di-monoubiquitination22,24. In addition, the FANCB-FANCL-FAAP100 complex is reported to enhance FANCD2 ubiquitination21. We propose that Ube2T activation could be augmented when FANCL is embedded in this complex. FANCI phosphorylation reportedly regulates substrate ubiquitination36 and may optimise FANCI’s target acidic patch. Moreover, we find efficient FANCI monoubiquitination relies on ubiquitinated FANCD2. Conversely, FANCI phosphorylation reportedly impedes activity of the cognate deubiquitinating enzyme USP135, thus favouring the ubiquitinated state. Evidently, the monoubiquitination status of FANCI-FANCD2 is regulated at multiple levels. Using the engineered E2-E3 pair we have designed a method49 that can readily generate natively monoubiquitinated FANCD2 and FANCI. This will enable characterisation of the ubiquitinated substrates, both in isolation and as a complex. Moreover, this approach has been instrumental in revealing how the site-specific ubiquitin signal on FANCD2 is removed by USP150. Similarly, the monoubiquitinated products could provide insights into their interactions with DNA, the temporal nature of di-monoubiquitination and facilitate discovery of downstream readers of the signal.
Online Methods
Plasmids, oligonucleotides and peptides
Human Ube2T and Ube2D3 (I.M.A.G.E. clone, Geneservice) were inserted using restriction cloning into a modified pET15 vector (Novagen) harbouring a 3C-cleavable 6xHis-tag sequence at the N-terminus. Human Ube2B (I.M.A.G.E. clone, Geneservice) was inserted via Gateway Cloning (Invitrogen) into a modified pDEST17 vector (Invitrogen), harbouring a TEV-cleavable 6xHis-tag sequence at the N-terminus. A synthetic human FANCL sequence (GeneArt) was used as template to insert FANCL (residue 1-375), FANCLUR (residues 109-375) and FANCLR (residues 289-375) coding regions into a pET SUMO vector (Invitrogen) using restriction-free cloning. All FANCL constructs harbour a 6xHis-smt3 tag at the N-terminus. A synthetic human Rad18 sequence (GeneArt) was inserted by restriction cloning into a modified pET28a (Novagen) harbouring a 6xHis-smt3 tag at the N-terminus. The Xenopus laevis FANCD2, FANCI (gifts from Prof. J.C. Walter, Harvard) and human Uba1 genes were inserted into modified pFBDM vectors harbouring an N-terminal 6xHis-TEV-HAtag, 6xHis-TEV-V5tag and 6xHis-TEV, respectively, via restriction cloning. Codon optimised synthetic FANCB, FANCL and FAAP100 genes (GeneArt) were inserted into a single pFBDM vector by sequential restriction cloning. A 6xHis-FLAGtag was inserted at the N-terminus of FANCB. Human PCNA (gift from Dr Svend Petersen-Mahrt, IFOM-FIRC Institute of Molecular Oncology, Milan), was inserted using restriction cloning into a pRSF Duet1 vector (Novagen), that harbours a 6xHis-tag at the N-terminus. Ubiquitin was inserted using restriction cloning into a modified pRSF Duet1 vector that harbours a 6xHis-TEV at the N-terminus. Silencer Select Negative Control No. 1 and Custom Silencer Select Ube2T siRNA duplexes (siT1 - 5’AGAGAGAGCUGCACAUGUUTT 3’, siT2 - 5’UCUAAGUUGCCUACCUUGATT 3’, siT3 - 5’UCUAAGUUGCCUACCUUGATT 3’) described previously11 were purchased from Life Technologies. A Ube2T gene with silent mutations to disrupt siRNA complementarity and a C-terminal 1D4-tag was synthesised (GeneArt) and inserted into a pcDNA5/FRT vector (Invitrogen). Various DNA cofactors for ubiquitination of the FANCD2-FANCI heterodimer have been previously described19. For this study, the following oligonucleotides were synthesised and obtained as PAGE purified duplex DNA (IDT technologies).
O10 - TTGATCAGAGGTCATTTGAATTCATGGCTTCGAGCTTCATGTAGAGTCGACGGTGCTGGGATCCAACATGTTTTCAATCTG
O11 - CAGATTGAAAACATGTTGGATCCCAGCACCGTCGACTCTACATGAAGCTCGAAGCCATGAATTCAAATGACCTCTGATCAA
All PCR reactions were carried out using Phusion High-Fidelity DNA polymerase (Thermo Scientific) except for mutagenesis which were generated using primer-based inverse PCRs using KOD Hot Start DNA polymerase (Novagen). Custom oligonucleotides for PCR and mutagenesis were obtained from Sigma Aldrich or IDT technologies. The coding regions of all constructs and mutants were verified by automated DNA sequencing using MRC PPU DNA Sequencing and Services or Eurofins Genomics DNA sequencing services. The biotinylated ubiquitin tail peptide (BiotinLRLRGG) was custom synthesised (GenScript) to >95% purity and trifluoroacetic acid exchanged with hydrochloric salt.
Expression and purification of recombinant proteins
The expression and purification of Ube2T and FANCL fragments (FANCLUR/FANCLR) has been previously described16,17,49. The 6xHis-smt3-Rad18 and 6xHis-MBP-rat RNF4 RING-RING (RNF4RR) proteins was purified as described elsewhere27,34. Bacmid preparation, baculovirus generation, expression and purification of FANCD2, FANCI and Uba1 proteins is as described elsewhere49. Bacmid preparation, baculovirus generation and expression of 6xHis-FLAGtag-FANCB-FANCL-FAAP100 were similar to FANCD2/FANCI. Purification of 6xHis-FLAGtag-FANCB-FANCL-FAAP100 involved Ni-NTA affinity, anion-exchange and gel-filtration chromatography. All purification steps were performed at 4°C and completed within 24-36 hours of cell lysis. Buffer solutions for protein purification are made using ultrapure water (resistivity of 18.2 MΩ cm at 25 °C) and kept cold. The constructs for ubiquitin, Ube2D3, Ube2B and PCNA were transformed into chemically competent BL21 (DE3) E. coli cells (Invitrogen) and cultured in Miller LB broth (Fisher) at 37°C until OD600 ~ 0.4 following which temperature was reduced to 16°C. At OD600 ~ 0.8, protein expression was induced by adding a final concentration of 500 μM Isopropyl-Beta-d-Thiogalactopyranoside (IPTG, Formedium) and allowed to proceed for a further 16-18h. Cell lysis and affinity purification of ubiquitin, Ube2D3, Ube2B and PCNA is similar to that described for Ube2T. The affinity tags were cleaved using GST-3C (for Ube2T and Ube2D3), 6xHis-Ulp1 (for FANCLUR/FANCLR) and 6xHis-TEV (for ubiquitin, RNF4 and Ube2B) proteases, respectively, that were made in-house. The 6xHis-tag on PCNA and the 6xHis-smt3 on RAD18 and FANCL (full-length) were retained. The ubiquitin, Ube2D3, Ube2B and 6xHis-PCNA proteins were subject to gel-filtration chromatography as described for FANCL 49. For all FPLC runs ӒKTA Pure chromatography systems (GE Life Sciences) were used. All purified proteins were concentrated using Amicon Ultra Centrifugal filters (Merck) with the desired Molecular Weight Cut Off. All proteins were flash-frozen in liquid nitrogen as single-use aliquots and stored at -80°C.
In vitro substrate ubiquitination assays
Frozen protein aliquots were thawed on ice and reactions were performed on the same day. All reactions were carried out in 50 mM HEPES pH 7.6, 100 mM NaCl, 1 mM Tris 2-carboxyethyl-phosphine (TCEP), 2 mM MgCl2 and 4% (v/v) glycerol buffer system at 30°C. For monitoring substrate ubiquitination, 20-100 μL reactions containing 50 nM 6xHis-Uba1, 5 μM Ubiquitin (or 6xHis Ub where indicated), 250 nM of Ube2T and FANCLUR (unless indicated otherwise) and 1 μM of xFANCD2 or xFANCI substrates in isolation. In case of the substrate complex, 1 μM of xFANCD2 and xFANCI were mixed with 2 μM of DNA and used for the reaction. Reaction mixes were prepared on ice, equilibrated to room-temperature and initiated by addition of Adenosine triphosphate (ATP) at a final concentration of 2.5 mM. At the desired time-points, reaction samples were terminated with 2xLDS sample buffer (Pierce) containing 7.5% (v/v) β-mercaptoethanol, and heated at 98°C for 3min. For visualizing reaction products, 3 μL of the terminated samples were resolved using Novex 4-12% Tris-Glycine Mini Gels, WedgeWell format (Invitrogen), stained overnight using InstantBlue Coomassie stain (Expedeon) and de-stained by extensive washes with distilled water. For immunoblotting, 1.5 μL of the terminated samples were resolved in the same gel system. Refer Immunoblots and Antibodies for transfer procedure and detection reagents. Coomassie stained gels and immunoblotted membranes were scanned by direct fluorescence monitoring using the Odyssey CLX Imaging System (Li-COR) and quantification were carried out using Image Studio software (v4.0.21).
For assays with the RNF4RR-Ube2D3 pair, 250 nM of the E3-E2 pair was used instead of FANCLUR-Ube2T. For assays with the biotinylated ubiquitin tail peptide (BiotinLRLRGG), 50 μL reactions contained 200 nM 6xHis-Uba1, 50 μM BiotinLRLRGG, 1 μM of Ube2T1-152,K91R and FANCLUR and 2 μM of xFANCD2 or xFANCD2 K562R. For PCNA ubiquitination assays, 30 μL reactions contained 50 nM 6xHis-Uba1, 10 μM Ubiquitin, 3 μM of Ube2B, 6xHis-smt3-Rad18 and 6xHis-PCNA. At the indicated time-points, reaction samples were mixed with 2xLDS sample buffer (Pierce) containing 7.5% (v/v) β-mercaptoethanol and terminated by heating at 98°C for 3min. Samples were resolved using NuPAGE 4-12% Bis-Tris Gels (Invitrogen) and analysed by immunoblotting
For single-turnover xFANCD2 ubiquitination assays, E2 charging reactions (30 μL) contained 150 nM 6xHis-Uba1, 12.5 μM Ubiquitin and 10 μM E2 and started with ATP (2.5 mM final). To prevent auto-ubiquitination, the Ube2T1-152,K91R variant was used and mutations of interest were made in this background. After 10 min at 30°C, 0.25 U of Apyrase (NEB) was added, the reaction diluted to 60 μL with assay buffer and left at room temperature for 5 min to hydrolyse the ATP. The E2~Ub charging efficiency was determined to be ~80%. The E2~Ub discharge mix (45 μL) contained ~1 μM of E2~Ub, 1 μM FANCLUR (unless indicated otherwise) and 1 μM of xFANCD2. At the indicated time-points, reaction samples were mixed with 3 μL of 3xLDS sample buffer (Pierce) containing 3 mM TCEP. Reaction products were resolved using NuPAGE 4-12% Bis-Tris Gels (Invitrogen) and visualised by InstantBlue Coomassie staining as above.
E2 charging, auto-ubiquitination and nucleophile reactivity assays
Frozen protein aliquots were thawed on ice and reactions were performed on the same day. All reactions were carried out in 50 mM HEPES pH 7.6, 100 mM NaCl, 1 mM Tris 2-carboxyethyl-phosphine (TCEP), 2 mM MgCl2 and 4% (v/v) glycerol buffer system at 30°C. E2 charging reactions (10 or 30 μL) contained 150 nM 6xHis-Uba1, 12.5 μM Ubiquitin and 10 μM E2. Reactions were commenced by addition ATP (2.5 mM final), maintained at 30°C and samples were taken at the indicated time-points. To terminate the reaction, samples were mixed with 3xLDS sample buffer (Pierce) containing 3 mM TCEP. E2 auto-ubiquitination reactions (10 μL) containing 100 nM 6xHis-Uba1, 12.5 μM Ubiquitin and 10 μM E2 and Ubiquitin were maintained at 30°C. At the indicated time-point, samples were mixed with 3xLDS sample buffer (Pierce) containing 7.5% (v/v) β-mercaptoethanol and heated at 98°C for 3min. To observe both E2 charging and auto-ubiquitination, 0.75-1 μg of each E2 was resolved using NuPAGE 4-12% Bis-Tris Gels (Invitrogen) and visualised by InstantBlue Coomassie staining as above. For nucleophile reactivity assays, the Ube2T1-152,K91R variant was used to prevent E2 auto-ubiquitination and desired mutations were made in this background. E2 charging reactions (50 μL) contained 150 nM 6xHis-Uba1, 7.5 μM E2 and 10 μM Ub and started with ATP (2.5 mM final). After 10 min at 30°C, 0.25 U of Apyrase (NEB) was added and left at room temperature for 5 min to hydrolyse the ATP. The E2~Ub discharge mix (50 μL) contained ~3 μM of E2~Ub in assay buffer containing 2 mM Hydroxylamine-HCl (Pierce) or 20 mM L-lysine (Sigma) or no nucleophile. The reactivity of each nucleophile was also assessed in the presence of 5 μM FANCLUR. The E2~Ub discharge reactions were carried out at 30°C and at the depicted time-points, reaction samples were terminated with 3xLDS sample buffer (Pierce) containing 3 mM TCEP. Reaction products were resolved using NuPAGE 12% (Supplementary Fig. 4d) or 4-12% (Fig. 5c) Bis-Tris Gels (Invitrogen) and visualised by InstantBlue Coomassie staining as above.
Cell based substrate ubiquitination assays
Human U2OS (osteosarcoma) cells (ATCC# HTB-96, Lot# 70008732) were purchased from the American Type Culture Collections and maintained in McCoy’s 5a medium (Sigma) with L-Glutamine, sodium bicarbonate and 10% (v/v) Foetal Bovine Serum (full-medium). All experiments were carried out with low-passage cells (<15) of good viability (>95%). siRNA transfections were performed in a 12-well dish using reverse transfection. In each well, 15 pmol of siRNAs and 4.5 μL Lipofectamine RNAiMAX (Invitrogen) were mixed in 100 μL of Opti-MEM I reduced serum medium and incubated for 20 min. To this mix, U2OS cells (0.2-0.25x106) in full-medium were added and final volume made up to 1 mL/well. After 24 hr, cells were washed with Dulbecco’s Phosphate Buffered Saline without CaCl2 and MgCl2 (Invitrogen) and full-medium was added. After 12 hr of the medium change, each well was transfected with 1 μg of vector using ViaFect (Promega) following manufacturer’s protocol. After 12 hr of vector transfection, the medium was replaced with 1 mL of full-medium containing 0.3 μM Mitomycin-C (MMC, Sigma) dissolved in dimethyl sulfoxide (DMSO, Sigma) or equal amount of DMSO. After 12hr of MMC/DMSO treatment cells were harvested and whole-cell lysates were prepared by lysing cells in 50 mM Tris-HCL pH 6.8, 2.5% SDS, 6% (v/v) β-mercaptoethanol and heating at 98°C for 10 min. Lysates (~25 μg) were resolved using Novex 4-12% Tris-Glycine Mini Gels, WedgeWell format (Invitrogen) and subject to immunoblotting. Refer Immunoblots and Antibodies for transfer procedure and detection reagents.
Immunoblots and Antibodies
All gels were transferred onto nitrocellulose membranes using the iBlot Gel Transfer Device (Invitrogen) set at P3 (20 V) and 8 min. Membranes were blocked using 1x Phosphate buffered saline (PBS, Fisher Bioreagents) containing with 5% (w/v) milk powder (Marvel) and 0.05% (v/v) Tween20 (Sigma). The membranes were incubated overnight at 4°C in blocking buffer containing the desired primary antibodies. Blots were washed 3x15 min with 1xPBS buffer with 0.05% v/v Tween20 and where required probed with IRDye 800CW or 680RD labelled secondary antibodies (Li-COR) at 1/10,000 dilution for 2 h at room-temperature. Blots were washed 3x15 min with 1xPBS buffer with 0.05% v/v Tween20 and scanned by direct fluorescence monitoring using Odyssey CLX Infrared Imaging System (Li-COR). The primary antibodies used in this study are as listed in Supplementary Table 3.
Isothermal titration calorimetry
ITC experiments were performed using MicroCal PEAQ-ITC (Malvern). All experiments were performed at 20°C, in duplicate, using freshly purified proteins. Proteins were buffer exchanged using 7K MWCO Zeba Spin Desalting Columns (Pierce) into 100 mM Tris-HCl, 100 mM NaCl, 0.4 mM TCEP buffer at pH 8.0 that was filtered and degassed. FANCLUR (ranging 22 to 34 μM) and FANCLR (~32 μM) was held in the cell, while Ube2T (ranging 400 to 600 μM) was present in the syringe. A total of 16 injections were carried out with the first injection of 0.3 μL/0.6 s followed by 15 injections of 1.5 μL/3 s. All injections were spaced by 120 s with mix speed set at 500 rotations per minute. Each experiment was controlled by an identical E2 into buffer run to account for the heat of dilution. All data were fitted with a single-site binding model using MicroCal PEAQ-ITC analysis software (v1.0).
Microscale Thermophoresis
MST measurements were performed using Monolith NT.115 (Nanotemper). All experiments were performed at 20°C, in triplicate, using freshly purified ligand protein (Ube2T1-152). The Ube2T1-152 fragment was used as it tolerates the high ligand concentrations required for interaction analysis. The target, 6xHis-HA-xFANCD2, was labelled in binding buffer (50 mM HEPES pH 7.5, 100 mM NaCl, 4 mM DTT, 1% (w/v) Ovalbumin) using His-Tag Labeling Kit RED-tris-NTA 2nd generation (Monolith) following the manufacturers guidelines. The ligand proteins were concentrated and buffer exchanged into binding buffer using 7K MWCO Zeba Spin Desalting Columns (Pierce). Ligands, at 3 mM, were diluted across a 16-point (2:1) dilution series in low binding tubes. Each dilution was mixed with the labelled target at a final concentration of 35 nM. The reaction mixture were loaded into premium capillaries (Monolith) and analysed at 100% LED power and 40% MST power. The Pre-MST, MST-acquisition and post-MST periods were 3 s, 15 s and 1 s, respectively. The resulting dose-response curves were fitted to a one-site binding model for Kd determination inbuilt in the MO.Affinity Analysis software (v2.3, Nanotemper). Fluorescence from the cold/pre-MST (-1.0 to -0.5 s) and hot/MST-acquisition (0.1 to 0.6 s) periods were used for Kd determination.
Residue Network Analysis
PDB files were first processed through the PDB-REDO webserver (https://pdb-redo.eu/) and Residue Interaction Networks (RINs) generated using the RIN generator webserver (http://protein.bio.unipd.it/ring/)51. Networks were generated for consecutive residues, excluding water molecules and hetero atoms, using relaxed (Ube2T) and strict (Ube2B) distance thresholds. Network policy was set to closest and all atoms of the residue pair are considered however, only the single most energetic interaction type was outputted. The output files were uploaded in Cytoscape (v3.6.1), and the RINs were compared using the RINalyzer (v2.0.0)52 using node identifier as matching attribute. In case of Ube2T, two copies of the RING bound state were individually compared to the unbound state using edge difference weighting. The resulting RIN comparison matrices were merged using the merge network tool in Cytoscape. Molecular figures prepared in PyMOL (v2.0.5).
Crystallization and Structure determination
The Ube2T E54R was concentrated to 24 mg/mL and used for sitting drop vapour diffusion crystallisation with various commercial screens. Crystals appeared in multiple conditions at 4°C. Diffraction quality crystals from the condition 0.1 M Tris-HCl pH 8.5, 20% v/v glycerol ethoxylate, 3% v/v poly(ethylene imine) were cryoprotected with 20% (v/v) glycerol and flash-frozen in liquid nitrogen. Native X-ray diffraction datasets were collected at 0.996 Å, 100 K on the ID30A-1/MASSIF-1 beamline at ESRF synchrotron (Grenoble, France) on a CMOS Hybrid Pixel detector (Dectris Pilatus3 2M) at phi scans of 0.15° at. Indexing and integration were performed using iMosflm(v7.2.2)53 and the reduced data scaled using AIMLESS(v0.7.3)54. The structure was solved using molecular replacement with Phaser(v2.8.2)55 with PDB ID 1yh2 as the search model. Manual model building and automated refinement were performed iteratively using COOT(v.0.8.9.2)56 and REFMAC5(v5.8.0238)57, respectively. No electron density observed for Ube2T C-terminus, residues 154-197, and are absent in the final model. The final model contained 98.1% of residues in favoured regions and 100% in allowed regions of the Ramachandran plot (no outliers).
Molecular Dynamics Simulations
All MD simulations (80 ns x 3) were performed using GROMACS (v5.1.1)58 using experimentally determined X-ray structures as starting point (PDB IDs: 4ccg, 1yh2, and the Ube2T E54R structure (6r75)). The all-atom force field AMBER99SB59 was used for the simulation. The starting structures were solvated using the TIP3P water model with water box extending 10Å from the outermost protein atom. The systems were neutralised with addition of Na+ and Cl- ions and equilibrated before and after insertion of ions. Production simulations were performed at constant temperature and pressure with periodic boundary conditions imposed in all directions. All bonds with hydrogen atoms were constrained by the LINCS algorithm and Coulomb interactions were computed with the PME method. For electrostatic and van der Waals interactions, 10Å cut off values were used. The simulations were run for 80 ns and the first 80 ps were removed prior to analysis. The analysis was performed using standard procedures (g_rmsf, g_dist) within the GROMACS package.
Statistical Analysis
All gels and membranes were scanned by direct fluorescence monitoring using the Odyssey CLX Imaging System (Li-COR) and quantification were carried out using the associated Image Studio software (v4.0.21). Custom shapes were used to quantify intensities of the total and ubiquitinated substrate. After average background subtraction, the trimmed signal intensities were exported into Microsoft Excel (2016) to calculate percentage of ubiquitination. Where reported, statistical significance of the difference from the FANCLUR-Ube2T reaction was assessed by one-way analysis of variance with Dunnett’s multiple-comparison test. For quantifying hydrolysis/aminolysis of the E2~Ub thioester, percentage of E2~Ub were fitted with one-phase decay mode using the equation Y= (Y0 - plateau)* e-kt + plateau. Y0 and plateau is percentage E2~Ub at 0 min and infinite time, respectively, k is the rate constant (%E2~Ub.min-1), t is time (min) and half-time (t1/2) = ln(2)/k (min). For quantifying ubiquitination of the FANCD2-FANCI heterodimer, percentage of ubiquitination were fitted with one-phase accumulation model using the equation Y= plateau(1 - e-kt). Plateau is percentage substrate ubiquitination at infinite time, k is the rate constant (%Ub.min-1), t is time (min) and half-time (t1/2) = ln(2)/k (min). All statistical analysis and graph generation were carried out in GraphPad Prism (v8.1.2).
Supplementary Material
Acknowledgements
We thank past and current members of the Walden laboratory for experimental suggestions, comments on the manuscript and their support. The pLou3 Rat RNF4 RING-RING DNA construct was a gift from R.T. Hay (University of Dundee). The X. laevis FANCD2 and FANCI coding genes were gifts from J.C. Walter (Harvard Medical School). The human PCNA cDNA template, was a gift from S. Petersen-Mahrt (IFOM-FIRC Institute of Molecular Oncology). All constructs are available from the MRC Protein Phosphorylation and Ubiquitylation Unit reagents Web page (http://mrcppureagents.dundee.ac.uk) or on reasonable request from the corresponding authors. We thank S.A.A Rehman (University of Dundee) for assistance during protein crystal harvest. Diffraction experiments were performed on beamline ID30A-1/MASSIF-1 at the European Synchrotron Radiation Facility (ESRF, Grenoble) and we are grateful to M. Bowler for providing assistance in using the beamline. We thank B. Smith (University of Glasgow) for access to computing resources for MD simulations. We thank J. Southall and S.M. Kelly (University of Glasgow) for assistance for the MST experiments. This work was supported by the Medical Research Council (MRC grant number MC_UU_12016/12); the EMBO Young Investigator Programme to H.W.; the European Research Council (ERC-2015-CoG-681582 ICLUb consolidator grant to H.W.
Footnotes
Data Availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 6r75. Other data and materials are available from the corresponding authors upon reasonable request or from the MRC Protein Phosphorylation and Ubiquitylation Unit reagents Web page (http://mrcppureagents.dundee.ac.uk).
Author contributions
V.K.C and H.W. conceived, designed and supervised the research; R.T and V.K.C generated various expression vectors and mutagenesis; C.A. and V.K.C established protein purification methodology and generated all recombinant material; V.K.C conducted biochemical and biophysical assays, cell biology experiments, residue network analyses and protein crystallization; M.L.R executed structure determination and validation; O.K. performed molecular dynamic simulations; V.K.C and H.W. wrote the manuscript with input from all authors.
Competing interest
Authors declare no competing interests.
References
- 1.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 2.Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17:626–42. doi: 10.1038/nrm.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. E2 enzymes: more than just middle men. Cell Res. 2016;26:423–40. doi: 10.1038/cr.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Hakim A, et al. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 2010;9:1229–40. doi: 10.1016/j.dnarep.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garaycoechea JI, Patel KJ. Why does the bone marrow fail in Fanconi anemia? Blood. 2014;123:26–34. doi: 10.1182/blood-2013-09-427740. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–62. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 7.Sims AE, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–7. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 8.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sareen A, Chaudhury I, Adams N, Sobeck A. Fanconi anemia proteins FANCD2 and FANCI exhibit different DNA damage responses during S-phase. Nucleic Acids Res. 2012;40:8425–39. doi: 10.1093/nar/gks638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joo W, et al. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science. 2011;333:312–6. doi: 10.1126/science.1205805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machida YJ, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–96. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 12.Walden H, Deans AJ. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys. 2014;43:257–78. doi: 10.1146/annurev-biophys-051013-022737. [DOI] [PubMed] [Google Scholar]
- 13.Alpi AF, Chaugule V, Walden H. Mechanism and disease association of E2-conjugating enzymes: lessons from UBE2T and UBE2L3. Biochem J. 2016;473:3401–3419. doi: 10.1042/BCJ20160028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meetei AR, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–70. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 15.Cole AR, Lewis LP, Walden H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat Struct Mol Biol. 2010;17:294–8. doi: 10.1038/nsmb.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodson C, et al. Structural analysis of human FANCL, the E3 ligase in the Fanconi anemia pathway. J Biol Chem. 2011;286:32628–37. doi: 10.1074/jbc.M111.244632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodson C, Purkiss A, Miles JA, Walden H. Structure of the human FANCL RING-Ube2T complex reveals determinants of cognate E3-E2 selection. Structure. 2014;22:337–44. doi: 10.1016/j.str.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32:767–77. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Longerich S, et al. Regulation of FANCD2 and FANCI monoubiquitination by their interaction and by DNA. Nucleic Acids Res. 2014;42:5657–70. doi: 10.1093/nar/gku198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato K, Toda K, Ishiai M, Takata M, Kurumizaka H. DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res. 2012;40:4553–61. doi: 10.1093/nar/gks053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajendra E, et al. The genetic and biochemical basis of FANCD2 monoubiquitination. Mol Cell. 2014;54:858–69. doi: 10.1016/j.molcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Twest S, et al. Mechanism of Ubiquitination and Deubiquitination in the Fanconi Anemia Pathway. Mol Cell. 2017;65:247–259. doi: 10.1016/j.molcel.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Hornbeck PV, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–20. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swuec P, et al. The FA Core Complex Contains a Homo-dimeric Catalytic Module for the Symmetric Mono-ubiquitination of FANCI-FANCD2. Cell Rep. 2017;18:611–623. doi: 10.1016/j.celrep.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugahara R, Mon H, Lee JM, Kusakabe T. Monoubiquitination-dependent chromatin loading of FancD2 in silkworms, a species lacking the FA core complex. Gene. 2012;501:180–7. doi: 10.1016/j.gene.2012.03.071. [DOI] [PubMed] [Google Scholar]
- 26.Zhang XY, et al. Xpf and not the Fanconi anaemia proteins or Rev3 accounts for the extreme resistance to cisplatin in Dictyostelium discoideum. PLoS Genet. 2009;5:e1000645. doi: 10.1371/journal.pgen.1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–20. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao B, et al. Inhibiting the protein ubiquitination cascade by ubiquitin-mimicking short peptides. Org Lett. 2012;14:5760–3. doi: 10.1021/ol3027736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheng Y, et al. A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol Cell Proteomics. 2012;11:329–41. doi: 10.1074/mcp.O111.013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y, et al. Modularized functions of the Fanconi anemia core complex. Cell Rep. 2014;7:1849–57. doi: 10.1016/j.celrep.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castella M, et al. FANCI Regulates Recruitment of the FA Core Complex at Sites of DNA Damage Independently of FANCD2. PLoS Genet. 2015;11:e1005563. doi: 10.1371/journal.pgen.1005563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen SK, Lima CD. Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer. Mol Cell. 2013;49:884–96. doi: 10.1016/j.molcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479–89. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 34.Hibbert RG, Huang A, Boelens R, Sixma TK. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc Natl Acad Sci U S A. 2011;108:5590–5. doi: 10.1073/pnas.1017516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung RS, et al. Ubiquitination-Linked Phosphorylation of the FANCI S/TQ Cluster Contributes to Activation of the Fanconi Anemia I/D2 Complex. Cell Rep. 2017;19:2432–2440. doi: 10.1016/j.celrep.2017.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishiai M, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–46. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ceccaldi R, Sarangi P, D'Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–49. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 38.Gallego LD, et al. Structural mechanism for the recognition and ubiquitination of a single nucleosome residue by Rad6-Bre1. Proc Natl Acad Sci U S A. 2016;113:10553–8. doi: 10.1073/pnas.1606863113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattiroli F, Uckelmann M, Sahtoe DD, van Dijk WJ, Sixma TK. The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat Commun. 2014;5 doi: 10.1038/ncomms4291. 3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–6. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Streich FC, Jr, Lima CD. Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature. 2016;536:304–8. doi: 10.1038/nature19071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–20. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 43.Middleton AJ, Day CL. The molecular basis of lysine 48 ubiquitin chain synthesis by Ube2K. Sci Rep. 2015;5:16793. doi: 10.1038/srep16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–20. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 45.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–81. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu W, et al. Dimeric Ube2g2 simultaneously engages donor and acceptor ubiquitins to form Lys48-linked ubiquitin chains. EMBO J. 2014;33:46–61. doi: 10.1002/embj.201385315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morreale FE, et al. Allosteric Targeting of the Fanconi Anemia Ubiquitin-Conjugating Enzyme Ube2T by Fragment Screening. J Med Chem. 2017;60:4093–4098. doi: 10.1021/acs.jmedchem.7b00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hewitt WM, et al. Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9. Angew Chem Int Ed Engl. 2016;55:5703–7. doi: 10.1002/anie.201511351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaugule VK, Arkinson C, Toth R, Walden H. Enzymatic preparation of monoubiquitinated FANCD2 and FANCI proteins. Methods Enzymol. 2019;618:73–104. doi: 10.1016/bs.mie.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 50.Arkinson C, Chaugule VK, Toth R, Walden H. Specificity for deubiquitination of monoubiquitinated FANCD2 is driven by the N-terminus of USP1. Life Sci Alliance. 2018;1:e201800162. doi: 10.26508/lsa.201800162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piovesan D, Minervini G, Tosatto SC. The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 2016;44:W367–74. doi: 10.1093/nar/gkw315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doncheva NT, Klein K, Domingues FS, Albrecht M. Analyzing and visualizing residue networks of protein structures. Trends Biochem Sci. 2011;36:179–82. doi: 10.1016/j.tibs.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 53.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–81. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–14. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–67. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abraham MJ, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 59.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.