Abstract
N-methyl-D-aspartate receptors (NMDARs), a subset of ligand-gated ionotropic glutamate receptors, are critical for learning, memory, and neuronal development. However, when NMDAR subunits are mutated, a host of neuropathological conditions can occur, including epilepsy. Recently, genetic variation within the GRIN2D gene, which encodes the GluN2D subunit of the NMDAR, has been associated with a set of early-onset neurological diseases, notably developmental and epileptic encephalopathy (DEE). Importantly, patients with GRIN2D variants are largely refractory to conventional anti-epileptic drug (AED) treatment, highlighting the need to further understand the distinctive characteristics of GluN2D in neurological and pathological functions. In this review, we first summarize GluN2D’s unique spatial and temporal expression patterns, electrophyslogical profiles, and contributions to both pre- and postsynaptic signaling. Next, we review thirteen unique case studies from DEE patients harboring ten different causal GRIN2D variants. These patients are highly heterogenous, manifesting multiple seizure types, electroencephalographic recordings, and neurological and developmental outcomes. Lastly, this review concludes by highlighting the difficulty in treating patients with DEE-associated GRIN2D variants, and stresses the need for selective therapeutic agents delivered within a precise time window.
Keywords: Autism, Channelopathy, Developmental and epileptic encephalopathy, Developmental delay, Epilepsy, Glutamate receptor, Intellectual disability, Precision medicine, Translational study
1. Introduction
N-methyl-D-aspartate receptors (NMDARs) are ligand-gated ionotropic glutamate receptors which constitute a calcium-permeable component of fast excitatory neurotransmission. These receptors are heterotetramers (Figure 1A), comprised of two GluN1 subunits (one gene, GRIN1) and two GluN2 subunits (four genes, GRIN2A-D). NMDARs are unique in that they require both co-agonist binding (glycine/D-serine on GluN1 and glutamate on GluN2) and membrane depolarization in order to flux ions.[1–3] At most resting membrane potentials, NMDARs are blocked by extracellular magnesium ions within the channel pore in a voltage dependent manner, which allows them to integrate neurotransmitter release with postsynaptic membrane depolarization.[1] This ‘coincidence detection’ and significant calcium permeation endows NMDARs with the ability to modulate synaptic plasticity, making them key regulators of neuronal excitability.[4] Moreover, NMDAR activity is critical for neurodevelopment, synaptogenesis, locomotion, spatial learning, memory formation, and general cognition.[5–10]
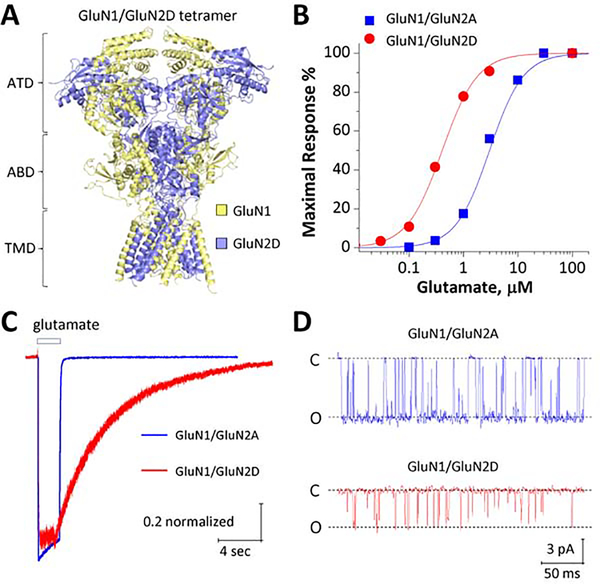
Figure-1. Distinct functional features of GluN2D-containing NMDARs.
A, Space-filled ribbon structure of a homology model of human GluN1/GluN2D receptor generated from GluN1/GluN2B crystallographic data (PDB-4PE5; Karakas and Furukawa, 2014; Lee et al., 2014). ATD: amino terminal domain, ABD: agonist-binding domain, TMD: transmembrane domain, comprised by M1, M2, M3, and M4. B, Concentration-response curves for glutamate (in the presence of 30 μM glycine) for GluN1/GluN2A (Blue) and GluN1/GluN2D (Red) generated from two-electrode voltage-clamp current recordings at VHOLD −40 mV. C, Representative traces of whole cell voltage clamp current recordings in response to 1 mM glutamate from human GluN1/GluN2A- and GluN1/GluN2D-transfected HEK293 cells at 60 mV. D, Representative unitary currents (activated by 1 mM glutamate and 50 μM glycine) from outside-out single channel patch clamp recordings from human GluN1/GluN2A- and GluN1/GluN2D-transfected HEK293 cells at VHOLD −80 mV. C, close; O, open. H.Yuan, unpublished data.
Given their importance in normal brain activity, it is not surprising to discover that mutations within the GRIN genes are among some of the least tolerant in the genome.[11–15] NMDAR mutations are associated with a host of neurological diseases, including schizophrenia, intellectual disabilities, autism, epilepsy, and attention-deficit/hyperactivity disorder (see XiangWei et al., 2018[13]). However, the focus of this review will be on developmental and epileptic encephalopathy (DEE), which are a group of devasting epilepsies that combine severe seizures with the presence of one or more developmental abnormalities. Patients with DEE are largely resistant to standard anti-epileptic drug (AED) therapies and tend to show poor recovery from secondary neurodevelopmental symptoms even after seizure cessation. Given the severity of DEE and its poor prognosis, there is a pressing need to further our understanding of the molecular underpinnings of DEE, with a particular focus on epileptogenesis.
All of the GRIN genes are capable of generating DEE via rare or de novo genetic mutations. The GRIN2A and GRIN2B genes clearly demonstrate a strong correlation between genetic variation and an epileptic phenotype, however, the functions of these genes are very well understood and have been extensively researched. For example, PubMed articles containing ‘NR2A/GRIN2A/GluN2A’ generated 2,966 hits and a search for ‘NR2B/GRIN2B/GluN2B’ generated 4,397 (accessed on 8/19/2019). However, there is also a strong epilepsy-associated component seen within the GRIN2D gene, as 63% (10/16) of patients with GRIN2D variants display some form of epilepsy, and more specifically DEE. This gene encodes the GluN2D NMDAR subunit, and its function within the nervous system is largely understudied, with PubMed searches for ‘NR2D/GRIN2D/GluN2D’ yielding only 406 papers (accessed 8/19/19). GluN2D-containing NMDARs (GluN2D receptors) exhibit unique expression patterns, cellular specificity, and electrophysiological profiles, which are unparalleled amongst the GRIN gene family. The combination of these features make unraveling the precise epileptogenic role of GRIN2D mutations complex, yet highly intriguing. The goal of this review is to highlight some of these distinguishing physiological features and conclude by presenting several case studies of pediatric DEE patients with genetic variation within the GRIN2D gene, with the hopes of putting epileptic mutations within this gene under the limelight.
2. Distinctive expression and functional features of GluN2D-containing NMDARs
2.1. GluN2D spatial and temporal expression patterns
The GRIN2D gene is located at 19q13.1-qter in the human genome, with its longest splice variant containing 1,336 amino acids.[1] The GluN2D subunit shares >80% sequence homology with the GluN2C subunit, >70% sequence homology with the GluN2A and GluN2B subunits, and about 60% sequence homology with all other ionotropic glutamate receptors, including the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) and the kainate receptor.[1, 16] For a more detailed description of transcription, translation, trafficking, etc. of ionotropic glutamate receptors, readers are invited to examine Traynelis et al. 2010.[1]
GluN2D mRNA expression begins in the latter portion of embryogenesis, not appearing until embryonic day (E)15–18 in rodents.[17] Levels of GluN2D mRNA continue to increase during neurodevelopment, reaching peak expression level at postnatal day (P)7–10.[17] After the neonatal phase, mRNA levels of GluN2D decrease and do not reach their relatively low steady-state expression level until late adolescence (P40–50).[17, 18] Although not as extensively studied, GluN2D mRNA expression in human brain largely follows a similar spatial and temporal pattern as described both above and below.[19, 20] However, most of this review in regards to functional data in native tissue will focus on rodent studies.
During the early phases of life, GluN2D has a ubiquitous expression pattern, appearing in the spinal cord (both dorsal and ventral horns)[21], midbrain nuclei such as the periaqueductal gray and reticular formation[21], all nuclei of the diencephalon (thalamus, hypothalamus, etc.)[18, 22], select nuclei within the basal ganglia (striatum, substantia nigra, and subthalamic nucleus), retina[23], cochlear/vestibular nerves[24], olfactory bulb[17], cerebellum[17], and cerebral cortex including the hippocampal formation[17, 18]. The widespread distribution of GluN2D during the embryonic and early neonatal phases of life give this subunit the opportunity to exert influence on circuit connectivity and function during a critical phase of neurodevelopment.
As the rodent ages, GluN2D expression remains in all of the regions listed above, but diminishes in transcript number and becomes more localized to particular cell types.[17, 18, 25, 26] The most notable examples of GluN2D cell-type specificity occur in the hippocampus and cerebellum.[27, 28] In juvenile hippocampus (P3–5), GluN2D receptors are believed to contribute to synaptic transmission in both glutamatergic pyramidal cells and GABAergic interneurons.[28] However, throughout maturation, NMDAR-mediated currents in hippocampal pyramidal cells lose hallmarks of GluN2D receptors, such as a prolonged synaptic decay and insensitivity to extracellular magnesium (see section below), while these characteristics are retained in GABAergic interneurons.[28] While the function of early GluN2D expression in excitatory pyramidal cells is not known, one plausible explanation is that the GluN2D subunit is necessary for neuronal maturation. Further work using GluN2C/D-specific NMDAR modulators has confirmed the presence of GluN2D in GABAergic hippocampal interneurons of both the medial ganglionic eminence (MGE; parvalbumin-expressing basket cells, etc.) and caudal ganglionic eminence (CGE; cholecystokinin-expressing basket cells, etc.) embryonic origins.[29–31] The sustained expression of GluN2D receptors on interneurons may indicate that this subunit is integral to maintain proper inhibitory drive to help control overall circuit function.
A similar change in GluN2D cell-type specificity occurs in the cerebellum. In early neurodevelopment, GluN2D has been found in Purkinje cells and in both stellate and Golgi cells.[17, 27, 32] After adolescence (>P40), Purkinje cells no longer express GluN2D and it becomes restricted to just stellate and Golgi cells.[17] This cell-type restriction during neonatal and preadolescent development may not be specialized to just the hippocampus and cerebellum, but could occur throughout all regions in which GluN2D is expressed during embryogenesis. Thus, a rare or de novo variant occurring within the GRIN2D gene, combined with the robust embryonic expression of GluN2D, would be able to impact nearly every facet of brain function. Empirical evidence for this hypothesis is presented later during this review, however, the neurological phenotypes of patients with GRIN2D variants are quite heterogeneous (see section 3.3).
2.2. Electrophysiological signatures of GluN2D-containing NMDARs
Glutamate (EC50 = 0.2–0.4 μM; Figure 1B) and glycine (EC50 = 0.1–0.2 μM) act with relatively high potency at GluN2D receptors [1]. Glutamate shows over5– 6-fold higher potency at the GluN2D receptor than at GluN2A- or GluN2B-containing NMDARs (Figure 1B).[1] As mentioned previously, NMDARs are unique in that they are blocked by extracellular magnesium at negative membrane potentials.[1] This allows NMDARs to function as coincidence detectors, producing an inward current in response to presynaptic glutamate release when the postsynaptic membrane is depolarized.[4] However, the potency of magnesium block on GluN2D receptors is 10-fold lower than that observed on GluN2A- and GluN2B-containing NMDARs.[33, 34] This indicates that neurons expressing GluN2D receptors may be more sensitive synaptically-released glutamate than those expressing GluN2A- or GluN2B-containing NMDARs alone. Conversely, uncompetitive NMDAR pore blockers, such as memantine (5-fold more potent) and ketamine (2-fold more potent), are more potent at GluN2D receptors than on GluN2A and GluN2B receptors.[1, 35].
Another unique electrophysiological characteristic of GluN2D receptors is their prolonged deactivation time. Using a rapid solution exchange system on transfected HEK cells, glutamate exposure can mimic a synaptic-like time course (1 ms exposure time).[36] In this paradigm, GluN2D receptors exhibit a weighted tau, a measure used to describe receptor deactivation following agonist removal, of over 5,000 ms.[18, 37, 38] This is over 10-fold slower than GluN2B and GluN2C receptors and ~100-fold slower than GluN2A receptors (Figure 1C).[1] Additionally, unlike GluN2A and GluN2B receptors, GluN2D-mediated current responses are virtually void of desensitization following prolonged agonist exposure.[18, 38–40] An exaggerated decay time with a near lack of desensitization makes GluN2D receptors capable of transferring a large quantity of ions across the plasma membrane, sustaining depolarization for a sufficient amount of time to initiate burst-firing (see sections 2.3 and 2.5). However, GluN2D receptors also have a much lower open channel probability (Figure 1D), around 0.01 – 0.04%, and a slightly lower calcium permeability than GluN2A and GluN2B receptors.[1, 41]
In addition to forming diheteromeric complexes, for example GluN1/GluN2D/GluN1/GluN2D, NMDARs can also be formed using two different GluN2 subunits. Multiple reports have demonstrated that both GluN1/GluN2A/GluN1/GluN2D and GluN1/GluN2B/GluN1/GluN2D triheteromeric assemblies are found in native tissue.[1] The presence of two different GluN2 subunits will alter receptor characteristics, typically observed as a blending of the two diheteromeric forms.[42] For example, the GluN2B/GluN2D triheteromeric receptor has a weighted tau of ~1,800 ms when transfected HEK cells are exposed to rapid synaptic-like glutamate exposure, whereas the weighted tau for diheteromeric GluN2B receptors is about 500 ms and the weighted taufor diheteromeric GluN2D receptors is >5,700 ms.[43] Other receptor kinetic parameters for GluN2B/GluN2D triheteromeric complexes are also an intermediate of diheteromeric GluN2B and GluN2D receptors, such as open channel probability, magnesium sensitivity, and efficacy of modulators like ifenprodil, ketamine, and memantine.[43] GluN2A/GluN2D triheteromeric assemblies have not been extensively evaluated for kinetic parameters, but it is plausible these receptors will also exhibit this unique intermediate blending phenotype.
2.3. Roles of GluN2D in postsynaptic neurotransmission
As mentioned previously, the GluN2D subunit is present in synaptic NMDARs in GABAergic interneurons in the hippocampus and cortex.[29, 44] However, the ratio of true GluN2D diheteromeric receptors to triheteromeric assemblies with GluN2A and/or GluN2B is still poorly understood. Regardless of stoichiometry, the presence of GluN2D within the receptor complex may endow interneurons with the unique ability to generate dendritic calcium spikes, integrate long-range signals, and synchronize large groups of interneurons in coordinated firing via glutamate spillover.[45–47] The prolonged synaptic decay, high glutamate potency, and modest magnesium block generated by the GluN2D subunit could allow interneurons to generate burst-firing, and thus could be partially responsible for their oscillatory and rhythmic nature.[48] It should also be noted that although the GluN2D subunit is found in both MGE- and CGE-derived interneurons, the relative contribution of NMDAR-mediated current observed on CGE-derived interneurons is much higher.[49] This suggests that NMDARs and the GluN2D subunit may not be the main driver for dendritic calcium spikes in MGE-derived interneurons. However, a recent study has shown that tonic activation of GluN2D receptors on developing cortical interneurons is necessary for proper intrinsic excitability, dendritic arborization, GABAergic synaptogenesis, and inhibitory tone onto excitatory pyramidal cells.[44]
In addition to GABAergic interneurons, the GluN2D receptor plays a pivotal role in postsynaptic signaling within the basal ganglia. Subthalamic nuclei (STN) neurons have been shown to have a high expression of dendritic GluN2D receptors.[50] The STN is the sole glutamatergic, excitatory nucleus within the basal ganglia, receiving excitatory drive from the thalamus and cortex, and providing an excitatory output to the globus pallidus interna and substantia nigra pars reticulata.[51] Thus, one important function of the STN is to modulate the strength of indirect pathway activity that reaches the thalamus and motor cortex. GluN2D receptors have also been reported on dopaminergic projection neurons in the substantia nigra pars compacta (SNc).[52] Here, both GluN2B and GluN2D receptors provide the excitatory NMDAR-mediated current to facilitate the release of dopamine onto medium spiny neurons within the dorsal striatum. Both the STN and SNc are hypoactive in Parkinson’s disease, and positive allosteric modulators specific for GluN2C/GluN2D receptors have shown some promise in rescuing motor symptoms in animal models.[53] In addition, the central role of GluN2D receptors in modulating GABAergic interneuron activity and coordinating the balance of direct versus indirect pathway activation within the basal ganglia make mutations within this subunit directly poised to have a substantial impact on circuit excitability and coordinated motor function.
GluN2D receptors have also been confirmed to play a central role in postsynaptic signaling within the spinal cord.[54] More specifically, GluN2D receptors have been found to mediate excitatory synaptic transmission in laminae I of the dorsal horn. The dorsal horn relays pain and temperature signals from sensory neurons in the periphery to the somatosensory cortex. The notion that the GluN2D receptors play a role in pain perception is supported by evidence using GluN2D-knockout mice, discussed more extensively below, as the GluN2D receptor was shown to be critical for the induction of AMPAR-mediated allodynia.[55]
2.4. Roles of GluN2D in presynaptic neurotransmission
In addition to serving as postsynaptic dendritic receptors, GluN2D receptors have also been reported in presynaptic sites. Presynaptic GluN2D receptors have mainly been studied in the hippocampus and cerebellum, although within these regions they serve different roles in circuit function. In the hippocampus, presynaptic GluN2D receptors have been suggested to mediate the induction of long-term depression (LTD) between the canonical CA3-CA1 pyramidal cell synapse.[56] More specifically, GluN2D receptors have been shown to be necessary for spike-time dependent LTD (t-LTD).[56] This form of plasticity helps to marry the temporal sequence of postsynaptic signal generation with action potential firing within the same cell.[57] In the context of t-LTD, if dendritic postsynaptic signals occur just after a cell fires an action potential, subsequent postsynaptic signaling strength will be diminished.[57] Andrade-Talavera and colleagues report that t-LTD induction does not require any postsynaptic NMDAR activation, and that GluN2B and GluN2D receptors, along with postsynaptic metabotropic glutamate receptors, work in concert to generate this form of Hebbian synaptic depression.[56]
In the cerebellum, presynaptic GluN2D receptors on stellate cells help modulate GABA release onto Purkinje cells.[58] Here, researchers postulate that ‘spilled over’ glutamate from parallel fibers binds to presynaptic GluN2D receptors under times of rapid granule cell firing and increases stellate cell GABA release.[58] Although the GluN2D subunit has been reported in multiple other brain regions, the remaining areas not discussed in this section have only been confirmed via localization studies (in situ hybridization, Western blots, or antibody binding). Thus, their direct role in circuit function is not fully understood. Regardless, GluN2D receptors play a critical role in modulating circuit function via their direct involvement in plasticity induction and in the fine-tuning of presynaptic neurotransmitter release. It should also be noted that GluN2D receptors have not been reported on any glial cells.[59]
2.5. Physiological role of the GluN2D subunit via knockout mice
GluN2D-knockout (KO) mice are viable, reproducible normally, and have no overt changes in neuronal histology.[60] However, these mice do exhibit a unique locomotor phenotype. This locomotor phenotype is likely due to altered levels of monoamines, such as dopamine and norepinephrine, which were shown to be downregulated in KO mice in the hippocampus and striatum.[61] Accordingly, KO mice have a diminished propensity for spontaneous motor movements via open field assays, but display no deficits in general locomotor abilities measured via the rotarod test.[62] There were also no gross changes in mRNA levels of the other NMDAR subunits, however, expression within the synapse has not yet been described.[60] On the other hand, GluN2D-KO mice have a reduced pharmacologically-induced hyperlocomotor phenotype to ketamine[63] and phencyclidine[62], both of which are pan-NMDAR pore blockers. This indicates a key role of GluN2D receptors in motor control, as would be postulated given their expression patterns in the spinal cord and basal ganglia. Finally, GluN2D-KO mice show deficits in spatial memory and prepulse inhibition.[48]
GluN2D receptors are also partially responsible for the generation of high-frequency gamma oscillations (>60 Hz), as KO mice had a lower power threshold between 65–140 Hz.[48]These oscillations are crucial for information processing, proper cortical functions, and generation of working memory.[64–66] Gamma oscillations are generated via fast-spiking parvalbumin-positive GABAergic interneurons.[64, 66] Sapkota and colleagues reported a decrease in PV-immunoreactivity of GluN2D-KO mice in the substantia nigra pars compacta and in the basolateral amygdala.[48] Other brain regions, such as the hippocampus and frontal cortex, did not show changes in PV-immunoreactivity. However, the loss of GluN2D on PV-interneuron function as well as its impact on overall circuit function remain unexplored.[48] Given the robust expression of GluN2D, especially during early neurodevelopment, it seems likely that perturbations to GluN2D function will impart a multitude of changes in cellular biology, synaptic function, and network output.
3. Rare and de novo variants in the GRIN2D gene contribute to developmental and epileptic encephalopathy
Genetic variations within the GRIN2D gene cause severe forms of developmental and epileptic encephalopathy. Epilepsy is one of the world’s most prevalent neurological disorders, afflicting nearly 50 million people.[67] Developmental and epileptic encephalopathy (DEE) represents a group of devastating neurological diseases, which are difficult to treat and involve multiple brain abnormalities in conjunction with severe convulsive seizures.[68–70] DEE manifests early during infantile or adolescent development.[71–73] Common comorbidities of DEE include slowed or incomplete neurological development, aberrant fine motor control, aphasia, and intellectual disabilities.[68, 69] Children with DEE have a shortened life expectancy, and most patients find little to no relief from seizures with AEDs.[74–78] Additionally, the prognosis for secondary symptoms in DEE patients is poor, with some never developing proper intellectual, motor, and verbal function.[79–82] This incomplete recovery from secondary symptoms suggests an underlying problem in neuronal circuitry that may become hardwired past a certain stage during development. Furthermore, children with DEE often require around-the-clock care, placing a heavy social and financial burden on primary caregivers, highlighting an urgent need to understand its disease etiology and progression.[77, 83–86]
3.1. Overview and location of DEE-associated GRIN2D variants
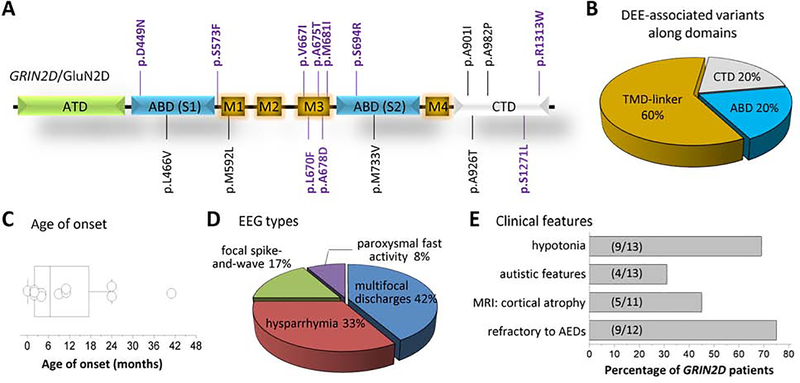
DEE can be caused by a variety of factors, including both genetic and environmental. Recently, thanks to advances in whole exome sequencing, genetically-derived DEE has gained much-deserved attention and causative gene mutations have now been identified. To date, 16 unique missense GRIN2D variants, which are absent in the gnomAD database (Genome Aggregation Database, Cambridge, MA, http://gnomad.broadinstitute.org/, evaluated on 07–31-2019), have been reported.[39, 87–89] Of these 16 variants in Table 1, 63% (10/16) are associated with DEE.[39, 88, 89] These DEE-associated GRIN2D missense variants are scattered through various domains of the GluN2D subunit, except for the extracellular amino-terminal domain (ATD; Figure 2A). Of the 10 DEE GRIN2D variants, the agonist-binding domain (ABD) contains 1 variant, the transmembrane domain and linker regions (TMD) contains 7 variants, and the carboxyl-terminal domain (CTD) contains 2 variants (Figure 2B).
Table 1.
Summary of missense mutations and rare variants in GRIN2D/GluN2D NMDARs
| Gene | Genotype | Protein | Location | Phenotype | References | |
|---|---|---|---|---|---|---|
| 1 | GRIN2D | c.1345G>A | p.D449N | ABD (S1) | Epi, ID | Tsuchida et al., Clinical Genetics 2018 |
| 2 | GRIN2D | c.1396C>G | p.L466V | ABD (S1) | SCZ | Tarabeux et al., Transl Psychiatry 2011 |
| 3 | GRIN2D | c.1718C>T | p.S573F | pre-M1 | Epi; ID | XiangWei et al., Brain 2019 |
| 4 | GRIN2D | c.1774A>C | p.M592L | M1 | ASD | Tarabeux et al., Transl Psychiatry 2011 |
| 5 | GRIN2D | c.1999G>A | p.V667I | M3 | Epi; ID; 3 pts | Li et al., Am J Hum Genet. 2016; XiangWei et al., Brain 2019 |
| 6 | GRIN2D | c.2008C>T | p.L670F | M3 | Epi, ID; 2 pts | XiangWei et al., Brain 2019 |
| 7 | GRIN2D | c.2023G>A | p.A675T | M3 | Epi; ID | XiangWei et al., Brain 2019 |
| 8 | GRIN2D | c.2033C>A | p.A678D | M3 | Epi; ID | XiangWei et al., Brain 2019 |
| 9 | GRIN2D | c.2043G>C | p.M681I | M3 | Epi, ID | Tsuchida et al., Clinical Genetics 2018 |
| 10 | GRIN2D | c.2080A>C | p.S694R | ABD (S2) | Epi, ID | Tsuchida et al., Clinical Genetics 2018 |
| 11 | GRIN2D | c.2197A>G | p.M733V | ABD (S2) | SCZ | Tarabeux et al., Transl Psychiatry 2011 |
| 12 | GRIN2D | c.2701G>A + c.2702C>T | p.A901I | CTD | SCZ | Tarabeux et al., Transl Psychiatry 2011 |
| 13 | GRIN2D | c.2776G>A | p.A926T | CTD | ASD | Tarabeux et al., Transl Psychiatry 2011 |
| 14 | GRIN2D | c.2944G>C | p.A982P | CTD | SCZ | Tarabeux et al., Transl Psychiatry 2011 |
| 15 | GRIN2D | c.3812C>T | p.S1271L | CTD | Epi; ID | XiangWei et al., Brain 2019 |
| 16 | GRIN2D | c.3937C>T | p.R1313W | CTD | Epi, ID | XiangWei et al., Brain 2019 |
ASD: Autism Spectrum Disorder, Epi: epilepsy/seizures, ID: Intellectual Disability, SCZ: Schizophrenia
ATD: amino-terminal domain, ABD: agonist-binding domain, TMD: transmembrane domains (M1, M2, M3, M4), CTD: carboxy-terminal domain
Figure-2. Developmental and epileptic encephalopathy (DEE)-associated human GRIN2D/GluN2D missense variants.
A,B, Locations of DEE-associated GRIN variants (indicated by BOLD/PURPLE). S1,S2 comprise the ABD; TMD-link: transmembrane domains (comprised by M1, M2, M3, and M4) and linker regions (S1-M1 linker, M1-M2 linker, M2-M3 linker, M3-S2 linker, and S2-M4 linker); see Fig-1 for domain organization; References are in the text. C, Box-and-whisker plot for the age of onset of 12 patients harboring 9 unique DEE-associated GRIN2D variants. Mean age of onset is 11 ± 3.6 months, with a median age of onset of 6.5 months. Onset data for Patient #8 was not available. D, Different types of electroencephalographic (EEG) patterns were observed from 12 patients harboring 9 unique DEE-associated GRIN2D variants. EEG type was not available for Patient #8. The most common EEG recording was multifocal discharges (5/12), followed by hysparrhymia (4/12), focal spike-and-wave discharges (2/12), and paroxysmal fast activity (1/12). E, Various clinical features observed in DEE-associated GRIN2D variant-containing patients. Besides the unifying phenotypes of DEE (epilepsy and developmental delay/intellectual disability), 69% of patients are hypotonic, 31% of patients display autistic-like features and behaviors, 45% of patients experienced cortical atrophy in brain MRI, and 75% are refractory to typical AED therapy. Magnetic resonance imaging (MRI) data was not available for Patient #4 and Patient #8. Patient #2 has not started any formal drug therapy and was not included in the calculation.
The ATD is furthest away from the plasma membrane and harbors sites for allosteric modulator binding and controls electrophysiological characteristics such as agonist potency and open channel probability.[1, 90] However, some studies have shown that NMDAR complexes are still functional without the ATD, albeit they are not comparable to wild-type receptor function, potentially explaining why no variants have been found within this domain. The ABD is a highly conserved region of the NMDAR, and helps to convert agonist binding, such as glutamate at GluN2 subunits, into molecular motion for pore opening.[1] The TMD of the receptor is also a critical region, harboring twelve transmembrane alpha helices (three per subunit) in addition to four reentrant loops arranged in a circle to form the receptor pore and ion selectivity filter.[1] Thus, it is not surprising that the majority of disease-associated GRIN2D variants lie within the TMD. Recent studies have shown the pre-M1 and M1 regions of the TMD are intolerant to genetic variation in a healthy population.[12] That is, if a mutation occurs in these regions, some form of neurological disease will likely manifest. Lastly, the CTD contains sites for protein-protein interactions, interacts with a host of intracellular proteins, and facilitates subunit trafficking from the endoplasmic reticulum to the plasma membrane.[1] Although the majority of GRIN2D variants are centered around the TMD, similar mapping studies with GRIN2A variants have shown that epilepsy-associated mutations occur throughout all domains of the NMDAR, including the ATD.[13, 14, 91]
3.2. Functional analysis of GRIN2D variants in heterologous systems
Of the 10 DEE-associated GRIN2D variants, 7 have undergone functional testing using heterologous expression systems.[39, 89] The outcome of this testing has been summarized in Table 2, and includes data on: S573F, V667I, L670F, A675T, A678D, S1271L, and R1313W. The use of these transient expression systems provides a multitude of electrophysiological and biological data, increasing our understanding of how each variant may be impacting synaptic transmission and neuronal viability. Using this data, these variants may be classified as gain-of-function (GoF, increased function compared to wild-type) or loss-of-function (LoF, diminished/decreased function compared to wild-type). There is no association between domain location along the receptor and functional outcome as both GoF and LoF variants are found throughout all domains of both GRIN2A and GRIN2B.[13] Surprisingly, 57% (4/7) – S573F, A675T, S1271L, and R1313W – of DEE-associated GRIN2D variants might be considered LoF. GoF variants – V667I and L670F – make up 29% of those tested, while the A678D variant likely needs to undergo further testing before a proper GoF or LoF assignment can be made. Additionally, it should be noted that all of the GoF or LoF designations assigned above are only approximations based off previous work that involved more complete functional analyses and calculations.[11]
Table-2.
Summary of DEE-related GRIN2D variants and patient’s phenotypes
| Patient-1 | Patient-2 | Patient-3 | Patient-4 | Patient-5 | Patient-6 | Patient-7 | Patient-8 | Patient-9 | Patient-10 | Patient-11 | Patient-12 | Patient-13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GRIN2D variants (NM_000836) | c.1345G>A:p.(Asp449Asn) | c.1718C>T: p.(Ser573Phe) | c.1999G>A: p.(Val667Ile) | c.1999G>A:p.(Val667Ile) | c.1999G>A:p.(Val667Ile) | c.2008C>T: p.(Leu670Phe) | c.2008C>T: p.(Leu670Phe) | c.2023G>A: p.(Ala675Thr) | c.2033C>A: p.(Ala678Asp) | c.2043G>C:p.(Met681Ile) | c.2080A>C:p.(Ser694Arg) | c.3812C>T: p.(Ser1271Leu) | c.3937C>T: p.(Arg1313Trp) |
| Variant location | ABD | pre-M1 | M3 | M3 | M3 | M3 | M3 | M3 | M3 | M3 | ABD | CTD | CTD |
| Gender | male | female | male | female | female | male | Female | N/A | Female | male | female | male | female |
| Age at onset | 3 days | 2mo | 9mo | 4mo | 2mo | < 1yo | 4mo | N/A | 3y, 5mo | 2yo | 2yo | 2mo | <1yo |
| Seizure types and EEG | startling seizure; EEG: intermittent multifocal discharges at 15 mon old | focal motor to GTCS; EEG: slow background, multifocal spikes | atypical absence; EEG: bilateral central spikes; hypsarrhythmia | complex partial seizure; EEG: frequent spikes and waves during awake and hypsarrhythmia during sleep | generalized seizure; EEG: sharp and spike wave discharges | epileptic spasms; EEG: diffuse paroxysmal abnormalities | focal motor to GTCS; EEG: continuous hypsarrhythmia with bilateral synchrony | epileptic spasms; EEG: N/A | focal motor → GTCS; EEG: frequent multifocal spikes; runs of faster alpha activity, multifocal spikes | febrile convulsion, afebrile generalized or unilateral convulsions, myoclonic seizure; EEG: Frequent multifocal spikes and diffuse spwc and sharp waves | tonic seizures, atonic seizures, atypical absence; EEG: Diffuse spwc. Focal spikes. Continuous diffuse spwc during sleep | epileptic spasms, myoclonic jerks; EEG: modified hypsarrhythmia; sporadic focal epileptic activity, runs of faster beta activity | GTCS, focal clonic, myoclonic, epileptic spasms; EEG: frequent/almost continuous sharp-waves with high amplitude |
| Developmental delay and other neurological features | no head control, no rolling over, no speech; hypotonia, hyperreflexia; Disturbed sleep pattern | walked at 2yo; poor eye contact, autistic behaviors | severe DD; mild hypotonia, dyskinetic and choreiform movements; cerebral visual impairment, oculomotor apraxia, changing tone, periodic breathing pattern | sitting alone at 3.5yo, walking with support at 3.5yo, speaking single words at 6.5yo; axial hypotonia, mild appendicular hypertonia; facial dysmorphisms, bilateral mild fifth finger clinodactyly, mild pes planus; poor sleep | rolling over at 9 mon old, unable to sit alone; truncal and orofacial hypotonia, appendicular hypertonia, athetoid limb movements, autistic features; microcephaly, severe pes planus | severe DD; severe hypotonia; cerebral visual impairment, feeding difficulties | severe DD; hypotonia, dyskinetic and choreiform movements; visual impairment with inconstant fixation | DD | walked at 2yo; single words at 2.5yo; hypotonia; poor eye contact, autistic behaviors | sitting alone at 18 mon old, DQ < 10 at 7 yo; myoclonus, ballismus, autism | Walking alone at 14 mon old, speaking two-word sentences at 36 mon old, IQ 64 at 7yo, full-scale IQ 58 at 11yo; ADHD | Severe DD; severe axial hypotonia, continuous movements; cerebral visual impairment, pyramidal signs with abnormal plantars (2yo), failure to thrive | severe; severe hypotonia, tetraplegia; wheelchair user, scoliosis, cerebral visual impairment, amaurosis, feeding difficulties |
| Brain MRI | loss of white matter, thin corpus callosum at 7.5 mon old | Normal | mild cerebral atrophy | N/A | normal | cortical atrophy | mild cerebral atrophy | N/A | Normal | normal at 6yo | normal at 3yr | normal | mild cerebral atrophy |
| Response to AEDs | PB, CZP, VPA: partially effective. CLB, Vit. B6: not effective | no formal therapy | seizure free on memantine, IVIG, oral steroids and Mg | Intractable, ACTH, LEV, Memantine, Mg, ketamine, pentobarbital, MDZ | intractable; seizure free on memantine, sultiame, lamotrigine, and valproic acid | no response | mild amelioration of EEG on steroids – no clinical overt seizures | no response | seizure free on VPA, LEV, and clonazepam | Intractable, VPA, CLB, LTG, LEV, LCM | ESM: effective for atonic seizures and atypical absence. CBZ, VPA, CZP: not effective | relatively controlled by VPA, TPM and ELF (in combination with VNS) | no response |
| Functional consequences | N/A | ↑glu/D-serine potency, ↑Mg inhibition, ↓pH inhibition, ↓Popen, ↓current amplitude, ↓receptor cell surface expression | ↑glu/gly potency, ↓Mg/pH/zinc inhibition, ↑Popen, ↑deactivation tau, causes neuronal cell death, dendritic swelling | ↑glu/gly potency, ↓Mg/pH/zinc inhibition, ↑Popen, ↑deactivation tau, causes neuronal cell death, dendritic swelling | ↑glu/gly potency, ↓Mg/pH/zinc inhibition, ↑Popen, ↑deactivation tau, causes neuronal cell death, dendritic swelling | ↑glu/gly/D-serine potency, ↓pH inhibition, ↑Popen, ↑deactivation tau, ↑charge transfer, ↓receptor cell surface expression | ↑glu/gly/D-serine potency, ↓pH inhibition, ↑Popen, ↑deactivation tau, ↑charge transfer, ↓receptor cell surface expression | ↑glu/gly/D-serine potency, ↓pH inhibition, ↓current amplitude, ↓receptor cell surface expression | ↑glu/gly/D-serine potency, ↑Popen, ↓current amplitude, ↓receptor cell surface expression; a significant ↓ in neuronal viability | N/A | N/A | ↓gly potency, No effects on Mg/pH inhibition, no effects on Popen and current amplitude, ↓receptor cell surface expression | ↓gly potency, No effects on Mg/pH inhibition, no effects on Popen and current amplitude, ↓receptor cell surface expression |
| Source | Tsuchida et a., 2018 | XiangWei et al., 2019 | XiangWei et al., 2019; Li et al., 2016 | Li et al., 2016 | Li et al., 2016; XiangWei et al., 2019 | XiangWei et al., 2019 | XiangWei et al., 2019 | XiangWei et al., 2019 | XiangWei et al., 2019 | Tsuchida et a., 2018 | Tsuchida et a., 2018 | XiangWei et al., 2019 | XiangWei et al., 2019 |
AED, anti-epileptic drug; DD, developmental delay; GTCS, generalized tonic clonic seizures; LEV, levetiracetam; NA, not available; TPM, topiramate; VGB, vigabatrin; VNS, vagal nerve stimulator; ELF, ethylloflazepate; VPA, valproate.
The S573F mutation lies within the pre-M1 region, a critical linker within the TMD that has profound impact on receptor gating.[1] The pre-M1 linker helps transfer molecular motion within the ABD to movement of the membrane-spanning helices within the TMD, allowing for receptor opening and ion flux across the membrane. Not surprisingly, the S573F variant impacts multiple aspects of receptor function including agonist potency, efficacy of magnesium block, current amplitude, and open channel probability.[89] Although this variant shows a modest increase in agonist potency, this may be considered a LoF mutation as magnesium block is increased, while current amplitude, open channel probability, and surface expression are all diminished.[89]
The V667I and L670F variants are both strong GoF mutations, found within the M3 helix of the TMD, and are just upstream of the highly conserved SYTANLAAF region which is also critical for receptor gating.[1, 39] Both variants display an increased agonist potency and open channel probability, as would be expected for mutations within the region of the receptor.[39, 89] Additionally, both V667I and L670F have prolonged deactivation times compared to wildtype GluN2D.[39, 89] This feature makes these variants unique, as all other DEE-associated GRIN2D mutations show either no change in receptor deactivation or an accelerated (e.g. a shortened) deactivation time course, which is a measure of how long the receptor active open following agonist removal. Thus, both the V667I and L670F variants will flux more ions into the neuron, potentially leading to calcium-induced excitotoxicity and cell death.[92] Indeed, cell viability assays with cultured cortical neurons transfected with a GluN2D-V667I plasmid revealed substantial neuronal swelling and extensive cell death.[39]
The A675T and A678D variants also reside within the M3 helix of the TMD. The A675T variant shows an increased agonist potency together with a strong decrease in current amplitude, therefore this variant likely represents a LoF mutation.[89] In fact, both the A675T and A678D variants had such small current amplitudes that measuring deactivation time was not possible. In both cases, the peak current amplitude measured in transfected HEK cells was less than 10% of a wildtype GluN2D-containing NMDA receptors.[89] This intriguing finding suggests that both mutations severely impact the ability of the receptor to open in response to agonist binding. In addition, the A678D variant displayed an increase in open channel probability, which makes classifying this variant more difficult. Either way, the profound decrease in current amplitude indicates that neurons harboring these variants will likely experience deficits in calcium influx. In agreement with these findings, transfected cultured neurons containing a GluN2D-A678D plasmid showed a significant decrease in viability.[89]
The S1271L and R1313W variants are located within the CTD. This domain of the receptor has not been extensively studied but has been shown to be important for surface trafficking and a localization site for protein-protein interactions. Interestingly, both the S1271L and R1313W variants showed a decreased agonist potency, despite being localized far away from the ABD. However, this change in agonist potency was not robust enough to elicit changes in other aspects of receptor function as open channel probability, current amplitude, and deactivation time were all unchanged compared to wild-type. This combination of decreased agonist potency and decreased surface expression likely indicate that both S1271L and R1313W are LoF mutations.[89] The finding that mutations within the CTD can generate DEE may be suggestive of the importance of protein-protein interactions facilitated by the GluN2D subunit in controlling circuit excitability.
In fact, unique to all variants tested for functional activity was a decrease in surface expression. Changes in surface expression have been reported previously with epilepsy-associated variants in the GRIN2A and GRIN2B genes, where most of which also showed a reduction in surface expression.[11] The idea that any perturbation to the amino acid composition of the GluN2D subunit diminishes surface expression is interesting, perhaps suggesting that biology has precisely tuned surface expression for its maximal level. On the other hand, mutations that increase surface expression of GluN2D may be lethal and thus might not be represented within our current findings. Changes in surface expression may also impact the subunit composition and ratio of NMDAR assemblies. As neuronal circuits grow and develop, this alteration combined with the modification to receptor function, may invoke a multitude of maladaptive homeostatic plasticity mechanisms. Examples of this phenomenon can be shown in the vast range of neurological and developmental phenotypes seen in patients with GRIN2D variants.[39, 87–89]
Overall, it seems that a loss of GluN2D function within the brain is sufficient to promote DEE, likely via hypofunction of GABAergic interneurons. The two GoF variants found, V667I and L670F, may also be indirectly diminishing interneuron function by initiating calcium-induced excitotoxicity. Future characterization of these variants using knock-in mouse models is needed to better understand the impact of these mutations on circuit function and interneuron viability.
3.3. Clinical symptoms and electroencephalographic signatures of patients with GRIN2D variants
Although DEE-associated GRIN2D variants may share some of the same functional characteristics, the seizure types, electroencephalographic (EEG) patterns, and developmental phenotypes they cause are largely heterogeneous. A surprising finding was that two variants, V667I (n = 3) and L670F (n = 2), were found in more than one patient, with a total of 13 individuals containing a DEE-associated GRIN2D variant (Table 2).[39, 89] The mean age of onset is 11 ± 3.6 months, with a median age of onset of 6.5 months (Figure 2C). GRIN2D-related DEE does not appear to be gender-related as the male to female ratio is 5:7, with gender unknown for the A675T patient.[89]
There appears to be little commonality in the type of seizures elicited by GRIN2D mutations, even amongst individuals carrying the same GRIN2D variant. All three V667I patients display different seizure patterns, with one having atypical absence seizures, one having partial complex seizures, and another having generalized seizures.[39] The same is true of the two patients harboring the L670F variant: one has epileptic spasms while the other has generalized tonic-clonic (GTC) seizures.[89] The heterogeneity of seizure types amongst patients with the same GRIN2D variant indicates a highly complex nature of epileptogenesis. Unfortunately, this complexity and disease heterogeneity amongst individuals containing the same mutation appears to be common, as individuals with Dravet’s syndrome – a genetic epilepsy caused by a haploinsufficiency in the SCN1A gene – also exhibit multiple different seizure phenotypes.[93] While a full list of the different seizure types caused by GRIN2D variants can be found in Table 2, two of the most common are focal motor to GTC (n = 4) and epileptic spasms (n = 4).
DEE-associated GRIN2D patients also exhibit diverse types of electrical activity recorded via EEG, although EEG patterns are more uniform than seizure type (Table 2). The majority (75%, 9/12; EEG pattern for Patient #8 is unavailable) of patients exhibited multifocal epileptiform activity, indicating that more than one brain region may be affected. This finding is not surprising given the expression patterns of GluN2D throughout the brain and their ability to regulate inhibitory tone via GABAergic interneurons. The two most common EEG patterns were multifocal spikes (n = 5) and hypsarrhythmia (n = 4; Figure 2D). The other three patients – Patient #5-V667I, Patient #6-L670F, and Patient #13-R1313W – all had focal epileptiform activity that presented as spike-and-wave discharges or paroxysmal fast activity.[12, 39] Again, patients containing the same GRIN2D variant did not exhibit similar EEG recordings, as one of the three V667I patients and one of the two L670F patients presented focal interictal EEG recordings.[39, 89]
Furthermore, patients with GRIN2D mutations express a host of neurological and developmental abnormalities. As mentioned previously, the GluN2D subunit is heavily involved in motor control likely via robust expression in the spinal cord and basal ganglia. Thus, it is not surprising that 69% (9/13) of DEE-associated GRIN2D variants are associated with hypotonia and poor motor control (Figure 2E). However, the type and location of hypotonia experienced spans the gamut between minor hypotonia with dyskinetic and choreiform movements (Patient #3) to severe hypotonia with tetraplegia (Patient #13). Approximately 45% (5/11; data not available for Patient #4 and Patient #8) experienced some form of cortical atrophy when overall brain morphology was assessed via magnetic resonance imaging (Figure 2E). Only 38% (5/13) GRIN2D DEE-associated patients have cerebral visual impairment, indicating an improper relay between the information sensed by the retina and that received by the visual cortex. These results are somewhat surprising given the strong GluN2D expression in rod bipolar cells and in interneurons in the cerebral cortex, as described earlier.[23, 59]
Autism and the presence of autistic-like features and behaviors are also fairly common, as 31% (4/13) patients with DEE-associated GRIN2D variants are on the autism spectrum. Most present with poor eye contact and social withdrawal. 23% (3/13) of patients have plantar deformities, with two having pes planus and one having ‘abnormal plantars’. Other behavioral difficulties manifest as problems with sleeping (n = 2), feeding (n = 2), breathing (n = 1), and speaking (n = 1). As with seizure type and EEG pattern, neurological and developmental symptoms are highly varied among individuals with the same GRIN2D variant.
3.4. Treating patients with GRIN2D variants
Unfortunately, another unifying characteristic of these 13 DEE GRIN2D patients is a resistance to typical AED therapy. Specifically, 75% (9/12; Patient #2 has not reported any formal drug therapy) of these patients were either refractory or only saw partial recovery from seizure burden on AEDs (Figure 2E).[39, 88, 89] Patient #7 showed a modest improvement of EEG recordings on steroids, whereas Patient #9 and Patient #11 were seizure free using typical AEDs. Additionally, these data only reflect information on seizure burden and EEG readouts in response to drug therapy as data regarding recovery from secondary behavioral, neurological, and developmental symptoms are not available. However, given the severity of DEE, it seems unlikely that any conventional drug therapy would be able to ameliorate all the GRIN2D variant-inflicted symptoms.
The three patients containing the V667I mutation are unique in that all three were exposed to targeted drug therapies that were specific for NMDARs. After exome sequencing, all three patients were placed on a combination therapy of memantine and magnesium.[39, 89] Memantine, as mentioned previously, is an NMDAR pore blocker that is efficacious at all NMDAR subunits, and shows an increase in potency at GluN2D receptors.[35] Additionally, magnesium was chosen as this divalent cation can block the NMDAR pore at polarized membrane potentials. Of the three patients receiving the memantine/magnesium cocktail, only one (Patient #3) responded positively. One patient did not have a response, whereas the other patient worsened and saw an increased seizure burden. When examining age of onset, Patient #3 was diagnosed and placed on this targeted treatment earlier than the other two patients.[39, 89] This suggests that not only is precision medicine needed but perhaps there exists a critical time window in which therapeutics may be most beneficial. While the exact mechanism behind the exacerbated seizures for the V667I patient on memantine/magnesium is unclear, this study provides critical information needed to combat the severe group of neurological diseases generated via GRIN2D variants.
4. Conclusions
The confluence of the advances in genetics, structural biology, and functional analysis will provide an opportunity to gain insight into the relationships between human disease and GRIN2D/GluN2D NMDA receptors and provide new insight into new strategy of personalized medicine to improve diagnosis and effective treatment. In turn, the evaluation of the disease associate GRIN2D variants will provide new insight into GluN2D NMDAR function.
Acknowledgements
The authors thank Dr. Stephen F. Traynelis, Dr. Subhrajit Bhattacharya, Dr. Guojun Zhang, and Britton R. Barbee for their critical reading and comments.
Funding sources
H.Y. is supported by the National Institutes of Health (NIH/NICHD; R01HD08237).
Footnotes
Conflict of interest
None of the authors have any conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Traynelis SF, et al. , Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev, 2010. 62(3): p. 405–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karakas E and Furukawa H, Crystal structure of a heterotetrameric NMDA receptor ion channel. Science, 2014. 344(6187): p. 992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee CH, et al. , NMDA receptor structures reveal subunit arrangement and pore architecture. Nature, 2014. 511(7508): p. 191–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuste R, et al. , Mechanisms of calcium influx into hippocampal spines: heterogeneity among spines, coincidence detection by NMDA receptors, and optical quantal analysis. J Neurosci, 1999. 19(6): p. 1976–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endele S, et al. , Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet, 2010. 42(11): p. 1021–6. [DOI] [PubMed] [Google Scholar]
- 6.Gambrill AC and Barria A, NMDA receptor subunit composition controls synaptogenesis and synapse stabilization. Proc Natl Acad Sci U S A, 2011. 108(14): p. 5855–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlsson M and Carlsson A, The NMDA antagonist MK-801 causes marked locomotor stimulation in monoamine-depleted mice. J Neural Transm, 1989. 75(3): p. 221–6. [DOI] [PubMed] [Google Scholar]
- 8.Sakimura K, et al. , Reduced hippocampal LTP and spatial learning in mice lacking NMDA receptor epsilon 1 subunit. Nature, 1995. 373(6510): p. 151–5. [DOI] [PubMed] [Google Scholar]
- 9.Bauer EP, Schafe GE, and LeDoux JE, NMDA receptors and L-type voltage-gated calcium channels contribute to long-term potentiation and different components of fear memory formation in the lateral amygdala. J Neurosci, 2002. 22(12): p. 5239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krystal JH, et al. , Subanesthetic effects of the noncompetitive NMDA antagonist, ketamine, in humans. Psychotomimetic, perceptual, cognitive, and neuroendocrine responses. Arch Gen Psychiatry, 1994. 51(3): p. 199–214. [DOI] [PubMed] [Google Scholar]
- 11.Swanger SA, et al. , Mechanistic Insight into NMDA Receptor Dysregulation by Rare Variants in the GluN2A and GluN2B Agonist Binding Domains. Am J Hum Genet, 2016. 99(6): p. 1261–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogden KK, et al. , Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology. PLoS Genet, 2017. 13(1): p. e1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.XiangWei W, Jiang Y, and Yuan H, De Novo Mutations and Rare Variants Occurring in NMDA Receptors. Curr Opin Physiol, 2018. 2: p. 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan H, et al. , Ionotropic GABA and Glutamate Receptor Mutations and Human Neurologic Diseases. Mol Pharmacol, 2015. 88(1): p. 203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traynelis J, et al. , Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res, 2017. 27(10): p. 1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burger PB, et al. , Mapping the binding of GluN2B-selective N-methyl-D-aspartate receptor negative allosteric modulators. Mol Pharmacol, 2012. 82(2): p. 344–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akazawa C, et al. , Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. J Comp Neurol, 1994. 347(1): p. 150–60. [DOI] [PubMed] [Google Scholar]
- 18.Monyer H, et al. , Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron, 1994. 12(3): p. 529–40. [DOI] [PubMed] [Google Scholar]
- 19.Scherzer CR, et al. , Expression of N-methyl-D-aspartate receptor subunit mRNAs in the human brain: hippocampus and cortex. J Comp Neurol, 1998. 390(1): p. 75–90. [DOI] [PubMed] [Google Scholar]
- 20.Ritter LM, Unis AS, and Meador-Woodruff JH, Ontogeny of ionotropic glutamate receptor expression in human fetal brain. Brain Res Dev Brain Res, 2001. 127(2): p. 123–33. [DOI] [PubMed] [Google Scholar]
- 21.Tolle TR, et al. , The differential expression of 16 NMDA and non-NMDA receptor subunits in the rat spinal cord and in periaqueductal gray. J Neurosci, 1993. 13(12): p. 5009–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Hara BF, et al. , GABAA, GABAC, and NMDA receptor subunit expression in the suprachiasmatic nucleus and other brain regions. Brain Res Mol Brain Res, 1995. 28(2): p. 239–50. [DOI] [PubMed] [Google Scholar]
- 23.Wenzel A, et al. , N-methyl-D-aspartate receptors containing the NR2D subunit in the retina are selectively expressed in rod bipolar cells. Neuroscience, 1997. 78(4): p. 1105–12. [DOI] [PubMed] [Google Scholar]
- 24.Niedzielski AS and Wenthold RJ, Expression of AMPA, kainate, and NMDA receptor subunits in cochlear and vestibular ganglia. J Neurosci, 1995. 15(3 Pt 2): p. 2338–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wenzel A, et al. , Developmental and regional expression of NMDA receptor subtypes containing the NR2D subunit in rat brain. J Neurochem, 1996. 66(3): p. 1240–8. [DOI] [PubMed] [Google Scholar]
- 26.Dunah AW, et al. , Regional and ontogenic expression of the NMDA receptor subunit NR2D protein in rat brain using a subunit-specific antibody. J Neurochem, 1996. 67(6): p. 2335–45. [DOI] [PubMed] [Google Scholar]
- 27.Momiyama A, Feldmeyer D, and Cull-Candy SG, Identification of a native low-conductance NMDA channel with reduced sensitivity to Mg2+ in rat central neurones. J Physiol, 1996. 494 (Pt 2): p. 479–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Engelhardt J, et al. , GluN2D-containing NMDA receptors-mediate synaptic currents in hippocampal interneurons and pyramidal cells in juvenile mice. Front Cell Neurosci, 2015. 9: p. 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perszyk RE, et al. , GluN2D-Containing N-methyl-d-Aspartate Receptors Mediate Synaptic Transmission in Hippocampal Interneurons and Regulate Interneuron Activity. Mol Pharmacol, 2016. 90(6): p. 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelkey KA, et al. , Hippocampal GABAergic Inhibitory Interneurons. Physiol Rev, 2017. 97(4): p. 1619–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanger SA, et al. , A Novel Negative Allosteric Modulator Selective for GluN2C/2D-Containing NMDA Receptors Inhibits Synaptic Transmission in Hippocampal Interneurons. ACS Chem Neurosci, 2018. 9(2): p. 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Misra C, et al. , Slow deactivation kinetics of NMDA receptors containing NR1 and NR2D subunits in rat cerebellar Purkinje cells. J Physiol, 2000. 525 Pt 2: p. 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuner T and Schoepfer R, Multiple structural elements determine subunit specificity of Mg2+ block in NMDA receptor channels. J Neurosci, 1996. 16(11): p. 3549–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke RJ and Johnson JW, NMDA receptor NR2 subunit dependence of the slow component of magnesium unblock. J Neurosci, 2006. 26(21): p. 5825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kotermanski SE and Johnson JW, Mg2+ imparts NMDA receptor subtype selectivity to the Alzheimer’s drug memantine. J Neurosci, 2009. 29(9): p. 2774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clements JD, et al. , The time course of glutamate in the synaptic cleft. Science, 1992. 258(5087): p. 1498–501. [DOI] [PubMed] [Google Scholar]
- 37.Vicini S, et al. , Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. J Neurophysiol, 1998. 79(2): p. 555–66. [DOI] [PubMed] [Google Scholar]
- 38.Wyllie DJ, Behe P, and Colquhoun D, Single-channel activations and concentration jumps: comparison of recombinant NR1a/NR2A and NR1a/NR2D NMDA receptors. J Physiol, 1998. 510 (Pt 1): p. 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li D, et al. , GRIN2D Recurrent De Novo Dominant Mutation Causes a Severe Epileptic Encephalopathy Treatable with NMDA Receptor Channel Blockers. Am J Hum Genet, 2016. 99(4): p. 802–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vance KM, et al. , Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nat Commun, 2011. 2: p. 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegler Retchless B, Gao W, and Johnson JW, A single GluN2 subunit residue controls NMDA receptor channel properties via intersubunit interaction. Nat Neurosci, 2012. 15(3): p. 406–13, S1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen KB, et al. , Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron, 2014. 81(5): p. 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi F, et al. , Functional and pharmacological properties of triheteromeric GluN1/2B/2D NMDA receptors. Journal of Physiology, 2019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanson E, et al. , Tonic Activation of GluN2C/GluN2D-Containing NMDA Receptors by Ambient Glutamate Facilitates Cortical Interneuron Maturation. J Neurosci, 2019. 39(19): p. 3611–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szapiro G and Barbour B, Multiple climbing fibers signal to molecular layer interneurons exclusively via glutamate spillover. Nat Neurosci, 2007. 10(6): p. 735–42. [DOI] [PubMed] [Google Scholar]
- 46.Tremblay R, Lee S, and Rudy B, GABAergic Interneurons in the Neocortex: From Cellular Properties to Circuits. Neuron, 2016. 91(2): p. 260–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francavilla R, et al. , Calcium Dynamics in Dendrites of Hippocampal CA1 Interneurons in Awake Mice. Front Cell Neurosci, 2019. 13: p. 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sapkota K, et al. , GluN2D N-Methyl-d-Aspartate Receptor Subunit Contribution to the Stimulation of Brain Activity and Gamma Oscillations by Ketamine: Implications for Schizophrenia. J Pharmacol Exp Ther, 2016. 356(3): p. 702–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matta JA, et al. , Developmental origin dictates interneuron AMPA and NMDA receptor subunit composition and plasticity. Nat Neurosci, 2013. 16(8): p. 1032–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanger SA, et al. , NMDA Receptors Containing the GluN2D Subunit Control Neuronal Function in the Subthalamic Nucleus. J Neurosci, 2015. 35(48): p. 15971–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolam JP, et al. , Synaptic organisation of the basal ganglia. J Anat, 2000. 196 (Pt 4): p. 527–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearlstein E, et al. , Glutamatergic synaptic currents of nigral dopaminergic neurons follow a postnatal developmental sequence. Front Cell Neurosci, 2015. 9: p. 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hallett PJ and Standaert DG, Rationale for and use of NMDA receptor antagonists in Parkinson’s disease. Pharmacol Ther, 2004. 102(2): p. 155–74. [DOI] [PubMed] [Google Scholar]
- 54.Hildebrand ME, et al. , GluN2B and GluN2D NMDARs dominate synaptic responses in the adult spinal cord. Sci Rep, 2014. 4: p. 4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minami T, et al. , Characterization of the glutamatergic system for induction and maintenance of allodynia. Brain Res, 2001. 895(1–2): p. 178–85. [DOI] [PubMed] [Google Scholar]
- 56.Andrade-Talavera Y, et al. , Presynaptic Spike Timing-Dependent Long-Term Depression in the Mouse Hippocampus. Cereb Cortex, 2016. 26(8): p. 3637–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feldman DE, The spike-timing dependence of plasticity. Neuron, 2012. 75(4): p. 556–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dubois CJ, et al. , Presynaptic GluN2D receptors detect glutamate spillover and regulate cerebellar GABA release. J Neurophysiol, 2016. 115(1): p. 271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alsaad HA, et al. , In the Telencephalon, GluN2C NMDA Receptor Subunit mRNA is Predominately Expressed in Glial Cells and GluN2D mRNA in Interneurons. Neurochem Res, 2019. 44(1): p. 61–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ikeda K, et al. , Reduced spontaneous activity of mice defective in the epsilon 4 subunit of the NMDA receptor channel. Brain Res Mol Brain Res, 1995. 33(1): p. 61–71. [DOI] [PubMed] [Google Scholar]
- 61.Miyamoto Y, et al. , Lower sensitivity to stress and altered monoaminergic neuronal function in mice lacking the NMDA receptor epsilon 4 subunit. J Neurosci, 2002. 22(6): p. 2335–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hagino Y, et al. , Essential role of NMDA receptor channel epsilon4 subunit (GluN2D) in the effects of phencyclidine, but not methamphetamine. PLoS One, 2010. 5(10): p. e13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamoto T, et al. , Role of the NMDA receptor GluN2D subunit in the expression of ketamine-induced behavioral sensitization and region-specific activation of neuronal nitric oxide synthase. Neurosci Lett, 2016. 610: p. 48–53. [DOI] [PubMed] [Google Scholar]
- 64.Sohal VS, et al. , Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature, 2009. 459(7247): p. 698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yizhar O, et al. , Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature, 2011. 477(7363): p. 171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Korotkova T, et al. , NMDA receptor ablation on parvalbumin-positive interneurons impairs hippocampal synchrony, spatial representations, and working memory. Neuron, 2010. 68(3): p. 557–69. [DOI] [PubMed] [Google Scholar]
- 67.Fiest KM, et al. , Prevalence and incidence of epilepsy: A systematic review and meta-analysis of international studies. Neurology, 2017. 88(3): p. 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Engel J Jr. and International League Against E, A proposed diagnostic scheme for people with epileptic seizures and with epilepsy: report of the ILAE Task Force on Classification and Terminology. Epilepsia, 2001. 42(6): p. 796–803. [DOI] [PubMed] [Google Scholar]
- 69.Khan S and Al Baradie R, Epileptic encephalopathies: an overview. Epilepsy Res Treat, 2012. 2012: p. 403592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mastrangelo M, et al. , Epileptic seizures, epilepsy and epileptic syndromes in newborns: a nosological approach to 94 new cases by the 2001 proposed diagnostic scheme for people with epileptic seizures and with epilepsy. Seizure, 2005. 14(5): p. 304–11. [DOI] [PubMed] [Google Scholar]
- 71.Loddenkemper T, Fernandez IS, and Peters JM, Continuous spike and waves during sleep and electrical status epilepticus in sleep. J Clin Neurophysiol, 2011. 28(2): p. 154–64. [DOI] [PubMed] [Google Scholar]
- 72.Helmstaedter C and Witt JA, Clinical neuropsychology in epilepsy: theoretical and practical issues. Handb Clin Neurol, 2012. 107: p. 437–59. [DOI] [PubMed] [Google Scholar]
- 73.Bhardwaj P, et al. , Acquired epileptic aphasia: Landau-Kleffner syndrome. J Pediatr Neurosci, 2009. 4(1): p. 52–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jain P, Sharma S, and Tripathi M, Diagnosis and management of epileptic encephalopathies in children. Epilepsy Res Treat, 2013. 2013: p. 501981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomson T and Forsgren L, Life expectancy in epilepsy. Lancet, 2005. 365(9459): p. 557–8. [DOI] [PubMed] [Google Scholar]
- 76.Vigevano F, et al. , Therapeutic approach to epileptic encephalopathies. Epilepsia, 2013. 54 Suppl 8: p. 45–50. [DOI] [PubMed] [Google Scholar]
- 77.Nariai H, Duberstein S, and Shinnar S, Treatment of Epileptic Encephalopathies: Current State of the Art. J Child Neurol, 2018. 33(1): p. 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McTague A and Cross JH, Treatment of epileptic encephalopathies. CNS Drugs, 2013. 27(3): p. 175–84. [DOI] [PubMed] [Google Scholar]
- 79.Matsumoto A, et al. , Long-term prognosis after infantile spasms: a statistical study of prognostic factors in 200 cases. Dev Med Child Neurol, 1981. 23(1): p. 51–65. [DOI] [PubMed] [Google Scholar]
- 80.Oguni H, Hayashi K, and Osawa M, Long-term prognosis of Lennox-Gastaut syndrome. Epilepsia, 1996. 37 Suppl 3: p. 44–7. [DOI] [PubMed] [Google Scholar]
- 81.Mikaeloff Y, et al. , Devastating epileptic encephalopathy in school-aged children (DESC): a pseudo encephalitis. Epilepsy Res, 2006. 69(1): p. 67–79. [DOI] [PubMed] [Google Scholar]
- 82.Pedespan JM, et al. , Surgical treatment of an early epileptic encephalopathy with suppression-bursts and focal cortical dysplasia. Epilepsia, 1995. 36(1): p. 37–40. [DOI] [PubMed] [Google Scholar]
- 83.Begley CE and Durgin TL, The direct cost of epilepsy in the United States: A systematic review of estimates. Epilepsia, 2015. 56(9): p. 1376–87. [DOI] [PubMed] [Google Scholar]
- 84.Li L, et al. , Considering economic reality in calculating the financial burden of epilepsy in China. Epilepsia, 2011. 52(2): p. 416–8. [DOI] [PubMed] [Google Scholar]
- 85.Cardarelli WJ and Smith BJ, The burden of epilepsy to patients and payers. Am J Manag Care, 2010. 16(12 Suppl): p. S331–6. [PubMed] [Google Scholar]
- 86.Gao L, et al. , Burden of epilepsy: a prevalence-based cost of illness study of direct, indirect and intangible costs for epilepsy. Epilepsy Res, 2015. 110: p. 146–56. [DOI] [PubMed] [Google Scholar]
- 87.Tarabeux J, et al. , Rare mutations in N-methyl-D-aspartate glutamate receptors in autism spectrum disorders and schizophrenia. Transl Psychiatry, 2011. 1: p. e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsuchida N, et al. , GRIN2D variants in three cases of developmental and epileptic encephalopathy. Clin Genet, 2018. 94(6): p. 538–547. [DOI] [PubMed] [Google Scholar]
- 89.XiangWei W, et al. , Heterogeneous clinical and functional features of GRIN2D-related developmental and epileptic encephalopathy. Brain, 2019. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuan H, et al. , Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci, 2009. 29(39): p. 12045–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burnashev N and Szepetowski P, NMDA receptor subunit mutations in neurodevelopmental disorders. Curr Opin Pharmacol, 2015. 20: p. 73–82. [DOI] [PubMed] [Google Scholar]
- 92.Arundine M and Tymianski M, Molecular mechanisms of calcium-dependent neurodegeneration in excitotoxicity. Cell Calcium, 2003. 34(4–5): p. 325–37. [DOI] [PubMed] [Google Scholar]
- 93.Chopra R and Isom LL, Untangling the dravet syndrome seizure network: the changing face of a rare genetic epilepsy. Epilepsy Curr, 2014. 14(2): p. 86–9. [DOI] [PMC free article] [PubMed] [Google Scholar]