SUMMARY
Biosynthesis of the pyrimidine nucleotide uridine monophosphate (UMP) is essential for cell proliferation and is achieved by the activity of convergent de novo and salvage metabolic pathways. Here we report the development and application of a cell-based metabolic modifier screening platform that leverages the redundancy in pyrimidine metabolism for the discovery of selective UMP biosynthesis modulators. In evaluating a library of protein kinase inhibitors, we identified multiple compounds that possess nucleotide metabolism-modifying activity. The JNK inhibitor JNK-IN-8 was found to potently inhibit nucleoside transport and engage ENT1. The PDK1 inhibitor OSU-03012 (also known as AR-12) and the RAF inhibitor TAK-632 were shown to inhibit the therapeutically-relevant enzyme dihydroorotate dehydrogenase (DHODH) and their affinities were unambiguously confirmed through in vitro assays and co-crystallization with human DHODH.
eTOC Blurb
Pyrimidine nucleotides are produced interchangeably by convergent metabolic pathways. Abt et al. develop and apply a phenotypic screen that leverages this redundancy to identify selective small molecule metabolism modifiers. Multiple compounds developed as protein kinase inhibitors were found to possess secondary targets within nucleotide metabolism.
INTRODUCTION
The redundant and plastic nature of metabolic networks represents a significant obstacle in the targeting of cancer metabolism. This redundancy manifests in two ways; the first is the expression of multiple enzymes that perform identical biochemical reactions, such as the hexokinase isozymes which each phosphorylate glucose (Xu et al., 2018); the second is the existence of convergent metabolic pathways that produce a common metabolite from unique precursors. Such convergent metabolic nodes have been noted in nucleotide (Le et al., 2017), lipid (cholesterol) (York et al., 2015) and amino acid (aspartate) metabolism (Garcia-Bermudez et al., 2018).
Despite these difficulties, the development of metabolism modifiers remains a robust area of research. One such therapeutically relevant target is pyrimidine nucleotide biosynthesis which consists of nucleoside salvage (NSP) and de novo (DNP) pathways that converge to generate uridine monophosphate (UMP), the common precursor for all pyrimidine nucleotides (Okesli et al., 2017). The NSP allows for the scavenging of uridine from the extracellular environment, shuttling it into the cell via nucleoside transporters where it is phosphorylated by uridine-cytidine kinases (UCKs) to produce UMP. UCK2 is thought to be the primary NSP kinase, given its 20-fold higher catalytic efficiency compared to UCK1 (Van Rompay et al., 2001). The DNP is a 6-step process that utilizes glutamine, aspartate, bicarbonate, and glucose to produce UMP through the action of 3 enzymes: trifunctional CAD, electron transport chain-linked dihydroorotate dehydrogenase (DHODH), and bifunctional UMP synthase (UMPS). Among DNP enzymes, DHODH in particular has emerged as a therapeutic target in multiple cancers including pancreatic ductal adenocarcinoma (PDAC) (Madak et al., 2019; Sykes et al., 2016; Santana-Codina et al., 2018). Additionally, over 90 patent applications involving DHODH inhibition have been filed in the last decade (Lolli et al., 2018).
In this study, we show that the pyrimidine NSP and DNP are interchangeable in their ability to sustain cancer cell proliferation and that a synthetic lethal phenotype can be achieved through their simultaneous inhibition. We leverage this observation to construct a metabolic modifier screen that allows for the identification of selective modulators of NSP and DNP pathways. In screening a library of protein kinase inhibitors, we identified multiple compounds with nucleotide metabolism-modifying activity. We show that the c-Jun N-terminal kinase (JNK) inhibitor JNK-IN-8 is a potent inhibitor of uridine transport which is vital for NSP function, and that the 3-phosphoinositide-dependent protein kinase 1 (PDK1) inhibitor OSU-03012 (also known as AR-12) and the pan-RAF inhibitor TAK-632 both bind and inhibit the pyrimidine DNP enzyme DHODH.
RESULTS
Design of a differential metabolic modifier screen for identification of modulators of pyrimidine nucleotide metabolism
While the UMP-DNP and -NSP are interchangeable in their ability to sustain cell proliferation, their relative activity at baseline (when both pathways are functional) is poorly defined. De novo pyrimidine biosynthesis is allosterically inactivated by its end product, UTP, which is also produced by the uridine (rU) NSP (Evans & Guy, 2004). This allosteric control functions at the level of the CPSase activity of the tri-functional protein CAD, which performs the first committed step in de novo pyrimidine biosynthesis (Figure S1A). To quantitatively evaluate the discrete activities of the pyrimidine de novo and salvage pathways, we modified and applied a LC-MS/MS assay previously used by our group to track the contribution of stable isotope-labeled glucose and deoxycytidine to newly replicated DNA (Le et al., 2017). In this assay, cells are cultured in the presence of [13C6]glucose (to track DNP activity) and 10 μM [13C9; 15N2]rU (to track NSP activity). Their DNA is then extracted and hydrolyzed and the abundance of stable isotope-labeled nucleosides is evaluated using LC-MS/MS in the multiple reaction monitoring (MRM) mode (Figure S1B). We applied this assay to a panel of cancer cell lines and observed a heterogenous degree of total labeling ([13C6]glucose + [13C9; 15N2]rU) in the deoxycytidine compartment of DNA (DNA-C) after 24 h (Figure S1C). Consistent with the aforementioned model in which UTP produced by uridine savage allosterically impairs de novo biosynthesis, we found that the fractional contribution of [13C9; 15N2]rU exceeded that of [13C6]glucose in all models tested (Figure S1D). Interestingly, we found heterogeneity in the relative contribution of [13C6]glucose and [13C9; 15N2]rU to DNA-C across the cell line panel, which likely reflects differential expression or regulation of the various transporters, kinases, nucleotidases, and phosphorylases involved in rU salvage. Importantly, we confirmed that the contribution of [13C6]glucose to DNA-C could be blocked by NITD-982, an established DHODH inhibitor (Bonavia et al., 2011), and likewise the contribution of [13C9; 15N2]rU could be prevented by the FDA-approved nucleoside transport inhibitor dipyridamole (DPA; Figure S1E) (Young et al., 2013). Collectively, these results indicate that, under the conditions tested, both the UMP-DNP and -NSP pathways are simultaneously, but not equally, active.
While redundant routes for UMP biosynthesis can complicate targeting, impaired proliferation resulting from simultaneous restriction of both de novo and salvage pathways can be leveraged for the identification of selective DNP or NSP activity modifiers (Figure S1F). A metabolic modifier screen was developed for the discovery of small molecule modulators of UMP production by leveraging this biosynthetic redundancy. This cell-based platform concurrently tests the effects of small molecule compounds on the proliferation of cells cultured in baseline (both NSP and DNP active), NSP-only, and DNP-only conditions (Figure 1A). Compounds which inhibit proliferation in baseline conditions are classified as non-specific inhibitors, those which selectively inhibit proliferation in NSP-only conditions are NSP inhibitors, while those that selectively inhibit growth in DNP-only conditions are DNP inhibitors. The screen design was validated using NITD-982 (which inhibits the DNP) and DPA (which inhibits the NSP), with Cell Titer Glo (CTG) utilized to evaluate proliferation impairment (Figure S1G) (Wang et al., 2011).
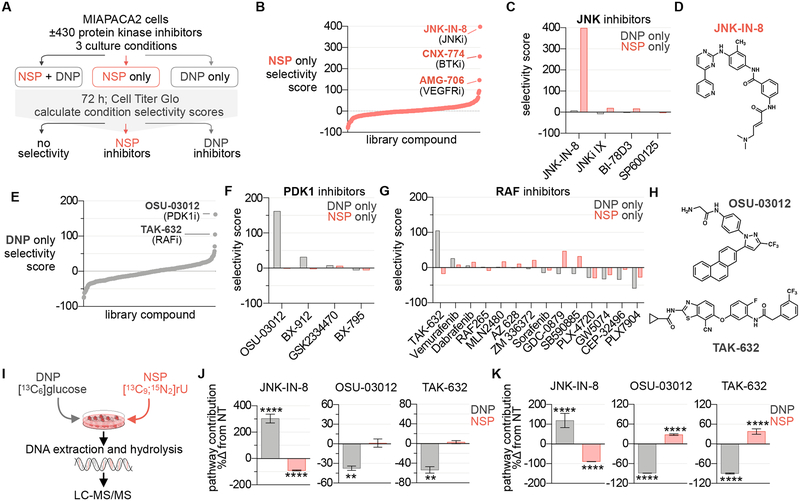
Figure 1 |. Identification of UMP-NSP and -DNP modulators in a protein kinase inhibitor library.
(A) Phenotypic screening strategy. The impact of 430 protein kinase inhibitors on cell proliferation was evaluated in MIAPACA2 cells plated in 3 distinct culture conditions; 1) NSP+DNP (media +10 μM uridine (rU)); 2) NSP only (media +10 μM rU +1 μM NITD-982); or 3) DNP only (media alone). % proliferation values were calculated using Cell Titer Glo (CTG) following 72 h treatment (7-point dose response; n=2). (B) Waterfall plot ranking library compounds based on NSP pathway selectivity score. (C) Summary of NSP and DNP selectivity scores across library compounds annotated as JNK inhibitors. (D) Structure of JNK-IN-8. (E) Waterfall plot ranking library compounds based on DNP pathway selectivity score. (F,G) Summary of NSP and DNP selectivity scores across library compounds annotated as PDK1 (F) or RAF inhibitors (G). (H) Structures OSU-03012 and TAK-632. (I) Experimental design to track contribution of UMP-DNP and -NSP to newly replicated DNA using stable isotope-labeled metabolite tracers. (J,K) LC-MS/MS analysis of [13C6]glucose (5.5 mM) and [13C9; 15N2] rU (10 μM) utilization for DNA-C replication in MIAPACA2 (J) or JURKAT (K) cells treated +1 μM JNK-IN-8 +5 μM OSU-03012 or +5 μM TAK-632 for 24 h (NT: not-treated; mean±SD; n=3; unpaired T-test; ** P < 0.01, **** P < 0.0001).
Cancer cell lines exhibited varying degrees of sensitivity to DHODH inhibition (as determined by doubling-time-normalized proliferation inhibition) and were all rescued by rU supplementation (Figure S2) (Hafner et al., 2016). MIAPACA2 PDAC cells were utilized for the screen due to their ability to maintain baseline proliferation levels in NSP-only or DNP-only conditions, while also exhibiting a significant decrease in proliferation upon simultaneous NSP and DNP inhibition (Figure S3A). A library of 430 protein kinase inhibitors was chosen for evaluation, the rationale being twofold. First, it was hypothesized that our synthetic lethality screen may identify compounds that indirectly target pyrimidine metabolism through regulatory signal transduction pathway inhibition. Second, because a substantial fraction of kinase inhibitors are ATP-mimetics and therefore resemble nucleotides, we predicted that protein kinase inhibitors may possess secondary, non-canonical targets within nucleotide metabolism. Consistently, several protein kinase inhibitors, specifically those exhibiting similarities with imatinib’s phenylamino pyrimidine (PAP) scaffold, and a subset of p38 MAPK inhibitors, exhibit activity against nucleoside transporters (Damaraju et al., 2016; Huang et al., 2002). This kinase inhibitor library was screened at 7-point dose response in duplicates. Composite NSP and DNP pathway selectivity scores were calculated for each compound as the sum of condition-specific anti-proliferative effects across the dose range (Data S1, Figure S3B). Phenotypic screen quality was monitored using the Z-factor metric (Zhang et al., 1999) (Figure S3C).
The JNK inhibitor JNK-IN-8, the BTK inhibitor CNX-774, and the VEGFR inhibitor AMG-706 were active in the NSP-only condition, exhibiting positive NSP-selectivity scores (Figure 1B). The selectivity of these hits for the NSP was unique among inhibitors of JNK (Figures 1C, 1D), BTK, and VEGFR (Figures S3D, S3E, S3F), indicating this phenotype likely did not result from on-target effects. The PDK1 inhibitor OSU-03012 (also known as AR-12) and the pan-RAF inhibitor TAK-632 elicited potent and selective inhibition of proliferation in the DNP-only condition (Figure 1E) (Zhu et al., 2004; Okaniwa et al., 2013). Among the four PDK1 inhibitors and 14 RAF inhibitors tested, OSU-03012 and TAK-632 were unique in their ability to selectively inhibit the DNP, suggesting that this effect is not the consequence of on-target activity (Figures 1F, 1G, 1H).
Microplate immunofluorescence microscopy nuclei scoring analysis of MIAPACA2 cells stained with Hoechst 33342 was performed as an orthogonal evaluation of hit compound activity. These studies confirmed the culture-condition selectivity of our hits and validated the results of the CTG-based screen (Figures S3G, S3H). Additionally, we performed CTG analysis and trypan-blue exclusion cell scoring in a second cancer cell line, JURKAT, to confirm hit selectivity (Figure S3I, S3J).
In addition to its non-redundant role in de novo pyrimidine nucleotide biosynthesis, DHODH functions as an electron donor in the mitochondrial electron transport chain (Fang et al., 2013). To exclude the possibility that the selective activity of JNK-IN-8 reflects an interaction with NITD-982 at the level of electron transport chain modulation, we synthesized and evaluated N-phosphonacetyl-L-aspartate (PALA), an inhibitor of CAD which functions upstream of DHODH (Collins et al., 1971; Peters et al., 2018). We determined that JNK-IN-8 inhibits JURKAT cell proliferation when both PALA and rU are present in the culture media, supporting that its selective activity results from the inhibition of uridine salvage (Figure S4A).
We next applied our LC-MS/MS stable isotope tracking approach to evaluate the impact of JNK-IN-8, OSU-03012, and TAK-632 on the incorporation of [13C6]glucose and [13C9; 15N2]rU into newly replicated DNA (Figure 1I). In MIAPACA2 cells, we found that JNK-IN-8 blocked the NSP contribution while triggering a compensatory upregulation of the DNP. Conversely, OSU-03012 and TAK-632 selectively impaired DNP contribution (Figure 1J). Similar selectivity was observed in JURKAT cells, where both OSU-03012 and TAK-632 blocked DNP contribution while inducing compensatory up-regulation of the NSP (Figure 1K).
JNK-IN-8 inhibits nucleoside uptake
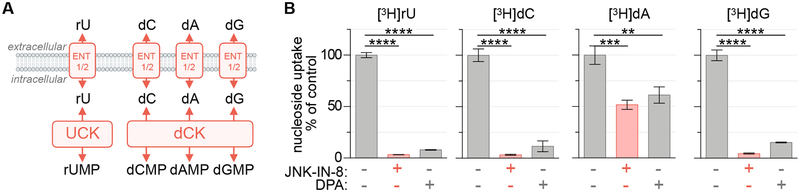
While three protein kinase inhibitors were identified as selective inhibitors of the pyrimidine NSP, JNK-IN-8 was exceptionally potent, with IC50 values in the low nanomolar range. We reasoned that the activity of JNK-IN-8 could arise from either the inhibition of nucleoside shuttling across the plasma membrane, which is achieved by nucleoside transporters, or through the inhibition of nucleoside phosphorylation by nucleoside kinases. To determine the level at which JNK-IN-8 is active, we determined the effects of JNK-IN-8 on the uptake of a panel of [3H]-labeled purine (dA, dG) and pyrimidine nucleosides (rU, dC) in MIAPACA2 cells. These nucleosides rely upon the same nucleoside transporters to enter the cell but require unique kinases for conversion into their respective monophosphate forms and intracellular accumulation. UCKs are required for the phosphorylation of rU while deoxycytidine kinase (dCK) is required for the phosphorylation of both purine and pyrimidine deoxyribonucleosides including dC, dA, and dG (Figure 2A) (Le et al., 2017). We found that JNK-IN-8 prevented the uptake of all nucleosides tested, but exhibited greater potency toward rU and dC. Importantly, JNK-IN-8 exhibits a selectivity pattern similar to the established ENT1 inhibitor DPA (Figure 2B). We confirmed JNK-IN-8 inhibited the uptake of both rU and dC with similar potency (33 nM and 31 nM, respectively), further suggesting that the compound inhibits nucleoside transport (Figure S4C). Additionally, JNK-IN-8 treatment prevented the anti-proliferative effects of gemcitabine (dFdC), a dCK-dependent nucleoside analog prodrug which relies upon nucleoside transporters for its activation, in a dose-dependent manner (Figure S4D) (Mackey et al., 1998). A similar pattern of dA, dG, rU and dC uptake inhibition by JNK-IN-8 and DPA was observed in a second cell line, the murine pancreatic cancer model KP4662 (Figures S4E, S4F)(Byrne et al., 2016).
Figure 2 |. JNK-IN-8 inhibits nucleoside uptake.
(A) Uridine salvage pathway activity can be prevented by inhibition of either nucleoside transporters or kinases. (B) Uptake of [3H]rU, [3H]dC, [3H]dA (+10 μM dCF) and [3H]dG (+1 μM BCX-1777) in MIAPACA2 cells following 2 h incubation ± 1 μM JNK-IN-8 or 1 μM dipyridamole (DPA; 18.5 kBq; mean±SD; n=3; one-way ANOVA corrected for multiple comparisons by Bonferroni adjustment; ** P<0.01; *** P<0.001; **** P<0.0001).
7 nucleoside transporters have been described and categorized into 2 families. Concentrative nucleoside transporters (CNT1–3; SLC28A1–3) are unidirectional inward transporters which co-transport Na+. Equilibrative nucleoside transporters (ENT1–4; SLC29A1–4) are bidirectional, energy-independent, and accept a broad range of purine and pyrimidine nucleosides (Young et al., 2013). We evaluated the expression of these transporters in MIAPACA2 and JURKAT cells and found that ENT1 (SLC29A1) is the predominantly expressed transporter in both models (Figure S4G) (Fernandez-Banet et al., 2016). ENT1 is an established transporter of a variety of nucleosides including natural purine and pyrimidine nucleosides as well as therapeutic analogs such as gemcitabine (Young et al., 2013). We next utilized the cellular thermal shift assay (CETSA), an approach that leverages the altered thermostability of proteins following ligand binding, to confirm ENT1 engagement by JNK-IN-8 (Figure S4H) (Martinez Molina & Nordlund, 2016). Collectively, these results indicate the JNK-IN-8 inhibits UMP-NSP activity by interfering with the transport of rU.
OSU-03012 and TAK-632 target de novo UMP biosynthesis and activate the DNA replication stress response pathway
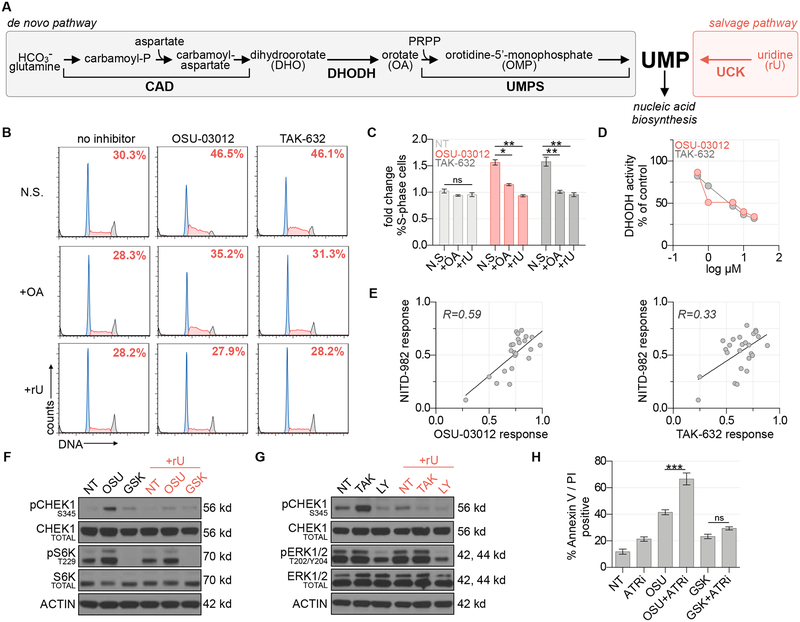
Two protein kinase inhibitors, TAK-632 and OSU-03012, were identified as potent and selective inhibitors of MIAPACA2 proliferation in the DNP-only culture condition (Figure 1E). We reasoned that these compounds could inhibit de novo pyrimidine biosynthesis by targeting either CAD, DHODH, or UMPS – the 3 enzymes essential for de novo UMP biosynthesis (Figure 3A). Both OSU-03012 and TAK-632 induced S-phase arrest in MIAPACA2 cells cultured in the DNP condition (Figure 3B). S-phase arrest is a phenotype associated with dNTP biosynthesis levels insufficient to sustain DNA replication and is the result of activation of intra-S phase cell cycle signaling checkpoints. This effect was rescued by orotate (the product of DHODH) supplementation and could be completely reversed by rU supplementation (Figures 3B, 3C). These data implicated DHODH as a likely target of both OSU-03012 and TAK-632. DHODH catalyzes one of three committed steps within the DNP and is an established druggable protein (Madak et al., 2019). Additionally, both OSU-03012 and TAK-632 posses fluorine substituents which have been shown to stabilize bioactive conformations of DHODH inhibitors (Bonomo et al., 2013; Baumgartner et al., 2006). In an in vitro colorimetric recombinant human DHODH activity assay, TAK-632 and OSU-03012 both inhibited DHODH activity in a dose-dependent manner (Figure 3D) (Baumgartner et al., 2006). Importantly, the response to TAK-632 or OSU-03012 correlated with the response to a known DHODH inhibitor in a panel of 25 pancreatic cancer cell lines (Figure 3E).
Figure 3 |. OSU-03012 and TAK-632 inhibit DHODH and activate the DNA replication stress response pathway.
(A) Schematic of UMP biosynthesis via the de novo and salvage pathways. (B) Propidium iodide cell cycle analysis of MIAPACA2 PDAC cells treated ±5 μM TAK-632 or ±5 μM OSU-03012 and supplemented with 50 μM orotate (OA) or 10 μM rU (N.S.: no supplement). Insert indicates % S-phase cells. (C) Summary of fold changes in S-phase cells from B (mean±SD; n=2; one-way ANOVA corrected for multiple comparisons by Bonferroni adjustment, ns: not significant; * P<0.05; ** P<0.01). (D) in vitro DHODH enzyme assay performed in the presence of OSU-03012 or TAK-632. (E) Correlation between DHODH inhibitor (1 μM NITD-982) and OSU-03012 (3.17 μM) or TAK-632 (3.17 μM) response across a panel of 25 PDAC cell lines determined using CTG following 72 h treatment. Response calculated as doubling time normalized proliferation inhibition. Pearson correlation coefficient is indicated. (F) Immunoblot analysis of MIAPACA2 cells treated ±1 μM PDK1 inhibitor GSK-2334470 (GSK) ±1 μM OSU-03012 (OSU) ±10 μM rU for 24 h. (G) Immunoblot analysis of MIAPACA2 cells treated ±10 μM RAF inhibitor LY3009120 (LY) ±10 μM TAK-632 (TAK) ±10 μM rU for 24 h. (H) Annexin V/PI flow cytometry analysis of MIAPACA2 PDAC cells treated ±1 μM OSU-03012 or 1 μM GSK-2334470 (GSK) ±500 nM VE-822 (ATRi) ±25 μM rU for 72 h (mean±SD; n=2; one-way ANOVA corrected for multiple comparisons by Bonferroni adjustment; ns: not significant; ** P<0.01; *** P<0.001).
OSU-03012 was recently reported to synergize with replication stress response kinase inhibitors in RSK-subtype mutant KRAS cancer models (Yuan et al., 2018). However, after confirming that OSU-03012 binds DHODH, we hypothesized that the observed synergy resulted from DHODH inhibition rather than PDK1 inhibition. Immunoblot analysis of S6K and S6 phosphorylation, PDK1 downstream targets, confirmed that GSK-2334470, a known PDK1 inhibitor, potently blocked PDK1 while OSU-03012 triggered S345 CHEK1 phosphorylation, a replication stress biomarker, only in the absence of rU (Figure 3F). Similarly, TAK-632 only triggered CHEK1 phosphorylation in the absence of rU whereas an established pan-RAF inhibitor, LY3009120 which does not exhibit paradoxical activation, down-regulated ERK1/2 phosphorylation but had no impact on CHEK1 phosphorylation (Figure 3G) (Peng et al., 2015). Consistently, we found that neither GSK-2334470 nor LY3009120 induced S-phase arrest at doses where we observed down-regulation of their target substrates, whereas a known DHODH inhibitor induced potent S-phase accumulation that was completely reversed by rU supplementation (Figures S4I, S4J). To compliment our evaluation of replication stress response biomarker induction we performed an assessment of DNA damage induced by OSU-03012, TAK-632, and the ATR inhibitor VE-822 as a positive control using γ-H2A.X flow cytometry. We found that while OSU-03012 and TAK-632 trigger activation of the replication stress response, they do not significantly induce γ-H2A.X. We hypothesize that activation of the replication stress response pathway by OSU-03012 or TAK-632 limits DNA double-strand breaks by preventing the collapse of stalled replication forks (Figure S4K) (Zeman & Cimprich, 2014).
To investigate the interaction between OSU-03012 and replication stress response inhibitors we treated MIAPACA2 cells with VE-822 (Le et al., 2017), an inhibitor of the proximal replication stress response kinase ataxia telangiectasia and Rad3-related (ATR), and either OSU-03012 or GSK-2334470 for 72 h. A synergistic increase in cell death was observed when OSU-03012 and an ATR inhibitor were combined, whereas the combination of GSK-2334470 and VE-822 demonstrated only a nominal increase in cell death as determined by AnnexinV/PI flow cytometry (Figure 3H). Taken together, these data indicate that replication stress triggered by OSU-03012 is the consequence of DHODH inhibition rather than inhibition of its canonical target.
Co-crystal structures of OSU-03012 and TAK-632 in complex with human DHODH
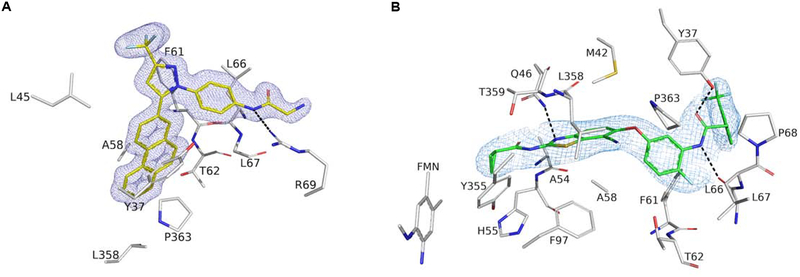
To determine the molecular interactions between the protein and its putative inhibitors, complete DHODH co-crystallization data sets were obtained and processed to 1.4 Å and 2.7 Å for OSU-03012 and TAK-632, respectively (Table S1). Both compounds bind in a hydrophobic channel composed by two N-domain α-helices through which ubiquinone travels, a mechanism consistent with previously identified DHODH inhibitors (Baumgartner et al., 2006). A long-range hydrogen bond between Arg 69 and OSU-03012 helps orient the molecule to the hydrophobic pocket where the phenanthrene moiety inserts, while the remainder of the molecule lies on the outer surface of DHODH, blocking the hydrophobic channel (Figures 4A). Three hydrogen bonds stabilize TAK-632 in the same hydrophobic pocket: two with Tyr 37 and Leu 66 help stabilize the inhibitor at the opening of the channel, while a third with Gln 46 helps pull the inhibitor deep into the pocket (Figures 4B).
Figure 4 |. OSU-03012 and TAK-632 bind DHODH.
(A,B) Crystal Structure of DHODH with compounds OSU-03012 (A) or TAK-632 (B). 2mFo-DFc electron density for OSU-03012 (carbons in yellow) or TAK-632 (carbons in green) contoured at 1 σ. Dashed black lines represent hydrogen bonds between DHODH and its ligands. Interacting residues as predicted by LigPlot+ are shown and labeled.
DISCUSSION
Our screening strategy expands upon previously described “nutrient-sensitized” genetic and small molecule cell-based screening approaches that leveraged the production of a proliferation-enabling metabolite by parallel and redundant metabolic networks to identify selective metabolism modifiers (Arroyo et al., 2016; Gohil et al., 2010). UMP biosynthesis (i.e. pyrimidine metabolism) proved to be compatible with this screening framework as UMP is produced by convergent (de novo and salvage) pathways, and UMP depletion triggers a quantifiable change in cellular proliferation.
JNK-IN-8, developed as an irreversible inhibitor of c-Jun N-terminal kinases 1, 2, and 3 with low-nanomolar affinity, was the most potent of three uridine salvage inhibitors identified (Zhang et al., 2012). Our data confirm that JNK-IN-8 also functions as a potent inhibitor of uridine and deoxycytidine transport and engages the nucleoside transporter SLC29A1. We conclude that JNK-IN-8 should not be used in conjunction with compounds which rely upon nucleoside transport, such as the anticancer agent gemcitabine, in research or therapy settings.
In addition to their role in pyrimidine salvage, equilibrative nucleoside transporters are well studied for their ability to regulate levels of the immuno-modulatory metabolite adenosine. ENT1 inhibitors increase extracellular adenosine levels which signal through the P1 purinergic receptor and are used clinically for the treatment of hypertension, among other disorders (Young et al., 2013). Thus, the development of potent and selective inhibitors of ENT1 is an active area of investigation.
The recently reported co-crystal structure of ENT1 in complex with two small molecule inhibitors (NBMPR and dilazep) provided new insight into the molecular mechanism of nucleoside transport and suggested that structurally diverse ENT1 inhibitors possess unique modes of inhibition (Wright & Lee, 2019). ENT1 contains 10 cysteine residues and ENT1-mediated uridine transport can be inhibited by covalent modification of Cys416 by N-ethylmaleimide (Yao et al., 2018). Intriguingly, the two highest scoring NSP inhibitors in our screen, JNK-IN-8 and the BTK inhibitor CNX-774, each contain a reactive acrylamide group and are cysteine-targeting drugs. Future work will explore the mechanism of ENT1 inhibition by JNK-IN-8 with a specific focus on the contribution of covalent interactions.
Positron emission tomography (PET) imaging is a powerful approach to monitor cellular metabolism in vivo, and several nucleoside analog PET probes have been developed including both pyrimidine ([18F]FAC, [18F]FLT) and purine analogs ([18F]CFA) (Radu et al., 2008; Shields et al., 1998; Kim et al., 2016). Interestingly, ENT1 knockout mice exhibit significantly higher plasma thymidine but also paradoxically higher levels of thymidine analog [18F]FLT uptake in the spleen and bone marrow compared to wild type controls (Paproski et al., 2010). Discrepancies between in vitro and in vivo findings could result from shifts in nucleoside gradients or differential expression of nucleoside transporters mediated by cytokine signaling. Future work will focus on exploring the utility of nucleoside analog PET as a pharmacodynamic biomarker for ENT inhibitor activity in vivo.
The structurally and functionally unrelated OSU-03012 and TAK-632 were identified as inhibitors of the pyrimidine DNP. A recent report described the ability of OSU-03012 and analogs to inhibit virus propagation via pyrimidine nucleotide biosynthesis inhibition, specifically implicating modulation of DHODH activity (Yang et al., 2018). Our work substantiates these findings and confirms engagement of DHODH by OSU-03012 and TAK-632 through crystallography studies. Notably, our studies show that OSU-03012 and TAK-632 bind in the same hydrophobic tunnel of DHODH as known inhibitors brequinar and teriflunomide (the active metabolite of leflunomide). This suggests that these two protein kinase inhibitors compete with ubiquinone, a redox partner of DHODH, which traverses this hydrophobic tunnel to regenerate FMN from FMNH2. By competitively inhibiting the binding of ubiquinone, these compounds prevent DHODH from completing its redox cycle and effectively abrogate its activity.
OSU-03012 has orphan drug designation in the European Union for treatment of tularaemia and cryptococcosis. We hypothesize that its effectiveness in these indications stems from its ability to inhibit DHODH, rather than from ‘on-target’ effects against PDK1. Indeed, DHODH inhibitors have demonstrated efficacy against viruses such as dengue virus and respiratory syncytial virus (Bonavia et al., 2011; Yang et al., 2018; Wang et al., 2011). In anticancer settings, OSU-03012 was recently demonstrated to synergize with CHK1 inhibitors in KRAS-mutant cancers (Yuan et al., 2018), which was initially attributed to its ability to inhibit PDK1. However, our data show that GSK-2334470, a PDK1 inhibitor more potent than OSU-03012, displayed little synergy with ATR inhibition. In light of our crystallographic data, we conclude that the synergy observed between OSU-03012 and ATR inhibition is likely a result of the DHODH-inhibitory ability of the former. Taken together, our data suggest that DHODH inhibitors may have utility in oncology, particularly if used in conjunction with ATR inhibitors or other DNA-damage response/replication stress response pathway inhibitors (Le et al., 2017).
In summary, we designed and applied a metabolic modifier screen which identified multiple protein kinase inhibitors as having non-canonical targets within pyrimidine metabolism. Similarly constructed phenotypic screens designed against other metabolic networks containing convergent nodes may find use in drug discovery campaigns or in repurposing screens using existing compounds.
METHODS
Experimental Model and Subject Details
Cell culture
Human cancer cells including MIAPACA2 (male) and JURKAT (male) were evaluated between passages 3 and 20 and maintained in antibiotic-free DMEM or RPMI +10% FBS at 37°C in 5% CO2. Mycoplasma contamination was monitored using the PCR-based Venor Mycoplasma kit. PDAC cell lines were acquired either from a commercial vendor (ATCC, DSMZ) or from collaborators (KP4662 from Dr. Vonderheide, UPenn, sex not available). Cell line identity was independently authenticated by PCR.
Drugs
Drug stocks were prepared in DMSO or H2O and diluted fresh in cell culture media for treatments. NITD-982 was synthesized as previously described (Bonavia, 2011). N-phosphonacetyl-L-aspartate (PALA) was synthesized as previously described (Morris & Cordi, 1997).
Method Details
Protein kinase inhibitor phenotypic screen
A library of 430 protein kinase inhibitors was arrayed in polypropylene 384-well plates at 200x concentrations covering a 7-point concentration range (corresponding to 1x concentrations: 5μM, 1.65μM, 550nM, 185nM, 61.5nM, 20.6nM, 6.85nM). 25μl per well of condition-specific growth media (DNP+NSP: media +10 μM rU; DNP: media alone; NSP: media +10 μM rU + 1 μM NITD-982) was plated in opaque-white 384-well plates using a BioTek multidrop liquid handler. Protein kinase inhibitors were added by 250 nL pin-tool transfer (BioMek FX, Beckman-Coulter) and inhibitor/media mixtures were incubated at room temperature for 30 m. 25 μL of a 40,000 cells/mL MIAPACA2 suspension (for 1000 cells/well) was subsequently added to each well. After 72 h, 50 μL of Cell Titer Glo reagent diluted 1:4 in dH2O was added to each well and luminescence was measured using a Wallac plate reader (Perkin Elmer). Each condition was assayed in duplicate (n=2) and % proliferation values were calculated by normalizing experimental wells to plate negative controls and averaging replicate values. Composite pathway selectivity synergy scores for each test compound were defined as the sum of the excess over additivity (% proliferation inhibition observed - % proliferation inhibition expected) between individual protein kinase inhibitor concentrations across the 7-point concentration range. Z factor scores for individual assay plates were calculated using eight positive and eight negative control wells on each plate as previously described (Zhang et al., 1999). All plates gave a Z factor > 0.5 (Figure S3C).
Cell Titer Glo viability analysis
Cells were plated at 1×103 cells/well in 50 μl in white opaque 384-well plates and treated as described. Following seeding or incubation for 72 h, 50 μl of Cell Titer Glo reagent (diluted 1:5 in dH2O) was added to each well, plates were incubated at room temperature for 5 m and luminescence was measured using a microplate luminescence reader. Proliferation rate normalized growth inhibition was calculated using the previously described GR metric (Hafner et al., 2016). Experimental CTG reading (CTGx) were normalized to vehicle treated control readings at seeding (CTGt0) or following 72 h of culture (CTGctrl).
Trypan blue exclusion cell viability
JURKAT cells were plated 1×105 cells/well in 6-well plates in 2 mL of media and treated with 10 μM rU, 1 μM NITD-982, 100 nM JNK-IN-8, 1 μM OSU-03012 or 1 μM TAK-632. Following incubation for 72 h, 500 uL of cell cultures were collected for trypan blue exclusion cell viability analysis using a ViCell analyzer. All conditions were tested in biological triplicate. Trypan blue-negative population counts are reported.
Microplate immunofluorescence microscopy cell scoring
MIAPACA2 cells were plated at 1000 cells/well in black-walled clear-bottom 384 well plates in 50 μL of media and treated as indicated with n=4 replicate wells per condition. After 72 h of drug exposure 50 μL of 10 μg/mL Hoechst 33342 dye diluted in culture media was added to microplate wells. Following a 30 m incubation at 37C images were acquired using a ImageXpress Micro Confocal High-Content Imaging System at 10x magnification and 1 image/well. Analysis was performed using the Cell Scoring Application Module in the MetaXpress analysis software. Nuclei counts for treatment groups were normalized to control wells.
Mass spectrometry
For analysis of stable isotope-labeled metabolite incorporation into newly replicated DNA, cells were cultured in glucose-free DMEM (for MIAPACA2 cells) or RPMI (for JURKAT cells) media supplemented with 10% dialyzed FBS, 4 mM glutamine, 1 g/L [13C6]glucose, 10 μM [13C9; 15N2]rU and treated as indicated.
Genomic DNA was extracted using the Quick-gDNA MiniPrep kit and hydrolyzed to nucleosides using the DNA Degradase Plus kit, following manufacturer-supplied instructions. In the final step of DNA extraction, 50 μL of water was used to elute the DNA into 1.5 mL microcentrifuge tubes. A nuclease solution (5 μL; 10X buffer/DNA Degradase PlusTM/water, 2.5/1/1.5, v/v/v) was added to 20 μL of the eluted genomic DNA in an HPLC injector vial. The samples were incubated overnight at 37 °C.
Hydrolyzed DNA was diluted 1/1 with solvent A (water/acetonitrile/formic acid, 95/5/0.1, v/v) and analyzed using a modified version of a previously reported method (Le et al., 2017; Nathanson et al., 2014) in which aliquots of the solution (15 μL) were injected onto a porous graphitic carbon column (Thermo Hypercarb, 100 × 2.1 mm, 5 micron particle size) equilibrated in solvent A and eluted (300 μL/min) with an increasing concentration of solvent B (acetonitrile/water/formic acid, 90/10/0.1). The HPLC timetable, in terms of min/%B, is the following: 0/0, 5/0, 12/20, 15/30, 17/50, 19/50, 20/0, 24/0. The effluent from the column was directed to Agilent Jet Stream connected Agilent 6460 QQQ operating in the positive ion MRM mode. After verification of retention times using authentic standards, the peak areas of the protonated nucleoside/protonated base fragment ion transitions for each of the nucleosides were recorded with instrument manufacturer-supplied software.
[3H]-labeled metabolite uptake assays
Radioactive probe uptake assays were conducted as previously described(Campbell et al., 2011). Briefly, cells were pretreated with JNK-IN-8 or DPA for 2 h before incubation with 18.5 kBq of [3H]-labeled probe for 2 h. For purine uptake assays, cells were cultured in the presence of 10 μM dCF (for [3H]-dA) or 1 μM BCX-1777 (for [3H]-dG) to prevent nucleoside catabolism. Following incubation, cells were washed with PBS and lysed. Cell lysate radioactivity was measured using a beta-counter (Perkin-Elmer).
Flow cytometry
All flow cytometry data were acquired on five-laser BD LSRII, and analyzed using FlowJo software.
AnnexinV/PI:
Treated PDAC cells were washed with PBS and incubated with AnnexinV and propidium iodide diluted in 1x Annexin binding buffer. 20,000 events were collected per sample.
Propidium iodide cell cycle analysis:
Treated PDAC cells were washed with PBS and suspended in propidium iodide cell cycle staining solution (100 μg/ml propidium iodide; 20 μg/ml Ribonuclease A). 10,000 events were collected per sample.
pH2A.XS139 flow cytometry:
Treated cells were collected by trypsinization, incubated with Cytofix/Cytoperm reagent for 15 m at 4C, washed with PBS and incubated in 100 μL PermWash buffer for 15 m at 4C. Cells were washed with 1 mL Perm/Wash buffer, resuspended in 50 μL of staining solution (1:800 dilution of FITC-conjugated pH2AXS139 antibody diluted in Perm/Wash buffer) and incubated for 20 m at 25C protected from light. Stained cells were washed and incubated in 500 μL of DAPI staining solution (1 μg/mL DAPI in PBS) before acquisition.
Gene cloning, protein expression, and purification of DHODH in E.coli cells
Primers were ordered to add NdeI (AGAGAACAGATTGGTGGTCATATGATGGCCACGGGAGATGAG) upstream of residue 29 (after the mitochondrial membrane associated loop) and BamHI (TCGGGCTTTGTTAGCAGCCGGATCCTTACCTCCGATGATCTGCTCC) after the stop codon to insert into N-terminal His-Sumo pET 14b vector. This clone, His-Sumo-DHODH 29–395 (subsequently referred to as DHODH) was successfully inserted into the vector in XL1-blue cells for vector propagation.
The vector was transformed into C41(DE3) cells for production. Cells were grown at 37 °C in 2xYT medium supplemented with 100 μg/mL ampicillin (Amp), treated with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at an OD600 nm of 0.6–0.8, and then cultured for an additional 18h at 18 °C. Cells were harvested by centrifugation, washed with 200 mM NaCl and 25 mM Tris pH 7.5, and pelleted at 5000 rpm for 20 minutes before storage at −20°C. 6.7g/L of cell pellet was obtained.
DHODH was purified according to known purification conditions (Baumgartner et al., 2006). The cell pellet was resuspended in lysis buffer (50 mM Tris pH 7.5; 600 mM NaCl; 0.33% w/v Thesit; 10% Glycerol; 1 mM PMSF) and lysed by sonication on ice. Lysed cells were centrifuged at 58,500 RCF for 45 minutes at 4°C, and the supernatant was filtered through a 0.45 μM filter and loaded onto a 5-mL His-Trap column pre-equilibrated with buffer A (50 mM Tris pH 7.4; 600 mM NaCl; 0.05% w/v Thesit; 10% Glycerol). The column was washed with buffer A for 70 mL, buffer A with 25 mM imidazole for 50 mL, and buffer A with 50 mM imidazole for 50 mL. The protein was eluted with buffer A with 250 mM imidazole. The eluted fraction was diluted 1:1 with Buffer A. Sumo protease was added and the protein was dialyzed overnight at 4°C against 1L of Buffer A. The dialyzed protein was loaded back onto the His-Trap column equilibrated with buffer A. The cut-DHODH was eluted with buffer A with 50 mM imidazole. The purified protein was concentrated to 5 mL and injected onto S-200 gel filtration column (GE Healthcare) equilibrated with: 50 mM HEPES pH 7.7, 400 mM NaCl, 10% Glycerol, 1 mM EDTA, 0.05 % w/v Thesit. Eluted fractions consistent with monomer size were collected, concentrated, flash frozen, and stored at −80°C.
Recombinant DHODH enzyme assay
Evaluation of DHODH inhibition was performed as previously described (Baumgartner et al., 2006). The standard assay mixture contained 50 μM decyclo-ubiquinone, 100 μM dihydroorotate, and 60 μM 2,6-dichloroindophenol (DCIP). The amount of DHODH was 337.4 ng/mL. Measurements were conducted in 50 mM TrisHCl, 150 mM KCl, 0.1% Triton X-100, pH 8.0, at 30 °C in a final volume of 1 mL. The components were mixed, and the reaction was started by adding dihydroorotate. The reaction was followed spectrophotometrically by measuring the decrease in absorption at 600 nm for 2.5 min at 30 second intervals. The assay was linear in time and enzyme concentration. Inhibitory studies were conducted in a standard assay with additional variable amounts of inhibitor.
Crystallization of DHODH with OSU-03012 and TAK-632 compounds
For co-crystallizaion of DHODH and OSU-03012, crystals were obtained using the same conditions reported in previously published DHODH structures (Lewis et al., 2016; Baumgartner et al., 2006; Das et al., 2013; Davies et al., 2009; Erra et al., 2011; Hurt et al., 2006; Ladds et al., 2018; Liu et al., 2000; McLean et al., 2010; Sainas et al., 2018; Walse et al., 2008), namely 1.6 – 2.6 M ammonium sulfate and 5–30% glycerol in the well in pH 4.5, with 20 mg/mL DHODH with 2 mM dihydroorotate (DHO), 20.8 mM dodecyldimethyl-N-amineoxide (DDAO), and 400 μM inhibitor. Protein was mixed 1:1 with mother liquor and hanging drops were used at room temperature. Crystals appeared after 48 hours and reached maximal size within one week. Molecular replacement used 4OQV (Deng et al., 2014) as the starting model. Interestingly, DHODH-TAK-632 crystals grown in similar conditions to those used for the DHODH-OSU-03012 complex did not show TAK-632 density. As a result, novel DHODH crystallization conditions were identified using commercial screens. For co-crystallization of DHODH with TAK-632, crystals were obtained in conditions of 1.4–1.6 M sodium phosphate, pH 8.2. Protein solution (20 mg/mL DHODH with 2 mM DHO, 20.8 mM DDAO, and 400 μM inhibitor) was mixed 1:1 with mother liquor and hanging drops were used at room temperature. Crystals appeared after 48 hours and reached maximal size within one week. The lack of density of the TAK-632 structure in the initial crystallography condition is most likely due to the difference in pH between the conditions, pH 4.5 and pH 8.2 for the OSU-03012 and the TAK-632 structure, respectively. There are multiple hydrogen bonds and potentially labile hydrogens on the TAK-632 structure that at low pH could be protonated and charged, potentially preventing their insertion into the hydrophobic tunnel. Ligplot+ (Laskowski & Swindells, 2011; Wallace et al., 1995) was used to determine hydrophilic and hydrophobic interactions between inhibitors and DHODH molecules.
Phasing/structural solution, refinement strategies and model validation
To phase both crystal structures, we used 4OQV as the starting model, including only Chain A without ligands. Phasing was done by molecular replacement using MOLREP included in the CCP4 software suite. Further refinement was done using restrained refinement using refmac, starting at a 4.0 Å cutoff, running 10 cycles. The cutoff was lowered by 0.5 Å each 10 cycles until highest resolution range was reached. The ligands were built in jLigand and added to the model using Coot. Final refinement was done using refmac. Model validation consisted of using Rampage and Procheck for outlier residues and positions, as well as the PDB validation service upon submission.
Immunoblot analysis
PBS-washed cell pellets were resuspended in cold RIPA buffer supplemented with protease and phosphatase inhibitors. Protein lysates were normalized using BCA assay, diluted using RIPA and 4x laemmli loading dye, resolved on 4–12% Bis-Tris gels and electro-transferred to nitrocellulose membranes. After blocking with 5% nonfat milk in TBS + 0.1% Tween-20 (TBS-T), membranes were incubated overnight in primary antibodies diluted (per manufacturers instructions) in 5% BSA in TBS-T. Membranes were washed with TBST-T and incubated with HRP-linked secondary antibodies prepared at a 1:2500 dilution in 5% nonfat dry milk/TBS-T. HRP was activated by incubating membranes with a mixture of SuperSignal Pico and SuperSignal Femto ECL reagents (100:1 ratio). Exposure of autoradiography film was used for detection.
CETSA
MIAPACA2 cells were cultured in 10 cm plates, washed with PBS, and harvested by cell scraping following addition of 4 mL of lysis buffer (100 mM ammonium sulfate, 400 mM NaCl, 10% glycerol, 0.5% DDM and 1x protease inhibitor cocktail). The cell lysate was collected in a 15 mL conical tube, incubated on ice for 20 m, centrifuged at 5,000×g for 20 m at 4C and protein content of the supernatant was measured. 30 μL of protein lysate was aliquoted into 1.5 mL Eppendorf tubes and treated with either DMSO, JNK-IN-8 or dipyridamole for 30 m on ice. Lysates were subsequently heated at the indicated temperatures using an Eppendorf Thermomixer for 6 m, cooled to room temperature for 3 m and transferred to ice. Heated lysates were centrifuged at 12,000×g for 40 m to pellet the insoluble protein fraction. Supernatants were processed for immunoblot analysis.
Quantification and Statistical analyses
Data are presented as mean ± SD with number of biological replicates indicated. Comparisons of two groups were calculated using indicated unpaired two-tailed Student’s t-test and P values less than 0.05 were considered significant. Comparisons of more than two groups were calculated using one-way ANOVA followed by Bonferroni’s multiple comparison tests, and P values less than 0.05/m, where m is the total number of possible comparisons, were considered significant.
Data and Code Availability
The full kinase inhibitor library phenotypic screen results related to Figure 1 are included as Data S1.
The atomic coordinates and structure factors for the OSU-03012:DHODH and TAK-632:DHODH co-crystal structures reported in this study are deposited to the Protein Data Bank under the accession codes PDB 6OC0 and PDB 6OC1, respectively.
Supplementary Material
Data S1. Full kinase inhibitor library phenotypic screen results, Related to Figure 1
Table S1. Crystallographic data collection and refinement statistics, Related to Figure 4
SIGNIFICANCE.
The pyrimidine nucleotide uridine monophosphate (UMP) is essential for cell proliferation and is produced by convergent de novo and salvage biosynthetic pathways which utilize unique metabolic precursors. This redundancy can be leveraged for the identification of selective de novo or salvage pathway inhibitors by employing a cell-based phenotypic metabolic modifier screen. In screening a library of 430 protein kinase inhibitors, multiple compounds which possess selective UMP metabolism-modifying activity were identified. This activity was subsequently shown to be unrelated to inhibition of the canonical targets of these inhibitors and is the consequence of their binding to and inhibition of nucleotide metabolism proteins, namely DHODH or ENT1. Herein, we describe a modular screening strategy which can be utilized for drug discovery and which can be adapted for the identification of small molecule inhibitors of additional convergent metabolic pathways.
Highlights.
Convergent UMP biosynthetic pathways interchangeably sustain cancer cell proliferation
Phenotypic screens can identify selective modulators of convergent metabolic networks
Multiple protein kinase inhibitors possess secondary targets within UMP metabolism
ACKNOWLEDGMENTS
We thank Bobby Tofig and Constance Yuen at the UCLA Molecular Shared Screening Resource for assistance performing the high-throughput screen. We thank Dr. Steven Mittelman for his critical review of this manuscript. We thank all members of the Ahmanson Translational Imaging Division at UCLA for their advice, technical expertise and support. This work was supported by the National Science Foundation Graduate Research Fellowship Program [Grant No. DGE-1650504 to E.W.R].
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information includes 4 figures, 1 table and 1 data file.
DECLARATION OF INTERESTS
The authors declare the following competing financial interest(s): C.G.R. and J.C. are co-founders of Sofie Biosciences and Trethera Corporation. M.E.J. is a co-founder of Trethera Corporation. They and the University of California (UC) hold equity in Sofie Biosciences and Trethera Corporation. The intellectual property developed by C.G.R., J.C. and M.E.J. and licensed by UC to Sofie Biosciences and Trethera Corporation was not used in this study.
LEAD CONTACT AND MATERIALS AVAILABILITY
Lead Contact
Reagent or resource requests should be submitted to the lead contact, Caius G. Radu (cradu@mednet.ucla.edu).
Materials Availability Statement
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
REFERENCES
- Arroyo JD, Jourdain AA, Calvo SE, Ballarano CA, Doench JG, Root DE et al. (2016). A Genome-wide CRISPR Death Screen Identifies Genes Essential for Oxidative Phosphorylation. Cell Metabolism, 24(6), 875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner R, Walloschek M, Kralik M, Gotschlich A, Tasler S, Mies J et al. (2006). Dual binding mode of a novel series of DHODH inhibitors. J Med Chem, 49(4), 1239–1247. [DOI] [PubMed] [Google Scholar]
- Bonavia A, Franti M, Pusateri Keaney E, Kuhen K, Seepersaud M, Radetich B, Shao J, Honda A, Dewhurst J, Balabanis K, Monroe J, Wolff K, Osborne C, Lanieri L, Hoffmaster K, Amin J, Markovits J, Broome M, Skuba E, Cornella-Taracido I, Joberty G, Bouwmeester T, Hamann L, Tallarico JA, Tommasi R, Compton T, Bushell SM (2011). Identification of broad-spectrum antiviral compounds and assessment of the druggability of their target for efficacy against respiratory syncytial virus (RSV). Proc Natl Acad Sci USA, 108(17), 6739–6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomo S, Tosco P, Giorgis M, Lolli M, Fruttero R (2013). The role of fluorine in stabilizing the bioactive conformation of dihydroorotate dehydrogenase inhibitors. J Mol Model, 19(3), 1099–1107. [DOI] [PubMed] [Google Scholar]
- Byrne KT, Leisenring NH, Bajor DL, Vonderheide RH (2016). CSF-1R–Dependent Lethal Hepatotoxicity When Agonistic CD40 Antibody Is Given before but Not after Chemotherapy. J.I, 197(1), 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DO, Yaghoubi SS, Su Y, Lee JT, Auerbach MS, Herschman H et al. (2011). Structure-guided Engineering of Human Thymidine Kinase 2 as a Positron Emission Tomography Reporter Gene for Enhanced Phosphorylation of Non-natural Thymidine Analog Reporter Probe. J. Biol. Chem, 287(1), 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins KD S. GR (1971). Aspartate Transcarbamylase: Interaction with the Transition State Analogue N-(phosphonoacetyl)-L-Aspartate. The Journal of Biological Chemistry, 6599. [PubMed] [Google Scholar]
- Damaraju VL, Weber D, Kuzma M, Cass CE, Sawyer MB (2016). Selective Inhibition of Human Equilibrative and Concentrative Nucleoside Transporters by BCR-ABL Kinase Inhibitors: Identification of Key hENT1 Amino Acid Residues for Interaction with BCR-ABL Kinase Inhibitors. J Biol Chem, 291(36), 18809–18817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P, Deng X, Zhang L, Roth MG, Fontoura BM, Phillips MA et al. (2013). SAR Based Optimization of a 4-Quinoline Carboxylic Acid Analog with Potent Anti-Viral Activity. ACS Med Chem Lett, 4(6), 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M, Heikkilä T, McConkey GA, Fishwick CW, Parsons MR, Johnson AP (2009). Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases. J Med Chem, 52(9), 2683–2693. [DOI] [PubMed] [Google Scholar]
- Deng X, Kokkonda S, El Mazouni F, White J, Burrows JN, Kaminsky W et al. (2014). Fluorine modulates species selectivity in the triazolopyrimidine class of Plasmodium falciparum dihydroorotate dehydrogenase inhibitors. J Med Chem, 57(12), 5381–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erra M, Moreno I, Sanahuja J, Andrés M, Reinoso RF, Lozoya E et al. (2011). Biaryl analogues of teriflunomide as potent DHODH inhibitors. Bioorg Med Chem Lett, 21(24), 7268–7272. [DOI] [PubMed] [Google Scholar]
- Evans DR, & Guy HI (2004). Mammalian pyrimidine biosynthesis: fresh insights into an ancient pathway. J Biol Chem, 279(32), 33035–33038. [DOI] [PubMed] [Google Scholar]
- Fang J, Uchiumi T, Yagi M, Matsumoto S, Amamoto R, Takazaki S et al. (2013). Dihydro-orotate dehydrogenase is physically associated with the respiratory complex and its loss leads to mitochondrial dysfunction. Biosci Rep, 33(2), e00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Banet J, Esposito A, Coffin S, Horvath IS, Estrella H, Schefzick S et al. (2016). OASIS: web-based platform for exploring cancer multi-omics data. Nature Methods, 13(1), 8–9. [DOI] [PubMed] [Google Scholar]
- Garcia-Bermudez J, Baudrier L, La K, Zhu XG, Fidelin J, Sviderskiy VO et al. (2018). Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat Cell Biol, 20(7), 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohil VM, Sheth SA, Nilsson R, Wojtovich AP, Lee JH, Perocchi F et al. (2010). Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat Biotechnol, 28(3), 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Niepel M, Chung M, Sorger PK (2016). Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods, 13(6), 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Wang Y, Collins M, Gu JJ, Mitchell BS, & Graves LM (2002). Inhibition of Nucleoside Transport by p38 MAPK Inhibitors. J. Biol. Chem, 277(32), 28364–28367. [DOI] [PubMed] [Google Scholar]
- Hurt DE, Sutton AE, Clardy J (2006). Brequinar derivatives and species-specific drug design for dihydroorotate dehydrogenase. Bioorg Med Chem Lett, 16(6), 1610–1615. [DOI] [PubMed] [Google Scholar]
- Kim W, Le TM, Wei L, Poddar S, Bazzy J, Wang X et al. (2016). [F]CFA as a clinically translatable probe for PET imaging of deoxycytidine kinase activity. Proc Natl Acad Sci USA, 113(15), 4027–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladds MJGW, van Leeuwen IMM, Drummond CJ, Chu S, Healy AR, Popova G et al. (2018). Publisher Correction: A DHODH inhibitor increases p53 synthesis and enhances tumor cell killing by p53 degradation blockage. Nat Commun, 9(1), 2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, & Swindells MB (2011). LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model, 51(10), 2778–2786. [DOI] [PubMed] [Google Scholar]
- Le TM, Poddar S, Capri JR, Abt ER, Kim W, Wei L et al. (2017). ATR inhibition facilitates targeting of leukemia dependence on convergent nucleotide biosynthetic pathways. Nat Commun, 8(1), 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis TA, Sykes DB, Law JM, Muñoz B, Rustiguel JK, Nonato MC et al. (2016). Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia. ACS Med Chem Lett, 7(12), 1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Neidhardt EA, Grossman TH, Ocain T, Clardy J (2000). Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure, 8(1), 25–33. [DOI] [PubMed] [Google Scholar]
- Lolli ML, Sainas S, Pippione AC, Giorgis M, Boschi D, & Dosio F (2018). Use of human Dihydroorotate Dehydrogenase (hDHODH) Inhibitors in Autoimmune Diseases and New Perspectives in Cancer Therapy. Recent Pat Anticancer Drug Discov, 13(1), 86–105. [DOI] [PubMed] [Google Scholar]
- Mackey JR, Mani RS, Selner M, Mowles D, Young JD, Belt JA et al. (1998). Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res, 58(19), 4349–4357. [PubMed] [Google Scholar]
- Madak JT, Bankhead A, Cuthbertson CR, Showalter HD, Neamati N (2019). Revisiting the role of dihydroorotate dehydrogenase as a therapeutic target for cancer. Pharmacology & Therapeutics, 195, 111–131. [DOI] [PubMed] [Google Scholar]
- Martinez Molina D, Nordlund P (2016). The Cellular Thermal Shift Assay: A Novel Biophysical Assay for In Situ Drug Target Engagement and Mechanistic Biomarker Studies. Annu. Rev. Pharmacol. Toxicol, 56(1), 141–161. [DOI] [PubMed] [Google Scholar]
- McLean LR, Zhang Y, Degnen W, Peppard J, Cabel D, Zou C et al. (2010). Discovery of novel inhibitors for DHODH via virtual screening and X-ray crystallographic structures. Bioorg Med Chem Lett, 20(6), 1981–1984. [DOI] [PubMed] [Google Scholar]
- Morris AD, Cordi AA (1997). A New, Efficient, Two Step Procedure for the Preparation of the Antineoplastic Agent Sparfosic Acid. Synthetic Communications, 27(7), 1259–1266. [Google Scholar]
- Nathanson DA, Armijo AL, Tom M, Li Z, Dimitrova E, Austin WR et al. (2014). Co-targeting of convergent nucleotide biosynthetic pathways for leukemia eradication. J Exp Med, 211(3), 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaniwa M, Hirose M, Arita T, Yabuki M, Nakamura A, Takagi T et al. (2013). Discovery of a selective kinase inhibitor (TAK-632) targeting pan-RAF inhibition: design, synthesis, and biological evaluation of C-7-substituted 1,3-benzothiazole derivatives. J Med Chem, 56(16), 6478–6494. [DOI] [PubMed] [Google Scholar]
- Okesli A, Khosla C, Bassik MC (2017). Human pyrimidine nucleotide biosynthesis as a target for antiviral chemotherapy. Current Opinion in Biotechnology, 48, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paproski RJ, Wuest M, Jans HS, Graham K, Gati WP, McQuarrie S et al. (2010). Biodistribution and uptake of 3’-deoxy-3’-fluorothymidine in ENT1-knockout mice and in an ENT1-knockdown tumor model. J Nucl Med, 51(9), 1447–1455. [DOI] [PubMed] [Google Scholar]
- Peng SB, Henry JR, Kaufman MD, Lu WP, Smith BD, Vogeti S et al. (2015). Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers. Cancer Cell, 28(3), 384–398. [DOI] [PubMed] [Google Scholar]
- Peters GJ (2018). Antipyrimidine effects of five different pyrimidine de novo synthesis inhibitors in three head and neck cancer cell lines. Nucleosides Nucleotides Nucleic Acids, 37(6), 329–339. [DOI] [PubMed] [Google Scholar]
- Radu CG, Shu CJ, Nair-Gill E, Shelly SM, Barrio JR, Satyamurthy N et al. (2008). Molecular imaging of lymphoid organs and immune activation by positron emission tomography with a new [18F]-labeled 2’-deoxycytidine analog. Nat Med, 14(7), 783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainas S, Pippione AC, Lupino E, Giorgis M, Circosta P, Gaidano V et al. (2018). Targeting Myeloid Differentiation Using Potent 2-Hydroxypyrazolo[1,5- a]pyridine Scaffold-Based Human Dihydroorotate Dehydrogenase Inhibitors. J Med Chem, 61(14), 6034–6055. [DOI] [PubMed] [Google Scholar]
- Santana-Codina N, Roeth AA, Zhang Y, Yang A, Mashadova O, Asara JM et al. (2018). Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis. Nat Commun, 9(1), 4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields AF, Grierson JR, Dohmen BM, Machulla HJ, Stayanoff JC, Lawhorn-Crews JM et al. (1998). Imaging proliferation in vivo with [F-18]FLT and positron emission tomography. Nature Medicine, 4(11), 1334–1336. [DOI] [PubMed] [Google Scholar]
- Sykes DB, Kfoury YS, Mercier FE, Wawer MJ, Law JM, Haynes MK et al. (2016). Inhibition of Dihydroorotate Dehydrogenase Overcomes Differentiation Blockade in Acute Myeloid Leukemia. Cell, 167(1), 171–186.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rompay AR, Linden K, Johansson M, Karlsson A (2001). Phosphorylation of Uridine and Cytidine Nucleoside Analogs by Two Human Uridine-Cytidine Kinases. Molecular Pharmacology, 59(5), 1181–1186. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM (1995). LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng, 8(2), 127–134. [DOI] [PubMed] [Google Scholar]
- Walse B, Dufe VT, Svensson B, Fritzson I, Dahlberg L, Khairoullina A et al. (2008). The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites. Biochemistry, 47(34), 8929–8936. [DOI] [PubMed] [Google Scholar]
- Wang QY, Bushell S, Qing M, Xu HY, Bonavia A, Nunes S et al. (2011). Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J Virol, 85(13), 6548–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright NJ, Lee SY (2019). Structures of human ENT1 in complex with adenosine reuptake inhibitors. Nat Struct Mol Biol, 26(7), 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Catapang A, Doh HM, Bayley NA, Lee JT, Braas D et al. (2018). Hexokinase 2 is targetable for HK1 negative, HK2 positive tumors from a wide variety of tissues of origin. J Nucl Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CF, Gopula B, Liang JJ, Li JK, Chen SY, Lee YL et al. (2018). Novel AR-12 derivatives, P12–23 and P12–34, inhibit flavivirus replication by blocking host de novo pyrimidine biosynthesis. Emerg Microbes Infect, 7(1), 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao SYM, Ng AML, Cass CE, Young JD (2018). Role of cysteine 416 in N-ethylmaleimide sensitivity of human equilibrative nucleoside transporter 1(hENT1). Biochem J, 475(20), 3293–3309. [DOI] [PubMed] [Google Scholar]
- York A, Williams K, Argus J, Zhou Q, Brar G, Vergnes L et al. (2015). Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling. Cell, 163(7), 1716–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Yao SY, Baldwin JM, Cass CE, Baldwin SA (2013). The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol Aspects Med, 34(2–3), 529–547. [DOI] [PubMed] [Google Scholar]
- Yuan TL, Amzallag A, Bagni R, Yi M, Afghani S, Burgan W et al. (2018). Differential Effector Engagement by Oncogenic KRAS. Cell Reports, 22(7), 1889–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA (2014). Causes and consequences of replication stress. Nat Cell Biol, 16(1), 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TDY, Oldenburg KR (1999). A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of Biomolecular Screening, 4(2), 67–73. [DOI] [PubMed] [Google Scholar]
- Zhang T, Inesta-Vaquera F, Niepel M, Zhang J, Ficarro SB, Machleidt T et al. (2012). Discovery of potent and selective covalent inhibitors of JNK. Chem Biol, 19(1), 140–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Huang JW, Tseng PH, Yang YT, Fowble J, Shiau CW et al. (2004). From the cyclooxygenase-2 inhibitor celecoxib to a novel class of 3-phosphoinositide-dependent protein kinase-1 inhibitors. Cancer Res, 64(12), 4309–4318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Full kinase inhibitor library phenotypic screen results, Related to Figure 1
Table S1. Crystallographic data collection and refinement statistics, Related to Figure 4
Data Availability Statement
The full kinase inhibitor library phenotypic screen results related to Figure 1 are included as Data S1.
The atomic coordinates and structure factors for the OSU-03012:DHODH and TAK-632:DHODH co-crystal structures reported in this study are deposited to the Protein Data Bank under the accession codes PDB 6OC0 and PDB 6OC1, respectively.