Abstract
Infection by the fungal pathogen Cryptococcus neoformans causes lethal meningitis, primarily in immune-compromised individuals. Colonization of the brain by C. neoformans is dependent on copper (Cu) acquisition from the host, which drives critical virulence mechanisms. While C. neoformans Cu+ import and virulence are dependent on the Ctr1 and Ctr4 proteins, little is known concerning extracellular Cu ligands that participate in this process. We identified a C. neoformans gene, BIM1, strongly induced during Cu limitation and which encodes a protein related to Lytic Polysaccharide Monooxygenases (LPMOs). Surprisingly, bim1 mutants are Cu deficient and Bim1 function in Cu accumulation depends upon Cu2+ coordination and cell surface association via a GPI anchor. Bim1 participates in Cu uptake in concert with Ctr1 and expression of this pathway drives brain colonization in mouse infection models. These studies demonstrate a new role for LPMO-like proteins as a critical factor for Cu acquisition in fungal meningitis.
Introduction
Fungal pathogenesis is a global public health issue, responsible for an estimated 1.5 million deaths each year largely in immune-suppressed individuals including those suffering from HIV-AIDS or tuberculosis, diabetics, cancer chemotherapy patients or immune suppressant-treated organ transplant recipients. Key to fungal virulence is the ability to colonize distinct host tissues and adapt to host environments with variations in pH, temperature, oxidants, and in the availability of nutrients and trace elements1. Trace elements such as zinc (Zn), iron (Fe), manganese (Mn) and copper (Cu) are essential for virtually all forms of life, where they perform structural, catalytic and signaling roles in processes such as transcription, protein degradation, respiration, reactive oxygen detoxification and a plethora of enzymatic activities2. At the host-microbial pathogen interface the availability of these trace elements that drive tissue colonization and virulence is often limited by a range of host processes that, together, are termed nutritional immunity3. For example, host proteins bind and sequester Fe, Zn and Mn from microbial pathogens, while host phagosomal membrane-associated metal transporters mobilize phagosomal Fe and Zn into the cytosol, further limiting their availability4,5.
Cryptococcus neoformans is among the most devastating fungal pathogens, with over 1 million infections reported annually and a high mortality rate6. Ubiquitously found in the environment on plants, in decaying wood and in bird guano, the initial route of infection occurs by inhalation of desiccated yeast cells or aerosolized spores. Successful lung colonization causes pneumonia and allows dissemination through the bloodstream to the brain, where it causes lethal meningitis7. Notably, one feature of C. neoformans that allows successful colonization of multiple locations is its ability to effectively adapt to distinct host Cu environments8-11. Central to C. neoformans Cu homeostasis is the unique Cu-sensing transcription factor, Cuf1, which regulates transcriptional responses to both high Cu and Cu-limitation10,11. Indeed, cuf1∆ strains are hypo-virulent in murine infection models12.
Upon pulmonary infection alveolar macrophages engulf C. neoformans and accumulate and compartmentalize Cu within the phagosome, where it is used for intoxication of microbial pathogens13,14. In response to these high Cu levels, the C. neoformans Cuf1 transcription factor activates genes encoding Cu-detoxifying metallothioneins (MT1 and MT2) and a mitochondrial ABC transporter (ATM1) that functions in delivery of additional Cu-sensitive FeS clusters to the cytosol8,10,15. Activation of Cu detoxification pathways is required for successful C. neoformans lung colonization and virulence in murine pulmonary models of cryptococcosis8. In contrast, C. neoformans senses a Cu-limiting environment in the brain, where Cuf1 activates expression of two cell surface Cu+ importers, Ctr1 and Ctr4, that are conserved in their overall structure and function from fungi to humans9,11. Both fungal and mammalian Cu+ importers are specific for Cu+ rather than Cu2+, and work in concert with coordinately regulated cell surface Cu2+ metallo-reductases10,11,16. However, the nature of the extracellular Cu source for high affinity Cu+ importers is not known. A C. neoformans ctr1∆ ctr4∆ strain is Cu-deficient, fails to support Cu-dependent virulence traits such as Fe acquisition, oxidative stress resistance, melanin production and respiration and is hypo-virulent in murine meningitis models via direct intra-cerebral administration9,11,12.
A recent study merging RNAseq and ChIPseq data defined the Cuf1-dependent C. neoformans Cu-regulon in response to both elevated Cu and Cu-limitation10. Surprisingly, there is a plethora of genes with previously uncharacterized roles in Cu metabolism, the transcription which are directly regulated by Cuf1 in response to alterations in Cu availability. Here we describe BIM1 (CNAG_02775, Uniprot ID J9VHN6), a gene that is strongly expressed under Cu-limitation that encodes a member of a new Lytic Polysaccharide Monooxygenase (LPMO)-like family. LPMO enzymes are secreted Cu-dependent enzymes that degrade recalcitrant carbohydrates through an oxidative mechanism17. We find that Bim1 binds Cu2+ in an atypical coordination environment for LPMOs that is reminiscent of the bacterial periplasmic Cu-binding protein CopC18, and unlike LPMOs, harbors a cell surface-tethering GPI anchor. Surprisingly, bim1∆ cells are Cu-deficient and genetic and biochemical studies demonstrate that Bim1functions in cellular Cu uptake via the Ctr1 importer. Bim1 has a critical role in fungal meningitis, suggesting that C. neoformans has exploited this unusual Cu2+-binding cell surface bound a new LPMO-like family member to facilitate Cu acquisition in the brain of mammalian hosts to drive Cu-dependent virulence traits.
Results
Bim1 is expressed and required during Cu-limited growth
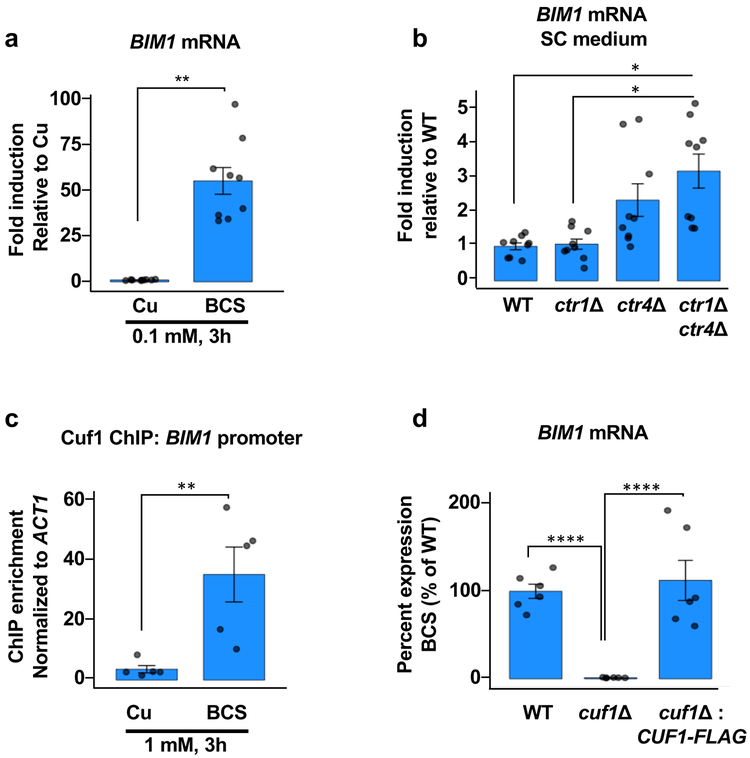
Many Cu-dependent processes are required for cryptococcal meningitis. As such, genes activated in environments with limited bioavailable Cu, such as the brain, may drive mechanisms required for adaptation for successful brain colonization. While genes encoding the high affinity Cu+ importers Ctr1 and Ctr4 are activated ~10- and ~400-fold, respectively in response to Cu-limitation by the Cu-specific chelator bathocuproine disulfonic acid (BCS), an additional transcript was the second most highly induced gene (CNAG_02775) under this condition10. The transcript encoding a protein termed Bim1 (BCS-inducible membrane protein), is elevated ~50-fold in C. neoformans cells grown under Cu-limiting conditions (BCS) compared to cells grown in the presence of CuSO4 (Figure 1a). BIM1 mRNA levels are also elevated during a genetically-induced Cu-deficiency, as shown by the increase in BIM1 transcript levels in cells lacking the Ctr1 and Ctr4 Cu+ importers as compared to wild type cells grown in synthetic complete (SC) medium (Figure 1b). The BIM1 promoter harbors three conserved Cu-Responsive Elements (CuRE)10, critical for Cuf1 binding and activation under Cu-limiting conditions, beginning at positions -239, -268 and -516. Consistent with their presence, binding of the Cuf1 Cu-sensing transcription factor to the BIM1 promoter is strongly induced under Cu-limiting conditions and expression of BIM1 under these conditions is Cuf1-dependent (Figure 1c,d).
Figure 1: C. neoformans BIM1 transcription is induced in Cu deficiency in a Cuf1-dependent manner.
(a) Mean (± SEM) values of BIM1 transcript levels by qRT-PCR analysis in C. neoformans wild-type H99 (DTY758) cells treated with BCS or CuSO4. n=3 biologically independent samples (3 technical replicates each), 2-tailed t-test, t=16.46, d.f.=4, P<0.0001 (*). (b) qRT-PCR analysis of mean (± SEM) relative BIM1 transcript levels in C. neoformans wild-type H99 (DTY758), ctr1∆ (DTY765), ctr4∆ (DTY763), and ctr1∆ ctr4∆ (DTY759) cells grown in SC medium. n=3 biologically independent samples (3 technical replicates each), 1-way ANOVA, F=3.6, d.f.=3, P<0.06, Fisher’s LSD test was performed for multiple comparisons. WT vs ctr1∆ ctr4∆ P=0.0275 (*), ctr1∆ vs ctr1∆ ctr4∆ P=0.0291 (*). (c) cuf1∆ cells complemented with Cuf1-FLAG (DTY758) were grown in either CuSO4 or BCS and the occupancy of Cuf1-FLAG protein on the BIM1 promoter was analyzed by ChIP-qPCR experiments. Mean (± SEM) fold enrichment in the immunoprecipitation relative to input material was calculated as described in the Online Methods. n=5 biologically independent samples, 2-tailed t-test, t=5.21, d.f.=8, P=0.0008 (*). (d) Cells from wild-type H99 (DTY758), cuf1∆ (DTY761), and cuf1∆ cells complemented with Cuf1-FLAG (DTY982) were grown in SC and treated with either 1 mM CuSO4 or 1 mM BCS for 3 h. Mean (± SEM) BIM1 transcript levels were normalized to 100% in the wild-type strain, and BIM1 expression levels in the other strains normalized to the wild-type. n=3 biologically independent samples (2 technical replicates each), 1-way ANOVA, F=86.63, d.f.=2, P<0.0001, Fisher’s LSD test was performed for multiple comparisons: WT vs cuf1∆ P<0.0001 (****), WT vs cuf1∆:CUF1-FLAG P = 0.084 and cuf1∆ vs cuf1∆:CUF1-FLAG P<0.0001 (****).
Annotation of the Bim1 open reading frame as a hypothetical protein in available databases prompted a detailed analysis of the putative Bim1 protein sequence. These analyses revealed the presence of an amino-terminal 19 amino acid signal sequence for secretion, several putative sites for O- and N-linked glycosylation and a predicted cleavage site between amino acids 190 and 191 for the addition of a glycosylphosphatidyl inositol (GPI) anchor (Supplementary Figure 1a). Together, these features suggest that Bim1 encodes a 218 amino acid glycoprotein that is anchored to the plasma membrane, and/or to polysaccharides in the fungal cell wall. To gain further insights into Bim1 protein structure and function, the Bim1 sequence was analyzed by the I-TASSER structural prediction server19 against all structures in the PDB library. Surprisingly, the 10 best alignments corresponded to proteins known to be Lytic Polysaccharide Monooxygenases (LPMOs) (Supplementary Table 1). LPMOs are secreted Cu-dependent enzymes that degrade recalcitrant polysaccharides and are characterized by a mono-nuclear Cu2+ active site formed by an internal histidine sidechain, an amino-terminal histidine imidazole and the amino-terminal amine20-23. LPMOs were first found secreted by fungi and bacteria that obtain their energy from recalcitrant polysaccharides, including cellulose or chitin, but are more broadly distributed in biology17. LPMOs have high binding affinity for Cu, and catalysis through the solvent-exposed Cu2+ binding site is dependent on the presence of appropriate reductants and oxidants such as O2 or H2O224.
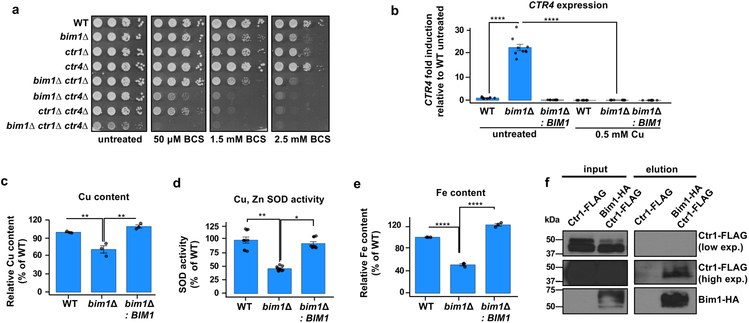
To gain insights into the potential rationale for C. neoformans robustly activating expression of an LPMO-like protein under Cu-limiting conditions, bim1∆ cells were generated and analyzed for growth during Cu deficiency. Interestingly, bim1∆ cells exhibit a growth defect on SC medium in the presence of BCS that is complemented by returning wild type BIM1 to the genome, and exacerbated on YPEG medium containing ethanol and glycerol as carbon sources that require Cu-dependent cytochrome oxidase function for their metabolism25 (Supplementary Figure 1b). The growth defect observed in bim1∆ cells on Cu deficient medium was not observed in other putative C. neoformans LPMOs (cazy.org database), such as the one encoded by CNAG_03405 (Uniprot J9VQT7) (Supplementary Figure 1c). To test potential functional interactions between Bim1, Ctr1 and Ctr4 in Cu-limited growth, a series of isogenic single or combined mutants were generated and evaluated for growth under increasingly Cu-deficient conditions using BCS. As previously observed, ctr1∆ or ctr4∆ single mutants have modest Cu-deficiency growth phenotypes, but loss of both high affinity Cu+ importers results in a severe growth phenotype on SC and on YPEG medium (Figure 2a). Moreover, bim1∆ ctr4∆ cells phenocopied ctr1∆ ctr4∆ Cu-deficiency growth defects, while bim1∆ ctr1∆ cells resemble bim1∆ mutants. These observations suggest that Bim1 and Ctr1 function in the same pathway to facilitate growth under Cu-limitation. Interestingly, the combined bim1∆ ctr1∆ ctr4∆ mutant has a more severe Cu-deficiency growth phenotype, suggesting that Bim1 has additional functions independent of Ctr1 and Ctr4 to foster growth under Cu-limiting conditions.
Figure 2: C. neoformans bim1∆ mutants have a Cu-deficiency growth phenotype.
(a) Cultures of strains wild-type H99 (DTY758), bim1∆ (DTY1000), ctr1∆ (DTY765), ctr4∆ (DTY763), bim1∆ ctr1∆ (DTY1002), bim1∆ ctr4∆ (DTY1003), ctr1∆ ctr4∆ (DTY759) and bim1∆ ctr1∆ ctr4∆ (DTY1004) were serially diluted and spotted on SC agar supplemented or not with BCS. (b) Mean (± SEM) relative CTR4 transcript levels in C. neoformans wild-type H99 (DTY758), bim1∆ (DTY1000), and bim1∆ complemented with BIM1 (DTY1001) cells grown in YPEG medium with or without CuSO4. n=3 biologically independent samples (3 technical replicates each), 2-way ANOVA, F=117, d.f.=2, P<0.0001, Fisher’s LSD test was performed for multiple comparisons: P<0.0001 (****). (c-e) Relative cell-associated Cu concentrations [(c), n=3 biologically independent samples, 1-way ANOVA, F=18.93, d.f.=2, P=0.0026, Fisher’s LSD test was performed for multiple comparisons: WT vs bim1∆ P=0.0037 (**), bim1∆ vs bim1∆:BIM1 P = 0.0011 (**), WT vs bim1∆:BIM1 P=0.2630], Cu, Zn superoxide dismutase activity [(d), n=3 biologically independent samples (2 technical replicates each), 1-way ANOVA, F=13.07, d.f.=2, P=0.0065, Fisher’s LSD test was performed for multiple comparisons: WT vs bim1∆ P=0.0025 (**), bim1∆ vs bim1∆:BIM1 P=0.0141(*), WT vs bim1∆:BIM1 P=0.1659] and relative cell associated Fe concentrations [(e), n=3 biologically independent samples, 1-way ANOVA, F=341.3, d.f.=2, P<0.0001, Fisher’s LSD test was performed for multiple comparisons: WT vs bim1∆ and bim1∆ vs bim1∆:BIM1 P<0.0001 (****), WT vs bim1∆:BIM1 P = 0.0013 (**)] in strains as in (b) grown in YPEG medium. (f) Bim1-HA co-immunoprecipitation with Ctr1-4FLAG (and Supplementary Figure 7). Protein extracts of strains bim1∆ complemented with Bim1-HA and expressing Ctr1-4FLAG tagged (DTY1016) and of bim1∆ expressing Ctr1-4FLAG tagged (DTY1015) were immunoprecipitated with HA-antibodies and analyzed for co-immunoprecipitation by immunoblotting with FLAG and HA antibodies. (a) and (f) were repeated independently three times with similar results.
To understand why bim1∆ mutants grow poorly under Cu-limitation, bim1∆ mutants were evaluated for biochemical hallmarks of Cu-deficiency10,11,26-29. In contrast to low levels of the CTR4 transcript in Cu-replete wild type cells, CTR4 transcript levels were strongly elevated in bim1∆ cells, which was suppressed by exogenous Cu or by restoration of a wild type BIM1 gene (Figure 2b). Moreover, bim1∆ cells showed a reduction in cell-associated Cu, in the enzymatic activity of Cu, Zn superoxide dismutase, in laccase-dependent melanin production (Figure 2c-d, Supplementary Figure 1d) and in the accumulation of cellular Fe, a well-established Cu-dependent process27,28 (Figure 2e). Given the genetic evidence that Bim1 functions in a Cu-accumulation pathway shared by Ctr1, Bim1 alleles were generated in which an HA-epitope was implanted within the Bim1 ORF starting at codon 36 or 92, to encode proteins (Bim1-HA) that functionally complement the Cu-deficiency bim1∆ phenotype (Supplementary Figure 2a-f). Bim1-HA was immunoprecipitated from a C. neoformans strain expressing a functional Ctr1-FLAG epitope fusion and the precipitate analyzed with anti-FLAG antibody by immunoblotting (Figure 2f). These experiments demonstrate that Bim1-HA forms a complex with Ctr1, either directly or indirectly, in vivo. Indeed, over-expression of Bim1-HA from the GAL7 galactose-inducible promoter restored Cu-limited growth in a cuf1∆ or a cuf1∆ ctr4∆ strain, presumably due significant constitutive expression of CTR111, but not in a cuf1∆ ctr1∆ strain or a ctr1∆ ctr4∆ strain (Supplementary Figure 3ab). Since Ctr1 transports Cu+ rather than Cu2+, we hypothesized that perhaps the C. neoformans metallo-reductase Fre4, which is induced in Cu-limiting conditions by Cuf110,11, functions in complex with Ctr1 and Bim1 to liberate Cu2+ bound to the Bim1 Cu binding site10,30. However, the protein Fre4-FLAG did not show an enrichment in a Bim1-HA immunoprecipitation experiment performed with a C. neoformans strain expressing a functional Fre4-FLAG epitope fusion (Supplementary Figure 3c). Taken together, these results demonstrate that bim1∆ cells are defective in Cu accumulation and Cu-dependent activities in multiple cellular compartments, providing a basis for the Cu-deficiency growth defect. While more experiments are required to understand the precise mechanisms for Bim1 function in copper acquisition, these experimental data evidence that Bim1 likely operates via a Ctr1-dependent pathway for Cu uptake.
Bim1 localizes to the cell surface and coordinates Cu2+
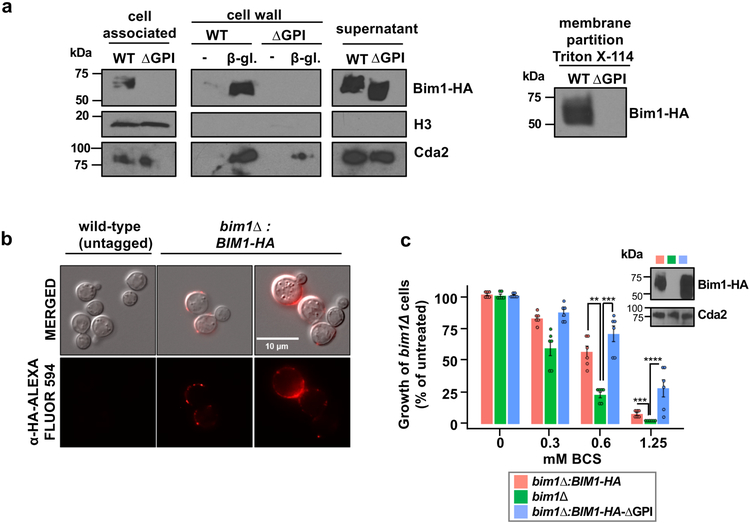
Bim1, related to LPMO enzymes at the primary sequence level, functions in C. neoformans Cu accumulation and is predicted to be a secreted, GPI-anchored Cu2+ binding protein. As fungal GPI anchors localize secreted proteins to both the plasma membrane and cell wall31, subcellular fractionation and indirect immunofluorescence microscopy experiments were used to localize Bim1-HA and Bim1-HA lacking the consensus GPI modification sequence (Bim1-HA-∆GPI) (Figure 3 ab, Supplementary Figure 4a). Bim1-HA fractionated with the plasma membrane, was released from the cell wall after treatment with fungal cell wall lytic enzymes and also partially fractionated in the culture supernatant (Figure 3a). While the Bim1-HA-∆GPI mutant protein was largely found in the culture supernatant (Figure 3a), indirect immunofluorescence microscopy validated cell surface association of Bim1-HA in intact C. neoformans cells (Figure 3b). Moreover, the Bim1-HA GPI anchoring sequence is required for complementation of the Cu-deficiency growth phenotype in a colony spot test on agar medium containing BCS (Supplementary Figure 4b). To test whether one role for the GPI anchor in Cu-limited growth may be to increase the cell surface concentration of Bim1, supernatants from bim1∆ cells expressing either Bim1-HA, Bim1-HA-∆GPI, or empty vector were concentrated and incubated with bim1∆ cells grown in SC medium in microtiter plates in the presence of increasing concentrations of BCS. Supernatants harboring Bim1-HA or Bim1-HA-∆GPI partially restored the growth of bim1∆ cells in Cu-limiting conditions, with a trend toward better complementation by Bim1-HA-∆GPI supernatant, which contains higher concentrations of Bim1 (Figure 3c).
Figure 3: C. neoformans Bim1 is a mannoprotein that is attached to the cell through a GPI anchor.
(a) Bim1-HA subcellular fractionation (and Supplementary Figure 8). Strains bim1∆ complemented with Bim1-HA (DTY1005) and bim1∆ complemented with Bim1-HA lacking the GPI signal sequence (DTY1015), were fractionated and Bim1-HA was probed for cell-association, cell wall attachment in untreated cells or cells treated with β-glycosidase, for abundance in cell culture supernatants and for membrane partitioning in Triton X-114 solubilized extracts as indicated. These experiments were repeated independently three (subcellular fractionation) and two (membrane partitioning) times with similar results. Immunoblotting was performed with antibodies specific for HA, histone H3 or Cda2. (b) Bim1-HA cellular localization assessed by indirect immunofluorescence. Cultures of strains wild-type H99 (DTY758) and bim1∆ complemented with BIM1-HA (DTY1005) were grown in SC medium with 0.5 mM BCS for 3 h. Cells were then fixed and processed for immunofluorescence microscopy. Top lane, merged differential interference contrast and fluorescence images (MERGE), bottom lane, fluorescence images after staining with α-HA antibody conjugated to Alexa Fluor 594. This experiment was performed once. (c) Culture supernatants from bim1∆ (DTY1000), bim1∆ complemented with Bim1-HA (DTY1005) and bim1∆ complemented with Bim1-HA-∆GPI (DTY1007) cells were collected and concentrated. bim1∆ (DTY1000) cells grown in SC medium were diluted to OD600 of 0.002 and grown in the presence of the indicated concentrated supernatants in 96-well plates and with the indicated concentrations of BCS. Protein concentration was evaluated in the concentrated supernatants by immunoblotting with HA and Cda2 antibodies (inset, and Supplementary Figure 9). n=3 biologically independent samples (2 technical replicates each), 2-way ANOVA, F=6.63, d.f.=6, P=0.0003, Fisher’s LSD test was performed for multiple comparisons: at 0.6 mM BIM1-HA vs bim1∆ P=0.0012 (**), bim1∆ vs BIM1-HA-∆GPI P=0.0001 (***), BIM1-HA vs BIM1-HA-∆GPI P=0.3804. At 1.25 mM BIM1-HA vs bim1∆ P=0.0010 (***), bim1∆ vs BIM1-HA-∆GPI P<0.0001 (****), BIM1-HA vs BIM1-HA-∆GPI P=0.0002 (****).
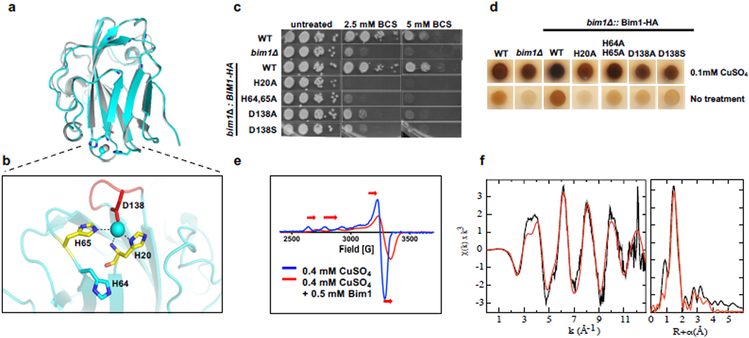
Bim1 shows high sequence homology (36% identity) to novel GPI-anchored LPMO-like proteins from the fungi Laetisaria arvalis and Laccaria bicolor32 . Given the conservation with these proteins of the Cu2+-coordinating histidines and a GPI anchor, a structural model for Bim1 was generated based on the LaX325 structure32 (Figure 4a, Supplementary Table 2). The threaded structural model of Bim1 displays the putative histidine brace Cu2+- coordinating ligands found in all LPMOs (H20 and H65) and a potential aspartic acid Cu2+ ligand (D138) found within a glycine-rich loop of LaX325 (Figure 4b). To ascertain if histidine H20, H64,65 an aspartate D138 are important for Bim1-HA function, these residues were independently converted to alanine (H20A, H64,65A or D138A) or to serine (D138S) and expressed in bim1∆ cells (Supplementary Figure 4a, c, d). The mutants were tested for complementation of bim1∆ cells by growth on Cu limiting medium and melanin production (Figure 4cd). Of note, H64 was also mutated as a potential Cu2+-ligand in Bim1, as these predictions come from a homology model rather than from an experimental structure. While wild type Bim1-HA complements bim1∆ phenotypes, expression of neither the Bim1-HA H20A, H64,65A nor D138A/S mutants was able to restore growth on Cu-limiting medium. All Bim1-HA putative Cu binding site variants showed a hypo-melanization phenotype similar to that observed for the bim1∆ strain. In all analyzed strains the hypo-melanization phenotype was restored to a similar level as that of the WT strain when 0.1 mM CuSO4 was added to the medium (Figure 4d). In a trans-complementation assay, although the Bim1-HA H64,65A mutant expressed comparable levels of secreted protein to Bim1-HA, it failed to restore growth of the bim1∆ mutant in Cu-limiting conditions (Supplementary Figure 4e).
Figure 4: Bim1 binds Cu2+ via His and Asp ligands.
(a) Bim1 homology structural model (cyan) aligned with the PDB structure corresponding to the LaX325 structure (grey) used as a template, with copper shown as a sphere and coordinating residues as sticks in the Bim1 model. (b) Close-up view of the Cu2+ coordination environment in the Bim1 structural model. The His brace residues (H20 and H65) are shown in yellow, a conserved Gly rich loop, including a predicted Cu-binding Asp residue (D138), is labelled in red. The additional His64 residue mutated in this study is also shown. (c-d) Cell cultures of wild-type (DTY758), bim1∆ (DTY1000), and bim1∆ complemented with either Bim1-HA (DTY1005), Bim1-HA H20A (DTY1008), Bim1-HA H64,65A (DTY1009) and Bim1-HA D138A or D138S (DTY1023 or DTY1025) strains were grown in SC medium and spotted on SC agar supplemented or not with BCS (c) or spotted on a DOPA agar plate supplemented or not with CuSO4 for assessment of melanin production (d). Plates were incubated in the dark at 30°C for 2 (c) or 1.5 (d) days and photographed. (e) EPR spectrum of CuSO4 in solvent (blue line) and in the presence of purified Bim1 (red line) at the indicated concentrations. These experiments were performed independently three [(c) and (d)] and two times (e) with similar results. (f) Unfiltered k3-weighted extended X-ray absorption fine structure (EXAFS) spectrum (left frame, black line) and the corresponding non-phase corrected Fourier transform of the Cu2+ Bim1 protein (right frame, black line). Multiple scattering pathways were calculated from the active site crystallographic coordinates of an LPMO bound in a histidine brace and the Cu-His distance refines to 2.03 Å (details provided in Supplementary Figure 5c). An additional first-shell, low-Z ligand at 1.94 Å was required to best fit the data.
The histidine residues in Bim1 that are required for growth and melanin production under Cu deficient conditions correspond to the Cu2+-coordinating histidine brace conserved in LPMOs23. To ascertain whether Bim1 binds Cu2+, Bim1 was expressed in and purified from Pichia pastoris (Supplementary Figure 5a) and evaluated for Cu binding by Electron Paramagnetic Resonance spectroscopy (EPR), which detects Cu2+, but not Cu+. The EPR spectrum of Cu2+ alone was modified by titration of approximately equimolar Bim1, indicating that Bim1 binds Cu2+ with an estimated 1:1 stoichiometry (Figure 4e). The EPR spectrum for the Cu2+-loaded Bim1 complex is axial with gz= 2.26 and a Az= 168 G (0.0177 cm−1) (Supplementary Table 3). These spectral features place the Cu2+-Bim1 complex within the Peisach-Blumberg classification of a type 2 copper centre33. The spin Hamiltonian parameters for Cu2+-Bim1 are comparable to LPMOs, although the coupling constant (Az/G = 168) is relatively high.
More definitive confirmation of the involvement of a putative histidine brace coordinating Cu2+ is evident from the analysis of X-ray Absorption Spectroscopy (XAS). The X-ray absorption edge energy, and lack of a Cu+ 1s→4p transition at 8984 eV, confirms the Cu2+ oxidation state34 (Supplementary Figure 5b). Figure 4f shows the k3-weighted extended X-ray absorption fine structure (EXAFS) (black line) and corresponding Fourier transformation (FT) for a representative sample of Bim1. Both the k3-weighted EXAFS oscillations and the transformed data show the hallmark features of rigid-ring imidazole scattering from histidine ligands, a double hump at 4 Å−1 and multiple FT peaks between 2-4 Å, respectively35. The fit shown in Figure 4f (red line, details reported in Supplementary Figure 5c) used multiple scattering pathways calculated from the crystallographic coordinates of a histidine brace Cu center from the LPMO from H. jecorina (PDB 5O2W)36 and requires two histidine ligands with 2.03 Å Cu-N. The identity of additional Cu2+ ligands is difficult to discern with XAS, but the fit requires an additional low Z ligand with a shorter distance (1.94 Å), an observation consistent with the possibility of a fourth unique ligand derived from aspartate oxygen. Single scattering fits are not shown but refine to an average ligand distance of 1.97 Å or can likewise be fit with two poorly resolved distances in the first shell (1.92 Å and 2.03 Å). Taken together, these studies demonstrate that Bim1 has a mononuclear Cu2+ center that, based on the LaX325 structure, is modeled to utilize solvent accessible histidine ligands. Based on these data and the Bim1 mutagenesis results, the Bim1 His residues at positions H20 and H65, and D138 are likely the key residues that coordinate the Cu2+. These spectroscopic data provide important context for the proposed unique adaptation of the LPMO-like active site for Cu acquisition and virulence (see below). Given the similarity of Bim1 to bona fide LPMOs, Cu-Bim1 was assayed for catalytic activity on phosphoric acid swollen cellulose, a standard enzymatic assay in the LPMO field37. However, no cellulose cleavage products were detected (Supplementary Figure 5d). This observation does not exclude the possibility of Cu-Bim1 exhibiting activity on other yet unidentified substrates.
Bim1 role in Cu import is critical for fungal meningitis
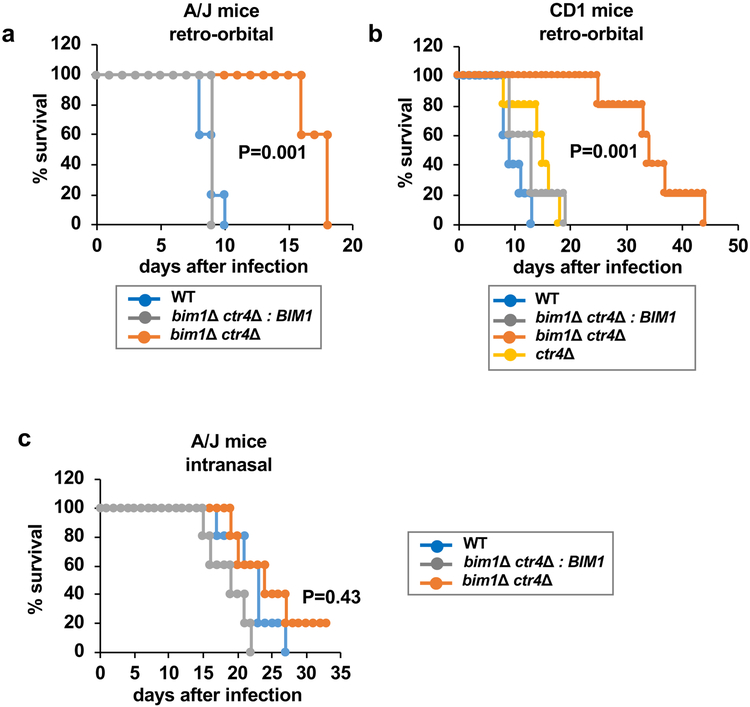
While expression of Cu detoxification pathways is induced during pulmonary infection and critical for C. neoformans survival within macrophages, these components are dispensable in mouse models of cryptococcal meningitis8. Conversely, the CTR1 and CTR4 genes are transcriptionally activated by Cuf1 in the low bioavailable Cu environment of the brain, and ctr1∆ ctr4∆ mutants, defective in Cu acquisition, are hypo-virulent in meningitis models9. Indeed, deletion of both Cu+ importers is required to observe a defect in mouse brain infection models. Given the similar Cu-deficiency phenotype of ctr1∆ ctr4∆ cells and bim1∆ ctr4∆ cells, experiments were conducted to ascertain whether Bim1 functions in the infectious niche of the brain. To bypass the lungs, retro-orbital mouse infections were conducted and mouse survival was assessed. The A/J inbred mouse strain infected with a bim1∆ ctr4∆ C. neoformans mutant survived nearly twice as long as mice infected with wild type cells or bim1∆ ctr4∆ cells complemented with wild type BIM1 (Figure 5a). As A/J mice are inbred, a similar experiment was conducted in the genetically diverse outbred mouse strain, CD1. Similar results were obtained in the CD1 retro-orbital infection model, demonstrating that Bim1 plays an important role in virulence in genetically diverse mouse models of cryptococcal meningitis (Figure 5b). Importantly, loss of both Cu acquisition pathways was required to observe this virulence defect, as loss of only Ctr4 (Figure 5b) or Bim1 (Supplementary Figure 6) displayed virulence in retro-orbital infections that were similar to that of wild type C. neoformans. Consistent with a requirement for deployment of Cu detoxification mechanisms in the high Cu environment of the lung, and lack of BIM1 expression in high Cu, inactivation of the Bim1 Cu acquisition pathway had no observable impact on virulence in an A/J mouse pulmonary infection model (Figure 5c).
Figure 5: The Bim-Ctr1 Cu uptake pathway is a virulence determinant in mouse models of cryptococcal meningitis.
(a) Retro-orbital infection of A/J mice with 5,000 cells of the following strains cultured in SC medium: wild-type H99 (DTY981), bim1∆ ctr4∆ (DTY1020), and bim1∆ ctr4∆ complemented with BIM1 (DTY1021). (b) Retro-orbital infection of outbred CD1 mice with 5,000 cells of the following strains cultured in SC medium: wild-type H99 (DTY981), bim1∆ ctr4∆ (DTY1020), ctr4∆ (DTY1019) and bim1∆ ctr4∆ complemented with BIM1 (DTY1021). (c) Intranasal infection of inbred A/J mice with 500,000 cells of the following strains cultured in SC: wild-type H99 (DTY981), bim1∆ ctr4∆ (DTY1020), and bim1∆ ctr4∆ complemented with BIM1 (DTY1021). Experiments a-c were performed once with 5 mice infected per strain. In all experiments the survival average of mice infected with the wild-type was compared to the survival average of mice infected with any of the mutants by the two-sided log rank statistical test. In (a) and in (b) statistical differences were found between mice infected with wild-type vs mice infected with bim1∆ ctr4∆. P values are indicated in the figures.
Discussion
The ability to rapidly adapt to changing environments is critical for colonization of infectious niches by microbial pathogens. During infection C. neoformans shifts from a Cu detoxification mode to actively acquiring Cu as it disseminates from the lungs to the brain8,9,12. This switch, orchestrated by the Cuf1 Cu-sensing transcription factor, is critical for colonization and pathology in both tissues10-12. While it is unclear how the brain, the most Cu-dependent organ on a tissue weight basis, limits pathogen access to Cu, C. neoformans senses a Cu-limiting environment, strongly activates expression of Ctr1 and Ctr4 and requires this Cu uptake machinery to drive a number of Cu-dependent virulence functions. What is striking from the current work is that under Cu limitation BIM1 is among the top two genes robustly induced by Cuf110. Bim1 binds Cu2+, has a critical function in Cu acquisition and the Bim1-Ctr1 pathway for Cu uptake plays a key role in cryptococcal meningitis.
What is the role played by Bim1 in Cu uptake? The Ctr1 and Ctr4 plasma membrane proteins are conserved in their overall topology and function from fungi to humans. These proteins support the permeation of Cu+ through membranes, generated through the action of cell surface metallo-reductases and coordinated via methionine thioether ligands through the membrane pore16,38. Currently it is unclear how eukaryotic Cu+ transporters, or the Cu2+ metallo-reductases, obtain Cu from the extracellular environment. Recent work demonstrates that albumin and the multi-copper oxidase ceruloplasmin39, which exists as both a secreted form in the serum and attached to the surface of specific cells via a GPI linkage, are sources of extracellular Cu in mammals. While in vitro studies show that the amino-terminal peptide of human Ctr1 can collect Cu2+ from albumin40, ceruloplasmin can provide Cu to both Ctr1-dependent and Ctr1-independent Cu uptake mechanisms in cultured cells39. Additionally, some forms of CopC, a periplasmic protein found in bacteria, bind a single Cu2+ atom using His and Asp ligands18 similar to that identified in the LaX325 protein32 and conserved in Bim1. Indirect evidence suggests that CopC could function in an analogous fashion to Bim1-Ctr1, in concert with the inner membrane protein CopD to import Cu18,41.
Given the genetic and biochemical evidence supporting a role for Bim1 in Ctr1-mediumted Cu+ uptake, it is possible that Bim1 serves as an extracellular Cu ligand in a fashion similar to mammalian ceruloplasmin or bacterial CopC. In fact, mutation of the putative Bim1 Cu binding His and Asp residues, provoked by the Bim1 structural model, compromised the ability of Bim1 mutant proteins to complement the growth and melanization phenotypes of bim1∆ growth on Cu limiting medium. We hypothesized that perhaps the C. neoformans Fre4 and Fre7 metallo-reductases, whose expression is also coordinately induced under Cu-limiting conditions by Cuf110,11, function together with Ctr1 and Bim1, to liberate the Cu2+ bound to the Bim1 Cu binding site10,30. For example, the reduction of Bim1-bound Cu2+ to Cu+ could reduce binding affinity for Bim1 but increase affinity for Ctr1. We did not see an enrichment in Fre4 protein levels in a Bim1-HA co-immunoprecipitation experiment, however this negative result could be solely due to a potentially very transient interaction as a result of the highly solvent-exposed nature of the Cu2+ center in Bim117. Moreover, as a bim1∆ ctr1∆ ctr4∆ mutant has a Cu-deficiency phenotype more severe than a ctr1∆ ctr4∆ mutant, Bim1 could also function with an as yet uncharacterized Cu importer, or through the oxidative degradation of the C. neoformans cell wall or capsule, both of which are reported to sequester metal ions7,42,43. While no obvious LPMO activity was detected in our experiments, current efforts are underway to ascertain if Bim1 has LPMO activity and to identify potential physiological substrates.
Bim1 is the first reported eukaryotic LPMO-like protein to function in Cu acquisition, thus expanding the repertoire of potential functions for this growing family of proteins. Interestingly, under Zn limiting conditions the pathogenic fungus Candida albicans induces expression and secretion of Pra1, a catalytically inactive member of a metalloprotease family which augments Zn uptake via interactions with the cell surface Zrt1 Zn importer44. Thus, organisms may generally exploit related enzyme family members that tightly and specifically bind metals to facilitate metal transfer, somewhat analogous to the structural resemblance between the intracellular CCS Cu chaperone for Cu, Zn superoxide dismutase (Sod1), which lacks enzymatic activity, and Sod1 itself29,45. It is unclear why, like Pra1 in C. albicans, Bim1 is also found in the culture supernatant. Perhaps within the brain Bim1 that is shed in a spontaneous or regulated fashion acquires Cu from host Cu proteins and returns to the cell surface in a manner similar to siderophores. Based on results demonstrating Bim1 in the culture medium can complement the Cu-deficiency phenotype of bim1∆ cells, perhaps the released Bim1 also functions in Cu acquisition in a cell non-autonomous manner, but the GPI anchor strongly improves this function.
While Bim1 plays a critical role in cryptococcal meningitis, at least in part through its role in Cu acquisition, it could play additional roles in C. neoformans during Cu limitation or as a means to invade and colonize host tissues in the brain by compromising host carbohydrate integrity. LPMO family members such as those from Vibrio cholerae and Listeria monocytogenes are virulence factors in mouse infection models46,47. Also, an insect virus uses an LPMO for infection and dissemination and several plant pathogens activate expression of a large number of LPMO coding genes during their necrotrophic life cycle48. While the mechanisms might be unrelated, there is still much to be discovered on how LPMOs and LPMO-like proteins function in virulence during infection by microbial pathogens. In this regard, the Cryptococcus genome encodes for three other putative LPMOs from families AA9, AA11 and AA14 (cazy.org database), the expression of which is independent of Cu and Cuf1, but which could have virulence roles in plant or animal hosts.
Online Methods
Generation of Cryptococcus neoformans mutants
DNA was introduced into C. neoformans by biolistic transformation. Yeast peptone dextrose (YPD) medium, supplemented with 1.5% agar and 100 μg ml-1 of nourseothricin (NAT), 200 μg ml-1 of neomycin (G418) or 200 μg ml-1 of hygromycin B (HYG) were used for colony selection after biolistic transformation. For Cu-import deficient mutants, YPD or YPD-sorbitol agar medium was supplemented with 40 μM CuSO4. Strains, plasmid descriptions and primers used are described in Supplementary Dataset 1. For cell growth assays C. neoformans cells were grown to stationary phase, diluted, plated with a pinner and incubated on untreated or treated SC-glucose, SC-galactose, or yeast peptone ethanol glycerol (YPEG) agar medium as indicated in figure legends.
RNA isolation and qRT-PCR
C. neoformans overnight cultures grown in synthetic complete (SC) medium (MP Biomedicals) or YPEG medium, as indicated in figure legends, were diluted to OD600 of 0.3, grown for 2 h and treated as indicated. C. neoformans mRNA isolation and expression analysis was performed as previously described10. Primers used for BIM1, CTR4 and ACT1 qRT-PCR are described in Supplementary Dataset 1.
Chromatin Immunoprecipitation
Overnight cultures of C. neoformans cuf1∆ cells complemented with Cuf1-FLAG (Supplementary Dataset 1) were diluted to OD600 of 0.2, grown for 3 h and then treated with 1 mM Cu or 1 mM BCS for 3 h. ChIP-PCR analysis was performed as previously described10. Specific primers used for detecting the BIM1 and TUB1 promoters by qPCR are described in Supplementary Dataset 1.
In silico analysis of the BIM1-encoded open reading frame
Bioinformatic analysis of CURE sites was performed with the motif-based sequence analysis tool provided by the Meme Suite platform49. The BIM1 coding sequence was analysed in SignalP 4.150, big-PI predictor51, NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/), NetOGlyc 4.052 and I-TASSER19 for predictions of amino-terminal signal peptide, GPI anchoring, N- and O-glycosylation and protein structure, respectively.
Inductively coupled plasma mass spectrometry (ICP-MS)
C. neoformans cells were grown in YPEG medium to stationary phase, diluted to OD600 of 0.2 and then grown for 3 h. Cells were harvested, pellets washed once with 1xPBS containing 5 mM EDTA, once with 1xPBS and twice with ultra-pure water. Pellets were digested by boiling overnight with 200 μl 100% HNO3. ICP-MS analyses were performed by the Environmental and Agricultural Testing Service, Department of Soil Science, North Carolina State University, Raleigh. Metal concentrations were obtained according to a standard curve and normalization between samples was performed by dividing the metal concentration of each sample to its phosphorous concentration, also determined by ICP-MS.
Enzyme activity assays
C. neoformans cells were grown in the indicated medium to stationary phase, diluted to OD600 of 0.2 and then grown for 3 h. Cells were pelleted, washed once with 1xPBS and protein extracts were obtained at 4°C with 50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1% Triton X-100, 1 mM EDTA and fungal specific protease inhibitors (Sigma). Cu, Zn SOD activity measurements were performed according to the SOD1 assay kit (Sigma). Data was normalized to protein concentration, determined with the Pierce BCA assay (Thermo Fisher). For melanin assays C. neoformans cells were grown in SC medium to stationary phase, diluted to an OD600 of 2.5 and plated with a pinner on L-3,4-dihydroxyphenylalanine (L-DOPA) agar medium (7.6 mM L-asparagine monohydrate, 5.6 mM glucose, 22 mM KH2PO4, 1 mM MgSO4.7H2O, 0.5 mM L-DOPA, 0.3 μM thiamine-HCl, 20 nM biotin, pH 5.6). L-DOPA plates were incubated at 30°C for 2 days in the dark and photographed.
C. neoformans subcellular fractionation
Protein analysis of Cryptococcus subcellular fractions and culture supernatant was performed as previously described with minor modifications53. C. neoformans cells were grown in YPD agar lawns for 72 h, cells harvested and resuspended in secretion buffer (10 mM imidazole, 2% glucose, pH 5.0) (ratio of 5 ml/plate) and incubated at 30°C for 17 h with shaking. After incubation, cells were collected by centrifugation and supernatants kept for protein content analysis (secreted fraction). Pelleted cells were washed once with 1xPBS, once with morpholineethanesulfonic (MES) acid-buffered saline (MBS; 25 mM MES, 150 mM NaCl, 2 mM EDTA, pH 6.5), resuspended in 500 μl of ice-cold MBS containing 0.1% (vol/vol) Triton X-100 and fungal specific protease inhibitors (Sigma) and lysed by bead beating for 6 cycles of 1 min at 4°C. Lysates were centrifuged at 3,500 g for 10 min at 4°C, supernatants were collected, set on ice and pellets resuspended in 1 ml of the same buffer and further disrupted by probe sonication for 5 cycles of 10 s ON, 10 s OFF, with at an output of 4. Samples were centrifuged at 3,500 g for 10 min at 4°C, and supernatants combined with those from the previous spin. Pellets from this spin, which contain the cell wall fraction, were washed twice with 1xPBS (with 1 min centrifugation after each wash), once for 1 h with 1% SDS at 37 °C with shaking, twice with 1 M NaCl and once with 1xPBS. Washed pellets were split into two tubes, one resuspended in 1 ml water and 100x fungal specific protease inhibitors (Sigma), the other resuspended in 1 ml of lytic enzymes from Trichoderma harziarum (Sigma) (prepared at 10 mg/ml in water and 100x fungal specific protease inhibitors). Both tubes were incubated for 1 h at 37 °C, and the supernatant, containing cell-wall released proteins, was collected by centrifugation (14,000 g, 15 min, 4°C). Those supernatants previously set aside on ice, were ultra-centrifuged at 135,000 g for 1 h at 4 °C to separate the pellet (crude membranes) from the supernatant (cell associated fraction). Approximately 100 μl of crude membranes per strain were resuspended in 200 μl of ice-cold 50 mM Tris-HCl pH 7.5, 0.5 mM EDTA and fungal specific protease inhibitors. Resuspensions were probe sonicated for 3 cycles of 5 seconds ON and OFF, with an output of 4. 120 μl of TritonX-114 were added to the samples mixed and chilled on ice for 1 h. After 1h, samples were centrifuged at 14,000 rpm for 10 min at 4 °C in a microcentrifuge to remove insoluble debris. The supernatant was heated to 37 °C for 30 min to achieve phase separation. The aqueous phase was removed from the lower (detergent) phase by centrifugation for 5 min at 14,000 rpm in a microcentrifuge. To remove residual aqueous material, the detergent phase was washed three times by mixing the detergent phase with 200 μl of 50 mM Tris-HCl pH 7.5, 0.5 mM EDTA followed by 5 min at 14,000 rpm centrifugation and supernatant removal. For the membrane partition fraction, proteins were recovered by trichloroacetic acid (TCA) precipitation addition to a final 10% concentration, followed by 20 min centrifugation at 14,000 rpm at 4 °C. Pelleted proteins were washed once with ice-cold acetone and resuspended in 50 μl of 100 mM Tris-HCl, pH 8.3, 1 mM EDTA, 1% SDS. Proteins from the different fractions were analyzed by immunoblotting with anti-HA (Y-11, polyclonal, Santa Cruz), anti-H3 (D1H2, polyclonal, Cell Signaling) and anti-Cda2 (a generous gift from J.K. Lodge) antibodies.
Bim1 trans-complementation assays
C. neoformans strains bim1∆ (DTY1000), bim1∆ complemented with either Bim1-HA (DTY1005), Bim1-HA (∆GPI) (DTY1007) and Bim1-HA H64A H65A (DTY1009) were grown in YPD agar lawns for 72 h (2 plates per strain). Cells were harvested and resuspended in secretion buffer (10 mM imidazole, 2% glucose, pH 5.0) (ratio of 5 ml/plate) and incubated at 30 °C for 17 h with shaking. After incubation, cells were pelleted and supernatants from 10 ml of overnight secretions (2 plates) were filtrated through 0.45 μm pore size filters (to facilitate supernatant concentration) and concentrated to approximately 200 μl (~ 6 μg/μl of total protein) in Amicon® Ultra 15 ml centrifugal filter devices (Millipore) with a 10,000 NMWL. After supernatant concentration, protein from all strains was normalized to 5 μg/μl in 200 μl of medium. 15 μl were used for immunoblotting analysis and the rest was diluted into 7 ml of SC medium. The 7 ml of medium, containing secreted proteins (including secreted Bim1-HA or mutants of Bim1-HA), were filter sterilized using 0.2 μm pore size filters. C. neoformans bim1∆ cells grown overnight in SC medium were diluted to an OD600 of 0.004 in 2 ml of each of the filter sterilized SC medium containing the secreted proteins from the different strains. Quantitative liquid growth assays were performed as described below.
Liquid growth assays
To perform growth analysis in liquid cultures, all wells of a 96-well plate were filled with 50 μl of SC medium. With a multichannel pipette, 50 μl of SC medium containing 20 mM BCS were added to each well of column number 12 in the plate and mixed with the previously added medium. With a multichannel pipette, 50 μl of SC medium from column number 12 (with 10 mM BCS) were transferred each well of column number 11, and this process was repeated until reaching column number 3 (column 1 was kept as a blank, and column 2 was the untreated control). After filling the plate with medium containing BCS, three rows of wells from column 1 to column 12 (2 technical replicates per experiment) were filled with 50 μl of the analyzed cells previously diluted to OD600 of 0.004 in SC medium. Plates were covered with a semipermeable membrane and incubated at 30 °C with shaking at 900 rpm in a Finstruments shaker instrument. Growth graphs were generated by plotting the OD600 readings versus the compound concentrations at the 48 h time point. Three biological replicates were performed.
Immunoprecipitation
C. neoformans cells were grown in 25 ml precultures overnight in SC medium at 30°C. Precultures were diluted to OD600 of 0.3 in 100 ml of SC, grown for 2 h and treated with 0.5 mM BCS for 3 hours. Formaldehyde (crosslinker) was added to the medium at 0.8% final concentration followed by 7 min incubation at room temperature with occasional shaking. After incubation, crosslinking was quenched with glycine at a final concentration of 125 mM followed by 5 min incubation at room temperature. Cells were collected by centrifugation and washed once in 125 mM glycine in 1xPBS and twice with 1 x PBS. Cell pellets were then resuspended in 450 μl of lysis buffer (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 5% glycerol, 1% NP-40, 0.5% foscholine-14 and protease inhibitors) and lysed by 6 cycles of 1 min ON, 1 min OFF bead beating. Cell debris was separated from lysates by 5 min centrifugation at 14,000 rpm at 4 °C. Protein concentrations were measured in lysates by the Pierce BCA protein concentration kit (Thermo Fisher) and normalized between the strains. A fraction of the protein lysates was kept frozen and loaded as input for the immunoblotting analysis, and the rest of the lysates were incubated overnight with rocking with 30 μl of anti-HA agarose resin slurry (Pierce), prewashed once with 500 μl TBST. After overnight incubation, the resin was washed twice with 1% Triton-X100, once with TBST and once with 50 mM ammonium bicarbonate. Proteins were eluted from the beads by 10 min boiling with 50 μl of 100 mM Tris-HCl, pH 8.3, 1 mM EDTA, 1% SDS. Input and eluted fractions were analyzed by immunoblotting with anti-FLAG (monoclonal, HRP-conjugated, Sigma) and anti-HA (Y-11, polyclonal, Santa Cruz) antibodies.
Mouse infection experiments
Inbred female A/J mice (Jackson Laboratories) and CD1 mice (Charles River Laboratories), aged 4 to 6 weeks, were used for virulence experiments. Purchased mice were acclimated for one week. For infections all C. neoformans strains were grown in SC medium overnight and washed 3 times with sterile 1 x PBS. Infections were performed intranasally or retro-orbitally with C. neoformans inocula as indicated in figure legends. Mouse infection experiments were performed in compliance with all ethical regulations. The mice protocol was revised and approved by the Duke University Institutional Animal Care and Use Committee (DUIACUC). Protocol approved documents were signed by Herman Staats, PhD (Chair, IACUC). Experimental endpoints were considered a 15 % reduction in body weight.
Protein extraction and immunoblotting
TCA-precipitated protein extracts were used to analyze Bim1-HA expression in wild type Bim1-HA or Bim1-HA mutants. For all figures, except Supplementary Figure 4c, overnight cultures of C. neoformans cells were diluted to an OD600 of 0.3 in 5 ml of fresh SC medium, left untreated or treated with the indicated concentration of BCS for the indicated times. For the data in Supplementary Figure 4c, overnight cultures of C. neoformans cells were diluted to an OD600 of 0.3 and grown to an OD600 of 3. After each time point TCA was added to a final concentration of 10%, cells pelleted, collected with 20% TCA and further processed as described. Supernatants from experiment in Supplementary Figure 4c were directly treated with loading buffer before SDS-PAGE and immunoblotting analysis. TCA protein extracts and supernatants were analyzed by immunoblotting with anti-HA (Y-11, polyclonal, Santa Cruz or SG-77, polyclonal, Invitrogen) and anti-H3 (D1H2, polyclonal, Cell Signaling) antibodies.
Indirect immunofluorescence
C. neoformans strains WT (DTY758) and bim1∆ complemented with Bim1-HA (DTY1006) were grown in SC to stationary phase. Precultures were then diluted to OD600 of 0.3 in 25 ml of SC medium. Cells were treated with 0.5 mM BCS and grown for 3 hours. After treatment, formaldehyde was added to a 3.7% final concentration followed by 20 min incubation at room temperature with shaking. After incubation, crosslinking was quenched with glycine at a final concentration of 125 mM followed by 5 min incubation at room temperature. Cells were collected by centrifugation and washed once in 125 mM glycine in 1xPBS and twice with 1 x PBS. Pellets were resuspended in 2 ml of 100 mM Tris-HCl pH 9.5 containing 10 mM DTT and incubated at 30°C for 20 min with 70 rpm shaking. Cells were pelleted at 3000 g during 5 min and pellets were resuspended in 5.2 ml of resuspension buffer (0.1 M sodium citrate, 1.1 M sorbitol pH 5.5) containing 10 mg/ml of lytic enzymes (Sigma, L1412) and halt protease inhibitors (Roche) and incubated for 2 h at 30°C with 70 rpm shaking. After incubation, spheroplasts were pelleted by centrifugation (8 min at 2200 g) and rinsed once with 10 ml of ice-cold resuspension buffer. Spheroplasts were collected by centrifugation (8 min at 2200 g) and resuspended in 2 ml of incubation buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% BSA and 0.02% sodium azide). 100 μl aliquots were incubated over night with 1:10 dilution of anti-HA (Y-11, polyclonal, Santa Cruz) antibody at 4°C. After the incubation, spheroplasts were washed three times with PBS containing 1% BSA. Spheroplasts were then resuspended in 100 μl PBS containing 1% BSA and 2 μg/ml of secondary antibody Alexa Fluor 594 and incubated at room temperature for 1 hour in the dark with 70 rpm shaking. The secondary antibody was three times with PBS containing 1% BSA and cells were mounted in a slide containing a 2% low melting point agarose pad prepared with SC medium. Slides were sealed with nail polish and cells observed with a Zeiss Axio Imager Microscope.
Homology modelling of Bim1
A homology model of Bim1 was created using the one to one threading option in Phyre254, based on the tetragonal crystal structure of LaX32532, PDB code 6IBJ. Bim1 and LaX325 share 36% sequence identity. Local alignment and secondary structure scoring (with a weight of 0.1) were used for producing a model with 100% Phyre2 confidence (this just indicates the confidence that the two sequences aligned truly represent two homologous proteins). Phyre2 aligns Hidden Markov Models (HMMs) of query and target. Based on this alignment, Phyre2 builds a crude backbone model based on the template, then fills missing loops using a fragment database from known structures. Finally, side chains were built in their most probable rotamer while avoiding clashes. In order to improve the geometry of the model, two Ramachandran outliers were corrected in COOT55, and 20 cycles of structure idealization carried out in REFMAC556 after adding coordinates for Cu2+ based on the template structure. The final model had good geometry as evaluated using the Molprobity server57. The Cα RMSD between the Bim1 model and the template was 0.679 Å.
Expression of Bim1 in Pichia pastoris
RNA from an exponentially growing C. neoformans cell culture was isolated with the Qiagen RNeasy mini kit, and DNAse treated with the turbo-DNA free kit (Roche). cDNA was synthesized with the SuperScript® III First Strand Synthesis System (Invitrogen), using oligo (dT) nucleotides and following the manufacturer instructions. A PCR with OLSG-120 and OLSG-192 on C. neoformans cDNA was performed and the PCR product was EcoRI/NotI digested and cloned into EcoRI/NotI digested pPIC3.5k (DTP1932) (Invitrogen). The reverse primer (OLSG-192) was designed to generate the expression of Bim1 attached to a TEV protease cleaving site and a 6 Histidine tag (Supplementary Dataset 1). The resulting plasmid, was digested with SalI and transformed by electroporation in the P. pastoris KM71 strain following the Pichia expression kit (Invitrogen) electroporation and colony selection protocols. A P. pastoris 25 ml culture of a Bim1-TEV-6His expressing cells (DTY1022) were grown until saturation in pH 6 buffered BMGY medium. This preculture was diluted into a 1 l of pH 6 buffered BMGY medium and grown overnight. Cells were pelleted and transferred to 4 baffled flasks, with 1.6 l each of pH 6 buffered BMMY medium, containing 1% methanol. Buffered BMGY and BMMY medium were prepared as described in the Pichia expression kit (Invitrogen). Bim1-TEV-6xHis expression was induced for 4 days by adding 1% methanol to the cultures once a day (or 0.5% methanol twice a day, with no difference in the outcome). At day 4 cells were harvested and cleared supernatants were used for protein purification. 40 ml of nickel-nitrilotriacetic acid-agarose (Ni-NTA) slurry were distributed into three 2.5 × 20 cm glass chromatography columns (BioRad). The Ni-NTA slurry was washed with 50 ml of a buffer containing 50 mM phosphate buffer pH 8, 300 mM NaCl and 10 mM imidazole. The washed beads were added to three different flasks, each containing 2.1 l of the collected supernatants. The supernatants were kept shaking with a magnetic stirrer at room temperature for 1 h. After 1 h, supernatants and Ni-NTA beads were poured through three glass chromatography columns (as above) and then washed with 50 ml of washing buffer (50 mM sodium phosphate, pH 8, 300 mM NaCl and 40 mM imidazole). For eluting the protein from the beads, beads were incubated 1 × 15 min with elution buffer (50 mM sodium phosphate pH 8, 300 mM NaCl and 40 mM imidazole) followed by other 2 × 5 min incubations with elution buffer. The eluted fractions were collected by gravity flow in all cases. In order to remove the Histidine tag, the elution buffer was exchanged to TEV cleavage buffer (50 mM Tris-HCl pH 8, 150 mM NaCl) until reaching an imidazole concentration below 0.25 mM, by using Amicon® Ultra 15 ml centrifugal filter devices (Millipore) with a 10,000 NMWL. After buffer exchange, the protein was diluted to a 1 mg/ml, checked using the Pierce BCA protein concentration kit (Thermo Fisher), and incubated for 17 h with 0.5 U of Histidine tagged turbo TEV protease (Eton Bioscience) per μg of purified Bim1 in TEV cleavage buffer containing 1 mM final concentration of DTT. After the 17 h incubation, the protein-turbo TEV protease mix was further diluted to buffer concentrations of 50 mM Tris-HCl, pH 8, 150 mM NaCl, 0.2 mM DTT and 10 mM imidazole. This diluted sample was incubated for 1 h with previously washed Ni-NTA beads. After 1 h, the flow through from the beads, containing purified Bim1 without the His tag, was collected by gravity flow (the turboTEV protease and the removed His-tag from Bim1 remained stacked to the Ni-NTA beads). In order to N-deglycosylate Bim1, the previously collected flow through was buffer exchanged to 50 mM sodium acetate, pH 6 by using Amicon Ultra 15 ml centrifugal devices, as described above. Bim1 was diluted to 1 mg/ml in 50 mM sodium acetate buffer and incubated for 17 h at 25 °C with 0.02 U of Endo H (NEB) per μg of Bim1 protein. Immediately after deglycosylation, Bim1 was further purified by size exclusion chromatography on a Superdex-200 column (GE Healthcare) at a constant flow rate of 1.3 ml/min in 50 mM sodium acetate buffer pH 6 using an Akta FPLC (GE Healthcare) system. Fractions containing pure Bim1 were collected, pooled and concentrated.
X-band Electron Paramagnetic Resonance (EPR) and X-ray Absorption Spectroscopy (XAS)
Purified Bim1 was used for X-band EPR analysis and X-ray absorption spectroscopy. EPR data were acquired using quartz EPR tubes and a Bruker EMX spectrophotometer with the following parameters; frequency 9.44 GHz, modulation 0.4 mT, center field 3400, sweep width 3000, sweep time 30 s, receiver gain 30 dB, microwave power 10.02 mW. Samples contained a 0.4 mM solution of CuSO4 in the presence or absence of 0.5 mM Bim1 at pH 6.0 (50 mM sodium acetate, 30% (v/v) glycerol) and spectra were collected at liquid nitrogen temperature. Cu2+ stock solutions were prepared by dissolving copper sulfate (CuSO4, Sigma) in nano-pure water and standardized by EDTA titration in an ammonium buffer to a murexide end point. X-ray absorption data on two replicate samples of the C. neoformans lytic polysaccharide monooxygenase homologue Bim1 were measured at the Stanford Synchrotron Radiation Laboratory on beamline 9-3. Bim1 protein samples were 0.4 mM in Cu and prepared in 50 mM sodium acetate buffer at pH 6 with 30% glycerol added as a cryoprotectant. Samples were loaded into a lucite sample cell wrapped in Kapton tape before flash freezing in liquid nitrogen. The SSRL storage ring was operating at 3 GeV with a maximum current of 500 mA. Beamline 9-3 uses a Rh-coated focusing mirror for harmonic rejection and was equipped with Si(220) monochromator crystals for energy selection. Copper fluorescence was detected with a Canberra 100- or 30-element germanium detector. Samples were maintained at 8K in an Oxford liquid helium continuous flow cryostat. A 6-micron nickel filter with a Soller-slit assembly was placed between the cryostat window and the detector to reduce scatter. The energy was calibrated by simultaneously measuring the edge spectrum of a copper foil, with the energy of the rising inflection point being assigned as 8980.3 eV. Spectra represent the average of 4 scans for the 100 element detector data (each scan on a fresh area of the sample) and 11 scans for the 30 element detector data (each set of 2-3 scans on a fresh area of the sample). The data were fit over a k range of 1-12.8 Å-1 without the use of any Fourier filtering or smoothing. Data were analyzed using EXAFSPAK (http://www-ssrl.slac.stanford.edu/exafspak.html) interfaced with FEFF7.058 for theoretical phase and amplitude functions. Multiple scattering pathways were calculated using the coordinates of a histidine brace active site in H. jecorina (PDB 5O2W) as shown in Supplementary Figure 5c36. Double and single scattering paths less than 4.5 Å with three or two legs were included in the fits. This scattering unit was then used as the basis for modeling the data allowing R and σ2 to vary, where σ2 is the Debye Waller factor, the mean square deviation in R. Coordination numbers for histidine ring atoms were scaled by two to model a second histidine. ∆E0 refined to −11 eV for the primary Cu-N interaction in the Histidine ring but was held constant in the other paths. The goodness of fit was evaluated using the F factor (a least-square deviation between data and fit) corrected for the number of variable parameters used in the fit (http://www-ssrl.slac.stanford.edu/exafspak.html). The Cu-ligand distance, R, and the Debye-Waller factor were freely varied for the primary Cu-N interaction but the other pathways were linked to maintain the rigid model system’s geometry. Metric parameters for the representative fit shown in Figure 4f are given in Supplementary Figure 5c.
Bim1 activity
Bim1 activity was tested in a cellulose saccharification assay by comparing its activity to that of the archetypical Thermoascus aurantiacus cellulolytic LPMO59. Briefly, 2 mL microcentrifuge tubes were filled with 0.5mL 0.3% phosphoric acid swollen cellulose, 25 mM citrate phosphate buffer pH 6.0, 2mM ascorbic acid and 12μM CuCl2. The assay was started by adding either 10μM Bim1 or 10μM TaAA9A or no-enzyme as a control. Samples were incubated for 50 hours in a thermomixer at 40°C and shacked at 550 rpm. The samples were clarified on a 0.45 μm filter and subjected to anion exchange chromatography60.
Statistical analysis
For all data error bars represent statistical errors of the means (SEM) of results from a number of biological replicates (N), as indicated in figure legends. Before statistical analysis was conducted, data from all experiments was log transformed for comparison of proportions. Statistical analysis was performed with GraphPad Prism software v7. The statistical tests chosen for each experiment and their results (i.e. P values) are indicated in figure legends. Asterisks in figures correspond to statistical significance as follows: ****, P < 0.0001; ***, P = 0.0001 to P < 0.001; **, P = 0.001 to P < 0.01; *, P = 0.01 to P < 0.05; ns (not significant), P > 0.05.
Supplementary Material
Acknowledgements
This work was partially supported by funds from the United States National Institutes of Health (NIH) (GM041840, D.J.T; GM084176, K.J.F., GM127390, N.V.G.), the Welch Foundation I-1505 to N.V.G., a Postdoctoral Fellowship from German Research Foundation grant PR 1727/1-1 to C.P., a fellowship support from NIH (GM100678-02) to R.A.F., NIH Molecular Mycology and Pathogenesis Training program (5T32a1052080) to A.D.S., the Novo Nordisk Foundation Grant NNF17SA0027704 to K.S.J., travel support from the School of Science and Math at the College of Charleston to P.R.G., fellowship support from NIH (GM084146-S1) and Duke University BioCoRE (R25-GM103765) to S.E.C. We thank Jennifer Lodge (Department of Molecular Microbiology, Washington University School of Medicine, St. Louis, Missouri, USA) for providing the anti-Cda2 antibody, Yudong Song, and Michael Hoy for technical assistance, and Jean-Guy Berrin for sharing information prior to publication. Use of the Stanford Synchrotron Radiation Light source, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the NIH, National Institute of General Medical Sciences (including P41GM103393). We thank authors of works that could not be appropriately cited in this work due to space limiting constrictions. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Köhler JR, Casadevall A & Perfect J The Spectrum of Fungi That Infects Humans. Cold Spring Harbor Perspectives in Medicine 5, doi: 10.1101/cshperspect.a019273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maret W The Metals in the Biological Periodic System of the Elements: Concepts and Conjectures. Int J Mol Sci 17, doi: 10.3390/ijms17010066 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hood MI & Skaar EP Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol 10, 525–537, doi: 10.1038/nrmicro2836 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer LD & Skaar EP Transition Metals and Virulence in Bacteria. Annu Rev Genet 50, 67–91, doi: 10.1146/annurev-genet-120215-035146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerwien F, Skrahina V, Kasper L, Hube B & Brunke S Metals in fungal virulence. FEMS Microbiol Rev 42, doi: 10.1093/femsre/fux050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajasingham R et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17, 873–881, doi: 10.1016/S1473-3099(17)30243-8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR & Casadevall A Cryptococcus: from Human Pathogen to Model Yeast. (American Society of Microbiology, 2011). [Google Scholar]
- 8.Ding C et al. Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe 13, 265–276, doi: 10.1016/j.chom.2013.02.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun TS et al. Reciprocal functions of Cryptococcus neoformans copper homeostasis machinery during pulmonary infection and meningoencephalitis. Nat Commun 5, 5550, doi: 10.1038/ncomms6550 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Santamarina S et al. Genome-wide analysis of the regulation of Cu metabolism in Cryptococcus neoformans. Mol Microbiol 108, 473–494, doi: 10.1111/mmi.13960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding C et al. The copper regulon of the human fungal pathogen Cryptococcus neoformans H99. Mol Microbiol 81, 1560–1576, doi: 10.1111/j.1365-2958.2011.07794.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waterman SR et al. Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans. J Clin Invest 117, 794–802, doi: 10.1172/JCI30006 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladomersky E et al. Host and Pathogen Copper-Transporting P-Type ATPases Function Antagonistically during Salmonella Infection. Infect Immun 85, doi: 10.1128/IAI.00351-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner D et al. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol 174, 1491–1500 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Santamarina S, Uzarska MA, Festa RA, Lill R & Thiele DJ Iron-Sulfur Protein Biogenesis Machinery Is a Novel Layer of Protection against Cu Stress. MBio 8, doi: 10.1128/mBio.01742-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith AD, Logeman BL & Thiele DJ Copper Acquisition and Utilization in Fungi. Annu Rev Microbiol 71, 597–623, doi: 10.1146/annurev-micro-030117-020444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsberg Z et al. Polysaccharide degradation by lytic polysaccharide monooxygenases. Curr Opin Struct Biol 59, 54–64, doi: 10.1016/j.sbi.2019.02.015 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Arnesano F, Banci L, Bertini I, Mangani S & Thompsett AR A redox switch in CopC: an intriguing copper trafficking protein that binds copper(I) and copper(II) at different sites. Proc Natl Acad Sci U S A 100, 3814–3819, doi: 10.1073/pnas.0636904100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12, 7–8, doi: 10.1038/nmeth.3213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaaje-Kolstad G et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222, doi: 10.1126/science.1192231 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Quinlan RJ et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci U S A 108, 15079–15084, doi: 10.1073/pnas.1105776108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horn SJ, Vaaje-Kolstad G, Westereng B & Eijsink VG Novel enzymes for the degradation of cellulose. Biotechnol Biofuels 5, 45, doi: 10.1186/1754-6834-5-45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen KS Lytic Polysaccharide Monooxygenases: The Microbial Power Tool for Lignocellulose Degradation. Trends Plant Sci 21, 926–936, doi: 10.1016/j.tplants.2016.07.012 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Forsberg Z et al. Structural and Functional Analysis of a Lytic Polysaccharide Monooxygenase Important for Efficient Utilization of Chitin in Cellvibrio japonicus. J Biol Chem 291, 7300–7312, doi: 10.1074/jbc.M115.700161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsukihara T et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Walton FJ, Idnurm A & Heitman J Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol 57, 1381–1396, doi: 10.1111/j.1365-2958.2005.04779.x (2005). [DOI] [PubMed] [Google Scholar]
- 27.Askwith C et al. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76, 403–410 (1994). [DOI] [PubMed] [Google Scholar]
- 28.Dancis A et al. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell 76, 393–402 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Culotta VC et al. The copper chaperone for superoxide dismutase. J Biol Chem 272, 23469–23472 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Saikia S, Oliveira D, Hu G & Kronstad J Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infect Immun 82, 839–850, doi: 10.1128/IAI.01357-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferguson MAJ, Hart GW & Kinoshita T in Essentials of Glycobiology (eds rd et al.) 137–150 (2015). [Google Scholar]
- 32.Labourel A et al. A fungal family of lytic polysaccharide monooxygenase-like copper proteins (Nature Chemical Biology (submitted), 2019). [DOI] [PubMed] [Google Scholar]
- 33.Peisach J & Blumberg WE Structural implications derived from the analysis of electron paramagnetic resonance spectra of natural and artificial copper proteins. Arch Biochem Biophys 165, 691–708 (1974). [DOI] [PubMed] [Google Scholar]
- 34.Kau LS, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO & Solomon EI X-ray absorption edge determination of the oxidation state and coordination number of copper. Application to the type 3 site in Rhus vernicifera laccase and its reaction with oxygen. 109, 6433–6442, doi: 10.1021/ja00255a032 (1987). [DOI] [Google Scholar]
- 35.Vu VV, Beeson WT, Span EA, Farquhar ER & Marletta MA A family of starch-active polysaccharide monooxygenases. Proc Natl Acad Sci U S A 111, 13822–13827, doi: 10.1073/pnas.1408090111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansson H et al. High-resolution structure of a lytic polysaccharide monooxygenase from. J Biol Chem 292, 19099–19109, doi: 10.1074/jbc.M117.799767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westereng B, Arntzen M, Agger JW, Vaaje-Kolstad G & Eijsink VGH. Analyzing Activities of Lytic Polysaccharide Monooxygenases by Liquid Chromatography and Mass Spectrometry. Methods Mol Biol 1588, 71–92, doi: 10.1007/978-1-4939-6899-2_7 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Pope CR, Flores AG, Kaplan JH & Unger VM Structure and function of copper uptake transporters. Curr Top Membr 69, 97–112, doi: 10.1016/B978-0-12-394390-3.00004-5 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Ramos D et al. Mechanism of Copper Uptake from Blood Plasma Ceruloplasmin by Mammalian Cells. PLoS One 11, e0149516, doi: 10.1371/journal.pone.0149516 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stefaniak E et al. The N-terminal 14-mer model peptide of human Ctr1 can collect Cu(ii) from albumin. Implications for copper uptake by Ctr1. Metallomics 10, 1723–1727, doi: 10.1039/c8mt00274f (2018). [DOI] [PubMed] [Google Scholar]
- 41.Lawton TJ, Kenney GE, Hurley JD & Rosenzweig AC The CopC Family: Structural and Bioinformatic Insights into a Diverse Group of Periplasmic Copper Binding Proteins. Biochemistry 55, 2278–2290, doi: 10.1021/acs.biochem.6b00175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nimrichter L et al. Self-aggregation of Cryptococcus neoformans capsular glucuronoxylomannan is dependent on divalent cations. Eukaryot Cell 6, 1400–1410, doi: 10.1128/EC.00122-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brady D, Stoll AD, Starke L & Duncan JR Chemical and enzymatic extraction of heavy metal binding polymers from isolated cell walls of Saccharomyces cerevisiae. Biotechnol Bioeng 44, 297–302, doi: 10.1002/bit.260440307 (1994). [DOI] [PubMed] [Google Scholar]
- 44.Citiulo F et al. Candida albicans scavenges host zinc via Pra1 during endothelial invasion. PLoS Pathog 8, e1002777, doi: 10.1371/journal.ppat.1002777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamb AL, Torres AS, O'Halloran TV & Rosenzweig AC Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat Struct Biol 8, 751–755, doi: 10.1038/nsb0901-751 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Loose JS, Forsberg Z, Fraaije MW, Eijsink VG & Vaaje-Kolstad G A rapid quantitative activity assay shows that the Vibrio cholerae colonization factor GbpA is an active lytic polysaccharide monooxygenase. FEBS Lett 588, 3435–3440, doi: 10.1016/j.febslet.2014.07.036 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Chaudhuri S et al. Contribution of chitinases to Listeria monocytogenes pathogenesis. Appl Environ Microbiol 76, 7302–7305, doi: 10.1128/AEM.01338-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Connell RJ et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet 44, 1060–1065, doi: 10.1038/ng.2372 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
References Online Methods
- 49.Bailey TL et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–208, doi: 10.1093/nar/gkp335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen TN, Brunak S, von Heijne G & Nielsen H SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8, 785–786, doi: 10.1038/nmeth.1701 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Eisenhaber B, Bork P & Eisenhaber F Prediction of potential GPI-modification sites in proprotein sequences. J Mol Biol 292, 741–758, doi: 10.1006/jmbi.1999.3069 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Steentoft C et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 32, 1478–1488, doi: 10.1038/emboj.2013.79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilbert NM et al. KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol Microbiol 76, 517–534, doi: 10.1111/j.1365-2958.2010.07119.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJ The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858, doi: 10.1038/nprot.2015.053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murshudov GN et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67, 355–367, doi: 10.1107/S0907444911001314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21, doi: 10.1107/S0907444909042073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ankudinov AL & Rehr JJ Relativistic calculations of spin-dependent x-ray-absorption spectra. Physical Review B 56, R1712–R1716, doi: 10.1103/PhysRevB.56.R1712 (1997). [DOI] [Google Scholar]
- 59.Quinlan RJ et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci U S A 108, 15079–15084, doi: 10.1073/pnas.1105776108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westereng B et al. Efficient separation of oxidized cello-oligosaccharides generated by cellulose degrading lytic polysaccharide monooxygenases. J Chromatogr A 1271, 144–152, doi: 10.1016/j.chroma.2012.11.048 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.