Abstract
Tumor cells must alter their antioxidant capacity for maximal metastatic potential. Yet the antioxidant adaptations required for ovarian cancer transcoelomic metastasis, which is the passive dissemination of cells in the peritoneal cavity, remain largely unexplored. Somewhat contradicting the need for oxidant scavenging are previous observations that expression of SIRT3, a nutrient stress sensor and regulator of mitochondrial antioxidant defenses, is often suppressed in many primary tumors. We have discovered that this mitochondrial deacetylase is specifically upregulated in a context-dependent manner in cancer cells. SIRT3 activity and expression transiently increased following ovarian cancer cell detachment and in tumor cells derived from malignant ascites of high-grade serous adenocarcinoma patients. Mechanistically, SIRT3 prevents mitochondrial superoxide surges in detached cells by regulating the manganese superoxide dismutase SOD2. This mitochondrial stress response is under dual regulation by SIRT3. SIRT3 rapidly increases SOD2 activity as an early adaptation to cellular detachment, which is followed by SIRT3-dependent increases in SOD2 mRNA during sustained anchorage-independence. In addition, SIRT3 inhibits glycolytic capacity in anchorage-independent cells thereby contributing to metabolic changes in response to detachment. While manipulation of SIRT3 expression has few deleterious effects on cancer cells in attached conditions, SIRT3 up-regulation and SIRT3-mediated oxidant scavenging are required for anoikis resistance in vitro following matrix detachment, and both SIRT3 and SOD2 are necessary for colonization of the peritoneal cavity in vivo. Our results highlight the novel context-specific, pro-metastatic role of SIRT3 in ovarian cancer.
Introduction
Epithelial ovarian cancer (EOC) remains the most deadly gynecological malignancy, with a five year survival rate of less than 29% for patients diagnosed with advanced stage metastatic disease [1]. During transcoelomic spread the peritoneal fluid facilitates the dissemination of detached ovarian cancer cells to peritoneal cavity organs, including the omentum [2, 3]. This is evident by malignant ascites accumulation in most advanced stage patients [3]. Thus evasion of anchorage-independent cell death (anoikis) upon detachment from the primary tumor is a likely critical step for ovarian cancer metastasis [4]. A feature of anoikis is the surge in oxidative stress elicited by cell detachment. Adaptations to oxidative stress are therefore necessary for successful hematological metastatic spread, as demonstrated in melanoma and breast cancer [5–7]. Moreover, administration of compounds that facilitate oxidant scavenging, such as N-acetyl-cysteine, can promote metastasis in vivo [8]. However, it remains largely unexplored if adaptations to oxidative stress are required by ovarian cancer cells for successful transcoelomic metastasis.
Contradicting the need of tumor cells for oxidant scavenging is the observation that expression of the nutrient stress sensor and regulator of mitochondrial antioxidant defenses, the Sirtuin deacetylase SIRT3 [9–12], is suppressed in many primary tumors [13–17]. Moreover, several studies have demonstrated that SIRT3 knock-down promotes proliferation and tumorigenesis in tumor models of breast [12, 18], mantle cell lymphoma [19] and liver cancer [16], promoting investigators to initially characterize SIRT3 as a tumor suppressor. However, it is becoming increasingly clear that the role of SIRT3 in tumor biology is complex [17, 20, 21]. Pro-tumorigenic properties of SIRT3 have conversely been reported in oral squamous cell carcinoma [22], diffuse large B cell lymphoma [23], and colorectal cancer [24], with increased SIRT3 expression being associated with poor outcome in colon and non-small cell lung cancer patients [17]. In addition, SIRT3 promotes glioblastoma multiforme (GBM) stem cell viability [25], and is an important component of the mitochondrial unfolded protein response (mtUPR) necessary for breast cancer metastasis [26, 27]. The latter function of SIRT3 is being attributed to its role as a regulator of the antioxidant response required for tumor cell survival and metastasis.
Although, previous reports have demonstrated that SIRT3 exerts anti-proliferative and anti-migratory effects on ovarian cancer cells [28, 29], the role of SIRT3 during ovarian cancer transcoelomic spread has not been investigated. Moreover, when and where SIRT3 is expressed during tumor progression remains unknown. We discovered that SIRT3 is upregulated in a context-dependent manner in ovarian cancer cells, and indeed has a specific pro-metastatic role, by supporting anchorage-independent survival. While SIRT3 expression is low in primary ovarian tumors and knock-down of its expression has no deleterious consequences in attached proliferating conditions, we demonstrate that SIRT3 activity and expression are specifically induced in response to anchorage-independence, and that this transient increase results in the activation of the mitochondrial antioxidant SOD2, which is necessary for anchorage-independent survival and peritoneal colonization in vivo. These findings provide important evidence of the function of SIRT3 in ovarian cancer, and clarify several contradictory findings associated with the role and expression of SIRT3 in cancer progression.
Results
SIRT3 expression increases in a context-dependent manner during ovarian cancer metastasis and is induced in response to matrix detachment
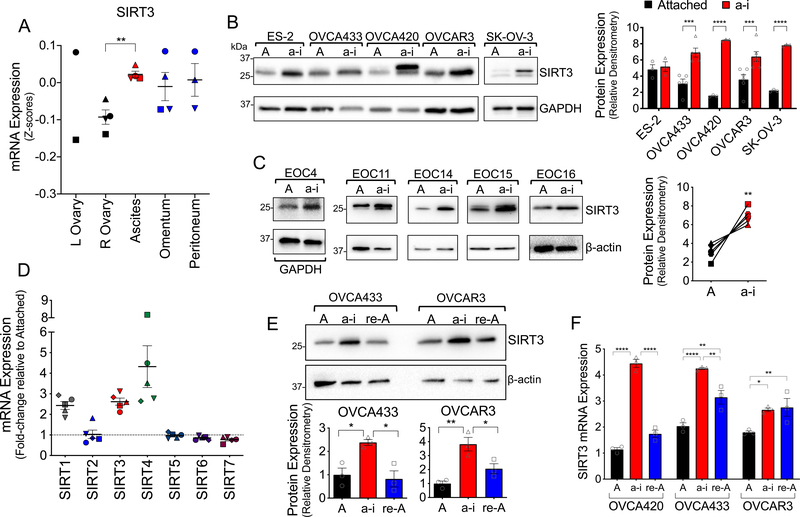
Expression analysis of primary ovarian tumors and matching cells derived from malignant ascites, and omental and peritoneal metastatic lesions from a publicly available data set (GEO:GSE85296) revealed that SIRT3 levels are heterogeneous in tissues representing different stages of transcolomic metastatic spread (Fig. 1A). While SIRT3 levels were lowest in primary ovarian tumors, highest SIRT3 expression was found in cells derived from malignant ascites. In two of four patient samples, the increase in SIRT3 expression was maintained in omental and peritoneal metastatic lesions, while in the other cases SIRT3 expression reverted to levels observed in primary tumors of the ovary. These data suggest that detachment induces transient increases in SIRT3 expression in malignant ascites.
Figure 1: Ovarian cancer SIRT3 expression is context-dependent, and increases in response to anchorage-independence.
A. SIRT3 mRNA expression in tumor samples from primary tumors of the ovary, matching ascites, and omental and peritoneal metastatic lesions (Geo:GSE85296, n=4, mean expression at each site per patient shown, repeated measures ANOVA P = 0.047, Dunnett’s multiple comparison test **P = 0.005). B. Ovarian cancer cell lines were cultured in anchorage-independent (a-i) conditions using ultra low attachment (ULA) plates for 72 h, and SIRT3 protein expression compared to cell cultures in attached (A) conditions using western blotting. Densitometry of SIRT3 bands is expressed relative to loading control GAPDH, and levels in a-i compared to attached conditions for each cell line (n=3–6; t-test ***P<0.001, ****P<0.0001). C. Epithelial ovarian cancer cells (EOCs) were derived from ascites of Stage III and IV high grade serous adenocarcinoma patients and cultured in attached or a-i conditions for 72 h. SIRT3 expression was assessed as in B (n=5, paired t-test **P=0.07). D. mRNA levels of sirtuin family members were assessed using U133 microarray after EOCs derived from patient ascites (n=5) were cultured in a-i conditions for 72 h as described in [47]. mRNA levels are expressed relative to cells grown in attached conditions. E. SIRT3 protein expression was assessed by western blotting of lysates from attached cell cultures (A), cells maintained for 24 h in ULA plates (a-i) and 24 hours following re-attachment (re-A). Densitometry of SIRT3 bands is expressed relative to loading control actin and normalized to attached conditions (n=3; one-way ANOVA, Tukey’s multiple comparison test *P<0.05, **P<0.01). F. SIRT3 mRNA expression was assessed by semi-quantitative real time RT-PCR following cell culturing as in E, and expressed relative to OVCA420 in attached conditions (n=3; two-way ANOVA, Tukey’s multiple comparison test *P<0.05, **P<0.001, ****P<0.0001).
We next tested if a loss of matrix attachment induces SIRT3 expression. Consistently, we found that SIRT3 expression increased when ovarian cancer cell lines (Fig. 1B), and ascites-derived primary epithelial ovarian cancer cells (EOCs, Fig. 1C) from Stage III and IV high grade serous adenocarcinoma patients were cultured under anchorage-independence in ultra-low attachment (ULA) dishes. RNA expression analysis of EOCs from a separate patient cohort revealed that SIRT3, SIRT1 and SIRT4 were the only members of the sirtuin gene family responsive to mRNA increases when maintained in anchorage-independent conditions (Fig. 1D). Interestingly, the transient increase in SIRT3 expression observed in patient ascites (Fig. 1A) could be recapitulated in 2/3 cell lines tested, where SIRT3 expression reverted to basal levels following cell re-attachment (Fig. 1E&F). These data highlight that SIRT3 expression in ovarian cancer is transient, and induced in a context-dependent manner in response to matrix detachment.
SIRT3 regulates mitochondrial reactive oxygen species scavenging by activating SOD2 in anchorage-independence
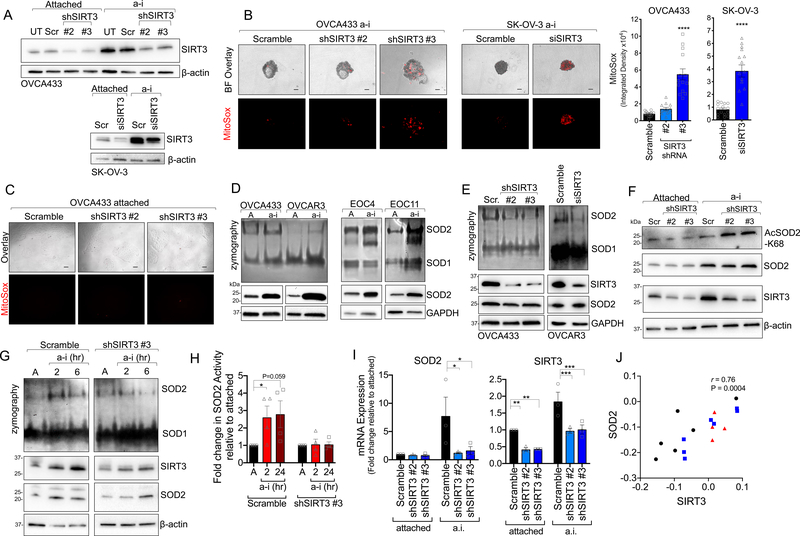
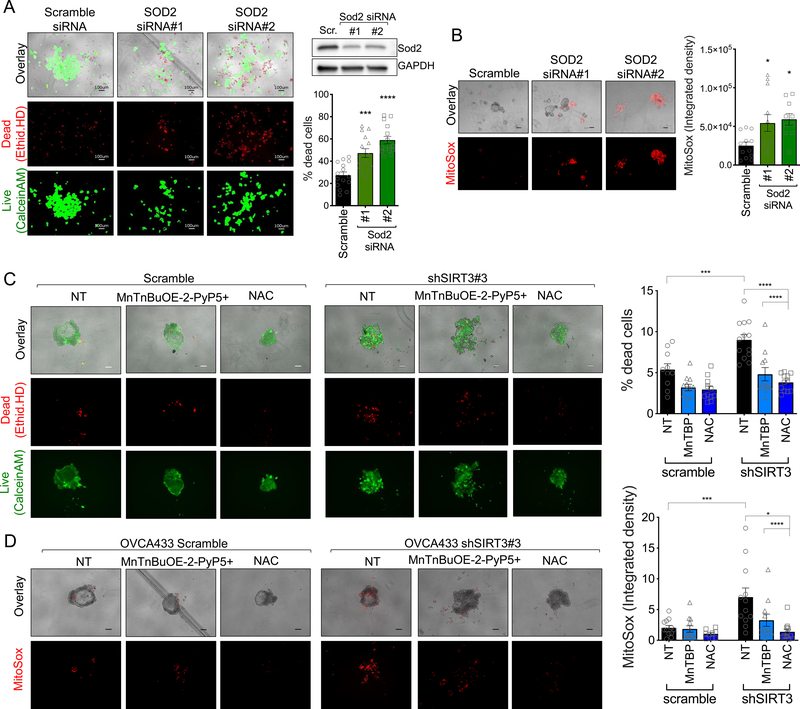
Matrix detachment elevates cytosolic [6] and mitochondrial reactive oxygen species [30], which are thought to contribute to anoikis of non-transformed epithelial cells [6]. To determine if SIRT3 protects cells from mitochondrial redox stress in anchorage-independence, we monitored MitoSox fluorescence in response to SIRT3 knock-down (Fig. 2). Inhibition of SIRT3 expression, using several independent si/shRNAs (Fig. 2A & Supp. Fig. 1A), increased MitoSox fluorescence in anchorage-independent conditions (Fig. 2B), indicating that SIRT3 is necessary to maintain low mitochondrial superoxide (O2.−) levels following detachment. Interestingly, SIRT3 knock-down had no effect on MitoSox fluorescence in attached conditions (Fig. 2C), suggesting that SIRT3 has a specific role in protecting against mitochondria oxidant stress during anchorage-independence.
Figure 2: SIRT3 maintains superoxide (O2.−) scavenging in anchorage-independent conditions by activating SOD2.
A. SIRT3 expression was inhibited using stable shRNA transfection of OVCA433 cells or transient delivery of siRNA in SK-OV-3 (UT, un-transfected; Scr, scramble control). B. SIRT3 knock-down increases oxidation and fluorescence of the mitochondrial O2.− probe MitoSox in ULA cultured OVCA433 (A) and SK-OV-3 (B) ovarian cancer cells. Quantification of MitoSox signal (n=12–14 ± SEM; OVCA433: one-way ANOVA P<0.0001, Tukey’s multiple comparisons test ****P<0.0001, **P=0.001; SK-OV-3: unpaired t-test ****P<0.0001, scale bar = 100μm). C. SIRT3 knock-down has no effect on mitochondrial O2.− in attached culture conditions, as assessed by MitoSox staining. D. Ovarian cancer cell lines and patient ascites-derived epithelial ovarian cancer cells (EOC) were cultured in anchorage-independent (a-i) conditions for 72 h using ultra low attachment (ULA) plates. SOD2 activity was assessed by in gel zymography and compared to cell cultures in attached (A) conditions. E. shRNA and siRNA-mediated SIRT3 knock-down inhibits SOD2 activity in ULA cultured (72 h) ovarian cancer cells, assessed by SOD zymography. F. SIRT3 knock-down increases SOD2 acetylation at lysine 68 in OVCA433 cells cultured in a-i conditions. G. SOD2 activity is induced within 2 h of matrix detachment in a SIRT3-dependent manner. SOD2 activity was assessed by zymography in attached (A) OVCA433 cells and cells cultured for 2 or 6 h in anchorage-independence (a-i). H. SOD2 activity, assessed by an in vitro SOD activity assay, increases in scramble transfected OVCA433 cells cultured for 2 and 24 h in a-i, while SIRT3 knock-down inhibits this a-I induced SOD2 activity (n=4 ± SEM; *P<0.05). I. SIRT3 knock-down decreases SOD2 mRNA levels in a-i. mRNA expression was assessed by semi-quantitative real time RT-PCR following cell culturing in ULA plates for 24 h. Data expressed relative to expression in scramble transfected cells in attached conditions (n=3; two-way ANOVA, Dunnett’s multiple comparison test *P<0.05, **P<0.01, ***P<0.001). J. Positive correlation between SIRT3 and SOD2 mRNA expression in tumor tissues derived from primary ovarian tumors ( ), ascites (
), ascites ( ), and peritoneal or omental lesions (
), and peritoneal or omental lesions ( ; Geo:GSE85296, Pearson correlation).
; Geo:GSE85296, Pearson correlation).
A major antioxidant target of SIRT3 is manganese superoxide dismutase 2 (SOD2), which is one of three superoxide dismutases in the cell, and the primary enzyme responsible for the dismutation of O2.− to hydrogen peroxide (H2O2) in the mitochondrial matrix. SIRT3 regulates SOD2 at both the transcriptional level, via deacetylaton and activation of the SOD2 transcription factor FOXO3a [26, 31], and by directly deacetylating and activating SOD2 dismutase activity [9–12]. Concomitant to SIRT3 increases, SOD2 activity and expression were strongly induced in response to detachment of ovarian cancer cell lines and patient ascites-derived cells (Fig. 2D), indicating that the SIRT3/SOD2 axis is an important adaptation for anchorage-independence. SIRT3 was directly responsible for enhanced SOD2 activity in detached cells, as evident by SIRT3 sh/siRNA mediated knock-down (Fig. 2E). This was accompanied by an increase in SOD2 acetylation at lysine 68, specifically in anchorage-independent conditions (Fig. 2F). We observed that increased SOD2 activity is an early response to matrix detachment, and that SOD2 activity rapidly increased within 2 hours of matrix detachment, prior to detectable changes in SOD2 mRNA transcript levels (Fig. 2G & H). Importantly, SOD2 activity in both short (2 h) and sustained anchorage-independence (24 h) were dependent on SIRT3, as cells with decreased SIRT3 expression failed to increase SOD2 activity in response to anchorage-independence (Fig. 2H).
In addition to early increases in SOD2 activity, we observed increased SOD2 mRNA expression after 6 hours following matrix detachment, and this was also significantly abrogated by SIRT3 knock-down (Fig. 2I). Notably, transcript levels of SIRT3 in patient specimens from GEO data set GSE85296 strongly correlated with SOD2 levels, where high expression was similarly observed in ascites-derived cells (Fig. 2J). These data demonstrate that the mitochondrial O2.− scavenger SOD2 is dually regulated by SIRT3 following detachment, and that the SIRT3/SOD2 axis is an early adaptation to anchorage-independence.
SIRT3 knock-down increases glycolysis in anchorage-independent cells
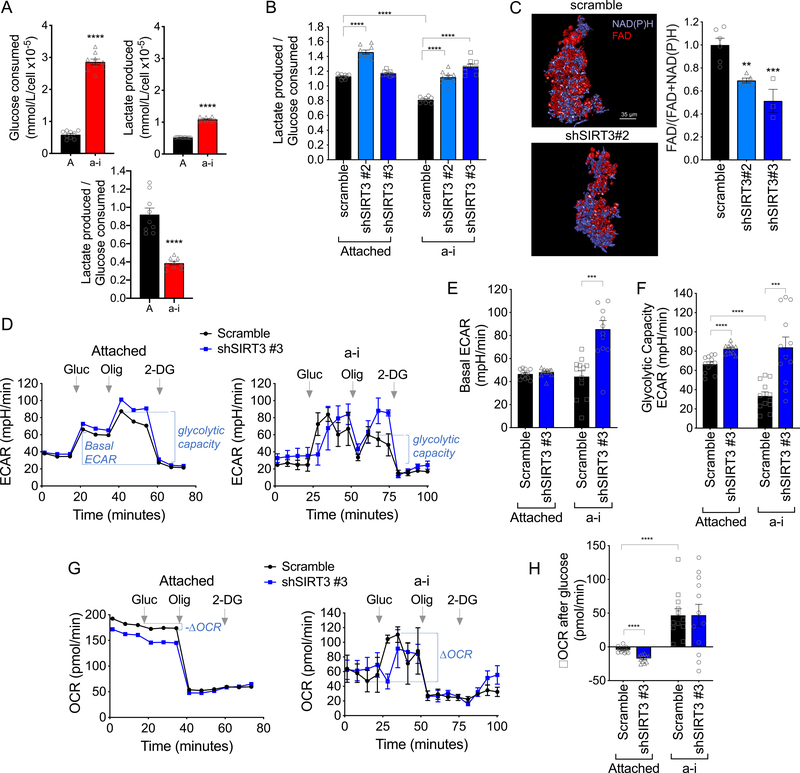
Given that SIRT3 has previously been associated with suppression of glycolysis [18, 32], we set out to determine if increased SIRT3 expression alters glycolytic flux following cellular detachment. First, we determined if glucose consumption and lactate production are altered in in detached cells, and found that overall glucose consumption was increased, while relative lactate production to glucose consumption was decreased, compared to attached cells (Fig. 3A). This suggests that cells in anchorage-independence re-route glucose consumption away from lactic acid production. SIRT3 knock-down significantly increased the ratio of lactate production to glucose consumption in detached cells (Fig. 3B & Supp. Fig. 2), and assessment of the optical redox ratio of the metabolic coenzymes FAD and NAD(P)H using multiphoton imaging [33] demonstrated a significant decrease in the FAD / [FAD + NAD(P)H] ratio with SIRT3 knock-down in anchorage-independent cells (Fig. 3C). To test if the above results indicate that increased SIRT3 expression switches glucose utilization away from lactate production towards Oxidative Phosphorylation, we examined changes in extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) using extracellular flux analysis. As expected, addition of glucose rapidly increased ECAR in both attached and detached cells (Fig. 3D). SIRT3 knock-down significantly increased basal glycolytic rate (basal ECAR) in detached cells following glucose addition, while it had no significant effect on basal ECAR in attached conditions (Fig. 3E). While cells in anchorage-independence displayed similar basal glycolytic rate, their maximal glycolytic capacity, stimulated by suppression of respiration with the mitochondrial ATP synthase inhibitor Oligomycin A, was significantly decreased compared to attached cells. Moreover, this was reversed when SIRT3 expression was suppressed (Fig. 3F). These data demonstrate that SIRT3 suppresses glycolytic capacity of tumor cells in anchorage-independent conditions.
Figure 3: SIRT3 suppresses the glycolytic capacity of anchorage-independent (a-i) cells.
A. OVCA433 a-i cultured cells (24 h) consume more glucose per cell compared to attached cells, but produce less lactate relative to glucose consumed (n=9 ± SEM; unpaired t-test ****P<0.0001). B. SIRT3 knock-down shifts OVCA433 cells towards enhanced lactate production relative to glucose consumption (n=9 ± SEM; one-way ANOVA P=0.01, Tukey’s multiple comparisons test ****P<0.0001). C. The optical redox ratio FAD / FAD + NAD(P)H is decreased in a-i conditions following SIRT3 knock-down in OVCA433 (n=3–5; unpaired t-test *P=0.04). D. Extracellular Acidification Rates (ECAR) were measured in attached and a-i OVCA433 cells using a Seahorse XFp extracellular flux analyzer. One representative experiment shown (n=3). 10mM glucose (Gluc), 1 μM Oligomycin A (Oligo) and 50mM 2-Deoxyglucose (2-DG) were added at indicated times. E. & F. SIRT3 knock-down increases basal ECAR/Glycolysis in response to glucose addition (E), and Glycolytic Capacity stimulated by Oligomycin A (F). G. Oxygen Consumption rate (OCR) was monitored simultaneously as in D. H. Attached cells display a decrease in OCR following glucose addition, which is further decreased with SIRT3 knockdown. Conversely, cells in a-i significantly increase their OCR following glucose addition. (E, F & H, n=12; ***P<0.001, *****P<0.0001)
As expected OCR was inhibited in response to glucose addition in attached conditions, as cells use glucose primarily for lactic acid production (Fig. 3G). Interestingly, extracellular flux analysis confirmed that detached cells do not utilize glucose primarily for lactate production, and also increase their oxygen consumption in response to glucose addition (Fig. 3G&H). While OCR was further suppressed by SIRT3 knock-down in attached cells, large variability in OCR readings between experimental replicates was unable to ascertain if SIRT3 directly inhibits Oxidative Phosphorylation in detached conditions.
SIRT3-dependent oxidant scavenging is necessary for ovarian cancer anchorage-independent survival
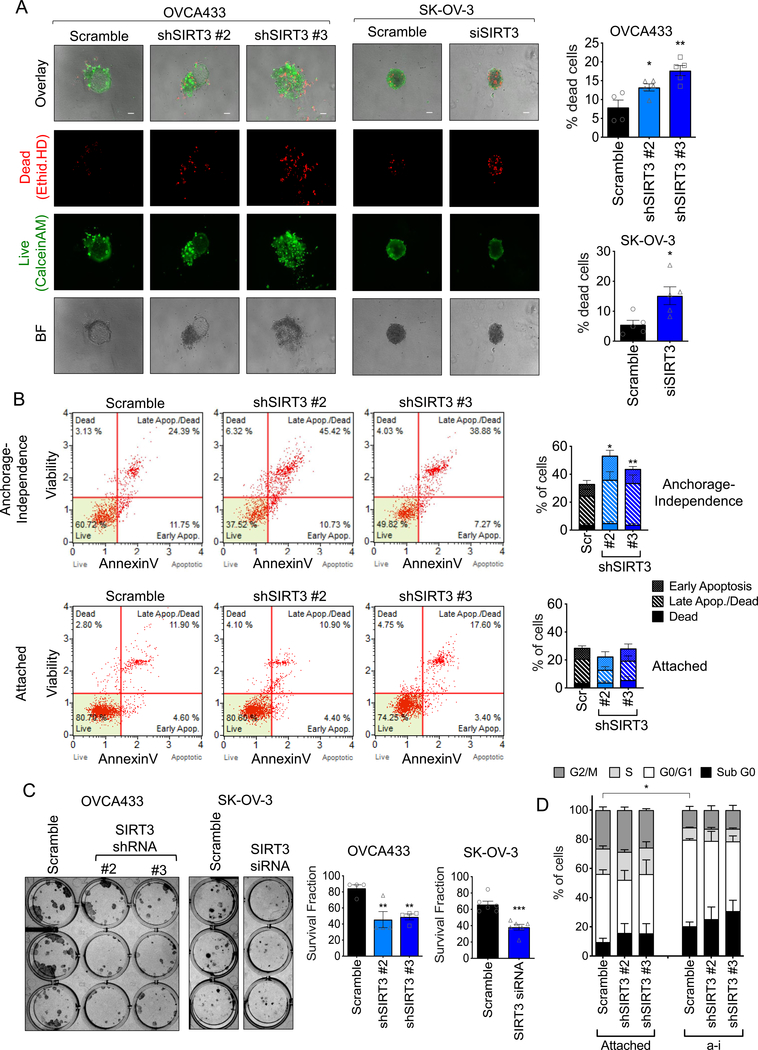
Since we found that SIRT3 is specifically increased upon cell detachment, we tested the necessity of SIRT3 for anchorage-independent survival. Knock-down of SIRT3 significantly increased the fraction of dead cells when OVCA433, SK-OV-3 and OVCAR3 cells were cultured in anchorage-independent conditions (Fig. 4A, Supp. Fig. 3A). This was accompanied by an inability of cells to rapidly aggregate into spheroid clusters within 6 hours following detachment (Supp. Fig. 3B), and resulted in loosely aggregated OVCA433 cells, and smaller and less uniform SK-OV-3 spheroids by 72 hours of anchorage-independence (Fig. 4A). Loss of SIRT3 expression increased the fraction of apoptotic cells following matrix detachment, while there was no effect of SIRT3 knock-down on apoptosis in attached conditions, suggesting that SIRT3 is inhibitory to anoikis (Fig. 4B). A pro-survival role for SIRT3 was also observed in single cell clonogenic assays. SIRT3 knock-down significantly inhibited colony number, but not average colony size, indicating that SIRT3 knock-down inhibits initial single cell survival and seeding, rather than proliferation (Fig. 4C). Accordingly, shRNAs to SIRT3 had no significant impact on cell cycle progression (Fig. 4D, Supp. Fig. 3C). Cells in detached conditions had fewer cells in S and G2/M phases of the cell cycle, suggesting that these cells slow their proliferation, as previously demonstrated [34], however this was unaffected by SIRT3 knock-down (Fig. 4D). These data demonstrate that SIRT3 is necessary for survival under anchorage-independence by inhibiting anoikis.
Figure 4: SIRT3 expression is required for anchorage-independent ovarian cancer cell survival.
A. SIRT3 knock down increases the dead cell fraction of cells in anchorage-independent (a-i) spheroid aggregates, when cultured in ULA plates for 72 h. Cells were stained with Ethidium homodimer (dead cells) and Calcein AM (live cells) and fractions of live and dead cells quantified (n=4; OVCA433: one-way ANOVA P=0.002, Tukey’s multiple comparison test *P=0.04, **P=0.001; SK-OV-3: unpaired t-test *P=0.019; scale bar = 100μm). B. SIRT3 knock-down increases the apoptotic fraction of cells cultured for 24 h in anchorage-independence, but not in attached conditions. Apoptosis was assessed by Annexin V (n=4–5 experimental replicates; repeated measures ANOVA of total dead cell fraction, a-i: P=0.008, Attached: not significant; Bonferroni’s multiple comparison test *P<0.05, **P<0.01). C. SIRT3 knock-down inhibits single cell clonogenic survival (n=4–6; OVCA433: one-way ANOVA P=0.005, Tukey’s multiple comparison test **P<0.01; SK-OV-3: unpaired t-test *P=0.0004). D. SIRT3 knock-down does not significantly affect cell cycle progression in either attached or a-i cultured OVCA433 cells. (a-i, 24 h, n=3 experimental replicates; Two-way ANOVA, Tukey’s multiple comparisons test *P=0.036, comparison of G0/G1).
As expected, SOD2, the target of SIRT3, was similarly necessary for anchorage-independent survival. SOD2 knock-down significantly increased the dead cell fraction and mitochondrial O2.− levels of detached ovarian cancer cells (Fig. 5A&B, Supp. Fig. 4). Cell death and MitoSox fluorescence induced by SIRT3 knock-down was successfully rescued with a mitochondrial SOD2 porphyrin mimetic MnTnBuOE-2-PyP5+ and the glutathione precursor N-acetyl-L-cysteine (NAC; Fig. 5C&D, Supp. Fig. 5), indicating that SIRT3-mediated oxidant scavenging is an important pro-survival mechanism for ovarian cancer cells following detachment.
Figure 5: SOD2 and oxidant scavenging are required for anchorage-independent survival.
A. SOD2 knock-down increases the dead cell fraction of OVCA433 cells in anchorage-independent (a-i) spheroid aggregates, when cultured in ULA plates for 72 h. Cells were stained with Ethidium homodimer (dead cells) and Calcein AM (live cells) and fractions of live and dead cells quantified (n=15; one-way ANOVA P<0.0001, Tukey’s multiple comparison test ***P=0.0003, ****P<0.0001). SOD2 expression was inhibited using siRNA in OVCA433 cells (Scr, scramble control) B. SOD2 knock-down increases oxidation and fluorescence of the mitochondrial O2.− probe MitoSox in ULA cultured OVCA433 cells (n=10–12; one-way ANOVA P=0.01, Tukey’s multiple comparisons test *P<0.05). C. Co-treatment of cells with 10μM MnTnBuOE-2-PyP5+ or 2mM NAC rescues OVCA433 cell viability following siRNA mediated SIRT3 knock-down; and D. results in decreased mitochondrial MitoSox oxidation (72 h a-i; n=12–15; ± SEM; OVCA433: one-way ANOVA P<0.0001, Tukey’s multiple comparisons test ****P<0.0001, **P=0.001; SK-OV-3: unpaired t-test ****P<0.0001).
SIRT3 and SOD2 are necessary for metastatic colonization of the peritoneal cavity
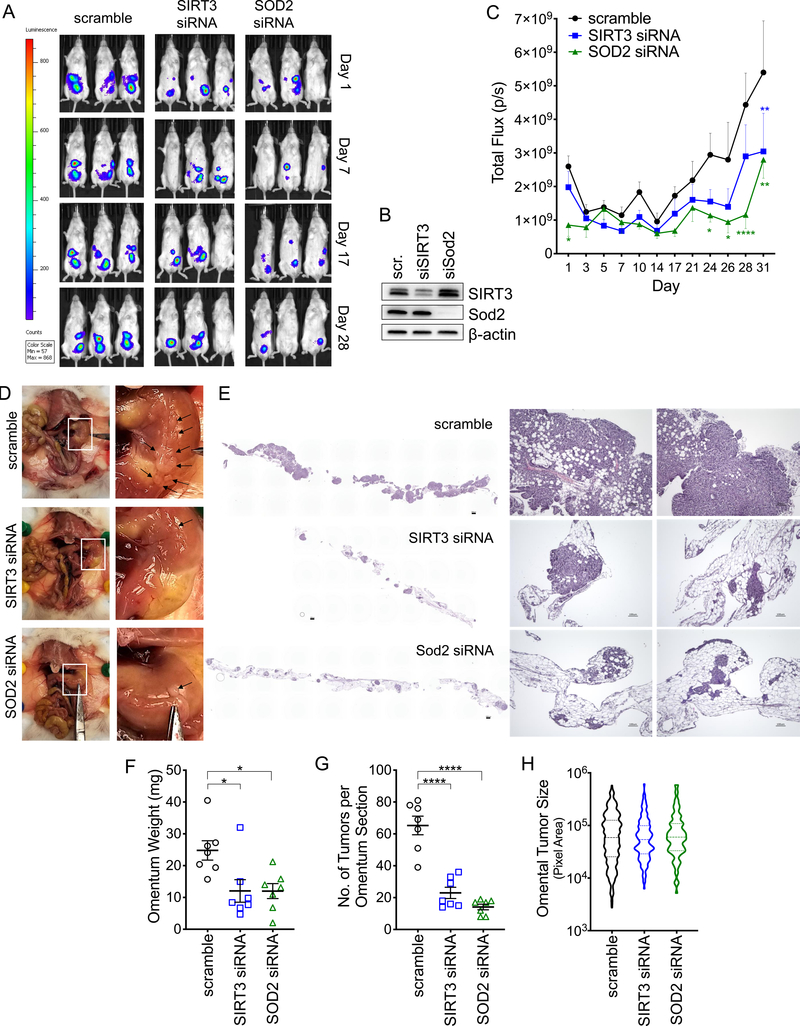
Since anchorage-independent survival is a critical step for successful transcoelomic metastasis of ovarian cancer cells in the peritoneal cavity [3, 4], we tested if the increases in SIRT3 and SOD2 specific to detachment are necessary for peritoneal tumor formation in vivo. To transiently decrease SIRT3 and SOD2 expression during the anchorage-independent phase, SK-OV-3-luc cells were transfected with siRNAs targeting either SIRT3, SOD2 or a scramble control. Twenty-four hours later cells were detached and 1×106 viable cells suspended in 150 μl PBS immediately injected into the peritoneal cavity of NSG mice (n=8), and tumor establishment monitored by bioluminescence imaging. Either SIRT3 or SOD2 knock-down significantly inhibited peritoneal tumor formation over time (Fig. 6A–C). Assessment of the omentum, a major target tissues of metastatic ovarian cancer [35], and a site where the majority of SK-OV-3 tumors were detected, revealed that control SK-OV-3 tumor cells replaced most of the adipocytes in this tissue. SIRT3 knock-down, and to a greater extent SOD2 knock-down, resulted in the establishment of fewer tumor nodules in the omentum, suggesting that the loss of these proteins leads to reduced seeding of viable tumor cells, likely as a consequence of increased anoikis during dissemination (Fig. 6D–H). The size of individual tumor nodules was not significantly different and varied widely in all experimental groups, although a trend in larger tumors was observed from control scramble siRNA transfected SK-OV-3 tumor cells (Fig. 6H). The above data demonstrate that SIRT3 and SOD2 are necessary for successful transcoelomic tumor formation in vivo.
Figure 6: SIRT3 and SOD2 are required for successful metastasis to the omentum.
A. Representative tumor luminescence images of NSG mice injected with SK-OV-3-luciferase cells transfected with either scramble siRNA, siRNA targeting SIRT3 or SOD2. B. Western blot demonstrating knock down of SIRT3 and SOD2 three days after transfection. C. Quantification of whole animal tumor luminescence over time (n=7–8; Mixed-design ANOVA P=0.004, Dunnett’s multiple comparisons test *P < 0.05, **P < 0.01, ****P < 0.0001). D. Assessment of the peritoneal cavity revealed that the majority of SK-OV-3 tumors (black arrows) were localized to the omentum. E. H & E staining demonstrates that SIRT3 and SOD2 knock down abrogate tumor burden in the omentum. F. SIRT3 and SOD2 knock down decreases omental weight and G. number of tumors per longitudinal section of omentum assessed by H & E (One way ANOVA, F: P = 0.01, G: P < 0.0001; Tukey’s multiple comparisons test *P < 0.05, P < 0.0001). H. Variability in individual omental tumor sizes between experimental groups (violin plot, median + interquartile range).
Discussion
Here, we present new evidence for the pro-metastatic role of SIRT3 in cancer, and demonstrate that the mitochondrial SIRT3/SOD2 stress response pathway is specifically upregulated in response to cellular detachment (Fig. 1), and specifically necessary for anchorage-independent survival and transcoelomic metastasis (Fig. 4&6).
Similar to several cancers previously reported [13–17], the expression of SIRT3 is low in primary tumors of high-grade serous adenocarcinoma samples from the Cancer Genome Atlas (TCGA). However, contrary to other tumor types, low SIRT3 expression was not associated with a decreased in overall patient survival (Supp. Fig. 6). The observation that a decrease in SIRT3 expression does not predict an unfavorable patient outcome somewhat contradicts previous reports of the anti-tumorigenic effects of SIRT3 in ovarian cancer cells [28, 29], and highlights that SIRT3 likely plays a dichotomous role during the progression of ovarian cancer. The context-dependent regulation of SIRT3 during ovarian cancer metastatic progression is evident by our findings that SIRT3 expression is significantly increased in cells derived from patient ascites, compared to matching primary ovarian tumor tissue (Fig. 1). We propose that this transient regulation addresses the seeming contradiction between SIRT3’s anti-proliferative properties and low primary tumor expression, and the role of SIRT3 as an important mitochondrial stress response gene important for cell survival.
Few studies have examined the context-specific regulation and function of SIRT3 in cancer. Most work demonstrating the anti-tumor role of SIRT3 was based on discoveries that SIRT3 expression is downregulated in a number of primary tumors, and that this decrease is associated with poor patient survival [13, 14, 16, 17]. The functional consequences of SIRT3 loss were mainly assessed in attached cell culture conditions or in vivo models testing the role of SIRT3 on primary tumor growth, without assessment of its function in metastatic progression [12, 18, 19]. However, our work and that of others is starting to unravel the complex role and regulation of SIRT3 during cancer progression. While we observed that manipulation of SIRT3 expression has few deleterious effects in attached cells, SIRT3 and its downstream target, the mitochondrial O2.− scavenger SOD2 were required for survival following matrix detachment (Fig. 4&5), and this was necessary for successful peritoneal tumor formation in vivo (Fig. 6). Importantly, we found that the SIRT3-dependent increase in SOD2 activity is an early response to matrix detachment and sustained under long term anchorage-independence through SIRT3-mediated SOD2 transcript regulation (Fig. 2), likely as a consequence of SIRT3-dependent FOXO3a deacetylation [26, 31]. Concurrent with our findings, SIRT3 has been implicated in anoikis resistance of oral squamous cell carcinoma cells [36], and for the maintenance of mitochondrial ROS scavenging in GBM stem cells [25]. In addition, the SIRT3/FOXO3a/SOD2 axis is upregulated as part of the mtUPR and necessary for breast cancer metastasis [26, 27, 37]. Moreover, we and others have demonstrated that SOD2 is required by metastatic cells as an adaptation to oxidative stress, for the maintenance of mitochondrial fidelity, and as a regulator of mitochondria redox signaling [20, 38–40].
Highlighting the dichotomous role of SIRT3 in cancer is the finding that the SIRT3-dependent regulation of SOD2 is also a previously described mechanism of SIRT3’s anti-tumor activity. We find that mitochondrial oxidant scavenging is necessary for anchorage-independent survival, and that both SIRT3 and SOD2 are required for this (Fig. 4&5). Conversely, increased oxidant production in Sirt3 knock-out mice was shown to induce tumor formation when Sirt3−/− MEFs were transformed with oncogenes Ras and Myc, as a result of increased DNA oxidation [12]. In addition, the anti-tumor effects of SIRT3 were demonstrated to be related to manipulation of tumor metabolism via SIRT3 inhibition of HIF-1α [18, 32]. The increase in mitochondrial O2.− levels in SIRT3 knock-down cells was shown to result in HIF-1α protein stabilization as a consequence of prolyl-hydroxylase inhibition, and the increased glycolytic phenotype of SIRT3 knock-down cells associated with this ROS-mediated increase in HIF-1α-signaling [18, 32]. While we similarly found that SIRT3 suppresses glycolysis, increased HIF-1α stabilization with SIRT3 loss was a phenotype limited to attached conditions (Supp. Fig. 7). HIF-1α levels remained unaltered following SIRT3 knock-down in anchorage-independence, indicating that the effects of SIRT3 on glycolysis are not coupled to HIF-1α suppression in this state of tumor metastasis. Several other metabolic enzymes are known SIRT3 targets. For example, a recent study showed that SIRT3 is important for the survival of Diffuse Large B cell Lymphoma cells by promoting glutaminolysis through regulation of glutamine dehydrogenase [23]. In addition, activation of IDH2 by SIRT3-dependent deacetylation can shift metabolism from glycolysis to oxidative phosphorylation [41], and the presence of IDH1 and IDH2 is important for survival of anchorage-independent cells by aiding in regeneration of NADPH for glutathione reduction [30]. Moreover, SIRT3 is a known activator of pyruvate dehydrogenase (PDH), resulting in enhance shuttling of pyruvate into the TCA cycle [42, 43].
It remains to be determined if these enzymes contribute to the suppression of glycolysis elicited by SIRT3 expression in anchorage-independence, or if alternate deacetylation targets of SIRT3 have functional roles in this stage of metastasis. As suggested by analysis of lactate levels and extracellular acidification rate measurements (Fig. 3), increased glucose uptake in anchorage-independent cells is more uncoupled from lactate production compared to attached cells. It is now well accepted that metabolic flexibility is a hallmark of cancer, allowing cells to cope with fluctuations in nutrient availability during different tumor stages and in response to changing tumor microenvironments. This is evident in anchorage-independent cancer spheroids, which are often enriched in cancer stem cells, and marked by the ability to readily switch their metabolism to induce the pentose phosphate pathway, TCA cycle and oxidative phosphorylation, based on nutrient availability and anabolic demands [6, 44]. As previously reported [34], we also observed a change in cell cycle in detached cells, suggesting that the switch from high lactate production to glucose utilization for other metabolic pathways may be a reflection of a switch from pro-proliferative energy demands in attached cells to pro-survival metabolism in anchorage-independence. The observed suppression of glycolytic capacity by SIRT3 in detached cells requires further attention, and could be an important mechanism for cancer cells to survive metastatic spread in new tumor environments with alternate carbon sources, including the ascites fluid and the adipocyte-rich omentum [45]. In addition, the suppression of glycolytic flux by SIRT3 may be a further survival mechanism to prevent lactic acid toxicity.
In conclusion, our data suggest that the rapid and sustained upregulation of SIRT3 following matrix detachment is necessary for SOD2-mediated mitochondrial oxidant scavenging to enhance anchorage-independent survival and peritoneal colonization of ovarian cancer cells. Additional consequences of the context specific increase of SIRT3 on metabolic changes likely further aid in transcoelomic spread as a response to changing nutrient environments. Our study highlights the context-dependent role and regulation of SIRT3 in cancer and has important implications for future targeting of this protein for anti-cancer therapies.
Materials and Methods
GEO data set
GEO data set GSE85296 was examined using GEO2R (NCBI) to determine SIRT3 and SOD2 expression in matching ovarian, peritoneal, omental and ascites specimens from four ovarian serous adenocarcinoma patients.
Cell lines and cell culture conditions
OVCA433 and OVCA420 cells were provided by Dr. S.K. Murphy (Duke University). ES-2 and NIH-OVCAR3 cells were purchased from American Type Culture Collection (CRL-1978, HTB-161; ATCC Manassas, VA). OVCA433, OVCA420 and OVCAR3 cells were cultured in RPMI media supplemented with 10% FBS. Luciferase expressing SK-OV-3-Luc cells were from the Japanese Collection of Research Bioresources Cell Bank and cultured in McCoy’s 5A medium with 10% FBS. All cells were maintained at 37°C with 5% CO2. Cell authentication was carried out by the commercial provider or in-house through STR genotyping, and cells routinely tested for mycoplasma contamination. In rescue experiments cells were co-treated with the Manganese Porphyrin O2•− scavenger ortho tetrakis(N-n-butoxyethylpyridinium-2-yl) porphyrin (MnTnBuOE-2-PyP5+), provided by Dr. Ines Batinic Haberle (Duke University), or with N-acetyl-L-cysteine (Sigma), for indicated times and doses.
Culturing cells in ULA plates
Cells were trypsinized, counted, and equal viable cells seeded in 6-well Ultra Low Attachment (ULA; Corning) plates (1×105 cells/2 mL/well) for RNA and protein extraction. For live/dead and MitoSOX staining, cells were seeded 1×103 cells/200 μL/well in 96-well round-bottom ULA plates.
EOC specimens from patient ascites
Ascites specimens from patients diagnosed with high grade serous adenocarcinoma (Stage III-IV) were collected after informed consent was sought at the Penn State Cancer Institute (Hershey, PA) and the London Health Sciences Centre (London, Ontario), with approval for the study granted from the Penn State College of Medicine and Western University Ontario institutional research ethics boards, respectively. EOCs were isolated from ascites, as previously described [46], and maintained in culture at 37°C, 5% CO2 in MCDB/M199 medium supplemented with 10% FBS and penicillin/streptomycin. Microarray analysis was carried out on total RNA extracted from EOCs obtained at Western University, as previously described [47], using the Ad-GFP transduced controls. RNA was extracted following culturing for 72 h in ULA plates or from controls grown in attached conditions. Affymetrix Human Genome U133A GeneChip analysis (Santa Clara, CA) was carried out at Precision Biomarker Resources Inc. (Evanston, IL).
Semi-quantitative real-time RT-PCR
RNA was extracted using the Direct-zol RNA MicroPrep (Zymo Research, R2062) and cDNA synthesized using the iScript cDNA kit (Bio-Rad, 1708891). Real-time RT-PCR was performed using the iTaq Universal SYBR Green Supermix (Bio-Rad) and Bio-Rad CFX96 Real-Time PCR System. Relative SIRT3 mRNA expression was normalized to the geometric mean of 4 housekeeping genes: HPRT1, TBP, 18S, and GAPDH, and data analyzed using the ΔΔCt method. The following primers were used: SIRT3-sense (S): 5’-AGCCCTCTTCATGTTCCGAAGTGT-3’; SIRT3-antisense (AS): 5’-TCATGTCAACACCTGCAGTCCCTT-3’; HPRT1-S: 5’-TGACCTTGATTTATTTTGCATACC-3’; HPRT1-AS: 5’-CGAGCAAGACGTTCAGTCCT-3’; TBP-S: 5’-TTGGGTTTTCCAGCTAAGTTCT-3’; TBP-AS: 5’-CCAGGAAATAACTCTGGCTCA-3’; 18S-S: 5’-AGAAACGGCTACCACATCCA-3’; 18S-AS: 5’-CACCAGACTTGCCCTCCA-3’
GAPDH-S: 5’-GAGTCAACGGATTTGGTCGT-3’; GAPDH-AS: 5’-TTGATTTTGGAGGGATCTCG-3’. SOD2-S: 5’-TCCACTGCAAGGAACAACAG-3’
SOD2-AS: 5’-CGTGGTTTACTTTTTGCAAGC-3’.
RNA interference
Short hairpin RNA (shRNA) with non-targeting scramble sequence (5’-GCACTACCAGAGCTAACTCAGATAGTACT-3’) or targeting SIRT3 sequences (shSIRT3_#1: 5’- GTACAGCAACCTCCAGCAGTACGATCTCC-3’; shSIRT3_#2: 5’-AACCAGAATATGTGAACTGAGTGGACACC-3’; shSIRT3_#3: 5’-TCTTCACTCTGCTGAAGCTCCTAATGGAA-3’; shSIRT3_#4: 5’-TCACATTCTGTTGACTCTCCATACTCAGC-3’) in pRS vector (Origene, TR309432) were used to stably transfect OVCA433 cells (Supp. Fig. 1A). Clones expressing shSIRT3_#2 and shSIRT3_#3 were used for all subsequent experiments. Scramble non-targeting SMARTpool control (D-001810–10-05), SIRT3-specific SMARTpool siRNA oligonuecliotides (L-004827–01-0005), SOD2-specific siRNA oligonucleotides (siSOD2_#1: 5’-AAGUAAACCACGAUCGUUA-3’, J-009784–06-0005 and siSOD2_#2: 5’-CAACAGGCCUUAUUCCACU-3’, J-009784–08-0005) were obtained from Life Technologies and 60pmol/1×105 cells transfected using Lipofectamine RNAiMax.
Immunoblotting
Lysates were prepared in RIPA buffer supplemented with protease and phosphatase inhibitors, and equal protein separated on tris-glycine SDS-PAGE, followed by transfer to PVDF membranes. Membranes were blocked in 5% milk/TBS/0.1%Tween-20, and incubated overnight at 4°C with the following antibodies: SIRT3 (5490S, Cell Signaling Technology); Histone H3 (9715S, Cell Signaling Technology); SOD2 (ab13533, Abcam); acetyl(K68)-SOD2 (ab137037, Abcam); GAPDH (AM4300, Invitrogen); β-actin (AM4302, Invitrogen); HIF1α antibody (610958, BD). HRP-conjugated secondary antibodies were obtained from GE Healthcare. Blots were visualized on a ChemiDoc MP system (Bio-Rad) using Femto ECL chemiluminescence substrate (Thermo Scientific).
Live/dead staining
Cell viability was determined by staining cells with 2μM Calcein AM and 4μM ethidium homodimer (Sigma) in PBS for 30 min at 37 °C, followed by imaging on a Keyence BZ-X700 fluorescence microscope and analysis using Image J.
Annexin V and Cell cycle Analysis
Apoptosis Annexin V and Cell Cycle analysis were performed using a Muse Flow cytometer (Sigma Milipore), according to manufacturer’s instructions.
Clonogenicity assay
Single cell survival clonogenicity assays were performed as previously described [48, 49]. Briefly, 45 cells/well were plated in 12-well cell culture dishes and colonies (≥50 cells/colony) visualizing with 0.05% crystal violet dye.
Mitochondrial Superoxide (O2.−) Detection
5 μM MitoSOX Red (Invitrogen) was added to cells in HBSS for 30 min followed by washing. Images were captured using a Keyence BZ-X700 fluorescence microscope and fluorescence signal quantified using Image J.
SOD Zymography
SOD activity was analyzed using zymography, as previously described [50, 51]. Briefly, cells were lyzed by sonication in potassium phosphate buffer (pH 7.8, 0.1 mM EDTA), and 50 μg of the proteins resolved by electrophoresis in a 10% non-denaturing polyacrylamide gel. Gels were incubated for 15 min in the dark with 2.5mM nitro blue tetrazolium, 30 mM TEMED, 0.028 mM riboflavine, 50 mM phosphate buffer, pH7.8, followed by washing in H2O, and visualization of SOD activity by light exposure.
SOD Activity assay
SOD activity was further quantified in an in vitro assay to monitor dismutation of superoxide radicals generated by xanthine oxidase and hypoxanthine (Cayman 706002). SOD2 activity was quantified in samples with addition of 1mM potassium cyanide to inhibit SOD1 activity. SOD activity was normalized against lysate protein concentrations.
Media Glucose and Lactate Quantification
Cells were seeded in 6-well adherent or ULA plates (1×105 cells/2 mL/well). After 24 h, media were collected by centrifugation and 200 μL analyzed for glucose and lactate content using the YSI 2900D Biochemistry analyzer (Xylem, Yellow Springs, OH), and values corrected for cell numbers.
Optical Redox Ratio imaging
Anchorage-independent spheroids were imaged using the Nikon A1 MP+ Multi-Photon Microscope system (Nikon Instruments, New York) to assess endogenous fluorescence signals produced by NAD(P)H and FAD, as detailed in Supplemental Methods.
Extracellular flux analysis
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a Seahorse XFp Analyzer (Agilent). 24 h prior to the assay cells were seeded at a density of 10,000 cells well into a XFp cell culture plate (attached conditions). Anchorage-independent cells were cultured for 24 h in 96 well round bottom ULA plates, followed by washing in assay media, and transfer of 10 spheroids into each assay well (1,000 cells per spheroid). Basal glycolysis was derived as the difference between ECAR following addition of glucose (10mM) and inhibition of glycolysis by 2-deoxy-glucose (50mM). Glycolytic capacity/maximal ECAR was determined following addition of the mitochondrial ATP synthase inhibitor Oligomycin (1μM).
Intraperitoneal in vivo xenografts
SK-OV-3-Luc cells were transfected with scramble, SOD2 or SIRT3 siRNA (Dharmacon, as above). Cells were detached 24 h after transfection, washed, counted and 1×106 viable cells resuspended in 150 μl of PBS before immediate IP injection into female Nod scid gamma mice (NSG, bred in house). Animals were randomized prior to tumor cell injection, and sample size (8 mice/group) chosen based on pilot experiments. Approval for animal studies was sought from the Penn State College of Medicine IACUC prior to study commencement. Luminescence imaging carried out every 2–3 days using an IVIS luminescence imaging system 10 min after mice were injected with 10 μl/g of body weight 15 mg/mL in vivo grade D-Luciferin (PerkinElmer). Mice were sacrificed by CO2 asphyxiation followed by cervical dislocation if they reached AAALAC-defined endpoints, or at day 31 post tumor cell injection. At necropsy organs were preserved in 10% buffered formalin, followed by paraffin embedding, sectioning, and staining with hematoxylin and eosin. Number and size of tumors per longitudinal section of each omentum were imaged by microscopy and quantified in a blinded manner using Image J.
Statistical analysis
All data are representatives of at least three independent experiments, unless otherwise noted. Data are expressed as mean ± SEM, and statistical analysis performed using GraphPad Prism Software v8, with statistical tests chosen based on experimental design, as described in figure legends. Differences between groups were assessed using two-tailed t-test. Analysis of variance (ANOVA) of multiple group comparisons were carried out using One or Two-way ANOVA and between group differences assessed by appropriate post-tests based on study design, as indicated in Figure legends. Paired analyses were performed by paired t-tests and repeated measures ANOVA.
Supplementary Material
Acknowledgements
We thank Usawadee Dier, Dr. LP Madhubhani Hemachandra, Dr. Sarah Engelberth, Larissa Suparmanto, Sadie Dierschke, and Gina Deiter for technical assistance. We thank Dr. Arati Sharma and Dr. David Claxton (Penn State) for providing PDX animals. We are grateful for Dr. Vonn Walter’s advice on TCGA data analysis. MnTnBuOE-2-PyP5+ was kindly provided by Dr. Ines Batinic Haberle (Duke University). OVCA433 and OVCA420 cells were a generous gift from Dr. Susan Murphy (Duke University). This work was supported by NIH grants R00CA143229 (N.H.), R01CA230628 (N.H. & M.K.), S100D018124 (T.A.), by the Rivkin Center for Ovarian Cancer (NH), an equipment grant from Seahorse/Agilent (N.H.), and the Penn State Cancer Institute Developmental Fund Award (N.H.). Collection of ascites was partially supported by DoD Pilot award W81XWH-16-1-0117 (N.H.).
Financial Support: This work was supported by NIH grants R00CA143229 (N.H.), R01CA230628 (N.H. & M.K.), S100D018124 (T.A.), by the Rivkin Center for Ovarian Cancer (N.H.), an equipment grant from Agilent (N.H.), and the Penn State Cancer Institute Developmental Fund Award (N.H.). Collection of ascites was partially supported by DoD Pilot award W81XWH-16-1-0117 (N.H.).
Footnotes
Conflict of Interest:
The authors declare no competing financial interests in relation to the work described.
Supplementary information is available at Oncomine’s website.
References
- 1.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018; 68: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sehouli J, Senyuva F, Fotopoulou C, Neumann U, Denkert C, Werner L et al. Intra-abdominal tumor dissemination pattern and surgical outcome in 214 patients with primary ovarian cancer. J Surg Oncol 2009; 99: 424–427. [DOI] [PubMed] [Google Scholar]
- 3.Tan DS, Agarwal R, Kaye SB. Mechanisms of transcoelomic metastasis in ovarian cancer. The lancet oncology 2006; 7: 925–934. [DOI] [PubMed] [Google Scholar]
- 4.Cai Q, Yan L, Xu Y. Anoikis resistance is a critical feature of highly aggressive ovarian cancer cells. Oncogene 2015; 34: 3315–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015; 527: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 2009; 461: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison CA, Durbin SM, Thau MR, Zellmer VR, Chapman SE, Diener J et al. Antioxidant enzymes mediate survival of breast cancer cells deprived of extracellular matrix. Cancer research 2013; 73: 3704–3715. [DOI] [PubMed] [Google Scholar]
- 8.Le Gal K, Ibrahim MX, Wiel C, Sayin VI, Akula MK, Karlsson C et al. Antioxidants can increase melanoma metastasis in mice. Science translational medicine 2015; 7: 308re308. [DOI] [PubMed] [Google Scholar]
- 9.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell metabolism 2010; 12: 662–667. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S et al. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO reports 2011; 12: 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Molecular cell 2010; 40: 893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer cell 2010; 17: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGlynn LM, McCluney S, Jamieson NB, Thomson J, MacDonald AI, Oien K et al. SIRT3 & SIRT7: Potential Novel Biomarkers for Determining Outcome in Pancreatic Cancer Patients. PloS one 2015; 10: e0131344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desouki MM, Doubinskaia I, Gius D, Abdulkadir SA. Decreased mitochondrial SIRT3 expression is a potential molecular biomarker associated with poor outcome in breast cancer. Hum Pathol 2014; 45: 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang B, Fu X, Shao L, Ding Y, Zeng D. Aberrant expression of SIRT3 is conversely correlated with the progression and prognosis of human gastric cancer. Biochem Biophys Res Commun 2014; 443: 156–160. [DOI] [PubMed] [Google Scholar]
- 16.Zhang CZ, Liu L, Cai M, Pan Y, Fu J, Cao Y et al. Low SIRT3 expression correlates with poor differentiation and unfavorable prognosis in primary hepatocellular carcinoma. PloS one 2012; 7: e51703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Cheng S, Chen S, Zhao Y. Prognostic and clinicopathological value of SIRT3 expression in various cancers: a systematic review and meta-analysis. Onco Targets Ther 2018; 11: 2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer cell 2011; 19: 416–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu W, Denu RA, Krautkramer KA, Grindle KM, Yang DT, Asimakopoulos F et al. Loss of SIRT3 Provides Growth Advantage for B Cell Malignancies. The Journal of biological chemistry 2016; 291: 3268–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YS, Gupta Vallur P, Phaeton R, Mythreye K, Hempel N. Insights into the Dichotomous Regulation of SOD2 in Cancer. Antioxidants (Basel) 2017; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. SIRT3 and cancer: tumor promoter or suppressor? Biochimica et biophysica acta 2011; 1816: 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D’Silva NJ et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer 2011; 117: 1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Chiang YL, Lyssiotis CA, Teater MR, Hong JY, Shen H et al. Non-oncogene Addiction to SIRT3 Plays a Critical Role in Lymphomagenesis. Cancer cell 2019; 35: 916–931 e919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Z, Song J, Wang G, Cui X, Zheng J, Tang Y et al. Deacetylation of serine hydroxymethyl-transferase 2 by SIRT3 promotes colorectal carcinogenesis. Nature communications 2018; 9: 4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park HK, Hong JH, Oh YT, Kim SS, Yin J, Lee AJ et al. Interplay between TRAP1 and sirtuin-3 modulates mitochondrial respiration and oxidative stress to maintain stemness of glioma stem cells. Cancer research 2019. [DOI] [PubMed] [Google Scholar]
- 26.Kenny TC, Hart P, Ragazzi M, Sersinghe M, Chipuk J, Sagar MAK et al. Selected mitochondrial DNA landscapes activate the SIRT3 axis of the UPR(mt) to promote metastasis. Oncogene 2017; 36: 4393–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. Journal of cell science 2011; 124: 1396–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang XY, Kang JS, Yang XC, Su J, Wu Y, Yan XY et al. SIRT3 participates in glucose metabolism interruption and apoptosis induced by BH3 mimetic S1 in ovarian cancer cells. Int J Oncol 2016; 49: 773–784. [DOI] [PubMed] [Google Scholar]
- 29.Dong XC, Jing LM, Wang WX, Gao YX. Down-regulation of SIRT3 promotes ovarian carcinoma metastasis. Biochem Biophys Res Commun 2016; 475: 245–250. [DOI] [PubMed] [Google Scholar]
- 30.Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 2016; 532: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangarajan P, Karthikeyan A, Lu J, Ling EA, Dheen ST. Sirtuin 3 regulates Foxo3a-mediated antioxidant pathway in microglia. Neuroscience 2015; 311: 398–414. [DOI] [PubMed] [Google Scholar]
- 32.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene 2011; 30: 2986–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou J, Williams J, Botvinick EL, Potma EO, Tromberg BJ. Visualization of Breast Cancer Metabolism Using Multimodal Nonlinear Optical Microscopy of Cellular Lipids and Redox State. Cancer research 2018; 78: 2503–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Correa RJ, Peart T, Valdes YR, DiMattia GE, Shepherd TG. Modulation of AKT activity is associated with reversible dormancy in ascites-derived epithelial ovarian cancer spheroids. Carcinogenesis 2012; 33: 49–58. [DOI] [PubMed] [Google Scholar]
- 35.Kenny HA, Lal-Nag M, White EA, Shen M, Chiang CY, Mitra AK et al. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nature communications 2015; 6: 6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamarajan P, Alhazzazi TY, Danciu T, D’Silva NJ, Verdin E, Kapila YL. Receptor-interacting protein (RIP) and Sirtuin-3 (SIRT3) are on opposite sides of anoikis and tumorigenesis. Cancer 2012; 118: 5800–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenny TC, Craig AJ, Villanueva A, Germain D. Mitohormesis Primes Tumor Invasion and Metastasis. Cell Rep 2019; 27: 2292–2303 e2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hempel N, Carrico PM, Melendez JA. Manganese superoxide dismutase (Sod2) and redox-control of signaling events that drive metastasis. Anti-cancer agents in medicinal chemistry 2011; 11: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemachandra LP, Shin DH, Dier U, Iuliano JN, Engelberth SA, Uusitalo LM et al. Mitochondrial Superoxide Dismutase Has a Protumorigenic Role in Ovarian Clear Cell Carcinoma. Cancer research 2015; 75: 4973–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connor KM, Hempel N, Nelson KK, Dabiri G, Gamarra A, Belarmino J et al. Manganese superoxide dismutase enhances the invasive and migratory activity of tumor cells. Cancer research 2007; 67: 10260–10267. [DOI] [PubMed] [Google Scholar]
- 41.Zou X, Zhu Y, Park SH, Liu G, O’Brien J, Jiang H et al. SIRT3-Mediated Dimerization of IDH2 Directs Cancer Cell Metabolism and Tumor Growth. Cancer research 2017; 77: 3990–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozden O, Park SH, Wagner BA, Yong Song H, Zhu Y, Vassilopoulos A et al. SIRT3 deacetylates and increases pyruvate dehydrogenase activity in cancer cells. Free radical biology & medicine 2014; 76: 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jing E, O’Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S et al. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes 2013; 62: 3404–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emmings E, Mullany S, Chang Z, Landen CN Jr., Linder S, Bazzaro M. Targeting Mitochondria for Treatment of Chemoresistant Ovarian Cancer. International journal of molecular sciences 2019; 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med 2011; 17: 1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepherd TG, Theriault BL, Campbell EJ, Nachtigal MW. Primary culture of ovarian surface epithelial cells and ascites-derived ovarian cancer cells from patients. Nat Protoc 2006; 1: 2643–2649. [DOI] [PubMed] [Google Scholar]
- 47.Peart TM, Correa RJ, Valdes YR, Dimattia GE, Shepherd TG. BMP signalling controls the malignant potential of ascites-derived human epithelial ovarian cancer spheroids via AKT kinase activation. Clinical & experimental metastasis 2012; 29: 293–313. [DOI] [PubMed] [Google Scholar]
- 48.Dier U, Shin DH, Hemachandra LP, Uusitalo LM, Hempel N. Bioenergetic analysis of ovarian cancer cell lines: profiling of histological subtypes and identification of a mitochondria-defective cell line. PloS one 2014; 9: e98479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Worley BL, Kim YS, Mardini J, Zaman R, Leon KE, Vallur PG et al. GPx3 supports ovarian cancer progression by manipulating the extracellular redox environment. Redox biology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 1971; 44: 276–287. [DOI] [PubMed] [Google Scholar]
- 51.Hempel N, Ye H, Abessi B, Mian B, Melendez JA. Altered redox status accompanies progression to metastatic human bladder cancer. Free radical biology & medicine 2009; 46: 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.