SUMMARY
Lipid droplets (LDs) provide a reservoir for triacylglycerol storage and are a central hub for fatty acid trafficking and signaling in cells. Lipolysis promotes mitochondrial biogenesis and oxidative metabolism via a SIRT1/PGC-1α/PPARα-dependent pathway through an unknown mechanism. Herein, we identify that monounsaturated fatty acids (MUFAs) allosterically activate SIRT1 towards select peptide-substrates such as PGC-1α. MUFAs enhance PGC-1α/PPARα signaling and promote oxidative metabolism in cells and animal models in a SIRT1 dependent manner. Moreover, we characterize the LD protein perilipin 5 (PLIN5), which is known to enhance mitochondrial biogenesis and function, to be a fatty acid binding protein that preferentially binds LD-derived monounsaturated fatty acids (MUFAs) and traffics them to the nucleus following cAMP/PKA-mediated lipolytic stimulation. Thus, these studies identify the first-known endogenous allosteric modulators of SIRT1 and characterize a LD-nuclear signaling axis that underlies the known metabolic benefits of MUFAs and PLIN5.
Keywords: ATGL, Fatty Acids, Lipid Droplets, Lipolysis, MUFA, Oxidative Metabolism, Olive Oil, PGC-1α, PLIN5, SIRT1
eTOC Blurb
Najt et al identify the first-known endogenous allosteric modulator of SIRT1 and characterize a lipid droplet-nuclear signaling axis that underlies the known metabolic benefits of monounsaturated fatty acids and PLIN5.
INTRODUCTION
During increased energy demand, fatty acids are hydrolyzed from triacylglycerol stored in cytoplasmic LDs to provide substrates for β-oxidation and oxidative phosphorylation. The hydrolysis of triacylglycerols (i.e. lipolysis) via adipose triglyceride lipase (ATGL), the major triacylglycerol lipase in most tissues, promotes the activation of the transcription factor/coactivator complex of PPAR-α/PGC-1α to upregulate mitochondrial biogenesis and, thus, couple oxidative capacity with the supply of fatty acid substrates (Haemmerle et al., 2011; Khan et al., 2015; Ong et al., 2011). While the supply of fatty acid ligands to activate PPAR-α may contribute to these effects (Haemmerle et al., 2011), we have shown that sirtuin 1 (SIRT1), which is known to deacetylate PGC-1α and promote its interaction with transcription partners, is activated in response to ATGL-catalyzed lipolysis and is required for ATGL-mediated upregulation of PPAR-α/PGC-1α signaling (Khan et al., 2015). Moreover, cAMP/PKA signaling, which promotes lipolysis and SIRT1, requires ATGL-catalyzed lipolysis for the induction of SIRT1 activity suggesting that ATGL is a key upstream regulator of SIRT1. A member of the sirtuin family of NAD+-dependent protein deacetylases, SIRT1 has a wide-range of biological functions including chromatin structure maintenance, cell cycle control, metabolism and the regulation of healthspan (Banks et al., 2008; Bordone et al., 2007; Houtkooper et al., 2012; Pfluger et al., 2008). In mice, SIRT1 promotes characteristics reminiscent of caloric restriction such as a decrease in the incidence of age-related diseases including diabetes, cardiovascular disorders and neurodegenerative diseases (Balasubramanian et al., 2017; Banks et al., 2008; Bordone et al., 2007; Chen et al., 2005; Pfluger et al., 2008). Numerous dietary small molecule activators of SIRT1, such as the polyphenol resveratrol and related compounds, have been identified and used to attenuate aging-related disease and improve lifespan (Hubbard and Sinclair, 2014; Kim et al., 2007; Lagouge et al., 2006; Sinclair and Guarente, 2014). Thus, SIRT1 plays a key role in sensing intracellular redox (i.e. NAD) and dietary phytochemicals to coordinate cellular function and disease resistance.
LD accumulation in non-adipose tissue is a hallmark and etiological factor of numerous diseases (Greenberg et al., 2011). Increased LDs in cells is commonly associated with lipotoxocity and altered metabolism that contributes to cellular dysfunction. Perilipin 5, a member of the perilipin (PLIN) family of LD proteins has been positively correlated with both triacylglycerol storage and fatty acid oxidation, and uncouples LD accumulation from lipotoxicity and metabolic dysfunction (Dalen et al., 2007; Gemmink et al., 2016; Kurmoto et al., 2012; Mohktar et al., 2016; Pollak et al., 2015; Wang et al., 2015; Wolins et al., 2006). Under basal conditions, PLIN5 directly interacts with and inhibits ATGL, but in response to lipolytic stimuli, such as cAMP/PKA signaling, it promotes triacylglycerol hydrolysis and fatty acid oxidation (Granneman et al., 2011; Granneman et al., 2009; Wang et al., 2015). While gain-and-loss of function studies have shown a connection between PLIN5 and fatty acid metabolism, the mechanism by which PLIN5 contributes to oxidative metabolism has remained largely unknown. Recent work providing insights into this mechanism demonstrate that PLIN5 interacts with PGC-1α and SIRT1 to promote PGC-1α/PPAR-α activity (Gallardo-Montejano et al., 2016).
Given that both ATGL and PLIN5 have been linked to SIRT1, we sought to elucidate the interplay between these two LD proteins and the mechanisms through which ATGL-mediated lipolysis promotes SIRT1 activity and downstream PGC-1α/PPAR-α signaling. Herein, we show that a specific class of fatty acids, MUFAs, bind and allosterically activate SIRT1 by reducing its Km for select peptide substrates. In addition, we identify PLIN5 to be a fatty acid binding protein that preferentially binds MUFAs derived from ATGL-catalyzed lipolysis and shuttles them to the nucleus for activation of SIRT1 following lipolytic stimulation.
RESULTS
MUFAs are allosteric activators of SIRT1 at nanomolar concentrations
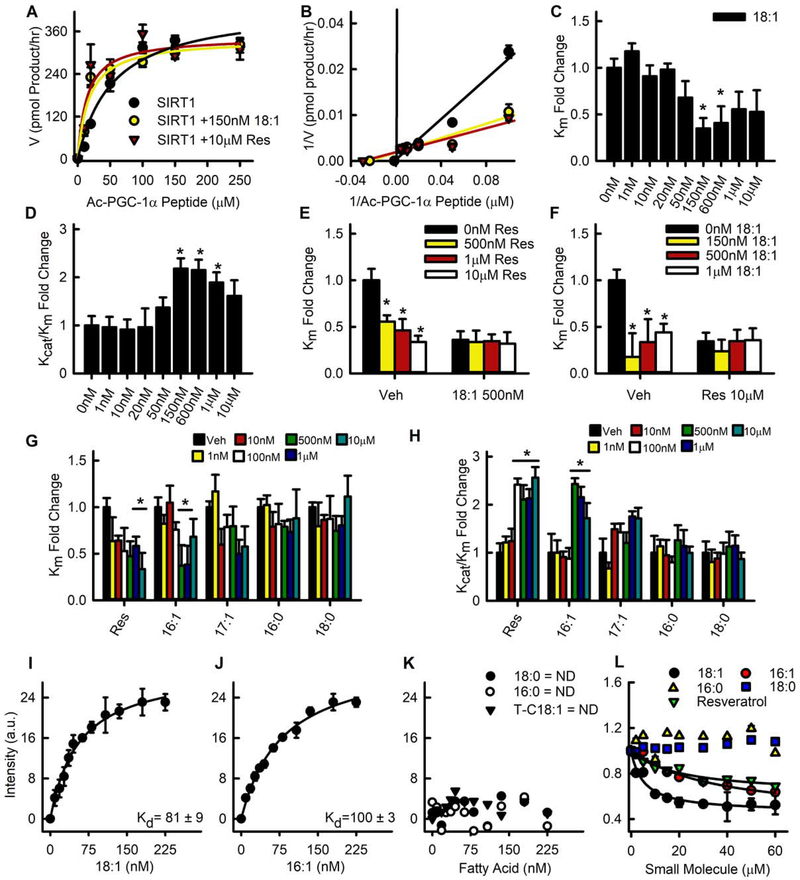
Given that ATGL promotes SIRT1 signaling, we explored if the products of ATGL-catalyzed lipolysis, fatty acids, could activate SIRT1. Indeed, SIRT1 has a hydrophobic pocket thought to be responsible for binding resveratrol and related sirtuin activating compounds (Borra et al., 2005; Cao et al., 2015; Kaeberlein et al., 2005). Using an MS-based Selective Reaction Monitoring method with recombinant SIRT1 (Fig. S1A–C), we found that the kinetics of PGC-1α peptide deacetylation were altered in the presence of the fatty acid 18:1 (Fig. 1A–D). The increase in SIRT1 catalytic efficiency (Kcat/Km) was due to a lowering of the Km of SIRT1 towards the PGC-1α peptide without a significant change in enzyme velocity. This effect was not additive to resveratrol, as co-addition of 18:1 and resveratrol did not alter the Km or catalytic efficiency when compared to addition of a single lipophilic compound (Fig. 1E–F) suggesting that 18:1 and resveratrol may activate SIRT1 through a common binding site. We next explored if other fatty acids had similar effects on SIRT1. While 17:1, 16:0, and 18:0 were unable to stimulate SIRT1 deacetylase activity, the addition of 16:1 also resulted in a lowering of the Km and an increase in catalytic efficiency towards the PGC-1α peptide comparable to the effects observed with 18:1 (Fig. 1G–H). For both even chain MUFAs, activation of SIRT1 was seen at concentrations of fatty acids ranging from 150 nM to 1 μM, but no deacetylase activation was observed at concentrations above 1 μM (Fig. 1D–H).
Fig. 1. The MUFAs 18:1 and 16:1 allosterically activate SIRT1 towards a PGC-1α substrate.
A) Saturation plot of the effect of fatty acids and resveratrol (Res) on human SIRT1 enzyme activity was measured by mass spectrometry (see supplemental methods) using a native peptide sequence of acetylated-PGC-1α. B) Lineweaver-Burk reciprocal plots were generated to determine Km, Vmax, and Kcat. C-D) Km and Kcat/Km fold change for each concentration of 18:1. E-F) Competition assays between 18:1 and resveratrol. G-H) Km and Kcat/Km fold change for each concentration of resveratrol and long chain fatty acids. I-K) SIRT1 binding affinity for fatty acids was determined by tryptophan fluorescence quenching assay (ND=not detected). L) Displacement of 1,8-ANS was used to determine the Ki of SIRT1 for fatty acids and resveratrol.
Next, we determined if fatty acid activation of SIRT1 was due to direct binding. Using tryptophan quenching assays, saturable binding curves for 18:1 and 16:1 were observed with Kd values of 81±9 nM, and 100±3 nM, respectively (Fig. 1I–J). No fatty acid binding was observed for 18:0, 16:0, or trans-18:1 (Fig. 1K) suggesting a preference for MUFAs. To further support these findings, fluorescence binding and displacement assays using 1,8-ANS were performed (Kane and Bernlohr, 1996). Displacement of the bound fluorophore using 18:1, 16:1, or resveratrol as a competing ligand revealed Ki values of 5.6±0.12, 12.5±0.06, and 16.7±0.07 μM respectively; displacement of 1,8-ANS was not observed with 18:0 and 16:0 (Fig. 1L and Table S1). Structure analysis using CD revealed that MUFAs elicited large changes in secondary structure with increased α–helical content of SIRT1 from 23% to 25.9% and 23% to 28.6% for 16:1 and 18:1, respectively (Fig. S1D and Table S2). The addition of 16:0 and 18:0 did not alter the shape of the CD spectrum of SIRT1 consistent with the lack of tryptophan quenching and ANS displacement showing no binding. These results suggest that MUFA-mediated allosteric activation of SIRT1 was due to direct fatty acid binding and subsequent conformational changes to the enzyme.
The activation of SIRT1 in response to resveratrol and related compounds is highly selective based upon the peptide substrate (Hubbard et al., 2013). Therefore, we tested if the ability of MUFAs to activate SIRT1 is also influenced by the acetyl peptide sequence. We chose peptide sequences from established SIRT1 targets FOXO3A and H3 (Fig. S1A). Similar to the results obtained with PGC-1α, 18:1 also increased SIRT1 activity towards the FOXO3a peptide through a reduced Km and increased catalytic efficiency comparable to what was observed with 10 μM resveratrol (Fig. 2A–F). In contrast, MUFAs were unable to increase SIRT1 activity towards the H3 peptide substrate (Fig. 2G–K). In fact, MUFA concentrations of 600 nM or more increased the Km and decreased Kcat/Km indicating inhibitory effects towards the H3 peptide. To further explore substrate selectivity, we used a competition assay with fixed amounts of PGC-1α, FOXO3a and H3 peptides and two doses of 18:1. The addition of either 150 or 600 nM 18:1 increased deacetylase activity towards FOXO3a and PGC-1α peptides, but decreased activity towards H3 (Fig. 2L). Taken together, these data show that MUFAs selectively target SIRT1 to specific peptide substrates.
Fig. 2. MUFAs selectively activate SIRT1.
A) Saturation plot of SIRT1 activity towards FOXO3a and the effects of 18:1 and resveratrol (n=4). B) Lineweaver-Burk reciprocal plots were generated to determine Km, Vmax, and Kcat for the FOXO3a peptide substrate. C-D) Km and Kcat/Km fold change for each concentration of 18:1 on FOXO3a. E) Kcat/Km fold change for resveratrol (Res; 10 μM). F) Saturation plot of SIRT1 activity towards H3 and the effects of 18:1 and resveratrol. G) Lineweaver-Burk reciprocal plots for the H3 peptide substrate. H-I) Km and Kcat/Km fold change for each concentration of 18:1 with H3. J-K) Kcat/Km fold change for each concentration of resveratrol and fatty acids for the H3 peptide substrate. L) Competition assay of SIRT1 activity towards FOXO3A, PGC-1α and H3 acetylated peptide substrates.
A hydrophobic residue at the +1 or +6 position upstream of the acetylated lysine is required for allosteric activation of SIRT1 (Hubbard et al., 2013). To determine if a similar requirement exists for MUFA activation, SIRT1 activity towards a p53 peptide was determined (Fig. S1A, and Fig. S2A–B). Lacking a hydrophobic residue at the +1 or +6 position resulted in a SIRT1 substrate that did not respond to allosteric activation via 18:1, resveratrol, or the SIRT1 activating compound SRT1720 (Fig. S2A–B). In contrast to the wild-type p53 peptide substrate, a mutant p53 peptide (p53-W) containing a tryptophan at the +6 position in replacement of alanine, was activated in response to 250 nM 18:1, 10 μM resveratrol, and 1 μM SRT1720 (Fig S2C–G). Examining the PGC-1 α, FOXO3a, and the H3 peptide substrates, both the PGC-1α and the FOXO3a substrates contained a hydrophobic residue at the +1 position, valine for PGC-1 α and tryptophan for FOXO3a, while the H3 substrate did not (Fig. S1A). Taken together, these data show that MUFAs selectively target SIRT1 to specific peptide substrates through the positioning of hydrophobic residues at either the +1 or the +6 position relative to the acetylated lysine similar to what has been reported for resveratrol and SRT1720.
18:1 increases PGC-1a transcriptional activity in an SIRT1 dependent manner
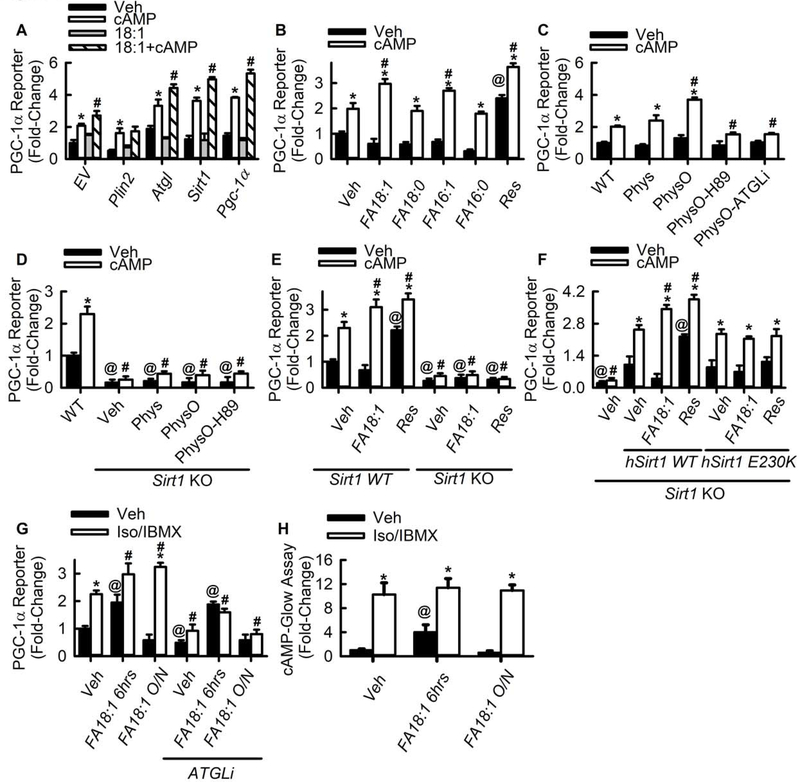
Since MUFAs bind and allosterically activate SIRT1, we tested the effects of lipolysis-derived 18:1 on SIRT1/PGC-1α signaling. As expected, cAMP treatment or Atgl, Sirt1, and Pgc-1α overexpression individually and synergistically increased PGC-1α activity (Fig. 3A). Preloading the hepatocytes with 18:1 further enhanced the response to cAMP and protein overexpression on PGC-1α activity indicating a synergistic effect of MUFA enrichment in LDs, cAMP and key proteins involved in LD-nuclear MUFA signaling; no effects of 18:1 were observed with Plin2 overexpression (Fig. 3A). To determine if the lipid loading effect was due to the presences of MUFAs rather than fatty acids in general, the experiments were repeated using individual fatty acids 18:1, 18:0, 16:1, and 16:0 (Fig. 3B), or a physiological mixture of the fatty acids that included or lacked 18:1 (Fig. 3C and Table S3). The individual fatty acids 18:1 and 16:1 synergized with cAMP to increase PGC-1α activity above non-loaded or 18:0/16:0 loaded cells (Fig. 3B). The physiological fatty acid mixture containing 18:1 increased PGC-1α activity above the physiological mixture lacking 18:1 and above cells not loaded with lipid in response to cAMP (Fig. 3C), a result similar to that of the individual fatty acids. Inhibition of PKA or ATGL negated the effects of 18:1 and cAMP. Taken together, the individual or physiological mixture of fatty acids experiments indicate the enhancing effect was due to the presences of MUFAs rather than fatty acids in general as saturated fatty acids or mixtures of polyunsaturated fatty acids lacking 18:1 did not enhance PGC-1α activity. Studies utilizing PKA or ATGL inhibitors revealed that PKA-stimulated lipolysis was required for 18:1 mediated activation of PGC-1α in contrast to more traditional SIRT1 activating compound resveratrol (Fig. 3B). This indicates MUFAs must be released from LDs by ATGL for activation to occur. In addition, studies utilizing mouse embryonic fibroblasts (MEFs) lacking Sirt1 revealed that SIRT1 was required for 18:1 mediated regulation of PGC-1α activity (Fig. 3D and E). Rescue experiments utilizing a GFP tagged human Sirt1 construct restored cAMP, 18:1, and resveratrol activation, while the Sirt1-E230K mutant, shown to block resveratrol binding (Dai et al., 2015; Hubbard et al., 2013; Sinclair and Guarente, 2014), restored basal and cAMP stimulated PGC-1α activity but did not restore MUFA or resveratrol mediated regulation of PGC-1α (Fig. 3F).
Fig. 3. Lipolytically-derived MUFA synergize with cAMP and signal via SIRT1 to activate PGC-1α.
A) PGC-1α luciferase reporter assays in primary hepatocytes transfected with the various overexpression plasmids (n=6–12). *p<0.05 vs. drug veh, #p<0.05 vs. cAMP alone. B) PGC-1α luciferase reporter assays in MEFs loaded with saturated fatty acids, MUFAs, or resveratrol (n=6–12). *p<0.05 vs. drug veh, #p<0.05 vs. lipid veh treated with cAMP. C) PGC-1α luciferase reporter assays in hepatocytes loaded with a physiological mix of fatty acids lacking 18:1 (Phys), or a physiological mix enriched in 18:1 (PhysO). ATGL inhibition was achieved by the addition of 30 μM ATGListatin (ATGLi). PKA inhibition was achieved by addition of 15 μM H89. Both drugs were administered for 1 hr followed by addition of 8-bromoadenosine 3’,5’-cyclic monophosphate (cAMP; 1mM). (n=6–12). *p<0.05 vs. drug veh, #p<0.05 vs. wild-type cells not loaded with lipid treated with cAMP. D) PGC-1α luciferase reporter assays in wild-type or Sirt1 knockout MEFs preloaded with as physiological mix of fatty acid and subsequently treated with inhibitors (n=6–12). *p<0.05 vs. drug veh, @P<0.05 vs wild-type, # p<0.05 vs. wild-type cells treated with cAMP. E) PGC-1α luciferase reporter assays in wild-type or Sirt1 knockout MEFs exposed to fatty acid or resveratrol preloading (n=6–12). *p<0.05 vs. wild-type treated with drug veh, @P<0.05 vs. lipid veh wild-type, #p<0.05 vs. lipid veh wild-type cells treated with cAMP. F) PGC-1α reporter assays from Sirt1 knockout cells transfected with human Sirt1 or human Sirt1 E230K mutant (n=8–12). *p<0.05 vs. drug veh, @P<0.05 vs. lipid veh treated hSirt1 expressing cells, #p<0.05 vs. lipid veh treated hSirt1 expressing cells treated with cAMP. G) PGC-1α reporter assays in MEFs loaded with 500 μM 18:1 acutely (6 hrs) or overnight (O/N - 16hrs). Lipolytic activation was achieved by the addition of 20 μM isoproterenol and 500 μM IBMX. ATGL inhibition was achieved by the addition of 30 μM ATGListatin (ATGLi) (n=6–12). *p<0.05 vs. drug veh, @P<0.05 vs. lipid veh, #p<0.05 vs. lipid veh treated with Iso/IBMX. H) Cellular cAMP levels were measured in MEF cells loaded acutely overnight with 500 μM 18:1 (n=12–16). Lipolytic activation was achieved by the addition of 20 μM isoproterenol and 500 μM IBMX.*p<0.05 vs. drug veh, @P<0.05 vs. lipid veh without Iso/IBMX, #p<0.05 vs. lipid veh treated with Iso/IBMX.
Acute exposure of cells to 18:1 has been shown to increase cellular cAMP as a means to activate SIRT1 (Lim et al., 2013). Therefore, we tested if alterations in cellular cAMP levels contributed to the effects of MUFA on SIRT1 activation in cells (Fig. 3G–H). Acute exposure (6 hrs) to 18:1 increased basal PGC-1α activity while chronic or overnight exposure (16 hrs) did not. Both acute and overnight 18:1 loading enhanced PGC-1α activity above non-loaded cells upon stimulation of β-andrenergic signaling. Inhibition of ATGL mediated lipolysis via ATGLstatin blocked the effects of β-andrenergic stimulation in 18:1 loaded cells. Cells acutely loaded with 18:1 still had elevated basal PGC-1α activity in the presence of ATGLstatin, however the stimulated response was blocked. Acute 18:1 exposure increased cellular cAMP levels similar to what was previously reported (Lim et al., 2013), however non-loaded, acutely loaded, and overnight loaded cells all exhibited similar levels of cellular cAMP upon treatment with isoproterenol/IBMX (Fig. 3H). Thus, in the experimental conditions where MUFAs and β-androgenic stimulation synergize to enhance PGC-1α activity in a SIRT1 dependent manner cAMP levels were not altered between non-loaded and 18:1 loaded cells. While these results are consistent our data showing ATGL-mediated activation of PGC-1α synergizes with cAMP/PKA, it should be noted that MUFAs also can signal acutely via regulation of cAMP independent of incorporation into and subsequent hydrolysis from LDs.
Olive oil diet increases oxidative metabolism in a SIRT1 dependent manner
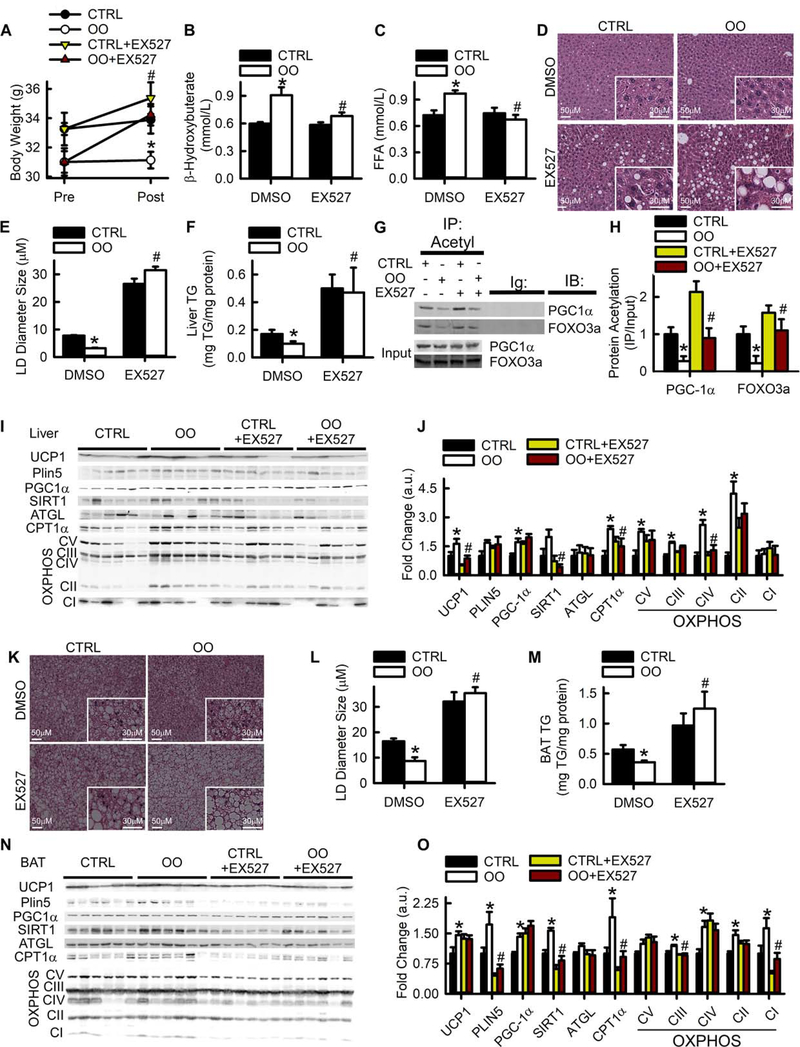
To investigate the effects of SIRT1 activating MUFAs in vivo, mice were fed diets enriched in lard/soybean oil (CTRL) or olive oil (OO), which contains ~75% 18:1 (Table S4), and were fasted overnight prior to sacrifice to stimulate lipolytic signaling. OO feeding decreased body weight over the course of 12 weeks due to a decrease in fat mass (Fig. S3A–C). Without effecting energy intake or locomotion, OO feeding increased oxygen consumption and heat production leading to increased energy expenditure (EE; Fig. S3D–N). To determine if the OO in the diet was exerting its physiological effects in a SIRT1 dependent manner, EX527, a potent and specific SIRT1 inhibitor, was administered over the course of three days prior to sacrifice (Fig. S4A). SIRT1 inhibition negated the decrease in body weight (Fig. 4A) and the increase in serum β-hydroxybutyrate and free fatty acids observed with OO feeding (Fig. 4B–C). The OO diet reduced white adipose tissue weights, an effect that was normalized in mice treated with EX527 (Fig. S4B–G). OO feeding decreased hepatic LD size and liver TAG content while SIRT1 inhibition ablated these effects (Fig. 4D–F). Acetylation of SIRT1 targets, PGC-1α and FOXO3a were decreased in OO fed mice, an effect that was attenuated by EX527 (Fig. 4G–H). To further test the importance of SIRT1 in MUFA-mediated signaling, we determined gene expression of key PGC-1α/PPARα oxidative genes (Fig. S5A). Consumption of the OO diet universally increased the expression of PGC-1α/PPAR-α targets genes, but these effects were ablated with EX527. The increased gene expression in OO fed mice corresponded to increased protein abundance of UCP1, PGC-1α, CPT1α, and various respiratory chain complex proteins in the liver (Fig. 4I–J and Fig. S5B). In addition to hepatic changes, histological examination of interscapular brown adipose tissue exhibited smaller LDs and decreased TAG, (Fig. 4K–M) indicative of enhanced thermogenesis. The smaller LDs corresponded to increased protein abundance of oxidative metabolism genes including UCP1, PLIN5, PGC-1α CPT1 α and complex I, II, III and IV of the respiratory chain (Fig. 4N–O and Fig. S5C). Similarly, OO feeding decreased LD size in inguinal white adipose tissue (Fig. S5D–E) along with increased protein abundance of UCP1, PGC-1α CPT1 α and respiratory chain complexes I, II and IV (Fig. S5F–H). Taken together, these findings define an ATGL-MUFA-SIRT1 axis that is critical for LD signaling to promote PGC-1α/PPAR-α and oxidative metabolism.
Fig. 4. MUFAs increases oxidative metabolism in vivo through SIRT1 activation.
A) Body weight of mice fed a control diet (CTRL) or a diet enriched in olive oil (OO). Three days prior to sacrifice mice were injected with 10mg/kg of EX527. Body weights were determined before and after EX527 treatment. B-C) Serum β-hydroxybutyrate and free fatty acid levels in C57Bl/6 mice were fed diets low in MUFAs (CTRL; black bars) or enriched in 18:1 (OO; white bars). A subset of mice were injected with 10 mg/kg daily of the SIRT1 inhibitor EX527 for 3 days prior to sacrifice. D-E) H&E staining of liver tissues from CTRL and OO fed mice. LD size was determined using 3–4 images from 2–3 mice per group. F) Quantification of TAG in liver samples was determined using 3–4 mice per group. G) Western blots of total and acetylated-PGC-1α and FOXO3a in livers from 3–4 mice. H) Quantification of immunoprecipitated acetylated-PGC-1α and FOXO3a. I-J) Relative protein expression levels of UCP1, PLIN5, PGC-1α, SIRT1, ATGL, CPT1α, and OXPHOS complex CI-V in liver were determined by Western blotting and quantified by densitometric analysis. K-L) H&E staining of brown adipose tissue (BAT) from CTRL and OO fed mice. LD size was determined using 3–4 images from 2–3 mice per group. M) Quantification of TAG in BAT samples was determined using 3–4 mice per group. N-O) Relative protein expression levels of UCP1, PLIN5, PGC-1α, SIRT1, ATGL, CPT1α, and OXPHOS complex CI-V in BAT were determined by Western blotting and quantified by densitometric analysis. *p<0.05 vs. CTRL diet, #p<0.05 vs. DMSO.
ATGL-mediated activation of PGC-1α requires PLIN5
PLIN5 co-localizes and interacts with ATGL suggesting and these two proteins may have bi- or unidirectional influence over one another (Granneman et al., 2011). Following PKA activation and its phosphorylation, PLIN5 translocates from LDs to the nucleus where it forms a complex with SIRT1 and PGC-1α to promote mitochondrial biogenesis in brown adipose tissue and muscle (Gallardo-Montejano et al., 2016). Given these links between ATGL, PLIN5, SIRT1 and PGC-1α, we investigated the role of PLIN5 in ATGL-mediated activation of SIRT1/PGC-1α. In hepatocytes, we found that ATGL preferentially colocalized with PLIN5-coated LDs (Fig. S6A) and translocated to the nucleus to directly interact with SIRT1/PGC-1α in response to fasting or cAMP signaling (Fig. S6B–D and Table S5). To test if PLIN5 is required for ATGL-mediated signaling, we used CRISPR/Cas9 to knockout Plin5 in mouse L-cells or antisense oligonucleotides (ASOs) to knockdown Plin5 in mouse primary hepatocytes (Fig. S6E–F). Cell-permeable cAMP (8-bromo-cAMP) and ATGL overexpression both individually and synergistically enhanced PGC-1α transcriptional activity in wild-type cells as expected, but ablation of Plin5 abrogated these effects (Fig. 5A–B). Rescuing the expression of PLIN5 in Plin5 knockout cells restored PGC-1α activity, but this restoration required ATGL lipolytic activity (Fig. 5A). Moreover, the increase in PGC-1α activity in response to PLIN5 overexpression and cAMP was blocked by chemical inhibition of ATGL or SIRT1 (Fig. 5B). Similarly, adenoviral-mediated ATGL overexpression in the livers of mice increased the expression of PGC-1α/PPAR-α target genes, however, ASO-mediated ablation of Plin5 negated these effects (Fig. 5C and Fig. S6F). In response to cAMP, PLIN5 undergoes PKA-mediated phosphorylation at Ser155 (Gallardo-Montejano et al., 2016; Pollak et al., 2015), which was verified with an antibody we generated specific for this phosphorylation site (Fig. S6G). Using PLIN5 phospho-mimetic (pM; S155E) and phospho-dead (pD; S155A) mutants we confirmed that this phosphorylation is both necessary and sufficient for nuclear translocation (Fig. S6H). Consistent with an important role of translocation, expression of the PLIN5-pD in the knockout cells was unable to restore the response to cAMP on PGC-1α activity (Fig. 5D). Expression of a PLIN5-pM increased basal PGC-1α transcriptional activity, but negated the response to cAMP (Fig. 5D). However, the increase in basal PGC-1α activity in the PLIN5-pM expressing cells required ATGL activity suggesting that ATGL-catalyzed lipolysis is critical for PLIN5-mediated signaling. To determine if PLIN5 translocation is dependent on ATGL-catalyzed lipolysis, we knocked-down Atgl in mouse primary hepatocytes and liver as described previously (Ong et al., 2011). PLIN5 was still able to translocate to the nucleus in response to cAMP (cells) or overnight fasting (livers) following Atgl knockdown (Fig. 5E–F). Thus, these data show that ATGL and PLIN5 are co-obligatory to increase PGC-1α activity and that ATGL inhibition does not influence PKA-mediated translocation of PLIN5 to the nucleus.
Fig. 5. ATGL-mediated activation of PGC-1α requires PLIN5.
A) PGC-1α luciferase reporter assays in wild-type or Plin5 knockout mouse L-cells transduced with control (Gfp) or Atgl adenoviruses. Rescue experiments were performed with overexpression of a plasmid harboring mCherry-Plin5. ATGL inhibition was achieved by the addition of 30 μM ATGListatin (ATGLi) (n=6). *p<0.05 vs. veh, #p<0.05 vs. wild-type, @p<0.05 vs. within treatment vehicle. B) PGC-1α luciferase reporters in primary hepatocytes transfected with control (Ctrl) or Plin5 ASOs. Treatment with EX527 (30 μM) was used to inhibit SIRT1 (n=6–12). *p<0.05 vs. veh, #p<0.05 vs Ctrl ASO, @p<0.05 vs within treatment vehicle. C) PGC-1α/PPAR α target gene expression in livers of mice treated with control or Plin5 ASOs and adenoviruses harboring Gfp or Atgl (n=6–8). *p<0.05 vs. GFP, #p<0.05 vs. Con ASO. D) PGC-1α luciferase reporters in wild-type or Plin5 knockout mouse L-cells transfected with an empty mCherry-vector (EV), mCherry-Plin5, mCherry-Plin5-pD, or mCherry-Plin5-pM (n=6). *p<0.05 vs veh, #p<0.05 vs. wild-type, @ p<0.05 vs. veh. treated Plin5-pM cells. E) Confocal imaging of mCherry-Plin5 transfected cells pretreated with vehicle or the PKA inhibitor H89 (15 μM) for 1 hr followed by addition of 8-bromoadenosine 3’,5’-cyclic monophosphate (cAMP; 1mM) for an additional hour. Cells were also transduced with control or shRNA adenoviruses (repeated with 3 individual hepatocyte isolations). F) Livers from 4 and 16 hr fasted mice were harvested and subjected to histological sectioning and immunostaining to detect PLIN5 (n=3).
PLIN5 binds fatty acids
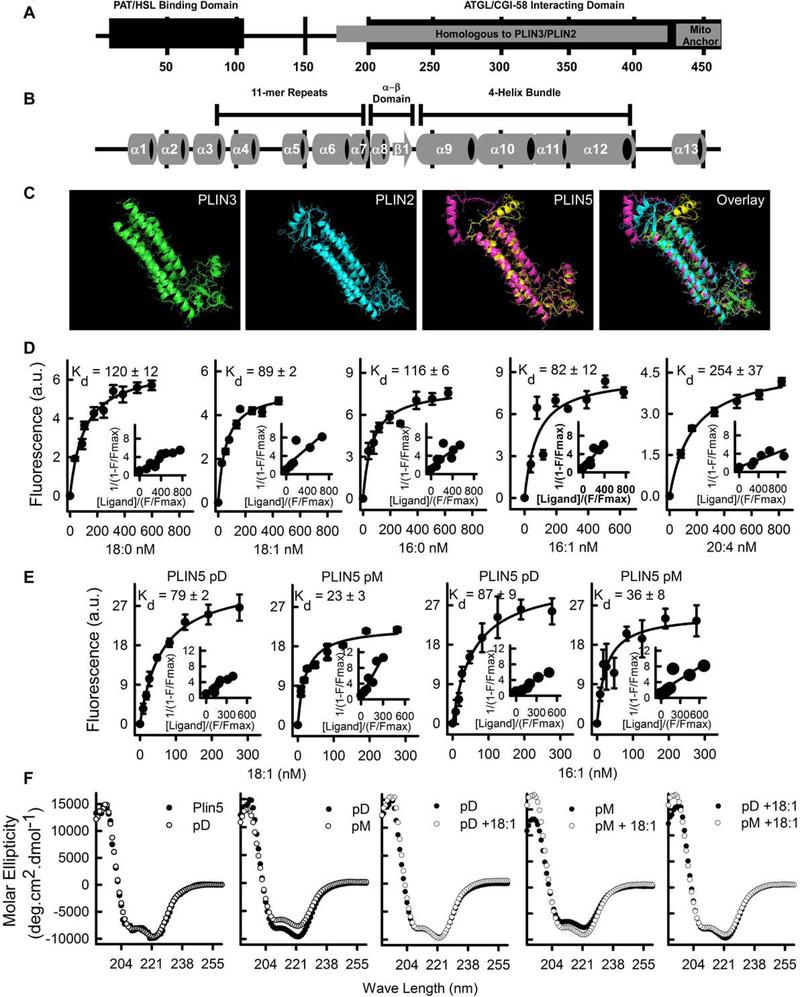
The above data suggest that an intrinsic function of PLIN5, independent of translocation, is critical for its signaling properties. Aligning the PLIN5 amino-acid sequence to its family members highlights a C-terminal region of PLIN5 that is homologous to PLIN2 and PLIN3 (Fig. 6A and Fig. S7A). The C-terminal region of PLIN2 and PLIN3 are α-helical and consist of one α–β domain, and a 4-helix bundle that comprise a hydrophobic pocket identified to bind fatty acids (Hickenbottom et al., 2004; Najt et al., 2014). We therefore carried out secondary structure analysis of PLIN5 using several prediction programs. Results from the algorithms predict that an α–β domain and 4-helix bundle found in PLIN2/3 exists in PLIN5 (Fig. 6B). We next constructed a homology 3D-model of the structure of C-terminal residues 164–390 of murine PLIN5 (Fig. 6C) aligned with murine PLIN3, which shares 42% sequence identity. Based on the structural model, PLIN5 contained a hydrophobic binding pocket of sufficient size and character to bind lipids similar to PLIN2 (Najt et al., 2014).
Fig. 6. PLIN5 is a fatty acid binding protein.
A) PLIN5 contains several domains of interest including the PAT/HSL binding domain, an ATGL/CGI-58 binding domain, a mitochondria anchor, and a region homologous to PLIN3/PLIN2. B) Based on prediction software (SABLE2, SAM and PsiPRED) and the known X-ray crystal structure of the homologous PLIN3 protein, the predicted secondary structure of PLIN5 contains 13 α-helices and 1 small β strand interconnected by random coils and unordered structure. C) The X-ray crystal structure of PLIN3 was used to homology model the C-terminal region of PLIN5. The crystal structure of PLIN3 is shown on the farthest left panel (residues 191–437, PDB entry 1SZI). PLIN2 homology model from (Najt et al., 2014) is shown in the second to the left panel, while the PLIN5 models are shown second from the right. Two structures, yellow and pink, were generated by the homology modeler Phyre2 each having a high-probability score. The region that differed between the two PLIN5 models was an α-helix connected to the 4-helix bundle by unordered structure. The structure contains a 4-helix bundle, which together with an α-β domain form the cleft, that when overlaid with the PLIN2 model aligns with the lipid binding pocket outlined in (Najt et al., 2014). D) The PLIN5 binding affinity for fatty acids was determined when recombinant protein was tittered with increasing amounts of ligand using a quenching of tryptophan fluorescence assay (n=4). E) PLIN5-pD (S155A) and PLIN5-pM (S155E) binding affinities for MUFAs were determined in a similar manner as PLIN5 (n=4). F) Circular dichroic analysis of PLIN5-pD and PLIN5-pM. Far ultraviolet (UV) circular dichroic (CD) spectra of PLIN5, PLIN5-pD and PLIN5-pM was shown in the presence or absence of ligand. Each spectrum represents an average of ten scans repeated in triplicate.
To determine if PLIN5 binds fatty acids, we employed tryptophan fluorescence assays with recombinant full-length murine PLIN5 (Fig. S7B). Saturable binding curves for stearic acid (18:0), oleic acid (18:1), palmitic acid (16:0), palmitoleic acid (16:1) and arachidonic acid (20:4) were observed (Fig. 6D). Kd values ranged from of 82 to 254 nM with the highest affinity determined with the monounsaturated fatty acids (MUFAs) 16:1 (82±12 nM) and 18:1 (89±2 nM). Similar analyses of the PLIN5 phospho-mutants with the two MUFAs were performed. PLIN5-pD exhibited a similar Kd to wild-type PLIN5, but the PLIN5-pM exhibited a 3.4- and 2.4-fold increase in binding affinity for 16:1 (23±3 nM vs. 79±2 nM) and 18:1 (36±8 nM vs. 87±9 nM), respectively (Fig. 6E). Fluorescence binding and displacement assays using (1,8,-ANS) as described above were conducted to further verify lipid binding (Kane and Bernlohr, 1996). Displacement of the bound fluorophore with natural ligands resulted in Ki values 4±0.13 μM, 9.9±0.07 μM, 17.7±0.04 μM, and 44.9±0.02 μM for 18:1, 16:1, 18:0, and 22:4, respectively (Fig. S7C and Table S6). To examine how phosphorylation affects function, the secondary structures of PLIN5 and the phosphorylation mutants, in the presence or absence of ligands, were analyzed by circular dichroism (CD). In absence of ligand, the CD spectrum for PLIN5-pD was not statistically different from PLIN5 (Fig. 6F). Analysis of CD spectra revealed that PLIN5-pM exhibited decreased helical content, and increased beta and random coil content over PLIN5-pD and wild-type suggesting that phosphorylation of PLIN5 causes the protein to undergo large conformational changes (Table S7). The addition of 18:1 robustly altered the shape of the CD spectrum of PLIN5-pM while more subtle changes were observed with the PLIN5-pD. These alterations were reflected by increases in both rigid and disordered helices while decreasing the percentage of β-sheets (Fig. 6F, Table S7). In summary, the CD results were consistent with the predicted secondary structure and indicated that the proteins were sensitive to fatty acid binding. Phosphorylation of PLIN5 alters the overall structure of the protein shifting from helical to β-sheet/β-turn, while addition of a fatty acid changes the overall fold of PLIN5-pM back to a more helical fold.
MUFA allosteric regulation of SIRT1/PGC-1α requires PLIN5
The link between lipolysis to changes in SIRT1/PGC-1α signaling and oxidative gene expression is enhanced in the presences of MUFAs while signaling between LDs and SIRT1/ PGC-1α requires PLIN5. We therefore tested the effects of PLIN5 deletion of MUFA activation of SIRT1. Studies utilizing L-Cells lacking Plin5 revealed that PLIN5 was required for 18:1 mediated regulation of PGC-1α activity (Fig. S7D). Transfection of the Plin5-pD mutant into the Plin5 knockout cells was unable to restore PGC-1α activity (Fig. S7E). Rescuing PLIN5 expression with transfection of the Plin5-pM plasmid restored basal PGC-1α activity, but the cells were unable to respond to cAMP and/or 18:1 loading suggesting that PLIN5 has to be present on the LD surface to acquire the fatty acid prior to nuclear translocation and SIRT1 activation (Fig. S7F).
DISCUSSION
Numerous studies have linked lipolysis, mediated through manipulation of ATGL or other LD proteins, to changes in PGC-1α/PPARα signaling and oxidative gene expression (Ahmadian et al., 2009; Haemmerle et al., 2011; Khan et al., 2015; Ong et al., 2011). This signaling is thought to play a key role in increasing the oxidative capacity of the cell to match the supply of lipolytic-supplied fatty acids. PLIN5 has been widely studied as a key LD protein that promotes oxidative metabolism and uncouples LD accumulation from lipotoxicity and insulin resistance (Bosma et al., 2013; Mason et al., 2014; Pollak et al., 2015; Sztalryd and Brasaemle, 2017; Wolins et al., 2006). Our data identify a novel role of PLIN5 in fatty acid binding and transport as an underlying mechanism that couples lipolysis to SIRT1/PGC-1α signaling (Fig. 7). In addition, PKA-mediated phosphorylation is a key event that both increases the ability of PLIN5 to bind fatty acids, preferentially MUFA, and trigger its translocation to the nucleus. These finding also implicate potential interactions between dietary lipids, PLIN5 expression and dietary or environmental stimuli, such as fasting, caloric restriction or exercise that increase cAMP/PKA signaling to promote lipolysis. Indeed, PLIN5 expression is induced by fasting, caloric restriction and exercise (Nogueira et al., 2012; Shepherd et al., 2013; Wolins et al., 2006). Taken together, these data unravel a novel mechanism through which PLIN5 elicits is protective effects against lipotoxicity and couples lipolysis to changes in oxidative metabolism (Fig. 7).
Fig. 7. Monounsaturated fatty acids traffic via PLIN5 to allosterically activate SIRT1.
A model describing lipid droplet derived monounsaturated fatty acids allosterically modulating SIRT1 via PLIN5.
SIRT1 has a wide-range of biological functions including chromatin structure maintenance, cell cycle control, metabolism and the regulation of healthspan (Banks et al., 2008; Bordone et al., 2007; Pfluger et al., 2008). Resveratrol and other naturally occurring polyphenols activate SIRT1 in a substrate dependent manner (Borra et al., 2005; Cao et al., 2015; Feldman et al., 2012) similar to what we observed with the selective activation of SIRT1 towards PGC-1α and FOXO3a, but not H3, in response to MUFAs. A previous study has shown that fatty acids do not modulate SIRT1 activity (Feldman et al., 2013). However, this study employed a fixed concentration of 18:1 (100 μM) and used the H3 peptide as a substrate. As we have shown (Fig. 1 and 2), MUFA do not activate SIRT1 at concentrations above 1 μM and do not enhance SIRT1 activity towards the H3 peptide. Our data suggest that in addition to sensing intracellular redox (NAD) and exogenous dietary compounds (e.g. resveratrol), SIRT1 also acts as a nutrient sensor to coordinate LD catabolism with downstream metabolic pathways responsible for the metabolism of fatty acids. The implications of these findings are widespread given the critical role of SIRT1 in many aging related disease and lifespan regulation directly linked to nutrient sensing. These studies may also provide a biologically feasible mechanism that underlies the health benefits of MUFAs (Fig. 7). MUFAs are common in many foods, but are enriched in a variety of foods including nuts, avocados, and olive oil. Evidence from model organism studies through clinical trials bare out the effects of MUFA and/or olive oil on improvements in oxidative metabolism and energy expenditure (Børsheim et al., 2006; Palou et al., 2002; Shin and Ajuwon, 2018) and in disease prevention and lifespan extension (Buckland and Gonzalez, 2015; Estruch et al., 2006; Han et al., 2017; Salas-Salvadó et al., 2011; Schwingshackl and Hoffmann, 2014a; Schwingshackl and Hoffmann, 2014b; Schwingshackl et al., 2011; Trichopoulou et al., 2005). Importantly, MUFA are regarded as one of the key components of the Mediterranean Diet, which is well-established to have wide-ranging health benefits including reduced aging related diseases and overall mortality (Sofi et al., 2010). The discovery that resveratrol, which is enriched red wine, activated SIRT1 was proposed as a mechanism through which a component of the Mediterranean Diet could promotes health benefits. However, doses of resveratrol needed to elicits its effects from diet alone far exceeds possible intake (Weiskirchen and Weiskirchen, 2016). While undoubtedly a plethora of components in the Mediterranean Diet contribute to its positive effects on health, the data presented herein provide at least one feasible biological mechanism that may underlay these well-established benefits.
In summary, these studies identify the MUFAs 18:1 and 16:1 as endogenous, non-substrate modulators of SIRT1 that can target the deacetylase to specific protein substrates. Additionally, these findings highlight the importance of LD composition and catabolism as a key regulatory node that integrates physiological inputs (dietary lipids and lipolytic stimuli) to coordinate cellular signaling and metabolism.
STAR METHODS AND KEY RESOURCES
LEAD CONTACT AND MATERIALS AVAILABILITY
The lead contact for this study is Douglas Mashek (dmashek@umn.edu). All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal protocols were approved by the University of Minnesota Institutional Animal Care and Use Committee. All cell lines used in this study were cultured in an atmosphere of 37°C, 5% CO2, 95% humidity. The cell lines are outlined in the legends and STAR methods table.
METHOD DETAILS
Mice and adenovirus administration
Male 6–8 week old C57Bl/6 mice were obtained from Harlan Laboratories and housed under controlled temperature and lighting (20–22°C; 14:10-h light-dark cycle). The mice were fed a purified control diet (TD 94045; Harlan Teklad Premier Laboratory) and acclimatized for 1 week before any experimental procedure.
Cell culture
Primary hepatocytes were isolated as described previously (Bu et al., 2009). Primary hepatocytes were cultured at 37°C under 5% CO2 in M199 media containing 23 mM HEPES, 26 mM sodium bicarbonate, 10% FBS, 50 IU/ml penicillin, 50 μg/ml streptomycin, 100 nM dexamethasone, 100 nM insulin, and 11 mM glucose and 1 mM carnitine. One hour before treatment with 1 mM of the cAMP analog 8-bromoadenosine 3’,5’-cyclic monophosphate, cells were washed twice with PBS, then allowed to incubate in the same M199 media minus serum and insulin with the addition of the lipase inhibitor ATGListatin (ATGLi; 30 μM), the cAMP-dependent protein kinase a (PKA) inhibitor H89 (15 μM), or the SIRT1 inhibitor EX527 (30 μM). Sirt1 knockout mouse embryonic fibroblasts (MEFs) were provided by Michael McBurney (University of Ottawa). Sirt1 knockout cells were cultured in DMEM supplemented with 10% FBS, 50 IU/ml penicillin, 50 μg/ml streptomycin. Prior to cAMP treatment, cells were cultured in DMEM without FBS. Plin5 CRISPR/Cas9 knockout L-cells were generated by the University of Minnesota Genome Engineering Shared Resource Center and validated in house (Fig. S1G). Plin5 knockout cells were cultured in DMEM supplemented with 10% FBS, 50 IU/ml penicillin, 50 μg/ml streptomycin. Prior to cAMP treatment, cells were cultured in DMEM without FBS. Adenoviruses to manipulate ATGL expression were generated and used as previously described (Khan et al., 2015; Ong et al., 2014; Sathyanarayan et al., 2017). For single adenovirus treatments, hepatocytes were treated with adenovirus 24–48 hrs prior to experimental set-up (AdAtgl or Ad-Gfp and control or Atgl shRNA adenoviruses).
PGC-1α reporter assay
Several cell lines were utilized for reporter assays including MEFs, L-cells, primary mouse hepatocytes, Sirt1 knockout MEFs, and Plin5 knockout L-cells. In all the reporter experiments cells were transfected with firefly luciferase reporter plasmids (TK-MH-UASluc), control Renilla luciferase (pRLSV40), and GAL4-Pgc-1a (pCMX-GAL4-Pgc-1a) using Targetfect Hepatocyte reagent (Targeting Systems) or Lipofectamine 3000 (ThermoFisher, Grand Island, NY). For overexpression or rescue experiments the following plasmids were co-transfected into cells; CFP-Plin2 (McIntosh AL et al., 2012) (Barbara Atshaves, Michigan State University), mCherry-Plin5 (GeneCopoeia, Rockville MD), eGFP-Atgl, eGFP-Sirt1, eGFP-Pgc-1α, mCherry-Plin5 (S155A; pD), mCherry-Plin5 (S155D; pM). Cells were stimulated with 1 mM cAMP analog for 6–8 hrs. Following treatments with indicated drugs, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Fitchburg, WI). Firefly luciferase activity was normalized to the co-expressed Renilla luciferase activity.
Site-directed mutagenesis
To generate mCherry-Plin5 S155A, S155E, S155D mutants, QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent; Santa Clara, CA) was used in accordance with the manufacturer’s protocol.
Antisense Oligonucleotides
Plin5 and control anti-sense oligonucleotides (ASO) were obtained from Ionis Pharmaceuticals (Carlsbad, CA). The ASO were both used at 40 mg/kg, prepared in sterile saline, and delivered via intraperitoneal injection twice a week for 3 weeks. Knockdown was confirmed through mRNA (RT-PCR) and protein (Western blot) analysis. For use in primary mouse hepatocytes, ASOs were complexed with Effectene transfection reagent (Qiagen; Venlo, Netherlands) following the manufacturer’s protocol.
RNA isolation and RT-PCR analysis
RNA was extracted with Trizol from liver tissues followed by reverse-transcription with SuperScript® VILO™ cDNA Synthesis Kit (Invitrogen) to generate cDNA. Gene expression was quantified as described previously (Khan et al., 2015; Najt et al., 2016; Ong et al., 2014).
Live cell and fluorescence resonance energy transfer (FRET) imaging
Fluorescence imaging and FRET experiments were performed with cells seeded at a density of 50,000 cells/plate on Mat-Tek cover-glass plates (Ashland, MA). The plasmid expressing mCherry-Plin5, eGFP-Pgc-1α, and eGFP-Sirt1 were purchased from GeneCopeia (Rockville, MD) and transfected into mouse primary hepatocytes using Targeting Systems (El Cajon, CA) Targefect Hepatocyte reagent. Digital images were acquired using a Nikon A1Rsi Laser Scanning Confocal Imaging System (LSCIS; Nikon, Melville, NY) equipped with 405 nm, 488 nm, 561 nm, and 640 nm laser, four channel GaSP detectors, and a 60x water immersion objective. To determine subcellular localization of PLIN5 under basal and stimulated conditions, mCherry-Plin5 overexpressing and control cells were cultured on glass bottom dishes using M199 media containing 23 mM HEPES, 26 mM sodium bicarbonate, 10% FBS, 50 IU/ml penicillin, 50 μg/ml streptomycin, 100 nM dexamethasone, 100 nM insulin, 11 mM glucose, 1 mM carnitine and Hoechst 33342 nuclear dye for 20 min. Cells were washed twice with PBS, then allowed to incubate in the same M199 media minus serum and insulin for 1 hr prior to stimulation. For probe excitation, the A1Rsi LSCIS utilized the 561 nm diode laser (mCherry), and the 405 nm laser line (Hoechst 33342) to acquire images of the cells by sequential excitation. The mCherry-Plin5 expressing cells were imaged showing PLIN5 localization at time zero, then treated with 1 mM of the cAMP analog 8-bromoadenosine 3’,5’-cyclic monophosphate and imaged every 10 min for 1.5 hrs. Image files were analyzed using NIS-Elements software. For colocalization experiments, NIS-Elements was used to identify mCherry-PLIN5 pixels colocalized with Hoechst 33342. Z-stack or multiple focal planes were imaged to ensure compartmentalization and localization.
To determine the molecular association between PLIN5, SIRT1, and PGC-1α in the nucleus, co-localization and FRET analysis was performed by acceptor photobleaching as described previously (McIntosh AL et al., 2012; Najt et al., 2014; Senthivinayagam et al., 2013). Briefly, primary hepatocytes were co-transfected with mCherry-Plin5 (acceptor) and eGFP-Sirt1 (donor) or eGFP-Pgc-1α (donor). Prior to imaging, cells were washed twice with PBS and placed in M199 media containing 23 mM HEPES, 26 mM sodium bicarbonate, 50 IU/ml penicillin, 50 μg/ml streptomycin, 11 mM glucose, and 1mM carnitine. Digital images were taken under basal conditions, then the cells were treated with 1 mM 8-bromoadenosine 3’,5’-cyclic monophosphate for 1 h. Images were acquired utilizing the 561 nm diode laser (mCherry) and the 488nm laser (eGFP) to acquire images of the cells by sequential excitation. Co-localization of the two probes was determined as described above. Upon establishing the two probes co-localized, acceptor photobleaching FRET experiments were performed to measure the increase in donor (eGFP) emission upon photobleaching of the acceptor (mCherry) as described elsewhere (McIntosh AL et al., 2012; Senthivinayagam et al., 2013). To calculate the FRET efficiency (E), representing the efficiency of energy transfer between donor and acceptor, the following equation was used: E = 1-(IDA/ID) where IDA is donor fluorescence intensity before acceptor photobleaching and ID is the donor fluorescence intensity after acceptor photobleaching. An average E value was calculated from eGFP fluorescence emission increase after photobleaching. The intermolecular distance R between PLIN5 and SIRT1 or PLIN5 and PGC-1α was calculated from the equation E = 1/(1-(R/Ro)6), where E is experimentally determined and Ro is the Foster radius for the eGFP-mCherry FRET pair. For the FRET efficiency images, analysis was performed in MetaMorph 7.5 (Molecular Devices, Sunnyvale, CA). Images were filtered to remove randomized noise by using a low pass filter. The filtered images of the donor emission before acceptor photobleaching were subtracted from the image after acceptor photobleaching. The resultant image was divided by the image of donor emission after acceptor photobleaching and multiplied by 100 to generate bar-scale FRET efficiencies.
Tissue histology
Tissue samples (25–75 mm3 segments) were fixed in a 10% buffered formalin solution at room temperature overnight, then stored in alcohol until embedded in paraffin, section (4–6m thickness). Immunohistochemistry was performed as described previously (Najt et al., 2014; Sathyanarayan et al., 2017). Sections were probed with anti-PLIN5 (Progen, Heidelberg Germany), anti-ATGL (CellSignaling Tech, Danvers MA), and anti-PLIN2 (prepared as previously described in (Atshaves et al., 1999)). Histological processing was done at the histopathology laboratory at University of Minnesota. Fluorescent imaging was performed on a Nikon A1Rsi Laser Scanning Confocal Imaging System (LSCIS; Nikon, Melville, NY). H&E slides were imaged on a Leica DM5500B microscope (Leica Microsystems) at 5x-20x magnification.
Western blotting
Cell lysates (30–50 μg protein) were separated on 10–12% tricine gels using a Mini-Protean II cell (Bio-Rad lab, Hercules, CA) system at constant amperage (30 mA per gel) for about 3 hrs. Proteins were then transferred onto PVDF membranes at constant voltage (90 V) for 1.5 hrs. Blots were stained with Ponceau S to confirm uniform protein loading (Aldridge et al., 2008; Willenborg et al., 2005) before blocking in 5% BSA in TBST (10 mM Tris-HCl, pH 8, 100 mM NaCl, 0.05% Tween-20) for 1 hr. Blots were incubated with specific poly- or monoclonal antibodies overnight and were developed with IRDye 800CW (LI-COR) or IRDye 680RD (LI-COR) secondary antibodies. To visualize the bands of interest, blots were scanned using the LI-COR Odyssey imaging system (Lincoln, NE). Protein bands were quantitated by densitometric analysis after image acquisition using NIH Scion Image to obtain relative protein levels expressed as integrated density. All values were normalized to β-actin expression or PonceauS staining. Antibodies were purchased or obtained from the following sources; Total-Plin5 (Progen; Heudelberg, Germany), Histone H3, SIRT1, Acetylated Lysine (Cell Signaling Technologies; Danvers, MA), PGC-1α (MilliporeSigma; Burlington, MA), phospho-PLIN5 (NeoBioLab targeting; Cys-LARRGRRW(pS)VELK), PLIN2 [Barbara Atshaves developed in (Atshaves et al., 1999)].
Cellular fractionation
Nuclear and cytoplasmic fractions from mouse tissues were prepared as previously described in (McIntosh et al., 2010; Muratore et al., 2018; Storey et al., 2012). Briefly, livers were excised, minced in homogenate buffer [10 mM Tris-base pH 7.0 with protease (Complete protease inhibitor cocktail, Roche, Basel Switzerland), phosphatase (PhosSTOP, Sigma-Aldrich, St. Louis, MO), and deacetylase inhibitors (De-acetylase Cocktail, MedChem Express, Monmouth Junction, NJ)], placed in a nitrogen cavitator and charged with nitrogen to 150 psi. The cavitator was submerged in ice allowed to lysis the tissue for 15 min. Liver lysates were harvested and spun at 1,000 × g for 10 min to obtain a post-nuclear supernatant. The pellet from this spin was suspended in nuclear isolation buffer (10 mM Tris-base pH 7.5, 10 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, with protease, phosphatase, and acetylase inhibitors). The re-suspended nuclear fraction was spun at 2,000 × g for 20 min, the supernatant was discarded and the pellet that contained nuclei kept. Nuclei were suspended in homogenate buffer described above supplemented with 10 mM KCL and 2 mM MgCl2. The post-nuclear supernatant was loaded onto a sucrose step gradient (4 ml 35% sucrose, 4 ml 25% sucrose) and centrifuged at 36,000 rpm in a Beckman SW41 swinging bucket rotor for 4 hrs. The LD fraction appeared as white film at the top of the tube, which was removed with a Pasteur pipette. Both the LD and nuclear fraction from fed and 16 hr fasted mice were snap frozen for latter analysis via western blotting.
PLIN5 structural prediction and analysis
The secondary structure of PLIN5 was predicted by the PredictProtein server https://www.predictprotein.org/. Secondary structural predictions generated by PredictProtein were consistent with further analysis by PSIPRED, SAM, and SABLE2. Similar to methods used to predict the lipid binding domain of PLIN2 (Najt et al., 2014) the PLIN5 C-terminal domain structure was modeled by homology to the structure of residues 209–431 in PLIN3 from Protein Data Bank entry 1SZ1 by using Modeler, as implemented in the ModWeb Web server (https://modbase.compbio.ucsf.edu/modweb/) and the online webserver Phyre2 (www.sbg.bio.ic.ac.uk/phyre2/).
Expression and purification of recombinant proteins in Escherichia coli cells
Recombinant PLIN5 was purified using the following procedure. Plin5 was cloned into pTEV6-HIS-MBP expression construct. Protein was overexpressed in E. coli host strains BL21 codon plus* (Agilent, Santa Clara, CA) and grown at 37°C in 1L cultures containing 2XYT sterile fermentation media 100 μg/ml Carbenicillin until OD600 = 0.6. Once the desired OD was obtained, the culture was cooled to 18°C and induced with IPTG. After 16 hrs, cells were harvested by centrifugation and re-suspended in buffer A (50 mM HEPES, 500 mM NaCl, 10% glycerol, pH 8.0). The re-suspended cells were cracked by sonication. Lysates were clarified by ultra-centrifugation (4°C for 60 min at 48,000 × g). The clarified lysate was applied to a amylose resin column (Thermo, Waltham, MA) equilibrated with buffer A at a flow rate of 0.5 ml/min. The column was washed with 20 column volumes of buffer B (50 mM HEPES, 300 mM NaCl, 10%glcerol 10 mM Maltose pH 8.0) at a flow rate = 1.0 ml/min. The protein was eluted using buffer C (50 mM HEPES, 300 mM NaCl, 250 mM Maltose, pH 8.0; flow rate = 1.0 ml/min) where the purified protein was identified by UV280 signal. The elution pool was diafiltered for 5 diavolumes of buffer D (25 mM HEPES, 100 mM NaCl, 5 mM DTT, 10% glycerol, pH 7.5) and then checked for purity by SDS-page analysis. The purified His-MBP-Plin5 protein was incubated with TEV-protease to cleave the His-MBP tag. The cleaved product was applied to Ni-NTA column and the flow through was collected. Purity of the un-tagged Plin5 protein was determined by SDS-page analysis. For binding and activity assays, purified protein was buffer exchanged to the appropriate buffer using a Ultrcel-50K (EMD Millipore, Darmstadt, Germany).
Recombinant SIRT1 was purified as previously described with some modifications (William C Hallows, Susan Lee, and John Denu, PNAS 2006). Briefly, proteins were overexpressed in E. coli host strains BL21 codon plus* (Agilent, Santa Clara, CA) and grown at 37°C in 15L cultures containing 2X YT sterile fermentation media 100 μg/ml Carbenicillin until OD600 = 3.0. Once the desired OD was obtained, the culture was cooled to 18°C and induced with IPTG. After 5 h, cells were harvested by centrifugation and re-suspended in buffer A (50 mM HEPES, 300 mM NaCl, 10 mM Imidazole, pH 8.0). The re-suspended cells were cracked by two passes through a microfluidizer (G30Z and H10Z interaction chambers) at 16,500 psi. Lysates were clarified by centrifugation (4°C for 30 min at 15,000 × g) and filtration through a 1μm filter. The clarified lysate was applied to a 20 ml Hispur-Ni2+ column (Thermo, Waltham, MA) equilibrated with buffer A at a flow rate of 5.0 ml/min. The column was washed with 10 column volumes of buffer B (50 mM HEPES, 300 mM NaCl, 20 mM imidazole pH 8.0) at a flow rate = 5.0 ml/min. The protein was eluted using buffer C (50 mM HEPES, 300 mM NaCl, 500 mM imidazole, pH 8.0; flow rate = 8.0 ml/min) where the purified protein was identified by UV280 signal. The elution pool was diafiltered for 5 diavolumes of buffer D (25 mM HEPES, 100 mM NaCl, 5 mM DTT, 10% glycerol, pH 7.5) and then checked for purity by SDS-page analysis. For binding and activity assays, purified protein was buffer exchanged to the appropriate buffer using a Ultrcel-50K (EMD Millipore, Darmstadt, Germany).
Intrinsic tryptophan fluorescence binding studies
The binding of fatty acids to PLIN5 and SIRT1 was examined by measuring the fluorescence quenching of PLIN5 and SIRT1 tryptophan residues after addition of ligand as described previously (Najt et al., 2014). In brief, the intrinsic tryptophan fluorescence of PLIN5 and SIRT1 (150 nM in 10 mM NaH2PO4, pH 7.5) was monitored from 300 to 400 nm after excitation at 295 nm (to minimize interference from tyrosine fluorescence) both before and after addition of increasing increments of fatty acids (fatty acids were dissolved in 200 proof spectroscopically clear ethanol) using a Cary Eclipse fluorescence spectrophotometer. Data was corrected for back ground scatter originating from the buffer and ligand without protein present. The intrinsic tryptophan fluorescence in the presence of different concentrations of ligand was plotted as the maximum fluorescence difference (ΔF = F₀-F) vs. ligand concentration to yield a saturation curve where F and F₀ were the measured fluorescence emission intensity of the protein solution in the presence and absence of ligand, respectively. The dissociation constant Kd was determined from the double reciprocal plot of the saturation curve. Linear regression of 1/[1-(F/Fmax)] versus [ligand]/(F/Fmax) yielded a slope = 1/ KD and ordinate intercept = nE₀/ Kd where F represented fluorescence intensity at a given concentration of ligand, Fmax was the maximal fluorescence. E₀ was the protein concentration, and n equaled the number of ligand binding sites.
Circular-Dichroic analysis of secondary structure
The far UV circular dichroic (CD) spectra of each protein was measured in phosphate buffer (10 mM NaH2PO4, pH 7.5 with 10 mM NaCl) in the presence and absence of ligand. PLIN5 was assayed at 3 μM and SIRT1 was assayed at 5 μM. Experiments were performed at 25°C in a 1 mm path length crystal cuvette using a JASCO J-815 CD spectrometer (JASCO Analytical Instruments, Easton, MD). Experiments with ligand were allowed to incubate 2–5 min prior to each scan to allow maximal protein-ligand interaction. CD spectra were recorded from 270 to 190 nm at a scan rate of 50 nm/min with a time constant of 1s and bandwidth of 2 nm. For each experiment, 10 iterations were performed in triplicate. Secondary structure analysis was carried out using the CDSSTR analysis program (Sreerama and Woody, 2000) with results reported as percentages of regular α-helices, distorted α -helices, regular β-strands, and distorted β-strands, turns and unordered structures.
1,8-ANS displacement assays for lipid binding
PLIN5 and SIRT1 lipid binding was measured using a 1-anilinonaphthalene 8-sulfonic acid (1,8-ANS) displacement assay as previously described (Kane and Bernlohr, 1996). 1,8-ANS was dissolved in absolute ethanol and its concentration determined spectrophotometrically (ε372 =8,000cm−1, M−1). Increasing amounts of protein were incubated with 500 nM 1,8-ANS in 50 mM sodium phosphate pH 7.5. The samples were mixed for 1 min under dim light and the fluorescence was measured in a Perkin Elmer 650–10S fluorescence spectrophotomer. Slit widths of 4 nm were used for both excitation and emission. Fluorescence intensity was plotted versus increasing protein concentration to generate a binding curve. Binding parameters were determined using Scatchard analysis. Upon establishing 1,8-ANS binding to PLIN5 and SIRT1, various lipids were assessed for their ability to displace the fluorescent probe. PLIN5 (1 μM) and SIRT1 (2.5 μM) were added to 50 mM sodium phosphate pH 7.5 mixed with 500 nM 1,8-ANS at 25°C and the fluorescence signal determined. Increasing concentrations of competitor lipids were added to the 1,8-ANS-protein complex, allowed to mix for 60s and the fluorescence signal recorded. The decay in normalized fluorescence as a function of the competitor concentration was used to determine the displacement curve and the I50. The apparent Ki was calculated using Ki = [I50]/(1+[L]/KD), where Ki is the apparent inhibitor constant, [L] is the free concentration of 1,8-ANS, and the KD is the apparent dissociation constant of a given protein for 1,8-ANS.
Peptide synthesis and purification
Peptides were synthesized using a Protein Technologies SymphonyX synthesizer and using 4-methylbenzydrylamine hydrochloride resin (Iris Biotech GMBH) (Perez et al., 2018). Standard Fmoc-protected amino acid (AA) coupling occurred in the presence of 95 mM HCTU (Iris Biotech GMBH) and 200 mM N-methylmorpholine (Gyros Protein Technologies; S-1L-NMM) over two 20-minute coupling cycles. Fmoc deprotection occurred in the presence 20% piperidine in dimethylformamide (DMF, Iris Biotech GMBH;) over two 5-minute cycles. The peptides were purified to >95% purity by preparative C18 reverse phase HPLC (Agilent 1200 series) over a water/0.1% TFA and acetonitrile/0.1% TFA gradient characterized using HPLC-MS (Agilent 6300 MSD). Peptide substrates were dissolved in mass spectrometry grade water. Absorbance measurements at 280 nm wavelength were used to determine the peptide concentration; peptide extinction coefficients were calculated using ExPASy Bioinformatics Resource portal (https://web.expasy.org/protparam/).
HPLC-MS/MS SIRT1 deacetylation assay
For the deacetylation assay, 1μM recombinant SIRT1 purified from E. coli was incubated with different concentrations of H3K9, PGC-1α, FOXO3a, and p53 acetylated peptides (0–250 μM) and fatty acids in 50 μL reaction mixture (25 mM Tris-base, pH 8.0, 50 mM NaCl, 1 mM DTT, 1 mM NAD+) at 37°C for 60 min. To quench the reactions, 50 μL ice cold acetonitrile was added into the reaction mixture. After centrifuging at 15,000 × g for 15 min, the supernatant was collected, transferred to a clean tube, and blown down under nitrogen. Samples were re-suspended in ultra-high purity MS/MS grade water and analyzed by HPLC-MS/MS Selective Reaction Monitoring (SRM) Analysis. Samples (10 μl) for SRM analysis were subjected to separation using an Shimadzu UFLCXR system coupled to an analytical Waters Acquity BEHc18 column (1.7um particle size, 2.1×50 mm) at 50°C, running a linear gradient of A: 15% acetonitrile/0.55% formic acid, and B: 55% acetonitrile/0.1% formic acid for 12 min at a column flow rate of 400 μl/min. The HPLC was connected to a Applied Biosystem 5500 iontrap fitted with a turbo V electrospray source run in positive mode with declustering potential and collision energies in Supplemental Table 5. The column was cleared with 95% acetonitrile for 2 min and then equilibrated to buffer A for 3 min. Transitions monitored as in Supplemental Table 5 were established using the instrument’s compound optimization mode with direct injection for each compound. The data was analyzed using MultiQuant™ software (ABI Sciex Framingham, MA) providing the peak area. A standard curve was constructed using synthetically produced product peptides from picomole to nanomole levels in 10 μl. Samples were run in triplicate and concentrations determined from the standard curve.
Dietary experiments
One week before the start of the feeding experiments, age-matched male (6–8 week old, 20–30 g) inbred C57Bl/6 mice obtained from Harlan Laboratories and placed on a control diet containing 15% fat derived primarily from soybean oil and lard (Envigo Teklad custom diet no. TD170820). After 1 week, half of the mice remained on the control diet, whereas the rest were switched to an isocaloric olive oil diet contain 15% fat derived primarily from olive oil (Envigo Teklad custom diet no TD170821). Mice were fed ad libitum for 12 weeks. At the end of the feeding study, mice were fasted for 16 h prior to tissue and serum collection. A second cohort of mice were placed on the same dietary regiment as described above with the following modifications; 3 days prior to tissue harvest, half the mice fed the control diet and half the mice fed the olive oil enriched diet were injected IP daily with 10 mg/kg of the SIRT1 inhibitor EX527 while the remaining mice were injected with equivalent amounts of DMSO.
Serum analysis
Non-esterified fatty acid levels in serum were determined using Wako Chemicals lipid assay systems (Wako Diagnostics, Richmond, VA). Colorimetric analyses of lipids were measured at 570 nm on an Omega FLOUstar 96-well plate reader from BMG labtech (Ortenberg, Germany). Serum β-hydroxybutyrate measurements were determined from mouse serum samples using a β-hydroxybutyrate LiquiColor kit (Stanbio Laboratory, Boerne, TX, USA) according to the manufacturer instructions.
Co-immunoprecipitation studies
The Nuclear Complex Co-IP system from Active Motif (Carlsbad, CA) was used for co-immunoprecipitation (co-IP) experiments following the manufacturers’ protocol. Briefly, nuclei from primary hepatocytes were isolated. Nuclear fractions were incubated overnight with kit reagents and anti-bodies (anti-acetyl-lysine, anti-FOXO3a, anti-PLIN5, anti-PGC-1α or anti-SIRT1) at 4°C with shaking. The next day, unbound fractions were separated by magnet, followed by washing and elution of the bound complex. Eluate proteins were analyzed by Western blotting. A parallel co-IP with the lysates using anti-rabbit IgG was performed to assess nonspecific binding (negative control). Input material was run in parallel to determine equal loading of sample. Immunoprecipitations performed to determine protein acetylation were carried out as described previously in (Khan et al., 2015).
cAMP-Glo Assay
The cAMP-Glo Assay from Promega (Promega, Fitchburg, WI) was used for cAMP detection according to the manufactures protocol. MEF cells were plated in a 96-well plate in complete media (DMEM 10% FBS, 1%P/S). Cells were treated for 6 or 16 hrs with 2% fatty acid free BSA or 500 μM 18:1 complexed to fatty acid free BSA. After being treated with complexed fatty acids, cells were washed 2x with PBS, then placed in fasting media (DMEM, 0.5% P/S) and treated with DMSO or 20 μM isoproterenol/500 μM IBMX for 20 minutes. Cells were lysed with cAMP-Glo lysis buffer at room temperature for 15 mins. Cell lysate was treated with cAMP Detection Solution, mixed and incubated at room temperature for 20 mins. The reaction was then treated with Kinase-Glo Reagent, mixed for 60s and incubated at room temperature for 10 minutes. Luminescence was then read on a Cary Eclipse fluorescence spectrophotometer (Agilent, Santa Clara, CA).
QUANTIFICATION AND STATISTICAL ANALYSIS
Values were expressed as the means ± SEM. In comparisons made between two groups, Student’s t-tests were performed using Graphpad Prism (San Diego, CA). When more than two groups were compared, analysis of variance (ANOVA) with Newman Keuls post-hoc test were performed. Values with p<0.05 were considered statistically significant.
DATA AND CODE AVAILABILITY
This study did not generate any unique dataset or code.
Supplementary Material
KEY RESOURCES TABLE.
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Guinea pig polyclonal anti-PLIN5 | Progen | Cat No. GP44 |
| Rabbit polyclonal anti-PLIN2 | (McIntosh AL et al., 2012) | N/A |
| Rabbit polyclonal anti-ATGL | Cell Signaling Technology | Cat No. 2138S |
| Rabbit polyclonal anti-FOXO3a | ThermoFisher Scientific | Cat No. PA5–27145 |
| Mouse monoclonal anti-PGC-1α | EMD Millipore | Cat No. ST1202 |
| Rabbit polyclonal anti-PGC-1α | Abcam | Cat No. ab54481 |
| Mouse monoclonal anti-Histone H3 | Cell Signaling Technology | Cat No. 14269 |
| Rabbit monoclonal anti-β Actin | LI-COR | Cat No. 926–42210 |
| Mouse Monoclonal anti-AcLysine | Novis Biologicals | Cat No. 15G10 |
| Mouse Monoclonal anti-AcLysine | Santa Cruz Biotechnology | Cat No. AKL5C1 |
| Mouse Monoclonal anti-AcLysine | Cell Signaling Technology | Cat No. 9681S |
| Mouse Monoclonal anti-AcLysine | Thermo Scientific | Cat No. 1C6 |
| Rabbit Polyclonal anti-Phos-PLIN5; CLARRGRRW(pS)VELK | NeoBioLab; This Paper | N/A |
| Rabbit Polyclonal anti-Total-PLIN5; CLARRGRRWSVELK | NeoBioLab; This Paper | N/A |
| Donkey anti-Guinea pig IRDye 800CW | LI-COR | Cat No. 926–32411 |
| Donkey anti-Guinea pig IRDye 680RD | LI-COR | Cat No. 926–68030 |
| Donkey anti-Rabbit pig IRDye 800CW | LI-COR | Cat No. 926–32213 |
| Donkey anti-Rabbit pig IRDye 680RD | LI-COR | Cat No. 926–68022 |
| Donkey anti-Mouse pig IRDye 800CW | LI-COR | Cat No. 925–32212 |
| Donkey anti-Mouse pig IRDye 680RD | LI-COR | Cat No. 926–68023 |
| Bacterial and Virus Strains | ||
| BL21 (DE3) | New England BioLab | Cat No. C2527H |
| BL21 (DE3) Codon+ | Fisher Scientific | Cat No. NC9122855 |
| XL1-Blue | Agilent | Cat No. 200150 |
| Ad-ATGL | (Miyoshi et al., 2008; Miyoshi et al., 2007) | N/A |
| Ad-shATGL | (Miyoshi et al., 2008; Miyoshi et al., 2007) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 8-bromoadenosine 39,59-cyclic monophosphate | Santa Cruz Biotechnology | Cat No. SC-217493A |
| Isoproterenol | Sigma-Aldrich | Cat No. I6504 |
| 3-isobutyl1-methylxanthine (IBMX) | Sigma-Aldrich | Cat No. I5879 |
| 1-anilinonaphthalene 8-sulfonic acid | Molecular Probes | Cat No. A47 |
| M199 media | Sigma-Aldrich | Cat No. M5017 |
| ATGL Statin (ATGLi) | Cayman Chemical | Cat No. 15284 |
| EX527 | Cayman Chemical | Cat No. 10009798 |
| H89 | Cayman Chemical | Cat No. 10010556 |
| Dynabeads Protein G | ThermoFisher Scientific | Cat No. 10004D |
| NucBlue | ThermoFisher Scientific | Cat No. R37605 |
| PGC-1α Peptide; KNSWSNETKVIAPNT | This Paper | N/A |
| Acetyl-PGC-1α; KNSWSNETK(Ac)VIAPNT | This Paper | N/A |
| Histone H3 Peptide; KWWGGTSKRATQK | This Paper | N/A |
| Acetyl-Histone H3; KWWGGTSK(Ac)RATQK | This Paper | N/A |
| FOXO3a Peptide; KDSPSQLSKWPGSPTS | This Paper | N/A |
| Acetyl-FOXO3a; KDSPSQLSK(Ac)WPGSPTS | This Paper | N/A |
| P53 Peptide; WEEKGQSTSSHSKSTEGAEE | This Paper | N/A |
| Acetyl-P53; WEEKGQSTSSHSK(Ac)STEGAEE | This Paper | N/A |
| P53-W Peptide; WEEKGQSTSSHSKSTEGWEE | This Paper | N/A |
| Acetyl-P53-W Peptide; WEEKGQSTSSHSK(Ac)STEGWEE | This Paper | N/A |
| SIRT1 | (Hallows et al., 2006) | N/A |
| PLIN5 | This Paper | N/A |
| Critical Commercial Assays | ||
| Dual-Luciferase Reporter Assay System | Promega | Cat No. E1960 |
| cAMP-Glo Assay | Promega | Cat No. V1501 |
| QuickChange Lightning Site-Directed Mutagenesis Kit | Agilent | Cat No. 210515 |
| Nuclear Complex Co-IP system | Active Motif | Cat No. 54001 |
| β-hydroxybutyrate LiquiColor kit | EKF Diagnostics | Cat No. 2440–058 |
| NEFA-Wako Chemicals lipid assay systems | Wako Chemicals | Cat No. 999–34691, 995–34791, 991–34891, 993–35191 |
| Targetfect Hepatocyte reagent | Targeting Systems | Cat No. Hep-01 |
| Lipofectamine 3000 | ThermoFisher Scientific | Cat No. L3000008 |
| Effectene | QIAGEN | Cat No. 301425 |
| SuperScript® VILO™ cDNA Synthesis Kit | Invitrogen | Cat No. 11754–250 |
| Deposited Data | ||
| N/A | N/A | N/A |
| Experimental Models: Cell Lines | ||
| Mouse Primary Hepatocytes | (Ong et al., 2011) | N/A |
| Mouse Embryonic Fibroblasts (MEF) | ATCC | Cat No. SCRC-108 |
| SIRT1 KO Mouse Embryonic Fibroblasts (SIRT1 −/− MEF) | (Di Sante et al., 2015) | N/A |
| Hep3B | ATCC | Cat No. HB-8064 |
| HepG2 | ATCC | Cat No. HB-8065 |
| L-Cells | (Atshaves et al., 2002; McIntosh AL et al., 2012) | N/A |
| Perilipin5 KO L-Cells (PLIN5 −/− L-Cells) | This Paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Male C57Bl/6 mice | Envigo | N/A |
| Oligonucleotides | ||
| Control Anti-Sense Oligonucleotide | Ionis Parmaceuticals; Mark Graham | N/A |
| Perilipin5 Anti-Sense Oligonucleotide | Ionis Parmaceuticals; Mark Graham | N/A |
| Recombinant DNA | ||
| pEZ-M98-eGFP-SIRT1 | GeneCopoeia | Cat. No EX-Mm12441-M98 |
| pEGFP-hSIRT1 | (Peng et al., 2011) | N/A |
| pEGFP-hSIRT1E230K | This Paper | N/A |
| pEZ-M98-eGFP-PGC-1α | This Paper | N/A |
| pEZ-M55-mCherry-PLIN5 | GeneCopoeia | Cat No. EX-Mm27089-M55 |
| pEZ-M55-mCherry-PLIN5-pD (S155A) | This Paper | N/A |
| pEZ-M55-mCherry-PLIN5-pM (S155E) | This Paper | N/A |
| pECFP-PLIN2 | (McIntosh AL et al., 2012; Senthivinayagam et al., 2013) | N/A |
| TK-MH-UASluc | (Finck et al., 2006; Puigserver et al., 1998) | N/A |
| pRLSV40 | (Finck et al., 2006; Puigserver et al., 1998) | N/A |
| pCMX-GAL4-PGC-1α | (Finck et al., 2006; Puigserver et al., 1998) | N/A |
| Software and Algorithms | ||
| SigmaPlot 11 | Systat Software, Inc | N/A |
| Prism8 | GraphPad | N/A |
| Canvas 11 | Canvas | N/A |
| PyMOL | PyMOL by Schrodinger | N/A |
| Image Studio v5 | LI-COR Biosciences | N/A |
| NIS-Elements 4 | Nikon | N/A |
| ZEN | Zeiss | N/A |
| Cary Eclipse Software | Agilent | N/A |
| CDSSTR | CDPro | N/A |
| MultiQuant | Sciex | N/A |
| Phyre2 | (Kelley et al., 2015; Kelly and Sternberg, 2009) | N/A |
| MetaMorph 7.5 | Molecular Devices | N/A |
| Other | ||
| Control Diet; 15% Fat Derived from Soybean Oil | Envigo: Teklad Custom Diet | Diet No. TD.170820 |
| Olive Oil Diet; 15% Fat Derived from Olive Oil | Envigo: Teklad Custom Diet | Diet No. TD.170821 |
Highlights.
MUFAs allosterically activate SIRT1 towards select substrates such as PGC-1α
MUFAs enhance PGC-1α signaling in vivo in a SIRT1-dependent manner
PLIN5 is a fatty acid binding protein that preferentially binds LD-derived MUFAs
PLIN5 mediates MUFA signaling to control SIRT1/PGC-1α
ACKNOWLEDGEMENTS
We would like to thank Candace Guerrero, Mitchell Fuller, Michael Autry, Colleen Forster, and Guillermo Marques for their technical assistance. We thank the University of Minnesota Imaging Center, Center for Mass Spectrometry and Proteomics, Clinical and Translational Science Biospecimen Support Center, and the Biophysical Technology Center for providing instrumentation and expertise. We thank Eduarado Chini for help with initial SIRT1 assays, Barbara Atshaves for antibodies and protocols, and Ann Hertzel for scientific discussions. Funding was provided for Charles Najt (NIH: T32DK007203 and T32AG029796), Timothy Heden (NIH: F32DK109556 and L30DK110338), Minervo Perez (NIH: R01CA182543-S1), Mallory Franklin (NIH: T32DK083250), David Bernlohr (NIH: R01DK053189 and the University of Minnesota E-0917-2), Lisa Chow (NIH: R01DK098203), and Douglas Mashek (NIH: R01AG055452, R01DK108790, R01DK114401 and the American Diabetes Association: 1-16-IBS-203).
Footnotes
DECLARATION OF INTERESTS
Authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahmadian M, Duncan RE, and Sul HS (2009). The skinny on fat: lipolysis and fatty acid utilization in adipocytes. Trends Endocrin Metab 20, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge GM, Podrebarac DM, Greenough WT, and Weiler IJ (2008). The use of total protein stains as loading controls: An alternative to high-abundance single-protein controls in semi-quantitative immunoblotting. J Neuroscience Methods 172, 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atshaves BP, Petrescu AD, Starodub O, Roths JB, Kier AB, and Schroeder F (1999). Expression and intracellular processing of the 58 KDa SCP-x/3-oxoacyl-CoA thiolase in transfected mouse L cells. J Lipid Res 40, 610–622. [PubMed] [Google Scholar]
- Atshaves BP, Storey SM, Petrescu AD, Greenberg C, Lyuksyutova OI, Smith R III, and Schroeder F (2002). Expression of Fatty Acid Binding Proteins Inhibits Lipid Accumulation and Alters Toxicity in L-cell Fibroblasts. Am J Phys 283, C688–C703. [DOI] [PubMed] [Google Scholar]
- Balasubramanian P, Howell PR, and Anderson RM (2017). Aging and Caloric Restriction Research: A Biological Perspective With Translational Potential. EBioMedicine 21, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutiérrez-Juárez R, Rossetti L, Gu W, and Accili D (2008). SirT1 Gain of Function Increases Energy Efficiency and Prevents Diabetes in Mice. Cell Metabolism 8, 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordone L, Cohen D, Robinson A, Motta MC, Van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, et al. (2007). SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell 6, 759–767. [DOI] [PubMed] [Google Scholar]
- Borra MT, Smith BC, and Denu JM (2005). Mechanism of Human SIRT1 Activation by Resveratrol. Journal of Biological Chemistry 280, 17187–17195. [DOI] [PubMed] [Google Scholar]
- Børsheim E, Kien CL, and Pearl WM (2006). Differential effects of dietary intake of palmitic acid and oleic acid on oxygen consumption during and after exercise. Metabolism 55, 1215–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma M, Sparks L, Hooiveld GJ, Jorgensen JA, Houten SM, Schrauwen P, Kersten S, and Hesselink MK (2013). Overexpression of Plin5 in skeletal muscle promotes oxidative gene expression and intrayocellular lipid content without compromising insulin sensitivity. Biochim Biophys Acta 1831, 844–852. [DOI] [PubMed] [Google Scholar]
- Bu SY, Mashek MT, and Mashek DG (2009). Suppression of Long Chain Acyl-CoA Synthetase 3 Decreases Hepatic de Novo Fatty Acid Synthesis through Decreased Transcriptional Activity. Journal of Biological Chemistry 284, 30474–30483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland G, and Gonzalez CA (2015). The role of olive oil in disease prevention: a focus on the recent epidemiological evidence from cohort studies and dietary intervention trials. British Journal of Nutrition 113, S94–S101. [DOI] [PubMed] [Google Scholar]
- Cao D, Wang M, Qiu X, Liu D, Jiang H, Yang N, and Xu R-M (2015). Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol. Genes & Development 29, 1316–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Steele AD, Lindquist S, and Guarente L (2005). Increase in Activity During Calorie Restriction Requires Sirt1. Science 310, 1641–1641. [DOI] [PubMed] [Google Scholar]
- Dai H, Case AW, Riera TV, Considine T, Lee JE, Hamuro Y, Zhao H, Jiang Y, Sweitzer SM, Pietrak B, et al. (2015). Crystallographic structure of a small molecule SIRT1 activator-enzyme complex. Nature Communications 6, 7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalen KT, Dahl T, Holter E, Arntsen B, Londos C, Sztalryd C, and Nebb HI (2007). LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1771, 210–227. [DOI] [PubMed] [Google Scholar]
- Di Sante G, Wang L, Wang C, Jiao X, Casimiro MC, Chen K, Pestell TG, Yaman I, Di Rocco A, Sun X, et al. (2015). Sirt1-deficient mice have hypogonadotropic hypogonadism due to defective GnRH neuronal migration. Molecular endocrinology (Baltimore, Md.) 29, 200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch R, Martínez-González MÁ, Corella D, Salas-Salvadó J, Ruiz-Gutiérrez V, Covas MI, Fiol M, Gómez-Gracia E, López-Sabater MC, Vinyoles E, et al. (2006). Effects of a Mediterranean-Style Diet on Cardiovascular Risk Factors: A Randomized TrialMediterranean Diet and Cardiovascular Risk Factors. Annals of Internal Medicine 145, 1–11. [DOI] [PubMed] [Google Scholar]
- Feldman JL, Baeza J, and Denu JM (2013). Activation of the Protein Deacetylase SIRT6 by Long-chain Fatty Acids and Widespread Deacylation by Mammalian Sirtuins. Journal of Biological Chemistry 288, 31350–31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Dittenhafer-Reed KE, and Denu JM (2012). Sirtuin Catalysis and Regulation. Journal of Biological Chemistry 287, 42419–42427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, and Kelly DP (2006). Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metabolism 4, 199–210. [DOI] [PubMed] [Google Scholar]
- Gallardo-Montejano VI, Saxena G, Kusminski CM, Yang C, McAfee JL, Hahner L, Hoch K, Dubinsky W, Narkar VA, and Bickel PE (2016). Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1α/SIRT1-dependent transcriptional regulation of mitochondrial function. Nature Communications 7, 12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmink A, Bosma M, Kuijpers HJH, Hoeks J, Schaart G, van Zandvoort MAMJ, Schrauwen P, and Hesselink MKC (2016). Decoration of intramyocellular lipid droplets with PLIN5 modulates fasting-induced insulin resistance and lipotoxicity in humans. Diabetologia 59, 1040–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Mottillo EP, Zhu Z, and Zhou L (2011). Interactions of perilipin-5 (plin5) with adipose triglyceride lipase. J Biol Chem 286, 5126–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HPH, Krishnamoorthy R, and Rathod M (2009). Perilipin control lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl). J Biol Chem 284, 34538–34544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, and Mashek DG (2011). The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest 121, 2102–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G, Moustafa T, Woelkart G, Büttner S, Schmidt A, van de Weijer T, Hesselink M, Jaeger D, Kienesberger PC, Zierler K, et al. (2011). ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nature medicine 17, 1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Lee S, and Denu JM (2006). Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proceedings of the National Academy of Sciences 103, 10230–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Schroeder EA, Silva-García CG, Hebestreit K, Mair WB, and Brunet A (2017). Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 544, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickenbottom SJ, Kimmel AR, Londos C, and Hurley JH (2004). Structure of a lipid droplet protein: The PAT family member TIP47. Structure 12, 1199–1207. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Pirinen E, and Auwerx J (2012). Sirtuins as regulators of metabolism and healthspan. Nature Reviews Molecular Cell Biology 13, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T, Riera TV, Lee JE, E SY, Lamming DW, et al. (2013). Evidence for a Common Mechanism of SIRT1 Regulation by Allosteric Activators. Science 339, 1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard BP, and Sinclair DA (2014). Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends in Pharmacological Sciences 35, 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, et al. (2005). Substrate-specific Activation of Sirtuins by Resveratrol. Journal of Biological Chemistry 280, 17038–17045. [DOI] [PubMed] [Google Scholar]
- Kane CD, and Bernlohr DA (1996). A Simple Assay for Intracellular Lipid-Binding Proteins Using Displacement of 1-Anilinonaphthalene 8-Sulfonic Acid. Analytical Biochemistry 233, 197–204. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJE (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10, 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly L, and Sternberg M (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols 4, 363–371. [DOI] [PubMed] [Google Scholar]
- Khan SA, Sathyanarayan A, Mashek MT, Ong KT, Wollaston-Hayden EE, and Mashek DG (2015). ATGL-Catalyzed Lipolysis Regulates SIRT1 to Control PGC-1α/PPAR-α Signaling. Diabetes 64, 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, et al. (2007). SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. The EMBO Journal 26, 3169–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurmoto K, Okamura T, Yamaguchi T, Nakamura TY, Wakabayashi S, Morinaga H, Nomura M, Yanase T, Otsu K, Usuda N, et al. (2012). Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J Biol Chem 287, 23852–23863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. (2006). Resveratrol Improves Mitochondrial Function and Protects against Metabolic Disease by Activating SIRT1 and PGC-1α. Cell 127, 1109–1122. [DOI] [PubMed] [Google Scholar]
- Lim J-H, Gerhart-Hines Z, Dominy JE, Lee Y, Kim S, Tabata M, Xiang YK, and Puigserver P (2013). Oleic Acid Stimulates Complete Oxidation of Fatty Acids through Protein Kinase A-dependent Activation of SIRT1-PGC1α Complex. Journal of Biological Chemistry 288, 7117–7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R, Mokhtar R, Matzarism M, Selathurai A, Kowalski G, Mokbel N, Meikle P, Bruce C, and Watt M (2014). Plin5 deletion remodels intracellular lipid composition and causes insulin resistance in muscle. Molecular Metabolism 3, 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AL, Senthivinayagam S, Moon KC, Gupta S, Lwande JS, Murphy CC, Storey S, and Atshaves BP (2012). Direct interaction of ADRP with lipids on the surface of lipid droplets: A live cell FRET analysis. Am J Physiol Cell Physiol 303, C728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AL, Storey SM, and Atshaves BP (2010). Intracellular lipid droplets contain dynamic pools of sphingomyelin: ADRP binds phospholipids with high affinity. Lipids 45, 465–477. PMCID: PMC3065392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Perfield II JW, Obin MS, and Greenberg AS (2008). Adipose triglyceride lipase regulates basal lipolysis and lipid droplet size in adipocytes. Journal of Cellular Biochemistry 105, 1430–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Perfield JW, Souza SC, Shen WJ, Zhang HH, Stancheva ZS, Kraemer FB, Obin MS, and Greenberg AS (2007). Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J.Biol.Chem. 282, 996–1002. [DOI] [PubMed] [Google Scholar]
- Mohktar RAM, Montgomery MK, Murphy RM, and Watt MJ (2016). Perilipin 5 is dispensable for normal substrate metabolism and in the adaptation of skeletal muscle to exercise training. American Journal of Physiology - Endocrinology and Metabolism. [DOI] [PubMed] [Google Scholar]
- Muratore KA, Najt CP, Livezey NM, Marti J, Mashek DG, and Arriaga EA (2018). Sizing lipid droplets from adult and geriatric mouse liver tissue via nanoparticle tracking analysis. Analytical and Bioanalytical Chemistry 410, 3629–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najt CP, Lwande JS, McIntosh AL, Senthivinayagam S, Gupta S, Kuhn LA, and Atshaves BP (2014). Structural and Functional Assessment of Perilipin2 Lipid Binding Domains. Biochemistry 53, 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najt CP, S S, Aljazi M, Fader K, Olenic S, Brock J, Lydic TA, Jones AD, and Atshaves B (2016). Liver-specific loss of Perilipin 2 alleviates diet-induced hepatic steatosis, inflammation, and fibrosis. Am J Physiol Gastro Liver Physiol 310, G726–G738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira LM, Lavigne JA, Chandramouli GVR, Lui H, Barrett JC, and Hursting SD (2012). Dose-dependent effects of calorie restriction on gene expression, metabolism, and tumor progression are partially mediated by insulin-like growth factor-1. Cancer Medicine 1, 275–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KT, Mashek MT, Bu SY, Greenberg AS, and Mashek DG (2011). Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology 53, 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KT, Mashek MT, Davidson NO, and Mashek DG (2014). Hepatic ATGL mediates PPAR-α signaling and fatty acid channeling through an L-FABP independent mechanism. Journal of Lipid Research 55, 808–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palou A, Picó C, Macarulla MT, Rodríguez VM, and Portillo MP (2002). Olive oil feeding up-regulates uncoupling protein genes in rat brown adipose tissue and skeletal muscle. The American Journal of Clinical Nutrition 75, 213–220. [DOI] [PubMed] [Google Scholar]
- Peng L, Yuan Z, Ling H, Fukasawa K, Robertson K, Olashaw N, Koomen J, Chen J, Lane WS, and Seto E (2011). SIRT1 Deacetylates the DNA Methyltransferase 1 (DNMT1) Protein and Alters Its Activities. Molecular and Cellular Biology 31, 4720–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M, Blankenhorn J, Murray KJ, and Parker LL (2018). High-throughput identification of FLT3 wild-type and mutant kinase substrate preferences and application to design of sensitive in vitro kinase assay substrates. Molecular & Cellular Proteomics, mcp.RA118.001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, and Tschöp MH (2008). Sirt1 protects against high-fat diet-induced metabolic damage. Proceedings of the National Academy of Sciences 105, 9793–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak NM, Jaeger D, Kolleritsch S, Zimmermann R, Zechner R, Lass A, and Haemmerle G (2015). The Interplay of Protein Kinase A and Perilipin 5 Regulates Cardiac Lipolysis. Journal of Biological Chemistry 290, 1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, and Spiegelman BM (1998). A Cold-Inducible Coactivator of Nuclear Receptors Linked to Adaptive Thermogenesis. Cell 92, 829–839. [DOI] [PubMed] [Google Scholar]
- Salas-Salvadó J, Bulló M, Babio N, Martínez-González MÁ, Ibarrola-Jurado N, Basora J, Estruch R, Covas MI, Corella D, Arós F, et al. (2011). Reduction in the Incidence of Type 2 Diabetes With the Mediterranean Diet. Results of the PREDIMED-Reus nutrition intervention randomized trial 34, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayan A, Mashek MT, and Mashek DG (2017). ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Reports 19, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwingshackl L, and Hoffmann G (2014a). Mediterranean dietary pattern, inflammation and endothelial function: a systematic review and meta-analysis of intervention trials. Nutri Metabol Cardiovasc Dis NMCD 24. [DOI] [PubMed] [Google Scholar]
- Schwingshackl L, and Hoffmann G (2014b). Monounsaturated fatty acids, olive oil and health status: a systematic review and meta-analysis of cohort studies. Lipids in Health and Disease 13, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwingshackl L, Strasser B, and Hoffmann G (2011). Effects of monounsaturated fatty acids on glycaemic control in patients with abnormal glucose metabolism: a systematic review and meta-analysis. Ann Nutr Metab 58. [DOI] [PubMed] [Google Scholar]
- Senthivinayagam S, McIntosh AL, Moon KC, and Atshaves BP (2013). Plin2 inhibitis cellular glucose uptake through interactions with SNAP23, a SNARE complex protein. PLoS ONE 8, e73696. doi: 73610.71371/journal.pone.0073696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd SO, Cocks M, Tipton KD, Ranasinghe AM, Barker TA, Burniston JG, Wagenmakers AJM, and Shaw CS (2013). Sprint interval and traditional endurance training increase net intramuscular triglyceride breakdown and expression of perilipin 2 and 5. The Journal of Physiology 591, 657–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, and Ajuwon KM (2018). Effects of Diets Differing in Composition of 18-C Fatty Acids on Adipose Tissue Thermogenic Gene Expression in Mice Fed High-Fat Diets. 10, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, and Guarente L (2014). Small-Molecule Allosteric Activators of Sirtuins. Annual Review of Pharmacology and Toxicology 54, 363–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofi F, Abbate R, Gensini GF, and Casini A (2010). Accruing evidence on benefits of adherence to the Mediterranean diet on health: an updated systematic review and meta-analysis. The American Journal of Clinical Nutrition 92, 1189–1196. [DOI] [PubMed] [Google Scholar]
- Sreerama N, and Woody RW (2000). Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem 287, 252–260. [DOI] [PubMed] [Google Scholar]
- Storey S, AL M, Huang H, Landrock KK, Martin GG, Landrock D, Payne HR, Atshaves B, Kier AB, and Schroeder F (2012). Intracellular cholesterol binding proteins enhance HDL-mediated cholesterol uptake in cultured primary mouse hepatocytes. Am J Physiol Gastrointest Liver Physiol 302, G824–G839 PMCID: PMC3355564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztalryd C, and Brasaemle DL (2017). The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1862, 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichopoulou A, Orfanos P, Norat T, Bueno-de-Mesquita B, Ocke MC, Peeters PH, van der Schouw YT, Boeing H, Hoffmann K, Boffetta P, et al. (2005). Modified Mediterranean diet and survival: EPIC-elderly prospective cohort study. BMJ 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhao Y, Gao X, Li L, Yuan Y, Liu F, Zhang L, Wu J, Hu P, Zhang X, et al. (2015). Perilipin 5 improves hepatic lipotoxicity by inhibiting lipolysis. Hepatology 61, 870–882. [DOI] [PubMed] [Google Scholar]
- Weiskirchen S, and Weiskirchen R (2016). Resveratrol: How Much Wine Do You Have to Drink to Stay Healthy? Advances in Nutrition 7, 706–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenborg M, Schmidt C, Braun P, Landgrebe J, von Figura K, Saftig P, and Eskelinen E-L (2005). Mannose 6-phosphate receptors, Niemann-Pick C2 protein, and lysosomal cholesterol accumulation. J Lipid Res 46, 2559–2569. [DOI] [PubMed] [Google Scholar]
- Wolins N, Quaynor B, Skinner J, Tzekov A, Croce M, Gropler M, Varma V, Yao-Borengasser A, Rasouli N, Kern P, et al. (2006). OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes 55, 3418–3428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique dataset or code.