Abstract
Chloramphenicol (CHL) and linezolid (LZD) are antibiotics that inhibit translation. Both were thought to block peptide bond formation between all combinations of amino acids. Yet recently, a strong nascent peptide context-dependency of CHL- and LZD-induced translation arrest was discovered. Here, we probed the mechanism of action of CHL and LZD by using single-molecule Förster resonance energy transfer spectroscopy (smFRET) to monitor translation arrest induced by antibiotics. The presence of CHL or LZD does not significantly alter dynamics of protein synthesis until the arrest-motif of the nascent peptide is generated. Inhibition of peptide-bond formation compels the fully accommodated A-site tRNA to undergo repeated rounds of dissociation and non-productive rebinding. The glycyl amino-acid moiety on the A-site Gly-tRNA manages to overcome the arrest by CHL. Our results illuminate the mechanism of CHL and LZD action through their interactions with the ribosome, the nascent peptide and the incoming amino acid, perturbing elongation dynamics.
During translation, the ribosome catalyzes peptide bond formation between chemically and structurally diverse substrates. To accomplish this task, the ribosome precisely positions the peptidyl-tRNA (pept-tRNA) in the P site and aminoacyl-tRNA (aa-tRNA) in the A site1. The formation of the new peptide bond results in the transfer of the nascent peptide from the P- to the A-site tRNA, extending the nascent peptide by a single amino acid. Upon peptidyl transfer, the 50S subunit rotates by 7°−10° relative to the 30S subunit2–6 and tRNAs assume the hybrid state, with their anticodons remaining in the P and A sites in the small subunit, but their acceptor ends moved to the P and E sites in the large subunit (P/E and A/P tRNA states)4,7. A subsequent translocation step moves the mRNA and tRNA anticodon stem-loops to the E and P sites with the ribosome ratcheting back to the non-rotated state. Various intermediate rotation8–10 and small-subunit conformational11 states are sampled during these transitions, although some of them might be too short-lived to be experimentally identified.
Many protein synthesis inhibitors act by sterically disrupting the association and/or positioning of the substrates in the PTC and thus, blocking peptide bond formation12–15. Two such small molecules are the antibiotics chloramphenicol (CHL) and linezolid (LZD) (Fig 1a). CHL is a long-known antibiotic initially isolated from Streptomyces venezuelae16, whereas LZD is a newer synthetic drug and the first representative of the oxazolidinone class of ribosome inhibitors approved for clinical use17. Despite their structural differences, both CHL and LZD bind to the same site within the PTC5,18–21, where they are expected to clash with the A-site aminoacyl-moiety on the aa-tRNA. Therefore, CHL and LZD were initially proposed to be nondiscriminatory inhibitors of peptide bond formation20,22, by occluding the correct positioning of any aa-tRNA aminoacyl-moiety in the PTC active site. However, recent in vivo (Ribo-seq) and in vitro (toeprinting) results showed that neither CHL nor LZD acts as universal inhibitors of peptide bond formation23. Rather than stopping the ribosome with equal efficiency at every codon, CHL and LZD allow for translation progression through some codons until the ribosomes synthesize nascent-peptides with specific features that facilitate translation arrest by the antibiotic. Ribo-seq and toeprinting analyses revealed that CHL and LZD most efficiently exert their inhibitory activity when the ribosome carries a nascent peptide whose penultimate amino acid is an Ala, Ser or Thr23. The acceptor substrate (the incoming aa-tRNA) also plays an important role: despite carrying a nascent peptide with a penultimate Ala (or Ser/Thr), a CHL-bound ribosome can still catalyze peptide bond formation if Gly-tRNAGly enters the A site23. Thus, instead of CHL and LZD being universal inhibitors of translation, the action of these drugs depends strongly on the identities of the ribosomal P- and A-site substrates.
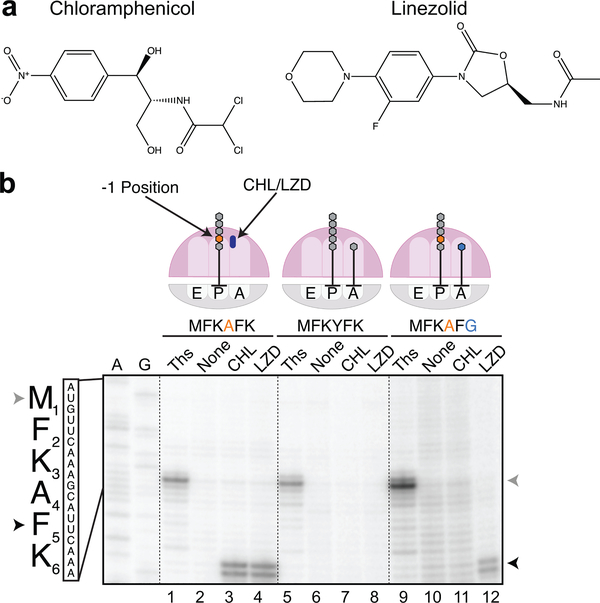
Figure 1. Context-specific inhibition of translation by chloramphenicol and linezolid.
a. Chemical structures of antibiotics chloramphenicol (CHL) and linezolid (LZD). b. In vitro toeprinting analysis of CHL- or LZD-induced translational arrest on three different mRNA constructs (complete sequences of the templates are shown in Supplementary Fig 1; independently repeated at least twice with similar results). The toeprint bands produced by CHL- or LZD-arrested ribosomes at the F5 codon of the mRNAs are indicated by black arrowheads. Gray arrowheads show the toeprint bands produced by ribosomes stalled at the start codons due to the presence of thiostrepton (Ths). Sequencing lanes A and G for the MFKAFK template are shown.
It remains unclear how the combined action of the nascent peptide and CHL or LZD induces translation arrest and which steps of translation leading to peptidyl transfer are affected. It is feasible that the acceptor substrate (aa-tRNA) is rejected at the early stages of its interaction with the ribosome, as has been observed with some other ribosome inhibitors24, or that tRNA fully accommodates in the A site but subsequent peptide bond formation is prevented. Furthermore, the mechanism of translation rescue by the A-site Gly-tRNAGly specific for CHL is unclear, as potentially both the glycine amino acid and the glycine-specific tRNA may contribute to overcoming the antibiotic action. Finally, it is unknown whether CHL and LZD stabilize specific conformational and compositional states of an actively translating ribosome conducive to translation arrest.
To understand the molecular mechanism underlying context-dependent protein synthesis inhibition by CHL and LZD, we combined biochemical and smFRET approaches to monitor CHL or LZD-induced translational arrests. We demonstrate that at the arrest site, the presence of CHL allows full accommodation of aa-tRNA body, but prevents the acceptor amino acid from participating in peptide bond formation with specific donor peptidyl-tRNA substrates. Drug-induced inhibition of the peptidyl-transfer reaction leads to multiple rounds of futile accommodation and dissociation of aa-tRNA without translation progression. We show that the ability of Gly-tRNAGly to overcome translation stall is determined by the nature of its aminoacyl moiety rather than the tRNAGly body, highlighting the exceptional properties of glycine as a peptidyl acceptor in the CHL-arrested ribosome.
Results
CHL/LZD context-specificity reproduced with model mRNA
Based on the results of our previous Ribo-seq analysis23 of the effects of CHL and LZD, we designed three model mRNAs encoding peptides with the sequences MFKAFKNIIRTRTL, MFKYFKNIIRTRTL and MFKAFGNIIRTRTL for use in smFRET experiments (see below) (for the further discussion of these model sequences we refer only to the underlined first 6 amino acids or the codons specifying them; full template sequences are shown in Supplementary Fig 1). These model mRNA constructs differ minimally in their nucleotide and amino acid sequences but are expected to vary drastically in their ability to promote CHL- and LZD-induced translation arrest at the F5 codon. The specificity of CHL- and LZD-induced ribosomal arrest was first validated using in vitro toeprinting analysis23,25 (Fig 1b). As expected, ribosomes synthesizing the MFKAFK sequence in the presence of either CHL or LZD were arrested with the F5 codon of the open reading frame (ORF) in the P site and the K6 codon in the A site, precisely when the nascent peptide contained an Ala residue in the penultimate position (Fig 1b, lanes 2–4). This result is consistent with the ribosome being unable to catalyze peptide bond formation between the C-terminal Phe of the MFKAF nascent peptide and the incoming Lys residue in the presence of CHL or LZD. In contrast, synthesis of the MFKYFK peptide, whose 4th residue is a Tyr instead of an Ala, proceeds unimpeded beyond the F5 codon despite the presence of inhibitory concentrations of CHL or LZD in the reaction (Fig 1b, lanes 6–8 and Supplementary Fig 2). Similarly, no CHL-induced translation arrest was observed with the mRNA encoding the MFKAFG peptide (Fig 1b, lanes 10,11). Therefore, ribosomes carrying the MFKAF peptide with a penultimate Ala residue, which typically favors blocking of aa-tRNA incorporation by CHL, successfully catalyze peptidyl transfer to Gly in spite of the presence of this drug. In line with our previous findings23, LZD is still able to inhibit peptide bond formation even with Gly as an acceptor (Fig 1b, lane 12). The results of the cell-free translation experiments with the model templates are fully consistent with and recapitulate the context-specific effects of CHL and LZD observed in vivo23 and justify the use of these designed ORFs for studying the mechanistic basis of action of these antibiotics in vitro.
Monitoring CHL/LZD-induced translation arrest in real time
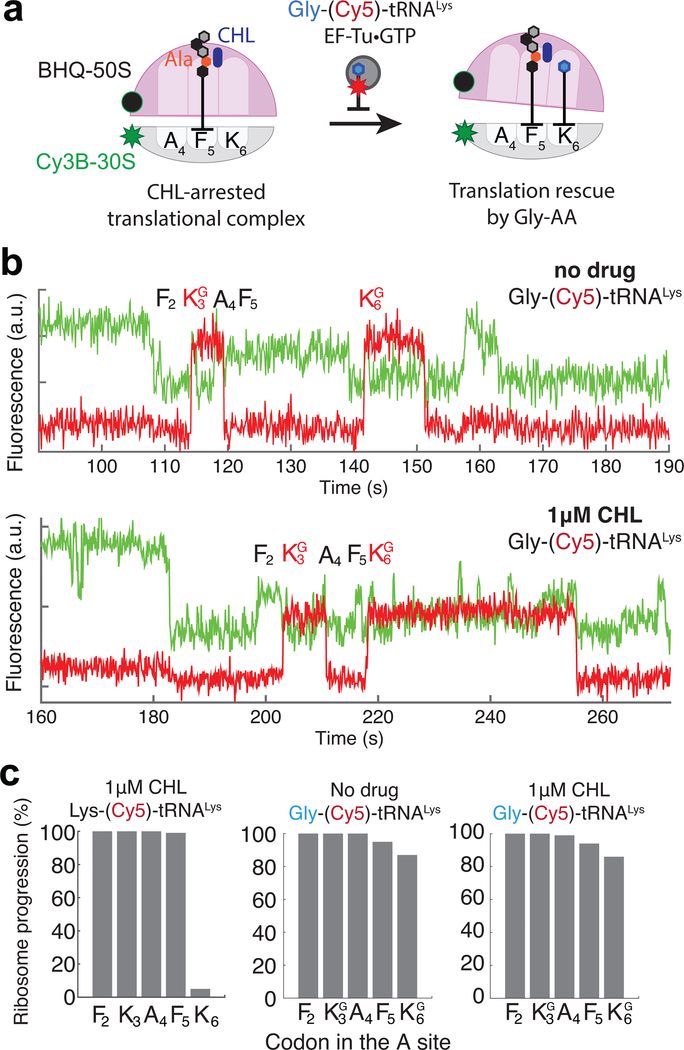
We next characterized CHL- and LZD-induced translation arrest using a smFRET-based translation assay26–28 (Fig 2a). To monitor multiple rounds of translation elongation that report CHL and LZD effects, we labeled E. coli ribosomal small subunits at helix 44 with Cy3B (Cy3B-30S) and labeled large subunits at helix 101 with the quencher BHQ-2 (BHQ-50S)27,29. The one-color FRET signal between the dyes allows the monitoring of ribosomal conformation changes during translation (Fig 2a, green trace). Before aa-tRNA binding, the ribosome assumes a non-rotated state, characterized by substantially quenched Cy3B fluorescence state because of its proximity to BHQ-2 on the large ribosomal subunit. Upon the aa-tRNA accommodation, the peptidyl-transfer reaction induces a transition to the rotated state, detected as a higher Cy3B intensity due to the increased distance between Cy3B and BHQ-2. Subsequently, translocation of the ribosome to the next codon resets the non-rotated state with the deacylated tRNA rapidly departing from the E site30, completing one cycle of translation elongation. To increase further the accuracy of the assignment of translation cycles based on monitoring the intersubunit rotation, we used the binding of fluorescently-labeled tRNAPhe labeled with Cy5 (at the naturally modified acp3U47 residue31; see Methods) at F2 and F5 codons28 (Fig 2a, red trace). The tRNA signal appears when aa-tRNA binds to the A site of the ribosome and persists after its translocation to the P site until the subsequent cycle of elongation places labeled tRNA in the E site for dissociation.
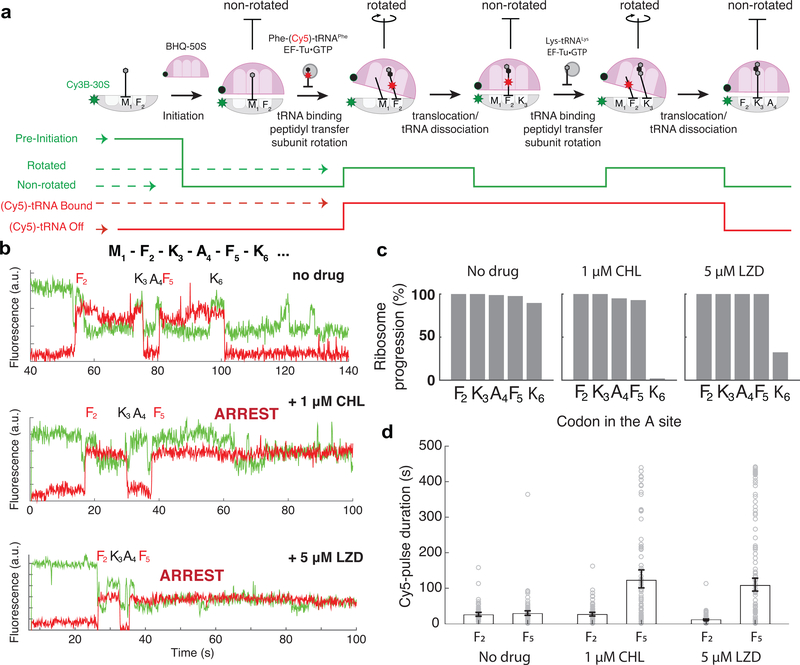
Figure 2. Monitoring the drug-induced translation arrest using smFRET-based assay.
a. Top: Diagram of smFRET-based assay to monitor ribosome structural changes coupled to translation elongation progression. Using Cy3B–BHQ-2 dye-quencher pair, non-rotated and rotated ribosome conformations are tracked. Binding of labeled specific tRNA (Phe-(Cy5)-tRNAPhe) is used to enhance fidelity of state transitions. Bottom: diagram of expected fluorescence intensities matching the structural states depicted immediately above. b. Representative traces for experiments without any antibiotics (similar results observed for n = 87, 100 and 139 for conditions “no drug”, “1μM CHL” and “5μM LZD”, respectively). c. Processivity of translation over the first six codons within the mRNA open reading frame at different conditions, measured as percentage of ribosomes that translated a particular codon over the entire population. d. Measurement of Cy5 pulse-durations from Phe-(Cy5)-tRNAPhe binding events at Phe codon 2 (F2) and Phe codon 5 (F5) for different conditions. Error bars represent 95% confidence interval from fitting the single-exponential distribution. Sample sizes for each conditions are identical in 2b-d.
Fluorescence intensity states corresponding to the non-rotated and rotated state for translating the first five codons were assigned as follows: a decrease in the Cy3B fluorescence intensity followed by a concurrent increase of both Cy3B and Cy5 intensities were assigned as a translation initiation event (binding of BHQ-50S to the surface-tethered mRNA–Cy3B-30S pre-initiation complex) and transition from non-rotated to rotated state for decoding of the first Phe (F2) codon and first peptide bond formation, respectively (Fig 2a,b). Observed non-rotated and rotated intensity levels observed during this translation of the first F2 codon were used to assign subsequent Cy3B fluorescence intensity changes between the non-rotated and rotated states for the first five codons (F2-K3-A4-F5-K6) following the start codon M1 in the MFKAFK mRNA construct. Concurrent decreases of the Cy3B and Cy5 intensities were used as a criterion for assigning the complete translation of two Lys codons (K3 and K6), placed following two Phe codons (F2 and F5). We have used this criterion to avoid the misassignment of Cy3B signal fluctuation as a rapid translation event. In previous studies, we have compared kinetic measurements from this smFRET setup with the ensemble kinetic measurements, and have observed a close agreement in detecting the effect of mRNA modifications to translation decoding kinetics between the two measurement schemes32,33.
By monitoring real-time translation elongation in the presence of 1 μM CHL or 5 μM LZD, we observed a strong arrest of translation at codon F5 of the MFKAFK template, when Ala is present at the penultimate position of the nascent peptide (Fig 2b, middle and bottom traces). An arrest does not occur in the absence of the inhibitors (Fig 2b, upper traces). In the presence of CHL or LZD, upon translocation to the arrest site (F5 codon in the P site), the ribosome does not transition to a stable rotated state, indicating that the aa-tRNA binding and/or peptidyl-transfer is inhibited. CHL inhibited translation more efficiently and at lower concentrations than LZD: at 1 μM CHL, 98 ± 2% of the translating ribosomes were halted at the arrest site, while the effect of LZD was only evident at 5 μM LZD (68 ± 4 % arrested ribosomes) (Fig 2c). The measured lifetime of ribosome occupancy by Phe-(Cy5)-tRNAPhe shows that the presence of CHL or LZD had little effect on the kinetics of tRNA transitioning through all three tRNA binding sites at the F2 codon of the MFKAFK template (Fig 2a,d). In contrast, the second Cy5 pulse lifetime corresponding to the F5 codon was substantially prolonged in the presence of the drugs. The measured Cy5 pulse lifetime of ~120 seconds at the F5 arrest codon (compared to the lifetime of ~25 seconds at the F2 non-arrest codon) is comparable to the Cy5 dye-lifetime before photobleaching34, suggesting that the actual duration of the translation arrest in our system could be even longer. Nonetheless, this result independently confirms the trapping of the ribosome by antibiotics after the F5 codon of the MFKAFK mRNA is placed in the P site.
When the penultimate alanine (A4) residue is changed to tyrosine (Y4) in the MFKYFK construct, neither CHL nor LZD prevents ribosomes from translating the MFKYFK sequence (Supplementary Fig 3a, b), confirming that our single-molecule assay recapitulates the specificity of the drug-dependent arrest. Furthermore, the presence of the drugs did not alter the kinetics of translation at any of the codons of the MFKYFK sequence (Supplementary Fig 3c), which indicates that CHL or LZD prevents the incorporation of the incoming amino acid predominantly when the ribosome carries nascent-peptides with specific sequences. The single-molecule data thus confirm and agree with prior biochemical results on the context-specific stalling in the presence of CHL or LZD. Since CHL was more active in our assays, most of the subsequent experiments were carried out with this drug.
Futile sampling of aa-tRNA to the CHL-arrested ribosome
To elucidate the state of the ribosome during the antibiotic-induced arrest, we monitored the binding of the A-site tRNA. Here we utilized Lys-(Cy5)-tRNALys instead of Phe-(Cy5)-tRNAPhe to cross-correlate elongation cycles deduced from the intersubunit FRET. In the presence of 1 μM CHL, the ribosome efficiently translated the first codons of the MFKAFK template (Fig 3a,b). However, when the F5 codon translocated to the P site of the CHL-bound ribosome, subsequent rounds of translocation were inhibited. Instead, we observed multiple bindings of Lys-(Cy5)-tRNALys to the ribosome directed by the K6 codon positioned in the A site. These tRNA sampling events were uncoupled from the ribosome transition from a non-rotated state to the fully rotated state but corresponded with an intermediate increase in Cy3B fluorescence intensity (referred to as the intermediate FRET state) (Fig 3b). About 30% of the time, sampling of the intermediate FRET state was observed without the appearance of the Cy5 signal (e.g., at 75–125 second in Fig 3b), which we attributed to binding of an unlabeled cognate Lys-tRNALys (present either in the preparation of Lys-(Cy5)-tRNALys and/or the bulk aminoacyl-tRNA despite using lysine-depleted amino acid mixture); alternatively this could be caused by binding of an unlabeled near-cognate tRNA after the miscoding event.
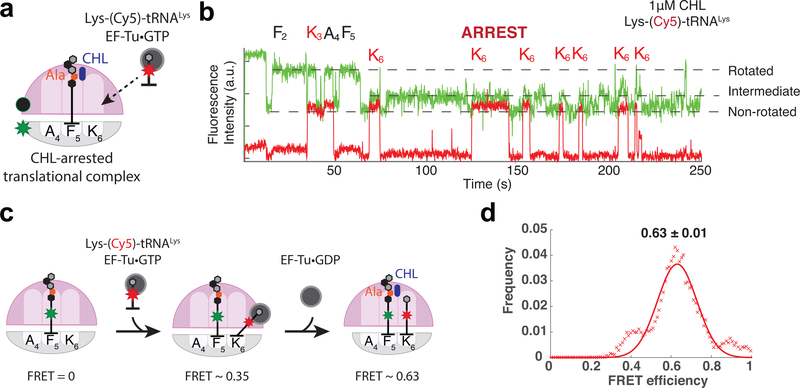
Figure 3. Monitoring accommodation of A-site tRNA during CHL-induced translational arrest.
a, b. Monitoring the binding of Lys-(Cy5)-tRNALys to the ribosome translating the MFKAFK template in the presence of CHL; a. schematics of the experiment, b. a representative trace. The multiple appearance of the intermediate FRET state coincides with the binding of Lys-(Cy5)-tRNALys. On average, Cy5 pulses were observed 3.5 times per translating ribosomes at codon 6 (n = 104 number of ribosomes) c. Schematics of tRNA-tRNA FRET experiment on CHL-arrested ribosome. FRET efficiencies report on the accommodation states of the A-site tRNA (Schematics and FRET efficiencies adapted from Polikanov et al.24). d. smFRET efficiencies (x axis) between the P-site tRNA (fMet-Phe-Lys-Ala-Phe-(Cy3)-tRNAPhe) on codon 5 (F5) and incoming A-site tRNAs (Lys-(Cy5)-tRNALys) on codon 6 (K6), and fitted normal distribution (red line) (n = 121 molecules or 186 individual FRET events).
We thus observed two unexpected features of the CHL-arrested translational complex: First, repeated cycles of A-site tRNA binding and dissociation occurring at the stall site and second, the transition of the ribosome to a state characterized by the intermediate FRET value that may be related to the cycles of tRNA binding.
aa-tRNA body accommodates into the CHL-stalled ribosome
To investigate the state of tRNAs during sampling of the CHL-arrested translational complex, we monitored smFRET between P- and A-site tRNAs, which reports their relative placement on the ribosome31,35. Before the accommodation and positioning of the acceptor aminoacyl residue in the PTC active site, aa-tRNA is delivered to the ribosome in the form of the EF-Tu·GTP·aa-tRNA ternary complex (TC). The TC on the ribosome undergoes conformational transitions that result in different tRNA-tRNA FRET values as the TC progresses from initial interaction with the ribosome to codon recognition, GTPase activation, and subsequent accommodation of the aa-tRNA body within the A site31,36,37. To follow the tRNA accommodation during the drug-induced arrest, the unlabeled ribosomes were stalled by CHL at the F5 codon of the MFKAFK mRNA with fMet-Phe-Lys-Ala-Phe-(Cy3)-tRNAPhe in the P site and the binding of Lys-(Cy5)-tRNALys encoded by the K6 codon was monitored following tRNA-tRNA FRET. (Fig 3c). The mean FRET efficiency of tRNA binding events on the arrested ribosome was 0.63 ± 0.01 (Fig 3d). The high FRET efficiency between two adjacent tRNA suggests that despite inhibiting the pentapeptidyl transfer to the Lys-tRNA, PTC-bound CHL allows for the full accommodation of the body of the incoming aa-tRNA in the A site during the sampling events. This resembles the effects of some other inhibitors of peptide-bond formation (e.g., hygromycin A or A201A) that also block aminoacyl-tRNA entry but apparently act at earlier steps in the tRNA selection pathway23.
A distinct FRET state is observed during aa-tRNA sampling
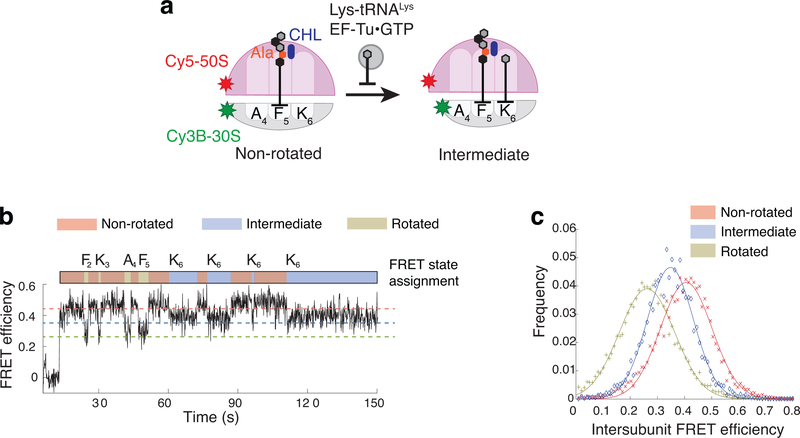
Being intrigued by a full, but non-productive, accommodation of the A-site tRNA, to the drug-stalled ribosome, we wanted to characterize in more detail the ribosome intermediate FRET state that we initially observed using the Cy3B-BHQ intersubunit FRET pair (Fig. 2b). To solidify and expand those observations, we followed the ribosomal conformation using the Cy3B–Cy5 donor–acceptor FRET pair whose anti-correlated signal is less ambiguous than that of Cy3B–BHQ-2 FRET signal38. Translation assays were carried out in the presence of unlabeled tRNAs with Cy5 on the large subunit (Cy5–50S) replacing BHQ-2 (Fig 4a). During translation of the first five codons of the MFKAFK mRNA in the presence of CHL, we observed transitions between high FRET (non-rotated) and low FRET (rotated) states, characterized by average FRET efficiency values of ~0.41 and ~0.26 respectively (Fig 4b and c, red and green lines), in agreement with previously reported results38. However, ribosomes that reached the arrest site (F5) do not acquire the fully rotated state. Instead, we observed repeated rounds of sampling of a state characterized by an average FRET value of ~0.35 (Fig 4b and c, blue line) before the Cy3B or Cy5 photobleaching. These data suggest that the non-productive accommodation of the aa-tRNA in the A site promotes the CHL-arrested ribosome to adapt the state characterized by the intermediate FRET value. Based on the FRET efficiency changes, it is expected that the distance between two labels (Cy3B on 30S subunit helix 44 and Cy5 on helix 101 of 50S subunit) increases by ~3 Å upon transitioning from the non-rotated to the CHL-dependent intermediate FRET state: less than half of the distance change (8 Å) from the non-rotated to fully rotated state measured in the same experiment. This state likely have resulted from a conformational change of the 30S subunit known as domain closure36,39,40 (Supplementary Fig 4), accompanying the full accommodation of the aminoacyl-tRNA in the A site (see Discussion).
Figure 4. Measuring FRET-efficiency for the intermediate FRET state during CHL-induced translational arrest.
a. Schematic diagram of smFRET assay using Cy3B–Cy5 pair to measure FRET efficiencies of non-rotated, rotated and intermediate FRET states of the ribosome during translation in the presence of CHL. b. Representative FRET trace and state assignments for the ribosome translating MFKAFK mRNA in the presence of 1 μM CHL (similar results observed for n = 94). c. FRET-efficiency histogram for each state (+,x,◊) and fitted normal distributions (line).
Incoming glycyl-tRNA relieves CHL-induced translation arrest
The presence of CHL (or LZD) prevents peptide bond formation with the incoming aminoacyl residue when Ala is the penultimate amino acid of the nascent chain, even though CHL (and likely LZD) does not prevent the accommodation of aa-tRNA body in the A site. The CHL-mediated translation block can be circumvented if Gly-tRNAGly serves as the acceptor substrate22 (Fig. 1b, compare lanes 3 and 11). In agreement with those findings, approximately 75% of the ribosomes translated past the arrest site of the MFKAFG mRNA construct in the presence of CHL in the smFRET assay (Supplementary Fig 3a). These data suggest that either the nature of the tRNA itself or of the glycyl moiety is responsible for Gly-tRNAGly rescue of the drug-mediated arrest. Therefore, we next sought to understand which feature of the A-site Gly-tRNAGly contributes to the rescue of the drug-stalled ribosome.
To distinguish the contribution of the tRNAGly body or the aminoacyl residue for circumventing the CHL-induced arrest, we prepared Gly-(Cy5)-tRNALys by mischarging Cy5-labeled tRNALys with glycine using the flexizyme reaction41,42 and then employed this substrate in the smFRET translation assay. Here, the Cy3B–BHQ-2 labeled ribosomes, and Gly-(Cy5)-tRNALys were used to translate MFKAFK mRNA constructs in the presence or absence of CHL (Fig 5a). Without CHL, the synthesized Gly-(Cy5)-tRNALys successfully participated in the translation of Lys codons, with 87% of ribosomes able to synthesize the MFGAFG peptide sequence (Fig 5b,c). Remarkably, even in the presence of 1 μM CHL, 86% of ribosomes translated past the arrest site using Gly-(Cy5)-tRNALys, in stark contrast to the only 2% of ribosomes that were able to incorporate lysine (K6) into the MFKAFK peptide when Lys-(Cy5)-tRNALys was used (Fig 5b,c). Contrary to what was observed with Lys-(Cy5)-tRNALys (Fig. 3b), we did not detect rounds of non-productive binding of Gly-(Cy5)-tRNALys to the CHL-bound ribosome. Thus, replacing the aminoacyl moiety of the Lys-tRNALys with Gly was sufficient for the bypass of the CHL-induced block of peptide bond formation. This agrees well with our observation of the full accommodation of the A-site tRNALys during the CHL-induced translational arrest (Fig 3d): when the body of tRNA is fully accommodated, the bulky lysine residue is likely displaced by CHL from the PTC active site, whereas glycine, that lacks the side-chain, achieves proper positioning to participate in the peptide bond formation reaction. Thus, our data demonstrate that not only penultimate amino acid of the nascent chain, but also the identity of the A-site amino acid critically contributes to defining the sites of CHL-induced translation arrest.
Figure 5. The glycyl residue of the A-site tRNA rescues CHL-arrested ribosomes.
a. Schematic diagram of experiments using Cy5-labeled tRNALys mischarged with Gly (Gly-(Cy5)-tRNALys). b. Representative fluorescence traces of translation of the MFKAFK mRNA in the reaction containing the mischarged Gly-(Cy5)-tRNALys in the absence (top) or presence (bottom) of CHL (similar results observed for n = 134 and 120 for top and bottom plots, respectively). c. Ribosome survival plots showing that the presence of Gly-tRNALys promotes bypassing of the CHL-induced translation arrest at the F5 codon of the MFKAFK template (n = 94, 134 and 120 from left to right).
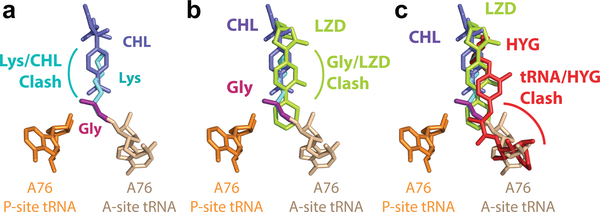
To understand how Gly acceptor aminoacid can rescue translation, we modeled the structure of the drug-bound PTC based on the available structures24,43–45. Because of the lack of side chain, the clash of Gly with the CHL molecule in the A site is greatly diminished (Fig 6a), thereby possibly allowing the Gly acceptor to reach the configuration required to participate in peptide bond formation. The avoidance of the clash is less likely when LZD is bound in the PTC (Fig 6b), possibly explaining why Gly acceptor amino acid rescues CHL-, but not LZD-induced arrest.
Figure 6. Modeling of relative placement of antibiotics and tRNA substrates in the PTC active site.
a. Overlay of the bound CHL molecule and the predicted position of the lysine amino-acid (Lys) on the A-site tRNA (A76 of both A-site and P-site tRNA shown; adapted based on PDB 1VY445 and amino-acid residues modeled). Lys is likely to clash with CHL (PDB 4V7T43), while the glycine (Gly) moiety can be properly accommodated without a direct steric clash. b. Overlay of the LZD (PDB 3CPW44) with CHL and tRNAs bound to the A site and P site of the ribosome. LZD is likely to clash with Gly on the A-site tRNA. c. Overlay of hygromycin A (HYG) with CHL and LZD (PDB 5DOY24). HYG is likely to clash directly with the A76 of A-site tRNA.
The context specificity of action of CHL and LZD has not been demonstrated for several other inhibitors that also bind in the PTC A site. One such example is hygromycin A, an antibiotic whose binding site overlaps with the sites of action of CHL and LZD23 (Fig 6c). Hygromycin A, that clashes not only with the aminoacyl-moiety, but also with the 3′-terminal adenosine of the aa-tRNA, can efficiently arrest the ribosomes at start codons of the genes by inhibiting the tRNA accommodation in the A site23, in contrast to CHL or LZD, both of which fail to inhibit the first peptide bond formation even when present at high concentrations23,25.
Discussion
The dynamic single-molecule fluorescence results presented here complement and extend the biochemical data and provide mechanistic insights into the drastic effect of CHL and LZD on elongation pathways within a given nascent-peptide chain context. As previously shown23, if a nascent peptide contains any amino acid but Ala (or Ser/Thr) in the penultimate position, then the following round of elongation proceeds relatively unimpeded despite the presence of the inhibitor. However, when the penultimate amino acid of the nascent peptide is an Ala (or Ser/Thr), translation is arrested.
We observed that at the site of the CHL-induced arrest, the ribosome can bind, and accommodate the body of the A-site aa-tRNA, but peptide-bond formation does not take place possibly due to the displacement of the incoming amino-acid and/or the nascent-peptide chain. Cognate codon-anticodon interaction in the decoding center, that must take place for the aa-tRNA ccommodation, results in the conformational change of the 30S subunit known as domain closure36,39,40. Such structural rearrangement of the small subunit relocates the placement of the tip of the small subunit helix 44 where our fluorophore was attached, relative to the large subunit. Therefore, the conformational change of the small subunit, induced by aa-tRNA ccommodation, could explain the intermediate intersubunit FRET state observed in our smFRET experiments during unproductive rounds of binding of aa-tRNA at the site of the drug-induced arrest. The same intermediate intersubunit FRET signal has been interpreted as a partially rotated state of the ribosome in our previous report30, caused by the presence of the E-site tRNA. However, in light of our new data, we favor the view that intermediate FRET state reports the 30S domain closure after the tRNA accommodation event and is the on-pathway rearrangement that, in the absence of antibiotic, would be followed by placement of the acceptor amino acid in the PTC A site, peptide bond formation, and intersubunit rotation. Normally this would be a transient state not observed by our smFRET assays, but in the presence of CHL, LZD or E-site tRNA, the duration of this domain-closed state is extended and detected. Further, in the case of CHL or LZD induced translation arrest, the inability of the drug-bound ribosome to catalyze the formation of the peptide bond prevents subsequent transition and instead results in tRNA dissociation and subsequent rounds of aa-tRNA sampling and 30S domain closure.
We can envision two compatible mechanisms that could account for the context specificity of CHL and LZD action defined by the nature of the penultimate amino acid of the nascent chain. First, the penultimate amino acid residue of a stalling nascent peptide may increase the affinity of CHL and LZD to the ribosome, facilitating their competition with the accommodating aa-tRNA. The bound antibiotic would not prevent the accommodation of the tRNA body but would preclude the proper placement of the acceptor substrate in the PTC active site, leading to translation arrest at codons that follow those specifying Ala, Ser or Thr. Second, CHL and LZD may also alter the positioning of the nascent peptide when its penultimate residue is Ala, Ser or Thr, displacing the ester group of the donor substrate from its optimal placement for the nucleophilic attack of the acceptor amino acid α-amino group. The conformational manipulation of both the donor and acceptor peptidyl-transfer substrates may be similar to inhibition mechanisms observed in the poly-proline track, where it has been assumed that not only the restricted acceptor substrate conformation but also maneuvering of the nascent peptide placement are required for translational arrest46. Determining whether either or both mechanisms above underlie antibiotic-induced translational arrest requires additional investigations.
Interestingly, both CHL and LZD have been demonstrated to induce miscoding47, and our work suggests a possible mechanism of how the context specificity of CHL and LZD could induce that effect. Unsuccessful samplings of the cognate tRNA without peptide bond formation open an opportunity for a near-cognate tRNA to deliver an amino acid (e.g., glycine) with a diminished clash with the antibiotic resulting in its incorporation and continued translation. As previously mentioned, this context specificity of action has not been demonstrated for several other inhibitors, such as hygromycin A that also bind in the PTC A site and efficiently arrest the first peptide-bond formation at the start codon23, suggesting a peptide-context independent mechanism of action. Such different context dependencies among antibiotics that inhibit peptidyl-transfer reaction may suggest that combinations of structural, genetic, and kinetic assays are needed to determine the precise mechanism of action for a given drug.
Online Methods
Generation of DNA templates for toeprinting analysis and smFRET experiments
DNA templates were generated by PCR reactions using AccuPrime DNA Polymerase (Thermo Fisher) and the appropriate 5-primer combinations of the oligonucleotides listed in Supplementary Table 1. For example, primers T7, NV1, SMFRET-Fwd, SMFRET-Rev and SMFRET-Mid(FKAFK) were used to generate the MFKAFK template. The structures of the resulting products are shown in Supplementary Fig 1. In the PCR reactions, the T7 and NV1 primers were at 100 μM while the primers SMFRET-Fwd and SMFRET-Rev were at 10 μM. The template specific “SMFRET-Mid” primers were at 10 μM.
Toeprinting assay
Toeprinting assays were conducted as previously described using the NV1 primer48,49. Briefly, translation reactions were performed in a total volume of 5 μl for 15 minutes at 37°C by using PURExpress system (New England Biolabs). After subsequent addition of AMV reverse transcriptase (New England Biolabs) primer extension was carried out for 10 min. When indicated, CHL (Fisher Scientific), LZD (Pharmacia) and thiostrepton (Sigma Aldrich) were present in the reaction at a 50 μM concentration; in these cases, the inhibitors were pre-dried at the bottom of the reaction tubes prior to setting up the translation reactions.
Preparation of biotinylated RNA templates for smFRET in vitro translation assay
The smFRET DNA templates generated by PCR for the toeprinting experiments (see above) were ligated into SmaI-cut pUC18 vector and the resulting plasmids were transformed into E. coli JM109 cells (Promega). Following verification by Sanger sequencing of the constructs, plasmids were cut with EcoRI and used for in vitro run-off transcription with T7 RNA polymerase. Transcription reaction was carried out in the presence of Biotin-GMP to generate 5’ biotinylated RNA.
smFRET experiments
Detailed methods regarding purification of ribosomal subunits, translation factors, tRNA, and specific reagent concentrations, buffer composition, and incubation times have been previously described30,32,33. Briefly, synthetic fluorescently labeled DNA oligonucleotides were annealed to the 30S and 50S ribosomal subunits isolated from E. coli to obtain Cy3B-30S, BHQ-50S or Cy5–50S. Aminoacylated Cy3 or Cy5 labeled tRNAs were prepared as previously described, where purified tRNA (Chem. Block. Ltd) was fluorescently-labeled via NHS-chemistry (Sulfo-Cyanine3 or Sulfo-Cyanine5 NHS ester from Lumiprobe Co., interacting with the acp3U47 residue), HPLC-purified, and charged with amino acid using aminoacyl-synthetase.
Single-molecule experiments were carried out as previously described. Briefly, Cy3B-30S were mixed with protein S1 prior to incubation with biotinylated mRNA (prepared as described above), IF2, and fMet-tRNAini to form pre-initiation complexes. In parallel, EF-Tu was charged with GTP by EF-Ts. Ternary complexes were formed by incubation of EF-Tu/GTP with both fluorescently labeled and bulk unlabeled tRNAs, where the bulk unlabeled tRNAs have been charged with a set of amino acids that lacks phenylalanine or lysine or both. Zero mode wavelength (ZMW) chips were prepared by pre-incubating the surface of the chip with Neutravidin (Thermo Scientific). Pre-initiation complexes containing biotinylated mRNA and Cy3B-30S were then immobilized onto the ZMW chip through Neutravidin-biotin interaction. Labeled 50S subunits (BHQ-50S or Cy5–50S; 65 nM), ternary complexes (50 nM), EF-G (50 nM), oxygen scavenging system (PCA and PCD; purchased from Pacific Bioscience), triplet state quencher (TSY; purchased from Pacific Bioscience) and, depending on the experiment, the inhibitors CHL and LZD (1–5 μM, as indicated), were mixed together and pipetted onto the ZMW chip containing pre-initiation complexes. Eight-minute videos at 10 frames per second of the translation reactions were recorded using PacBio RSII (Pacific Biosciences) while the fluorophores Cy3 (green) and Cy5 (red) were excited with 532-nm and 642-nm lasers, respectively.
Collected movie of fluorescence traces for each experiment was analyzed using previously described pipe-line. Briefly, fluorescence traces that contained high Cy3B and Cy5 signals were selected using a custom script, and manually inspected for correlated Cy3B and Cy5 signals expected for translating molecules. Selecting criteria for intersubunit FRET experiments involved quenching of Cy3B signal upon 70S formation (due to Cy3B–BHQ FRET), followed by Phe-(Cy5)-tRNAPhe binding to F2 codon correlated to non-rotated to rotated state transition. In case of tRNA–tRNA FRET and the intersubunit FRET experiments using Cy3 (or Cy3B) and Cy5 pair, selection criteria involved detecting anti-correlated signal between Cy3 and Cy5 signals. States were assigned in the selected traces, which were used to calculate the number of observable elongation events, Cy5 pulse durations, and the periods of non-rotated and rotated states. For non-rotated and rotated lifetime measurements or Cy5-pulse-duration, time spent in each state across entire population of molecules were collected and fitted with a single-exponential fit. For FRET efficiency measurements, single Gaussian function was fitted to collected frames in each state. For calculation of the ribosome progression, the number of ribosomes translating particular codon (showing both transition to rotated and non-rotated state) are divided with the number of ribosomes translating the first elongation codons (F2 or the second phenylalanine codon in all cases) and multiplied by 100 to calculate percentage. Similarly, the drug efficiency is calculated as:
and the error has been estimated assuming the binomial distribution:
For measuring tRNA-tRNA FRET efficiencies, experiments were carried out in the total internal reflectance (TIRF) microscope setup, for consistency with the previous reports. Briefly, pre-initiation complexes were prepared as outlined above, and mixed with the elongation factors (200 nM of EF-G and 6.7 μM of total tRNA TC charged without Phe-AA and 500 nM of Phe-(Cy3)-tRNAPhe) in the presence of 20 μM CHL, and incubated for 20 minutes in the room temperature, to form CHL-arrested elongation complexes. These were immobilized to the TIRF quartz slide that was functionalized with PEG/PEG-biotin prior to the experiment using biotin-Neutravidin interaction. Unbound complexes were washed out and the slide imaged in buffer containing PCA/PCD/TSY mix and 100 nM of Lys-(Cy5)-tRNALys for 5 minutes at 5 frames per second. The magnesium ion (Mg2+) concentration has been adjusted to 15 mM total MgCl2, to be consistent with the previous measurements.
Mischarging Cy5-tRNALys with glycine by the flexizyme reaction
Gly-(Cy5)-tRNALys was synthesized from flexizyme dFx-catalyzed reaction as previously described41,42,50. 3,5‐dinitrobenzyl ester of glycine (Gly-DBE) was generously provided by Prof. Hiroaki Suga (The University of Tokyo). The flexizyme (dFx) was prepared by run-off T7 transcription of the DNA template, followed by the FPLC-purification step. tRNALys (Chem. Block. Ltd.) has been labeled with Cy5 as described above. In the flexizyme reaction, final concentrations after mixing all the components are listed here: a mixture of 100mM HEPES-KOH pH7.5, 10 μM dFx and 8 μM Cy5-tRNALys was heated at 95°C for 2 min and slowly cooled to room temperature over 5 min. Then MgCl2 was added to the final concentration of 25 mM and the reaction was incubated at room temperature for another 5 min before cooling on ice for 3 min. Gly-DBE in DMSO was added to 5 mM from a 25 mM stock to initiate the charging reaction, which was incubated on ice for 5 hrs before quenching by the addition of final 300 mM NaOAc, pH 5.2. The charged tRNA was ethanol precipitated, resuspended in tRNA storage buffer (10 mM NaOAc pH 5.2 and 0.5 mM Mg(OAc)2) and further purified by running through a P6 spin column equilibrated with the same tRNA storage buffer. Final tRNA was stored at −80 °C after flash freezing with liquid nitrogen, and was used for making the ternary complex as described above.
Statistics and Reproducibility
Measurements from single-molecule fluorescence assay were resulting from specified number (n) of molecules from a single experiment.
Data Availability statement
All experimental data are available upon request.
Code Availability statement
All MATLAB scripts used in this study are available upon request.
Supplementary Material
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grants GM51266 to J.D.P. and R01 AI125518 to A.S.M. and N. V.-L.; by a Stanford Bio-X fellowship to J.C.; by Knut and Alice Wallenberg Foundation postdoctoral scholarships to J.Z. and to J.W. (2015.0406). We are grateful to Prof. Suga for providing the substrate for the flexizyme reaction and Prof. Innis for the advice and training in using the flexizyme-based procedures. We thank members of the Puglisi lab and the Mankin/Vázquez-Laslop lab for discussion and input.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Rodnina MV & Wintermeyer W Peptide bond formation on the ribosome: Structure and mechanism. Curr. Opin. Struct. Biol 13, 334–340 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Frank J & Agrawal RK A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406, 318–322 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Valle M et al. Locking and Unlocking of Ribosomal Motions. Cell 114, 123–134 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Agirrezabala X et al. Visualization of the Hybrid State of tRNA Binding Promoted by Spontaneous Ratcheting of the Ribosome. Mol. Cell 32, 190–197 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunkle JA et al. Structures of the Bacterial Ribosome in Classical and Hybrid States of tRNA Binding. Science (80-. ). 332, 981–984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ermolenko DN et al. Observation of Intersubunit Movement of the Ribosome in Solution Using FRET. J. Mol. Biol 370, 530–540 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Moazed D & Noller HF Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–8 (1989). [DOI] [PubMed] [Google Scholar]
- 8.Munro JB, Altman RB, O’Connor N & Blanchard SC Identification of Two Distinct Hybrid State Intermediates on the Ribosome. Mol. Cell 25, 505–517 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bock LV et al. Energy barriers and driving forces in tRNA translocation through the ribosome. Nat. Struct. Mol. Biol 20, 1390–1396 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Jamiolkowski RM, Chen C, Cooperman BS & Goldman YE tRNA Fluctuations Observed on Stalled Ribosomes Are Suppressed during Ongoing Protein Synthesis. Biophys. J 113, 2326–2335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voorhees RM & Ramakrishnan V Structural basis of the translational elongation cycle. Annu. Rev. Biochem 82, 203–236 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Polacek N & Mankin AS The ribosomal peptidyl transferase center: Structure, function, evolution, inhibition. Crit. Rev. Biochem. Mol. Biol 40, 285–311 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Wilson DN The A-Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol 44, 393–433 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Lin J, Zhou D, Steitz TA, Polikanov YS & Gagnon MG Ribosome-Targeting Antibiotics : Modes of Action, Mechanisms of Resistance, and Implications for Drug Design. Annu. Rev. Biochem 87, 451–478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arenz S & Wilson DN Bacterial Protein Synthesis as a Target for Antibiotic Inhibition. Cold Spring Harb Perspect Med 6, a025361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrlich J, Gottlieb D, Burkholder PR, Andierson LE & Pridham TG Streptomyces venezuelae, n. sp., the source of chloromycetin. J. Bacteriol. 56, 467–477 (1948). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brickner SJ, Barbachyn MR, Hutchinson DK & Manninen PR Linezolid (ZYVOX), the First Member of a Completely New Class of Antibacterial Agents for Treatment of Serious Gram-Positive Infections. J. Med. Chem 51, 1981–1990 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Schlunzen F et al. Structural basis for the interaction of antibiotics with the peptidyl transferase center in eubacteria. Nat. (London, United Kingdom) 413, 814–821 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Leach KL et al. The Site of Action of Oxazolidinone Antibiotics in Living Bacteria and in Human Mitochondria. Mol. Cell 26, 393–402 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Wilson DN et al. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci 105, 13339–13344 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulkley D, Innis CA, Blaha G & Steitz TA Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl. Acad. Sci 107, 17158–17163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pestka S in Antibitoics III: Mechanism of Action of Antimicrobial and Antitumor Agents (ed. Corcoran JW, Hahn FE) 370–395 (Springer-Verlag, 1975). [Google Scholar]
- 23.Marks J et al. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc. Natl. Acad. Sci. U. S. A 113, 12150–12155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polikanov YS et al. Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A. Mol. Cell 58, 832–844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orelle C et al. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob. Agents Chemother 57, 5994–6004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aitken CE & Puglisi JD Following the intersubunit conformation of the ribosome during translation in real time. Nat. Struct. Mol. Biol 17, 793–800 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorywalska M et al. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res 33, 182–189 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uemura S et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature 464, 1012–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Tsai A, Petrov A & Puglisi JD Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics. J. Am. Chem. Soc 134, 5734–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi J & Puglisi JD Three tRNAs on the ribosome slow translation elongation. Proc. Natl. Acad. Sci 114, 13691–13696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanchard SC, Jr RLG, Kim HD, Chu S & Puglisi JD tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol 11, 1008–1014 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Choi J et al. N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol 23, 110–115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi J et al. 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat. Struct. Mol. Biol 25, 208–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J et al. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc. Natl. Acad. Sci. U. S. A 111, 664–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wasserman MR, Alejo JL, Altman RB & Blanchard SC Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat. Struct. Mol. Biol 23, 333–341 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loveland AB, Demo G, Grigorieff N & Korostelev AA Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alejo JL & Blanchard SC Miscoding-induced stalling of substrate translocation on the bacterial ribosome. Proc. Natl. Acad. Sci. U. S. A 114, E8603–E8610 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall RA, Aitken CE & Puglisi JD GTP Hydrolysis by IF2 Guides Progression of the Ribosome into Elongation. Mol. Cell 35, 37–47 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fislage M et al. Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A. Nucleic Acids Res. 46, 5861–5874 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brilot AF, Korostelev AA, Ermolenko DN & Grigorieff N Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc. Natl. Acad. Sci. U. S. A 110, 20994–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami H, Ohta A, Ashigai H & Suga H A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat. Methods 3, 357–359 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Goto Y & Suga H Translation initiation with initiator tRNA charged with exotic peptides. J. Am. Chem. Soc 131, 5040–5041 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Dunkle JA, Xiong L, Mankin AS & Cate JHD Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci 107, 17152–17157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ippolito JA et al. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem 51, 3353–3356 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Polikanov YS, Steitz TA & Innis CA A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol 21, 787–793 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huter P et al. Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P. Mol. Cell 68, 515–527.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Thompson J, O’Connor M, Mills JA & Dahlberg AE The protein synthesis inhibitors, oxazolidinones and chloramphenicol, cause extensive translational inaccuracy in vivo. J. Mol. Biol 322, 273–279 (2002). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 48.Vazquez-Laslop N, Thum C & Mankin AS Molecular Mechanism of Drug-Dependent Ribosome Stalling. Mol. Cell 30, 190–202 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Orelle C et al. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 41, e144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Kwiatkowski M & Forster AC Kinetics of Ribosome-Catalyzed Polymerization Using Artificial Aminoacyl-tRNA Substrates Clarifies Inefficiencies and Improvements. ACS Chem. Biol 10, 2187–2192 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All experimental data are available upon request.