Abstract
Bile salt hydrolase (BSH) enzymes are widely expressed by human gut bacteria and catalyze the gateway reaction leading to secondary bile acid formation. Bile acids regulate key metabolic and immune processes by binding to host receptors. There is an unmet need for a potent tool to inhibit BSHs across all gut bacteria in order to study the effects of bile acids on host physiology. Here, we report the development of a covalent pan-inhibitor of gut bacterial BSH. From a rationally designed candidate library, we identified a lead compound bearing an alpha-fluoromethyl ketone warhead that modifies BSH at the catalytic cysteine residue. Strikingly, this inhibitor abolished BSH activity in conventional mouse feces. Mice gavaged with a single dose of this compound displayed decreased BSH activity and decreased deconjugated bile acid levels in feces. Our studies demonstrate the potential of a covalent BSH inhibitor to modulate bile acid composition in vivo.
Introduction
Bile acids, long relegated to the role of undistinguished detergents, have recently emerged as likely candidates for the molecular messengers that allow members of the human gut microbiome to modulate the physiology and behavior of their hosts.1,2 Investigating these potential roles in detail has been hampered by the lack of tools to regulate specific messages, and developing the appropriate tools has been hampered in turn by the complex biosynthesis of bile acids. Primary bile acids are produced in the liver from cholesterol and conjugated to taurine or glycine to produce primary conjugated bile acids (Fig. 1a). These molecules are stored in the gallbladder and released into the digestive tract where they aid in absorption of lipids and vitamins. Over 95% of bile acids are reabsorbed in the ileum and transported to the liver. The remaining ~5% pass into the colon, where the majority of gut bacteria reside. Gut bacteria then enzymatically modify these primary bile acids, producing a group of molecules called secondary bile acids (Fig. 1a). Roughly 50 secondary bile acids have been detected in human feces, and their concentrations can reach low millimolar levels.3,4
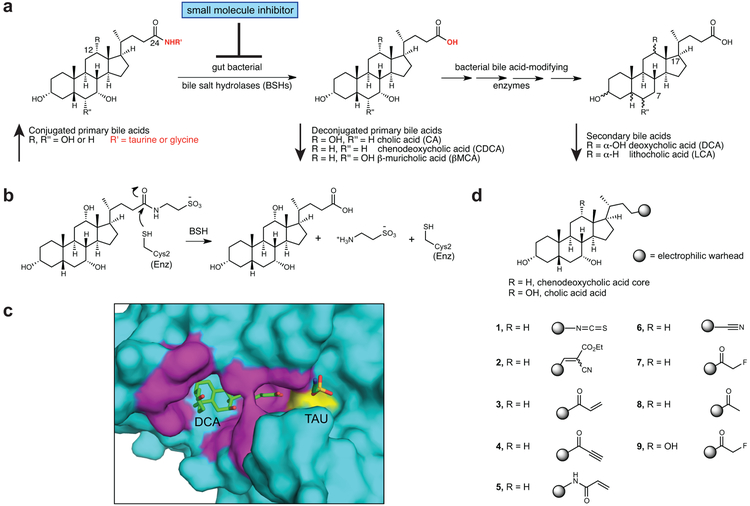
Figure 1. Rational design of gut bacterial bile salt hydrolase (BSH) inhibitors.
(a) BSHs are the gateway enzymes in the conversion of primary (host-produced) to secondary (bacterially produced) bile acids. Inhibition of BSHs should result in a decrease in deconjugated primary and secondary bile acids. (b) Mechanism of enzymatic amide bond cleavage by BSHs. (c) A co-crystal structure of the BSH from the Gram positive gut bacterium Clostridium perfringens (strain 13 / type A) and deconjugated tauro-deoxycholic acid (PDB 2BJF) guided inhibitor design. While hydrophobic interactions orient the bile acid core in the active site, the D-ring side chain is exposed to solvent (magenta residues are within 4Å of bile acid, Cys2 is yellow). (d) Library of synthesized inhibitors. Electrophilic warheads were appended to the chenodeoxycholic acid bile core in order to create broad-spectrum BSH inhibitors.
Bile acids, which share a carbon skeleton with steroids, can bind to host receptors, including nuclear hormone receptors (NhR) and G-protein coupled receptors (GPCRs). By acting as either agonists or antagonists for these receptors, bile acids regulate host metabolism, including energy expenditure and glucose and lipid homeostasis,2,5 and host immune response, including both innate and adaptive immunity.6,7 Dysregulated bile acid metabolism is thought to play a causal role in the pathophysiology of diseases including hypercholesterolemia, obesity, and cancer,2,8,9 further highlighting the biological importance of these molecules.
The key reaction in the conversion of primary into secondary bile acids is the hydrolysis (deconjugation) of the C24-amide bond of conjugated primary bile acids (Fig. 1a). This enzymatic conversion is performed exclusively by gut bacterial bile salt hydrolase (BSH) enzymes.1 BSHs (EC 3.5.1.24) are widespread in human gut bacteria. A recent study identified BSHs in gut species from 117 genera and 12 phyla.10 A non-toxic, small molecule pan-inhibitor of gut bacterial BSHs would provide a powerful tool to study how bile acids affect host physiology. Such a compound should limit bile acid deconjugation across the vast majority of gut strains without significantly affecting bacterial growth. The use of a pan-inhibitor in vivo would significantly alter bile acid pool composition, shifting the pool toward conjuated bile acids and away from deconjugated bile acids and secondary bile acids (Fig. 1a).
Herein, we report the development of a covalent inhibitor of bacterial BSHs using a rational design approach. Importantly, this compound completely inhibits BSH activity in conventional mouse feces, demonstrating its potential utility as a pan-BSH inhibitor. This chemical tool may allow researchers to investigate previously unanswered questions, including how primary and secondary bile acids differentially affect physiology in a fully colonized host.
Results
Rational design of covalent BSH inhibitors
Covalent inhibitors can inactivate their protein target with a high degree of potency and selectivity even in the presence of large concentrations of native substrate.11 The substrates for BSHs, conjugated bile acids, are found in high concentrations in the colon (1-10 mM),4 suggesting that covalent inhibition could be an effective strategy for targeting these enzymes. In addition, recent work has demonstrated that irreversible inhibitors of bacterial enzymes can be effective in the gut,12 thus further validating our approach.
While there is significant divergence in BSH protein sequence across gut strains, all BSHs possess a conserved active site that includes a catalytic cysteine (Cys2) (Fig. 1b).1,10 We reasoned that by designing compounds that targeted this conserved residue, we could develop pan-BSH inhibitors. A co-crystal structure of the Clostridium perfringens BSH and the substrate taurodeoxycholic acid showed that hydrophobic interactions engaged the bile acid core and oriented the amide toward Cys2, leaving the amino acid solvent-exposed (Fig. 1c).13 Furthermore, C. perfringens BSH tolerates a large degree of variability in the amino acid side chain, including longer chain conjugates.14 These results suggested that the C-17 bile acid side chain was a possible site for incorporation of an electrophilic group.
Based on this rationale, we designed a small library of potential inhibitors containing both a bile acid core motif and a pendant electrophilic warhead (Fig. 1d). While previous literature indicated that conjugated amino acid identity largely drives BSH specificity,1 sterol core configuration also affects BSH reactivity.15 Indeed, in recent work, we determined that some Bacteroidetes species cleave C12 = H but not C12 = OH primary bile acids (Fig. 1a).16 In order to target both Gram negative and Gram positive strains, we decided to utilize the steroidal portion of the human primary bile acid chenodeoxycholic acid (CDCA, C12 = H) as our scaffold.
For the electrophilic trapping groups, we chose warheads that have been successfully deployed in the development of protease and kinase inhibitors,17 including isothiocyanate (1),18 cyanoacrylate (2),19 α,β-unsaturated systems (3 and 4),20 acrylamide (5),21 and nitrile (6).22 We also synthesized an inhibitor with an α-fluoromethyl ketone warhead (FMK) (7). In contrast to the more electrophilic α-iodo-, α-bromo- and α-chloromethyl ketone warheads, the weak leaving group ability of fluorine renders the FMK warhead less reactive and hence more selective.23,24 As a result, FMK-based inhibitors have been shown to result in minimal off-target effects.23,25
Biochemical characterization of BSHs
Following inhibitor synthesis (Supplementary Note), we sought to evaluate the activity of inhibitors 1-9 against both Gram negative and Gram positive BSHs. We decided to use a selective Bacteroides BSH for inhibitor optimization, reasoning that the more limited substrate scope of this enzyme could make it more difficult to target. We heterologously expressed and purified the selective BSH (BT_2086) that we had previously identified in Bacteroides thetaiotaomicron VPI-5482 (B. theta) (Supplementary Table 1 and Supplementary Fig. 1).16 We then established kinetic parameters using a ninhydrin-based assay.26 Consistent with our previous results from B. theta cultures, purified B. theta BSH displayed a preference for tauro-ursodeoxycholic acid (TUDCA) deconjugation and did not deconjugate tauro-cholic acid (TCA) (Table 1 and Supplementary Fig. 1).16
We also cloned, expressed, and determined kinetic parameters for the BSH from the Gram positive strain Bifidobacterium longum SBT2928 BSH27 (Table 1 and Supplementary Fig. 1). The Km values for all of the recognized substrates are in the low millimolar range, which is approximately the concentration of these bile acids in the gut. While the kcat values are lower than the kcat reported for the Lactobacillus salivarius BSH, the Km values for these enzymes are similar to those of previously characterized BSHs.27-29
α-FMK compound 7 inhibits recombinant BSHs
We next evaluated the ability of the compounds in our library to inhibit B. theta and B. longum BSHs. We also tested riboflavin (10) and caffeic acid phenethyl ester (CAPE, 11), compounds that had been previously identified in a high-throughput screen for inhibition of a BSH from a Lactobacillus salivarius chicken gut isolate (Supplementary Fig. 1).30 To determine BSH inhibitory activity, we pre-incubated the B. theta BSH with each inhibitor (100 μM) for 30 minutes and then added a mixture of conjugated bile acids (100 μM final concentration). Because BSHs display varying reactivities toward different conjugated bile acids, we used an equimolar combination of two primary and two secondary conjugated bile acids that are predominant in the gallbladder and small intestine of conventional mice as our substrate mixture (tauro-β-muricholic acid (TβMC), TCA, TUDCA, and tauro-deoxycholic acid (TDCA)).31 Deconjugation of bile acids was monitored by Ultra Performance Liquid Chromatography-Mass Spectrometry (UPLC-MS) over 21 hours. Among the synthesized inhibitors, isothiocyanate (1) displayed modest inhibition. Other compounds containing Michael acceptor warheads (2-6) did not inhibit deconjugation. In contrast, incubation with α-FMK-based 7 resulted in almost complete inhibition of B. theta BSH activity for 21 hours (>98%, Fig. 2a, Supplementary Fig. 3-4, and Supplementary Table 2). In order to validate that the inhibitory activity of 7 was due to the presence of fluorine as a leaving group, we synthesized a methyl ketone analog (8).25 This analog did not display BSH inhibition against either recombinant protein or B. theta cultures, indicating that the α-fluoro group was necessary for activity (Fig. 2a, Supplementary Fig. 5, and Supplementary Table 2). Riboflavin did not display any inhibitory activity, while CAPE provided only moderate inhibition of B. theta BSH.
Figure 2. Identification of 7 as a potent, non-toxic, pan-BSH inhibitor.
(a,b) Screen of inhibitors versus B. theta BSH (a) and B. longum BSH (b) showing % deconjugation of tauro bile acids at 2 and 21 hours (for concentrations of substrates and products, see Supplementary Fig. 4; for % deconjugation of each bile acid see Supplementary Table 2). Inhibitor (100 μM) was incubated with 200 nM rBSH for 30 mins followed by addition of taurine-conjugated bile acids (tauro-β-muricholic acid, TβMCA; tauro-cholic acid, TCA; tauro-ursodeoxycholic acid, TUDCA; and tauro-deoxycholic acid, TDCA, 25 μM each). Deconjugation of substrate was followed by UPLC-MS. (c) Compound 7 inhibited BSH activity in growing cultures of Gram negative (B. theta VPI 5482, Bacteroides fragilis ATCC 25285, and Bacteroides vulgatus ATCC 8482) and Gram positive (Lactobacillus plantarum WCFS1, Clostridium perfringens ATCC 13124, and Bifidobacterium adolescentis L2-32) bacteria. Inhibitor (100 μM of 7 or CAPE) and taurine-conjugated bile acids (TβMCA, TCA, TUDCA and TDCA, 25 μM each) were added to bacterial cultures at OD600 0.1. Cultures were allowed to grow into stationary phase and percent deconjugation of tauro bile acids at 21h was determined by UPLC-MS (for concentrations of substrates and products, see Supplementary Fig. 8; for % deconjugation of each bile acid see Supplementary Table 2). (d) Compound 7 is not bactericidal. Bacterial strains were incubated with conjugated bile acids (as described in panel c) and compound (100 μM) and plated at 21h to assess strain viability. CAPE decreased the cell viability of the Gram negative strains tested. Red downward arrows indicate fold decrease compared to DMSO control. For (c) and (d), one-way ANOVA followed by Dunnett’s multiple comparisons test. (e) Compound 7 inhibited BSH activity in a fecal slurry. All assays were performed in biological triplicate, and data are presented as mean ± SEM.
We next evaluated the activity of 1, 7, and CAPE against the BSH from B. longum. Compound 7 was again the most active inhibitor, while CAPE was ineffective at inhibiting B. longum BSH at all timepoints (Fig. 2b and Supplementary Fig. 3-4, and Supplementary Table 2). Compound 7 inhibited both B. theta and B. longum BSHs in a dose-dependent fashion (IC50 values of 427 nM and 108 nM respectively, Supplementary Fig. 6). Taken together, these data indicate that compound 7 is a potent inhibitor of purified BSH protein from both a Gram negative and a Gram positive bacterial strain.
Compound 7 completely inhibited B. theta BSH, the more catalytically efficient of the two enzymes (Table 1), within 15 seconds at a concentration equimolar to substrate and without any preincubation of inhibitor with enzyme (Supplementary Fig. 7). In the presence of a large excess (~80-fold) of substrate, 7 entirely inhibited B. theta BSH activity within 15 minutes, the earliest measurable timepoint for product formation under these conditions. These results indicate that 7 is a kinetically efficient inhibitor of BSH activity.
Table 1:
Kinetic parameters for BSHs from Bactereroides thetaiotaomicron (B. theta) and Bifidobacterium longum (B. longum).
| BSH sourcea | Substrateb | kcat (min−1) | Km (mM) | kcat/Km
(min−1mM−1) |
|---|---|---|---|---|
| B. theta | TCAc | -- | -- | -- |
| TUDCA | 15.3 ± 0.8 | 8.2±1.0 | 1.9 ± 0.3 | |
| TDCA | 12.9 ± 0.6 | 3.4±0.6 | 3.8 ± 0.7 | |
| TCDCA | 4.3 ± 0.6 | 2.9±1.8 | 4.3 ± 0.9 | |
| B. longum | TCA | 6.9 ± 0.9 | 8.3±2.5 | 0.8 ± 0.3 |
| TUDCA | 0.9 ± 0.2 | 4.1±2.5 | 0.2 ± 0.1 | |
| TDCA | 3.5 ± 0.1 | 2.3±0.3 | 1.5 ± 0.2 | |
| TCDCA | 4.6 ± 0.6 | 7.0±2.3 | 0.6 ± 0.2 |
Characterization was performed using ninhydrin reagent and experiments were performed in PBS buffer at pH 7.5 and 37 °C.
Conjugated primary and secondary bile acid used as substrates were taurocholic acid (TCA), tauroursodeoxycholic acid (TUDCA), taurodeoxycholic acid (TDCA), taurochenodeoxycholic acid (TCDCA).
B. theta did not deconjugate TCA. n=3 biological replicates per condition. All data are presented as mean ± SEM.
Compound 7 inhibits BSHs in gut bacterial cultures
We next sought to evaluate the potency of 7 in growing bacterial cultures. To test the scope of BSH inhibition, we used three Gram negative and three Gram positive strains of BSH-containing human gut bacteria (Gram negative, B. theta, Bacteroides fragilis ATCC 25285, and Bacteroides vulgatus ATCC 8482; Gram positive, Lactobacillus plantarum WCFS1, Clostridium perfringens ATCC 13124, and Bifidobacterium adolescentis L2-32).1,16
Bacterial cultures were diluted to pre-log phase and both inhibitor (100 μM) and a mixture of conjugated bile acids (100 μM final concentration) were added simultaneously. Deconjugation was monitored over 21 hours using UPLC-MS. Strikingly, while all six bacterial strains deconjugated bile acids in the presence of vehicle control, we observed almost no deconjugation in any of the cultures grown in the presence of 7 (Fig. 2c, Supplementary Fig. 8, and Supplementary Table 2). We then incubated an isogenic BSH-deleted B. theta strain16 with either DMSO, 7, or CAPE. Under all three conditions, taurine-conjugated bile acids were recovered unmetabolized (Supplementary Fig. 9). These results suggest that BSH inhibition by 7 is not due to the effects of this inhibitor on other bile acid-utilizing processes. Compound 7 did not significantly affect the cell viability of the majority of the tested strains (Fig. 2d), indicating that the BSH inhibition observed was not due to bactericidal activity. We determined that the IC50 values of this inhibitor against B. theta and B. adolescentis were 1070 nM and 237 nM, respectively (Supplementary Fig. 10). These results indicate that 7 is a potent, broad-spectrum inhibitor of BSHs.
We observed no BSH inhibition in five out of the six bacterial strains grown in the presence of CAPE (Fig. 2c). Moreover, CAPE inhibited the cell viability of all three Gram negative bacterial strains tested (Fig. 2d). These results suggest that the dominant effect of CAPE on Gram negative bacteria is not inhibition of BSH activity but rather inhibition of growth.
Finally, to evaluate our hypothesis that C12=OH compounds would not be effective broad-spectrum inhibitors, we synthesized a compound in which we appended the α-FMK warhead to a C12=OH bile acid core, cholic acid (compound 9, Fig. 1d). Compound 9 displayed significantly reduced ability to inhibit BSH deconjugation in B. theta cultures compared to 7 (Supplementary Fig. 5). These results support the hypothesis that bile acid core structure, specifically C12 substitution, affects the ability of our probes to inhibit selective BSH. In addition, these results suggest that the α-FMK warhead is not broadly reactive but rather requires suitable positioning within the active site, a hypothesis that we later investigated using mass spectrometry and crystallography studies.
Compound 7 inhibits BSH activity in mouse feces
Previous literature had reported significant BSH activity in mouse feces.32 To further assess whether 7 is a pan-inhibitor of BSH, we tested its activity in resuspended feces from conventional mice. We reasoned that this fecal slurry should contain BSHs from nearly the entire bacterial community of the distal colon. Compounds 1, 7, and CAPE (20 μM) were added to a fecal suspension in buffer. After 30 minutes, the deuterated substrate glycochenodeoxycholic acid-d4 (GCDCA-d4) was added, and formation of deconjugated product was quantified after 18 hours using UPLC-MS. Strikingly, we observed that incubation with 7 completely inhibited BSH activity in feces (Fig. 2e). Consistent with our in vitro results, CAPE provided no inhibition of BSH activity in feces. These results demonstrate that 7 is a potent pan-inhibitor of BSH activity.
Compound 7 covalently modifies catalytic Cys2 residue
We next investigated the mechanism of inhibition of 7. The B. theta BSH contains two cysteine residues, Cys2 and Cys67. Analysis of an apo crystal structure of this enzyme revealed that both the cysteine residues are pointed towards the active site (PDB 3HBC). To confirm that 7 is a covalent inhibitor that modifies Cys2, we incubated purified B. theta BSH with an excess of this molecule. Analysis by mass spectrometry revealed a mass shift consistent with the addition of a single molecule of 7, confirming formation of a covalent bond (Supplementary Fig. 11). Subsequent top-down mass spectrometry analysis identified Cys2 as the modified residue (Supplementary Fig. 11).
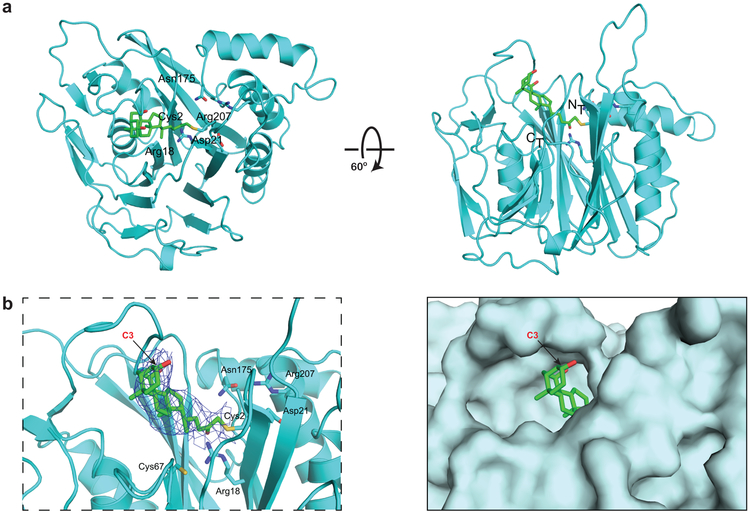
We then determined the structure of the B. theta BSH, first in its apo form to 2.7 Å resolution and then covalently bound to 7 to 3.5 Å resolution (Supplementary Table 3) (PDB 6UFY and 6UH4, respectively). The structure of the BSH-inhibitor complex contains four copies of the protein in the asymmetric unit. The electron density map is best resolved in two of the four subunits, and electron density is clearly visible for the inhibitor in one of these subunits covalently attached to Cys2 (Fig. 3a-b). Comparison with the apo structure also suggests that there is a loop (residues 127-138) which repositions to clasp the inhibitor in the active site in a solvent-exposed channel (Supplementary Fig. 12).
Figure 3. Compound 7 covalently modifies B. theta BSH at the active site cysteine residue.
(a) X-ray structure of 7 bound to B. theta BSH. The BSH (cyan) is shown in ribbon representation, with indicated side chains (cyan, with heteroatoms in CPK colors) rendered as sticks. Compound 7 (green, with heteroatoms in CPK colors) is rendered in stick form. There is electron density visible at the active site of one of the four subunits in the asymmetric unit, consistent with the conclusion that the inhibitor is covalently attached to Cys2. (b) Co-crystal structure of B. theta BSH and 7 shown in ribbon (left, with electron density of the compound shown as a blue net) and surface (right) representations. The A ring of 7, which includes the C3 hydroxyl group, is solvent-exposed. Panels a and b were prepared using PYMOL software (Schroedinger).
These data indicate that 7 selectively labels the B. theta BSH at Cys2. Furthermore, the co-crystal structure reveals that the C3-hydroxyl group is solvent-accessible, suggesting that this site might be amenable to further modification (Fig. 3b).
Compound 7 displays minimal off-target effects
Concerns have been raised that non-specific reactivity of covalent inhibitors could result in acute toxicity.11 Bile acids are ligands for the farnesoid X receptor (FXR) and the G protein-coupled bile acid receptor 1 (TGR5).2 An in vitro coactivator recruitment assay showed that 7 is neither an agonist nor an antagonist for FXR at physiologically relevant concentrations (Supplementary Fig. 13).31 Next, we evaluated the effect of compound 7 on TGR5 activation in a human intestinal cell line (Caco-2). Compound 7 neither agonized nor antagonized TGR5 over the range of concentrations tested (Supplementary Fig. 13). These results suggest that 7 does not induce off-target effects on either of these critical host receptors.
Bile acids are also known to be toxic due to their detergent properties.1,33 We tested the toxicity of this compound against human intestinal cells (Caco-2 and NCI-H716). No resultant toxicity was observed when these cells were incubated with up to 50 μM or 100 μM of compound 7, respectively (Supplementary Fig. 13). Because the IC50 values of 7 range from 237 to 1070 nM, these results suggest that it should be possible to achieve an effective, non-toxic in vivo dose. To test the effect of compound 7 on epithelial integrity, Caco-2 cells were differentiated in transwell inserts into a polarized monolayer with tight intercellular junctions.34 Compound 7 was incubated in the apical chamber of the transwells, and epithelial integrity was measured by passive diffusion of 4kDa FITC-dextran. No significant increase in fluorescence was observed in 7-treated cells compared to control-treated cells, indicating that 7 did not compromise epithelial monolayer integrity (Supplementary Fig. 14).
Despite these encouraging findings, it is important to understand the proteome-wide reactivity of 7.35 To assess target engagement and off-target interactions of compound 7, we synthesized a ‘clickable’ version of this inhibitor. We appended an α-azido moiety36 to 7 at the solvent-exposed C3 position to generate 7-N3 (compound 12, Fig. 4a). Like 7, 7-N3 potently inhibited BSH activity in mouse feces (Fig. 4b). These results demonstrate that azide substitution did not significantly perturb the BSH inhibitory activity of this molecule. To study on- and off-target effects in bacterial cells, cultures of B. adolescentis L2-32 were treated for 1 hour with 10 μM of 7-N3, a concentration at which 7 inhibited BSHs in bacterial culture (Supplementary Fig. 5). Lysed bacterial supernatants were then reacted with Fluor 488-alkyne under copper catalyzed azide-alkyne cycloaddition conditions, and proteins were visualized by in-gel fluorescence. Remarkably, only one fluorescent band was visible at a mass of ~35 kDa, the predicted mass of the annotated B. adolescentis BSH (Fig. 4c and Supplementary Fig. 15). To identify this protein, we clicked the clarified lysate with desthiobiotin-alkyne and performed streptavidin pulldown. Bound proteins from control and treated samples were resolved by SDS-PAGE and visualized by silver-staining (Fig. 4d and Supplementary Fig. 15). We observed a single silver-stained band at the predicted molecular weight (~35 kDa) of BSH. We excised this band along with the corresponding region of the control lane, digested both with trypsin, and performed LC-MS/MS. BSH was identified in the gel bands with high confidence, and a semi-quantitative analysis of these data indicated a 4.5-fold enrichment in 7-N3-versus vehicle-treated bacterial cultures (Supplementary Table 4). To assess off-target binding across the bacterial proteome, we digested streptavidin bead-bound proteins isolated from treated and control bacterial cultures. Label-free LC-MS/MS analysis identified BSH as 3.6-fold enriched in probe-treated cultures (Supplementary Tables 5-7). No other proteins exceeded a 2-fold enrichment threshold across biological triplicate experiments. Competition of 7 with 7-N3 showed dose-dependent labeling of the annotated B. adolescentis BSH (Fig. 4e and Supplementary Fig. 15), further confirming on-target activity of 7.
Figure 4. Compound 7 exhibits BSH engagement and minimal off-target effects.
(a) Structure of ‘clickable’ 7, 7-N3 (12), for on- and off-target studies. (b) 7-N3 displayed significant BSH inhibition in conventional mouse feces, showing that this probe retained its function as a BSH inhibitor. (c) Treatment of B. adolescentis L-32 culture with 7-N3 for 1 hour followed by cell lysis, click reaction with Fluor 488-alkyne, and visualization using in-gel fluorescence revealed labeling of only one protein ~35 kDa in size, the mass of the annotated B. adolescentis BSH. Proteins were loaded at a concentration of 1.5 mg/mL. B. theta recombinant BSH (BT BSH, 1 μM) was used as a positive control for the click reactions and as a standard to show 38 kDa. (d) Lysate from the treatment of B. adolescentis cultures with 7-N3 was reacted with desthiobiotin-alkyne, resolved by SDS-PAGE, and visualized by silver-staining. Arrow indicates a band in the probe-treated sample at the predicted molecular weight (~35 kDa) of BSH. (For full gels in triplicate, see Supplementary Fig. 15.) In-gel digestion followed by LC-MS/MS identified this band as BSH with high-confidence. Semi-quantitative analysis indicated a 4.5-fold enrichment in 7-N3-treated versus vehicle-treated bacterial cultures (see Supplementary Tables 4). (e) Treatment of B. adolescentis cultures with decreasing concentrations of 7 followed by treatment with 10 μM 7-N3 and click reaction with Fluor 488-alkyne resulted in a dose-dependent increase in fluorescence labeling of annotated B. adolescentis BSH. Experiment was repeated twice with similar results. (For full gels in triplicate, see Supplementary Fig. 15.) (f) One-hour treatment of NCI-H716 intestinal cells with 7-N3 followed by click reaction with Fluor 488-alkyne and visualization by in-gel fluorescence resulted in no significant labeling of proteins compared to control-treated cells (for triplicate gels, see Supplementary Fig. 16). For (b, c, d, and f), n=3 biological replicates per condition. For (b), data is presented as mean ± SEM.
We then profiled the off-target effects of compound 7 in mammalian intestinal cells (NCI-H716). These cells were also treated with 7-N3 and processed in the same manner as the bacterial cells. Click reaction with Fluor 488-alkyne showed no enrichment of any band by in-gel fluorescence (Fig. 4f and Supplementary Fig. 16). No proteins were enriched (≥2-fold) in probe-treated lysates based on label-free LC-MS/MS analysis (Supplementary Tables 8-10). Collectively, our data demonstrate on-target BSH binding of 7 and limited off-target activity against other bacterial proteins or mammalian proteins in intestinal cells.
Single dose of 7 inhibits BSH activity in vivo
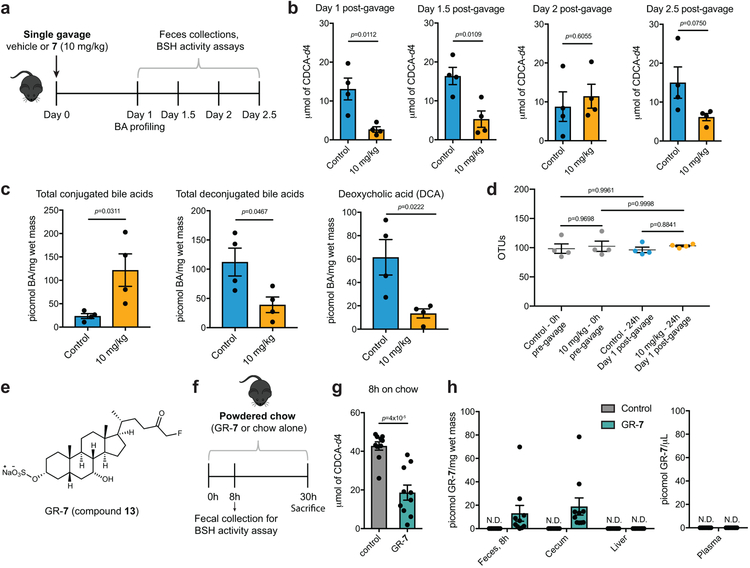
We next sought to evaluate the activity of this inhibitor in vivo. C57BL/6 mice were gavaged with a single dose of either 7 (10 mg/kg, see Online Methods for dose calculation) or vehicle control, and BSH activity in feces was monitored over time in half-day increments (Fig. 5a). As predicted, we observed a significant decrease in BSH activity in feces 1 day and 1.5 days post-gavage, while at subsequent timepoints, BSH activity recovered (Fig. 5b). We also observed a significant increase in fecal conjugated bile acids and a decrease in deconjugated bile acids 1 day-post gavage (Fig. 5c). 16S rDNA sequencing and plating of fecal samples from these mice indicated that compound 7 did not significantly affect gut bacterial OTUs, biomass, or community composition (Fig. 5d and Supplementary Fig. 17). Taken together, our results indicate that one dose of 7 can inhibit gut bacterial BSH activity and modulate the bile acid pool in vivo while not significantly affecting the gut bacterial community.
Figure 5. Compound 7 inhibits BSH activity in vivo and can be gut-restricted.
(a-c) Treatment of conventional mice with a single dose of 7 resulted in recoverable inhibition of BSH activity and a shift toward conjugated bile acids. n=4 mice per group, Student’s t test. (a) Design of in vivo BSH inhibition experiment. Adult male C57BL/6 mice were gavaged with a single dose of 7 (10 mg/kg) or vehicle control. (b) BSH activity was measured in half-day increments starting 1 day post-gavage. Resuspended fresh feces from inhibitor- or vehicle-treated groups were incubated with substrate (GCDCA-d4, 100 μM) for 25 min and formation of product was quantified by UPLC-MS. n=4 mice per group, two-tailed Student’s t test. (c) Fecal bile acid composition 1 day post-gavage. Deconjugated bile acids, including the secondary bile acid deoxycholic acid (DCA), were decreased in the inhibitor-treated group. n=4 mice per group, two-tailed Student’s t test. (d) Bacterial OTUs (operational taxonomic units) did not differ between the inhibitor- and vehicle-treated groups 1 day post-gavage. n=4 mice per group, one-way ANOVA followed by Tukey’s multiple comparisons test. (e) Structure of gut-restricted compound 7 (GR-7, 13). (f) Design of proof-of-concept in vivo study with GR-7. Adult male C57BL/6 mice were fed powdered chow containing 0.09% (w/w) GR-7 or powdered chow alone for 30 hours. Fecal pellets were collected 8 hours post-diet change. n =10 mice per group. (g) Resuspended fresh feces (20 mg/mL) from inhibitor- or control-treated mice were incubated with substrate (GCDCA-d4, 100 μM) for 25 min and formation of product was quantified by UPLC-MS. Significant inhibition of BSH activity was observed in the feces of GR-7-treated compared to control-treated mice. Student’s t test. n=10 mice per group, two-tailed Student’s t test. (h) Quantification of GR-7 in tissues and plasma. Inhibitor was detected in feces 8 hours post-diet change and in cecal contents at sacrifice. No GR-7 was detected in the liver or plasma. N.D. = not detected. n=10 mice per group. All data are presented as mean ± SEM.
Proof of concept of gut restriction of 7
To further minimize the likelihood that 7 would induce off-target effects, ideally, this molecule would be confined to the GI tract. As a proof of principle that derivatives of 7 could be restricted to the gut, we synthesized a 3-sulfated variant (gut-restricted 7 or GR-7, compound 13, Fig. 5e). Sulfation of endogenous bile acids in the liver detoxifies these compounds and increases their water solubility. As a result, sulfated bile acids largely remain in the gut.37 We reasoned that sulfation of 7 would also decrease its systemic absorption. We chose to modify the C3 position due to its solvent exposure (Fig. 3b).
Evaluation of GR-7 in mouse feces revealed that GR-7 remains a potent pan-BSH inhibitor (Supplementary Fig. 18). Because bile acids are secreted post-prandially, we hypothesized that feeding GR-7 in chow would ensure that the inhibitor was present when high concentrations of conjugated bile acids were also present. We fed C57BL/6 mice with either powdered chow containing 0.09% GR-7 (w/w) for 1 day or powdered chow alone (Fig. 5f). We observed significant inhibition of the BSH activity in the feces of inhibitor-treated mice 8 hours post-diet change (Fig. 5g). We detected GR-7 in feces collected at 8h, demonstrating that the inhibitor was being excreted at a rate consistent with mouse colonic transit time.38 We detected 20 picomol/mg wet mass (~20 μM) of this compound in cecal contents (mean value, Fig. 5h). This concentration was effective at BSH inhibition in the mouse feces assay and lower than the toxicity threshold of 100 μM for 7 (Supplementary Figs. 13 and 18). Moreover, GR-7 (60 μM) did not affect epithelial barrier integrity of Caco-2 cells, suggesting that this compound is relatively non-toxic (Supplementary Fig. 14). GR-7 also did not affect microbial biomass (Supplementary Fig. 18). Importantly, we did not detect any GR-7 in the serum and liver of inhibitor-treated mice (Fig. 5h). Collectively, these results provide proof of concept that 7 can be chemically modified to minimize absorption, and when fed in chow, a gut-restricted 7 derivative can inhibit BSH activity.
Discussion
To uncover the effects of bacterial metabolites on host health, tools are needed that control the levels of these compounds in conventional animals. In this work, we report the development of such a chemical tool, a potent, selective, pan-inhibitor of gut bacterial BSHs. We identified a lead inhibitor, 7, that effectively inhibits deconjugation in vitro and in vivo, and importantly, does not significantly affect the viability of gut bacteria.
Covalent inhibitors display several advantages compared to reversible compounds, and these benefits could be particularly advantageous in the context of BSH inhibition. First, covalent inhibitors demonstrate high potencies even in the presence of high concentrations of competing endogenous substrates, in this case, conjugated bile acids in the gut.11 Second, the duration of inhibition is dependent on protein resynthesis, making rapidly cleared compounds more acceptable and leading to lower systemic inhibitor exposure and fewer off-target effects.39 Indeed, we observed significant amounts of GR-7 in feces within 8 hours and undetectable levels of this molecule in circulation, suggesting that GR-7 is efficiently eliminated from the mouse gut at approximately the same rate as GI transit.38 Further studies that determine the rate of BSH turnover and protein resynthesis will aid in the establishment of optimal dosing strategies for BSH inhibitors. Third, targeting the active site cysteine residue with an irreversible binder has allowed us to develop a pan-inhibitor of BSHs. While in mammalian cells, the cross-reactivity of covalent inhibitors with other proteases has proved to be an impediment for the development of selective inhibitors,39,40 in this case, the conservation of the catalytic cysteine residue in BSHs across gut bacterial phyla has proved to be an asset.
Despite the potential benefits of covalent inhibitors, it is imperative to understand the target engagement and off-target interactions of all new covalent probes.35 To this end, we synthesized a ‘clickable’ version of our lead inhibitor, 7-N3. By incubating this probe with bacterial and mammalian intestinal cells, we showed that 7 displays on-target selectivity and limited off-target activity. In growing cultures of B. adolescentis, 7-N3 labeled only BSH to an appreciable extent. Likewise, 7-N3 did not appreciably label any proteins in intestinal NCI-H716 cells. In addition, the proof-of-principle development of GR-7 demonstrates that it is possible to confine a potent BSH inhibitor to the GI tract.
Nonetheless, it is possible that in the long-term, a non-covalent inhibitor of BSHs may prove to be the most effective tool for in vivo use. While covalent inhibition is the most effective strategy when complete inactivation of a protein target is required,41 it possible that long-term treatment with a covalent BSH inhibitor could result in the emergence of resistant bacterial strains in the microbiome community.39 In this case, a reversible inhibitor that results in partial BSH inhibition and thereby avoids the development of resistance mutations might be preferable.
Despite the potential draw-backs of covalent BSH inhibitors, our results suggest that 7 or derivatives thereof can be used as tools to study the biological roles of primary and secondary bile acids in conventional animals, including both wild-type and knock-out mouse strains. These investigations would broaden our understanding of how bile acids affect host immune and metabolic systems and reveal how these metabolites affect the composition and biogeography of the gut bacterial community.
In particular, BSH inhibitors could be used to better understand the effects of bile acids on host metabolism. Previous studies have reported conflicting results about how altering BSH activity in vivo may affect host metabolic responses.32,42 Moreover, in previous work, we demonstrated that mice colonized with a B. theta BSH knock-out strain gained less weight and displayed a decreased respiratory exchange ratio compared to mice colonized with the B. theta wild-type strain.16 However, these experiments were performed in monocolonized germ-free mice and do not reveal how limiting activity of all BSHs will affect the metabolism of conventional animals. Use of a non-toxic, pan-BSH inhibitor would enable investigation of how BSH activity directly affects metabolism in fully colonized hosts.
In addition, BSH inhibitors could enable investigation of how bile acids affect host immune response in the context of liver cancer. A recent study proposed a causal connection between the conversion of primary to secondary bile acids and a decrease in a tumor-suppressive environment in the liver mediated by the accumulation of NKT cells.43 Use of a BSH inhibitor in mouse models of liver cancer could further test this hypothesis by shifting the endogenous in vivo bile acid pool toward primary bile acids without significantly perturbing the enterohepatic system and the microbial community.
Looking ahead, if use of BSH inhibitors in vivo beneficially affects host physiology, these compounds could be developed as novel drug candidates. In this way, development of mechanism-based, non-toxic chemical probes for bacterial enzymes could lay the groundwork for potential therapeutic agents that target the microbiota.12,44
Online Methods
Reagents.
All bile acids and reagents for synthesis were commercially purchased from Steraloids Inc. and Sigma Aldrich. Stock solutions of all bile acids and inhibitors were prepared in molecular biology grade DMSO (Sigma Aldrich) at 1000X concentrations. Solvents used for preparing UPLC-MS samples were HPLC grade. New biological materials reported here are available from the authors upon request. 100 μM bile acid pool consisted of TCA, TβMCA, TUDCA and TDCA (25 μM each).
Chemical Synthesis.
See Supplementary Note.
Bacterial Culturing.
All bacterial strains were cultured at 37 °C in Cullen-Haiser Gut (CHG) media (which consists of brain heart infusion media (Bacto™ BHI, BD) supplemented with 1% BBL vitamin K1-hemin solution (BD), 1% trace minerals solution (ATCC), 1% trace vitamins solution (ATCC), 5% fetal bovine serum (Hyclone), 1 g/L cellubiose, 1 g/L maltose, and 1 g/L fructose) or BHI+ (Bacto™ BHI, BD, supplemented with with 5 mg/L hemin, and 2.5 uL/L Vitamin K1). All strains were grown under anaerobic conditions in a anaerobic chamber (Coy Lab Products Airlock) with a gas mix of 5% hydrogen and 20% carbon dioxide nitrogen. Escherichia coli was grown aerobically at 37 °C in LB medium supplemented with ampicillin to select for the pET21b plasmid.
UPLC-MS Analysis.
Bile acid profiling by UPLC-MS was performed using a published method.16 Correction factors for extraction efficiency were used and were determined by extraction of known concentrations of relevant bile acids from buffer or bacterial media and comparison to standard curves. The limits of detection for individual bile acids were determined using commercially available standards/ synthesized compounds solubilized in 1:1 MeOH/water and are as follows: βMCA, 0.03 picomol/μL; TβMCA, 0.01 picomol/μL; CA, 0.04 picomol/μL; TCA, 0.01 picomol/μL; UDCA, 0.04 picomol/μL; TUDCA, 0.01 picomol/μL; DCA, 0.04 picomol/μL; TDCA, 0.05 picomol/μL; GCDCA-d4, 0.1 picomol/μL; CDCA-d4, 0.1 picomol/μL; 7-oxo-CA, 0.5 picomol/μL; 7, 1.0 picomol/μL; GR-7, 0.05 picomol/μL.
Protein Expression and Purification.
B. thetaiotaomicron rBSH.
The gene encoding BT_2086 (without the leader sequence) was codon-optimized for E. coli and cloned into pET-21b(+) vector containing a C-terminal His6 tag (see Supplementary Table 1 for primers). The expression plasmid was then transformed into BL21(DE3)pLysS Escherichia coli (New England Biolabs) cells under ampicillin selection. Overnight cultures grown in LB media with ampicillin (50 μg/mL) were diluted 1:1000 in fresh LB media with ampicillin and grown at 37 °C. Expression was induced at an OD600 of 0.6-0.7 by the addition of 1 mM isopropyl-1-thio-D-galactopyranoside (IPTG) and further incubated at 18 °C overnight. The cells were pelleted by centrifugation at 7,000 g for 20 mins at 4 °C. The pelleted cells were then resuspended in PBS buffer (with 5% glycerol) containing 20 mM imidazole, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 0.25 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP). The resuspended cells were sonicated and pelleted by centrifugation at 16,000 g for 20 mins at 4 °C. The supernatant was then mixed with pre-formed Ni-NTA for 45 mins at 4 °C. The nickel-bound protein was eluted with gradually increasing concentration of imidazole in PBS buffer (with 0.25 mM TCEP and 5% glycerol). Collected fractions were tested for purity by SDS-PAGE. The pure fractions were combined and concentrated followed by dialysis using the storage buffer (PBS at pH 7.5 with 0.25 mM TCEP and 5% glycerol).
For crystallization purposes, the protein was further purified using S200 size exclusion column (from GE) on a BioRad FPLC in 50 mM tris(hydroxymethyl)aminomethane buffer with 300 mM NaCl, 0.25 mM TCEP and 5% glycerol at pH 7.5.
B. longum rBSH.
Recombinant BSH from B. longum SBT2928 was expressed and purified as above, except 0.25 mM IPTG was used for protein expression and 1 mM TCEP for protein purification.
Enzyme Kinetics.
The enzyme was characterized using a modified BSH activity assay.26 To 144.8 μL PBS buffer (containing 10 mM TCEP and 5% glycerol), 35.2 μL of rBSH was added to afford a final concentration of 6.2 μM and 7.0 μM for B. theta BSH and B. longum BSH, respectively. This solution was preheated to 37 °C in a water bath. 20 μL of a conjugated bile acid in DMSO at appropriate concentration was preheated to 37 °C in a water bath and added to the above solution. At every time interval, 15 μL of the mixture was quenched with 15 μL of 15% trichloroacetic acid. The cloudy solution was centrifuged at 4,200 g for 15 mins. 10 μL of the supernatant was added to 190 μL of ninhydrin mix (15 mL of 1% [wt/vol] ninhydrin in 0.5 M sodium citrate at pH 5.5, 36 mL glycerol and 6 mL 0.5 M sodium citrate buffer at pH 5.5) and the mixture was heated to 100 °C in a BioRad thermocycler for 18 mins. The obtained solution was cooled at 4 °C for 20 mins and absorbance was measured at 570 nm using a spectrophotometer (Molecular Devices).
Inhibitor Screen Using rBSHs.
200 nM rBSH was incubated with 100 μM inhibitor at 37 °C for 30 mins in 3 mL PBS buffer containing 0.25 mM TCEP and 5% glycerol at pH 7.5. Bile acid pool (100 μM) was added to the above solution and incubated at 37 °C. At timepoint intervals, 1 mL of the above buffer solution was acidified to pH = 1 using 6M HCl and extracted twice with 1 mL ethyl acetate. The combined organic layers were then dried using a Biotage TurboVap LV. The dried extracts were resuspended in 1:1 methanol:water and transferred to mass spec vials. Samples were analyzed as per the method described in “UPLC-MS analysis”. The obtained concentrations of bile acids were used to determine % deconjugation.
Equation for Calculating % Deconjugation.
% deconjugation = concentration of deconjugated bile acids detected / (concentration of deconjugated bile acids detected + concentration of conjugated bile acids detected)*100.
Compound 7 Kinetic Studies.
Assay was run in PBS buffer (containing 0.25 mM TCEP and 5% glycerol) and all reactants were incubated at 37 °C before reaction start time. B. theta BSH (200 nM) was added to a pool of 100 μM bile acid pool and 100 μM 7. 500 μL aliquots were removed at indicated time points and flash frozen in liquid nitrogen. After thawing the solution was acidified to pH = 1 using 6M HCl and then processed as per the method described in “Inhibitor Screen Using rBSHs”. Procedure was repeated with 8.2 mM TUDCA.
Determination of IC50 Values of Compound 7 against recombinant proteins.
200 nM rBSH was incubated with increasing concentrations of 7 at 37 °C for 1 h in 1 mL PBS buffer containing 0.25 mM TCEP and 5% glycerol at pH 7.5. 100 μM bile acid (TUDCA for B. theta BSH and TDCA for B. longum BSH) was added to the above solution and incubated at 37 °C for 2 h. The solution was acidified to pH = 1 using 6M HCl and then processed as per the method described in “Inhibitor Screen Using rBSHs”.
Inhibitor Screen in Bacteria.
Bacterial cultures were diluted to OD600 of 0.1 in 4 mL BHI+, containing 100 μM taurine conjugated bile acid pool 100 μM inhibitors. These cultures were then grown anaerobically at 37 °C. After 21 h, serial dilutions were plated on BHI+ agar to determine cell viability (CFU/mL). 1 mL of the entire bacterial culture was acidified to pH = 1 using 6M HCl followed by addition of 2 mL ethyl acetate and vortexed. The cultures were spun down in a centrifuge at 2,500 g for 5 mins to obtain better separation. The organic layer was then removed and the aqueous layer was extracted again using 2 mL of ethyl acetate. The dried organic extracts were resuspended in 1:1 methanol:water and transferred to mass spec vials and analyzed as per the method described in “UPLC-MS Analysis”. The obtained concentrations of bile acids were used to determine % deconjugation.
Determination of IC50 Values of Compound 7 in Bacterial Cultures.
Note that due to slow growth of B. longum, B. adolescentis was used for studies in growing bacteria. Overnight cultures of B. theta and B. adolescentis were diluted to an OD600 of 0.1 in 2 mL fresh CHG media (see “Bacterial Culturing”) containing 100 μM TUDCA or TDCA, respectively, and inhibitor 7 at increasing concentrations. B. theta and B. adolescentis deconjugated TUDCA and TDCA, respectively, to the greatest extent of any of the conjugated substrates in the Inhibitor Screen in Bacteria assay, and therefore these substrates were used to determine IC50 values. Cultures were then grown anaerobically at 37 °C for 24 h (B. adolescentis) or 48 h (B. theta). Longer incubation time was required for B. theta because for this bacterium, significant BSH activity was only observed during stationary phase. Cultures were extracted and analyzed as per the method described in “Inhibitor Screen in Bacteria”.
Screen of Inhibitors in Conventional Mouse Feces.
BSH activity in fecal pellets were quantified using a modified version of a published method.45 Fecal pellets (approximately 10-20 mg) were broken into fine particles in buffer (10% PBS, 90% sodium acetate at pH 5.2) to obtain a concentration of 1 mg/mL. Indicated concentration of inhibitors were added to the fecal slurry and the mixture was incubated at 37 °C for 30 mins. 100 μM glycochenodeoxycholic acid-d4 (GCDCA-d4) was added to the mixture and incubated at 37 °C for 18 h. The tubes were then frozen in dry ice for 5 mins and upon thawing were diluted with an equal volume of methanol. The slurry was centrifuged at 12,500 g for 10 mins. The supernatant was removed into a clean eppendorf tube and centrifuged again. The supernatant was transferred to MS vials and samples were analyzed as per the method described in “UPLC-MS Analysis”. The concentration of product detected from these assays was reported directly.
Crystallization, Data Collection, and Structure Determination.
Crystals of BSH and BSH in complex with 7 were grown in 24-well format hanging drops at room temperature. BSH crystals (5.0 mg/mL) grew from micro seeding after 3 days in 42% tacimate 100 mM Tris pH 7.4. The BSH-7 complex (5.0 mg/mL) crystals grew after 5 d in 21% PEG 3350 and 100 mM X Sodium citrate tribasic dihydrate pH 5.0. Crystals were cryoprotected by supplementing the mother liquor with 10% 2-methyl-2,4-pentanediol (v/v). Data collection was performed at Advanced Photon Source NE-CAT beamline 24 ID-C at 100 K using a wavelength of 0.979 Å. Diffraction images were processed and scaled using XDS. To obtain phases for the apo BSH structure, molecular replacement was performed in Phenix with Phaser46 using 3HBC as the search model. Iterative model building and reciprocal space refinement was performed in COOT and phenix.refine,47 respectively. The BSH-7 structure was phased using molecular replacement with apo BSH as a search model. Iterative model building and refinement for the BSH-7 grouped atomic B-factors and used an applied twinlaw of k h –l. Model quality for both structures was evaluated using composite omit density maps. In final cycles of model building, NCS restraints were removed. Final model quality was assessed using MolProbity.48 For 6UFY, 97% of residues were in favored regions of the Ramachandran plot, 3% were in allowed regions, and none were in outlier regions; for 6UH4, 89.3% of residues were in favored regions, 10.3% were in allowed regions, and 0.4% were outliers. All crystallographic data processing, refinement, and analysis software was compiled and supported by the SBGrid Consortium.49 Data acquisition and refinement statistics are presented in Supplementary Table 3. Figures were prepared using Pymol (Schrödinger).
Mass Spectrometry Analysis for Identifying Labeled Residue on BSH.
BSH protein was incubated with DMSO or a 10-fold molar excess of inhibitor 7 for 2 h at room temperature. Reactions were then analyzed by LC-MS using a Shimadzu LC and autosampler system (Shimadzu, Marlborough, MA) interfaced to an LTQ ion trap mass spectrometer (ThermoFisher Scientific, San Jose, CA).
To determine the site of modification, compound 7 modified protein was analyzed as described above, except that the LC system was interfaced to an Orbitrap Lumos Mass Spectrometer (ThermoFisher Scientific). The mass spectrometer was programmed to perform continuous cycles consisting of 1 MS scan (m/z 300-2000, profile mode, electron multiplier detection) followed by ETD MS/MS scans targeting the +41 charge state precursor of compound 7 modified protein (ETD reagent target = 200 ms, image current detection at 60K resolution, target value=2E6, ETD reaction time= 100 or 200 ms). Ion assignments were performed using mzStudio software.50
Effect of 7 on FXR.
LanthaScreen TR-FRET Coactivator Assay (Invitrogen, Carlsbad, CA) was used to test the effect of 7 on FXR according to the manufacturer’s instructions. Known FXR agonist GW4064 (Sigma, G5172) was used as a positive control (agonism assay) or added at its EC50 (50.3 nM, measured in this assay) (antagonism assay). Following 1 h incubation at room temperature, the 520/495 TR-FRET ratio was measured with a PerkinElmer Envision fluorescent plate reader using the following filter set: excitation 340 nm, emission 495 nm, and emission 520 nm. A 100 μsec delay followed by a 200 μsec integration time was used to collect the time-resolved signal.
Cell Culture.
Caco-2 cells and NCI-H716 cells were obtained from American Type Culture Collection (Manassas,VA). Caco-2 cells were maintained in Minimum Essential Medium (MEM) supplemented with GlutaMAX and Earle’s Salts, while NCI-H76 cells were maintained in Roswell Park Memorial Institute (RPMI) media (Gibco, Life Technologies, UK). All cell culture media were supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin (GenClone). Cells were grown in FBS- and antibiotic-supplemented ‘complete’ media at 37 °C in an atmosphere of 5% CO2.
Plasmids and Transient Transfections.
For luciferase reporter assays, vectors expressing human reporter constructs were used. The pGL4.29[luc2P/CRE/Hygro] plasmid (Promega Corporation) was transiently transfected in Caco-2 cells at a concentration of 2 μg/ml of media each for studying TGR5 activation respectively. The pGL4.74[hRluc/CMV] plasmid (Promega Corporation) was used as a transfection efficiency control at a concentration of 0.05 μg/ml of media. All plasmids were transfected using Opti-MEM (Gibco) and Lipofectamine 2000 (Invitrogen, Life Technologies, Grand Island, NY, USA) according to manufacturer’s instructions. Plasmid transfections were performed in antibiotic-free MEM media with 10% FBS. After overnight incubation, 7 and/or bile acids were added in complete media. 7 and/or bile acids were diluted in DMSO and the concentration of DMSO was kept constant. 10 μM of LCA was added along with 7 to study TGR5 antagonism and incubated overnight. Cells were harvested the next day for luciferase assay.
Luciferase Reporter Assay.
Luminescence was measured using the Dual-Luciferase Reporter Assay System (Promega Corporation) according to manufacturer’s instructions. Cells were washed gently with PBS and lysed in PLB from the kit. Luminescence was measured using a SpectraMax M5 plate reader (Molecular Devices, San Jose, CA) at the ICCB-Longwood Screening Facility at HMS. Luminescence was normalized to Renilla luciferase activity and percentage relative luminescence was calculated compared to DMSO control.
Cell Viability Assay.
Caco-2 and NCI-H716 cells were treated with indicated compounds diluted in DMSO in complete MEM and RPMI media respectively. The concentration of DMSO was kept constant and used as a negative control. Cells were incubated with compound overnight at 37 °C in an atmosphere of 5% CO2. The next day, cells were treated with 0.25% trypsin in HBSS (GenClone) for 10 min at 37 °C. Cell viability was measured in Countess II automated cell counter (Invitrogen). Percentage relative viability was calculated compared to DMSO control.
Epithelial Permeability Assay.
Undifferentiated Caco-2 cells were seeded in 24-well plate transwells (0.4 uM pore size, Costar) at 200,000 cells per transwell. Media was changed on days 4, 8, 12, 16, and 18 to differentiate Caco-2 cells in vitro.51 On day 21, fully differentiated and polarized cells were used for FITC-dextran permeability assay. Briefly, 7 and GR-7 were added in PBS at indicated concentrations to the apical chamber of the transwells containing differentiated Caco-2 cells and incubated for 6 or 12 h. The apical chamber of the transwells contained a volume of 100 ul PBS with compounds or DMSO control, while the basolateral chamber contained 500 uL of PBS. Caco-2 epithelial integrity was assayed by measuring passive diffusion of 4 kDa FITC-Dextran (Sigma Aldrich) added at a concentration of 5 uM to the apical chamber. Diffusion from the apical to basolateral side was measured by fluorescence reading in PBS on the basolateral side of the transwell system using a SpectraMax M5 plate reader (Molecular Devices, San Jose, CA) at the ICCB-Longwood Screening Facility at HMS. Fluorescence reading was normalized to the DMSO control.
Target Validation and Off-target Profiling in B. adolescentis using 7-N3.
Pilot studies with 7-N3 were performed using B. adolescentis (Gram positive) and B. theta (Gram negative). We chose to use B. adolescentis due to the stronger total fluorescent signal detected by-in gel fluorescence. B. adolescentis cultures were diluted to an OD600 of 0.1 in 6 mL fresh CHG media containing 100 μM taurine-conjugated bile acid pool. Cultures were allowed to grow anaerobically at 37 °C for 21 h. 10 μM 7-N3 (10 mM stock in DMSO) or 6 μL DMSO (to control tubes) was then added to the cultures and incubated anaerobically at 37 °C for 1 h. The cultures were centrifuged at 2,500 g at 4 °C for 15 mins. The media was decanted and cells were resuspended in PBS containing 1 mM TCEP and 1 mM PMSF and centrifuged 4,200 rpm at 4 °C for 15 mins. The buffer was decanted and the cells were suspended in 300 μL of fresh buffer and transferred to homogenizing tubes (Precellys lysing kit tough micro-organism lysing VK05 tubes) with ceramic beads. The suspension was then homogenized (5000 speed for 90 s*2, 6500 speed for 60 s) and spun down for 20 min at 15,000 g at 4 °C. The supernatant was removed and the concentration of proteins in the lysate was quantified by Bradford assay. The lysates were then subjected to click reaction as per “Click Chemistry for In-gel Fluorescence Imaging” for fluorescence imaging and “Click Chemistry for MS/MS on Bacterial Lysate” for mass spectrophotometer-based quantification and identification.
Dose-dependent Labeling of BSH in B. adolescentis via competition of 7 and 7-N3.
B. adolescentis cultures were diluted to an OD600 of 0.1 in 6 mL fresh CHG containing containing 100 μM taurine-conjugated bile acid pool. Cultures were allowed to grow anaerobically at 37 °C for 21 h. Decreasing concentrations of 7 were added to different tubes and the cultures were incubated anaerobically at 37 °C for 1 h. 10 μM 7-N3 was then added to the cultures and incubated anaerobically at 37 °C for an additional hour. The cultures were further processed as per the reported method in “Target Validation and Off-target Profiling in B. adolescentis using 7-N3” and “Click Chemistry for In-gel Fluorescence Imaging”.
Off-Target profiling in Mammalian Cells using 7-N3.
The human epithelial cell line NCI-H716 was used to study interactions with mammalian proteins. 10 μM 7-N3 (10 mM stock in DMSO) or 1 uL DMSO (for control) were added to ~ 8 × 106 cells in 1 ml DPBS (HiMedia) and incubated for 1 h. Cells were collected in 15 ml Falcon tubes and washed 2 times in 15 ml DBPS by centrifugation at 500 g for 5 mins. A 3rd wash by centrifugation was performed in 1 mM solution of cOmplete™ Protease Inhibitor Cocktail (Roche, Swtzerland) in DPBS. Cells were resuspended in 250 ul of DPBS with 1mM cOmplete™ Protease Inhibitor Cocktail and sonicated at 50% amplitude for 2 secs followed by 30 secs on ice for 3 cycles. The lysate was centrifuged at 15,000 g for 15 mins at 4 °C. The supernatant was removed and protein concentration was measured by Bradford assay. The lysates were then subjected to click reaction as per “Click Chemistry for In-gel Fluorescence Imaging” for in-gel fluorescence and “Click Chemistry for MS/MS on Mammalian Lysate” for mass spectrophotometer-based quantification and identification.
Click Chemistry for In-gel Fluorescence Imaging.
Click reactions were performed on 25 μL scale. Lysates (normalized to 1.5 mg/mL for both bacterial and mammalian cells) pretreated with 10 μM Compound 7-N3 were incubated with 100 μM fluor 488-alkyne (10 mM stock in DMSO), 100 μM CuBr (5 mM stock in DMSO), and 100 μM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (5 mM stock in 4:1 t-BuOH:DMSO) for 1 h at 37 °C in the dark. 10 μL of 2x Laemmli buffer (containing 5% β-mercaptoethanol) was added to the reactions and the tubes were heated at 95 °C for 10 mins. 15 μL of the protein samples were then resolved by 10% SDS-PAGE. The ladder was diluted 100-fold and 10 μL was loaded. Gels were destained for 30 mins using 40% methanol, 50% acetic acid, 10% water and visualized using Bio-Rad ChemiDoc MP Imaging System. Gels were stained for 20 mins in Coomassie blue and destained for 2 h prior to imaging.
Click Chemistry for MS/MS on Bacterial Lysate.
Click reactions were performed on 100 μL scale. Lysates (normalized to 1.3 mg/mL) pretreated with 10 μM 7-N3 were incubated with 100 μM desthiobiotin-PEG4-alkyne (10 mM stock in DMSO), 1 mM CuBr (50 mM stock in DMSO), and 1 mM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (50 mM stock in 4:1 t-BuOH:DMSO) for 1 h at 37 °C. The samples were then processed for further analysis as per “Proteomic Analysis of Click-Tagged Proteins”.
Click Chemistry for MS/MS on Mammalian Lysate.
Click reactions were performed on 100 μL scale. Lysates (1.5 mg/mL for mammalian cells) pretreated with 10 μM 7-N3 were incubated with 100 μM desthiobiotin-PEG4-alkyne (10 mM stock in DMSO), 100 μM CuBr (5 mM stock in DMSO), and 100 μM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (5 mM stock in 4:1 t-BuOH:DMSO) for 1 h at 37 °C. The samples were then processed for further analysis as per “Proteomic Analysis of Click-Tagged Proteins”.
Proteomic Analysis of Click-Tagged Proteins.
Pulldown of desthiobiotinylated proteins and on bead digestion was performed similar to a previously described protocol.52 After resuspending tryptic peptides in 5% acetonitrile with 0.1% formic acid, peptides were analyzed by nanoflow LC-MS/MS as described.53 Raw data were converted to .mgf using multiplierz54 and searched using Mascot 2.6.2 against forward reversed databases of either human or bifidobacterium adolescentis proteins (uniprot). Search results were downloaded from Mascot, converted to xls, and filtered to 1% FDR using multiplierz scripts. Normalized spectral abundance factors were derived as described.55 Data were filtered for proteins with more than 5 spectral counts (averaged across biological triplicates) for 7-N3 treated samples. In separate experiments, clicked bacterial lysate proteins were subjected to avidin enrichment and washed as described above. Proteins were then eluted with LDS loading buffer and subjected to SDS-PAGE and silver staining. Indicated bands were excised, subjected to in-gel digestion, and extracted peptides analyzed by nanoflow LC-MS/MS as described.53
Animal Studies.
C57BL/6 mice obtained from Jackson laboratories were maintained under a strict 12 h/12 h light/dark cycle and a constant temperature (21 ± 1 °C) and humidity (55–65%). All experiments were conducted on 8-9 week old male mice.
Single Gavage of 7.
Based on the efficacy of 7 at 10 μM to 100 μM in in vitro assays, our goal concentration of 7 in vivo was ~50 uM: (0.00005 M) x (~ 10 mL volume / 1 mouse GI tract) x (1 mmol compound 7 / 408 mg) = 0.2 mg / mouse x (1 mouse / ~0.02 kg) = 10 mg / kg.
Mice were maintained on a standard chow diet (LabDiet, catalog no. 5053) for the duration of the experiment. Mice were split into two groups of four mice each and were gavaged with either 200 μL of corn oil containing 5% DMSO (vehicle group) or with 200 μL of corn oil containing 7 at a concentration of 1.25 mg/mL (experimental group). For the fecal pellet collection, each mouse was transferred to a temporary cardboard cage for a few minutes until it defecated.
Feeding of GR-7 in Chow for One Day.
Mice were fed powdered standard chow diet (LabDiet, catalog no. 5053) for the duration of the experiment. Mice were split into two groups of ten mice each and were maintained on powdered chow (control group) or fed powdered chow containing 0.09% (w/w) GR-7 (experimental group). Feces from these mice were collected as described above at 8 h. Mice were euthanized after 30 hours of access to the powdered chow with or without GR-7 using carbon dioxide. Blood samples were collected by cardiac puncture and placed in EDTA-coated tubes on ice. The liver and cecal contents were then collected from each mouse, snap frozen in liquid nitrogen, and stored at −80 ˚C until further analysis. The blood samples were then centrifuged at 2500 g for 15 minutes at 4 ˚C. The resulting supernatant (plasma) was collected and stored at −80 ˚C until analysis.
BSH Activity in Feces.
BSH activity in fecal pellets were quantified using a modified version of a published method.45 Fecal pellets (approximately 10-20 mg) were suspended in buffer (10% PBS, 90% sodium acetate at pH 5.2) containing 100 μM (GCDCA-d4) to obtain a concentration of 20 mg/mL. The fecal pellets were broken into fine particles and the mixture was incubated at 37 °C for 25 mins. Samples were processed and analyzed as per the method described in “Screen of Inhibitors in Conventional Mouse Feces”. The concentration of product detected from these assays were reported directly.
Quantification of Bile Acids in Tissues and Plasma.
Bile acids from tissues and plasma that were collected from mouse experiments were extracted using a previously published method. 16
Determination of Microbial Biomass by Plating.
Frozen fecal pellets were used to determine colony forming units (CFU/g). Feces were suspended in PBS buffer in an anaerobic chamber. Serial dilutions were plated on CHG agar plates (see “Bacterial Culturing”) and incubated at 37 °C.
Isolation of Fecal Bacterial Microbiota and 16S rRNA Gene Sequencing Analysis.
Mouse fecal microbiota DNA was isolated by using ZymoBIOMICS 96 DNA Kit (ZymoBIOMICS™) according to the manufacturer’s instructions. The variable region 4 of the 16S rRNA genes was amplified using primers: Forward 5’- TATGGTAATTGTGTGCCAGCMGCCGCGGTAA-3’ Reverse 5’- AGTCAGTCAGCCGGACTACHVGGGTWTCTAAT-3’. PCR products were quantified using Quant-IT dsDNA high sensitivity assay (Invitrogen) according to the manufacturer’s instructions. Gel electrophoresis was used to check the success of PCR amplification. The concentration of the PCR product was measured by the Quan-IT dsDNA high sensitivity assay. ~120 ng of DNA of each PCR product was pooled together to generate an aggregated library for downstream processing. PCR DNA amplicons between 300-500bp were selected from the aggregated library on a targeted size selection platform (pippin prep 1.5% agarose cassette from Sage Sciences) according to the manufacturer’s instructions. The size of DNA amplicons was characterized on an Agilent Technologies 2100 bioanalyzer trace. DNA concentration of the aggregated library was measured by the Quant-IT dsDNA high sensitivity assay. The DNA in the library was denatured by NaOH and diluted to 7.5 pM with HT buffer provided in the Illumina kit. 600 ul of the denatured and diluted library with 20% phiX spike-in (120 ul, 7.5 pM of phiX) was loaded onto the MiSeq V2 reagent cartridge (Illumina) and was sequenced with paired-end 250bp reads using the custom primers described above. After MiSeq running, demultiplexed fastq files were generated by the Illumina MiSeq control software using default parameters and quality control was done by the pipeline at the Massachusetts Host-Microbiome Center. The resulting FASTQ sequences were then quality-filtered and analyzed by following QIIME_mothur_DADA2.56-59 Operational Taxonomic Units (OTUs) were picked with 97% sequence similarity. The phylogenetic affiliation of each OTUs were aligned to the Greengenes reference database and 99% ID.
Quantification of bacterial 16S rDNA copy number.
Bacterial DNA was isolated from mouse cecal contents using AllPrep Bact. DNA/RNA/Protein Kit (QIAGEN). The 16S rDNA was then amplified using 10 μM of the following pair of primers: Forward 5’-AGAGTTTGATCCTGGCTCAG −3’, Reverse 5’- CTGCTGCCTYCCGTA −3’. Amplification was performed using LightCycler 480 SYBR Green I Master on a QuantStudio 7 Flex Real-Time PCR System according to the provided qPCR protocol. The cycle threshold of each sample was compared to a standard curve, obtained from serial dilution of B. theta genomic DNA.60
Ethics.
All mouse experiments were performed under the approval of the Beth Israel Deaconess Medical Center IACUC.
Data availability statement
16S rDNA datasets analyzed in the manuscript are available throughs NCBI under accession number PRJNA574158. The coordinates for both the apo and covalently inhibited forms of the BSH are deposited in the PDB and have PDB ID codes 6UFY and 6UH4, respectively. Raw mass spectrometry data were deposited at MassIVE (massive.ucsd.edu). Native mass spectrometry data files are available for download from the MassIVE archive at UCSD (ftp://massive.ucsd.edu/MSV000084491/). All other data generated or analyzed during this study are included in this article and its supplementary information files.
Code availability statement
No custom code or mathematical algorithms were used in this study.
Supplementary Material
Acknowledgements
This research was supported National Institutes of Health (NIH) grant R35 GM128618 (A.S.D), R35 CA220340 (S.C.B.), R01 CA222218 (J.A.M.), an Innovation Award from the Center for Microbiome Informatics and Therapeutics at MIT (A.S.D), a grant from Harvard Digestive Diseases Center (supported by NIH grant 5P30DK034854-32) (A.S.D), a Karin Grunebaum Cancer Research Foundation Faculty Research Fellowship (A.S.D), a John and Virginia Kaneb Fellowship (A.S.D), a Quadrangle Fund for the Advancement and Seeding of Translational Research at Harvard Medical School (Q-FASTR) grant (A.S.D), and an HMS Dean’s Innovation Grant in the Basic and Social Sciences (A.S.D). L.Y. and S.N.C. acknowledge a Wellington Postdoctoral Fellowship and an American Heart Association Postdoctoral Fellowship, respectively. M.D.M. acknowledges an NSF Graduate Research Fellowship (DGE1745303). D.R. is supported by the Early postdoc mobility fellowship from the Swiss National Science Foundation. We are indebted to Nathanael Gray, David Scott, John M. Hatcher, Jinhua Wang, Jon Clardy, Matthew Henke, and members of the Clardy group for helpful discussions. We thank the ICCB-Longwood Screening Facility for use of their fluorescent plate reader.
Footnotes
Competing Interests Statement
A.S.D. is an ad hoc consultant for Kintai Therapeutics and HP Hood. S.C.B. serves on the SAB for Erasca, Inc., is a consultant on unrelated projects for Ayala Pharmaceutical and IFM Therapeutics, and receives funding from Novartis for an unrelated project. J.A.M. serves on the SAB of 908 Devices (Boston, MA). The other authors declare that no competing interests exist.
References
- 1.Ridlon JM, Kang D-J & Hylemon PB Bile salt biotransformations by human intestinal bacteria. J. Lipid Res 47, 241–259 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Fiorucci S & Distrutti E Bile Acid-Activated Receptors, Intestinal Microbiota, and the Treatment of Metabolic Disorders. Trends Mol Med 21, 702–714 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Setchell KD, Lawson AM, Tanida N & Sjövall J General methods for the analysis of metabolic profiles of bile acids and related compounds in feces. J. Lipid Res 24, 1085–1100 (1983). [PubMed] [Google Scholar]
- 4.Hamilton JP et al. Human cecal bile acids: concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol 293, G256–G263 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Modica S, Gadaleta RM & Moschetta A Deciphering the nuclear bile acid receptor FXR paradigm. Nucl Recept Signal 8, e005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vavassori P, Mencarelli A, Renga B, Distrutti E & Fiorucci S The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol 183, 6251–6261 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Pols TWH et al. Lithocholic acid controls adaptive immune responses by inhibition of Th1 activation through the Vitamin D receptor. PLOS ONE 12, e0176715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begley M, Hill C & Gahan CGM Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol 72, 1729–1738 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang JY Recent advances in understanding bile acid homeostasis. F1000Res 6, 2029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Z et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome 7, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strelow JM A Perspective on the Kinetics of Covalent and Irreversible Inhibition. SLAS Discov 22, 3–20 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Roberts AB et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med 24, 1407–1417 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossocha M, Schultz-Heienbrok R, Moeller von, H, Coleman JP & Saenger W. Conjugated bile acid hydrolase is a tetrameric N-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochem. 44, 5739–5748 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Huijghebaert SM & Hofmann AF Influence of the amino acid moiety on deconjugation of bile acid amidates by cholylglycine hydrolase or human fecal cultures. J. Lipid Res 27, 742–752 (1986). [PubMed] [Google Scholar]
- 15.Kawamoto K, Horibe I & Uchida K Purification and characterization of a new hydrolase for conjugated bile acids, chenodeoxycholyltaurine hydrolase, from Bacteroides vulgatus. J. Biochem 106, 1049–1053 (1989). [DOI] [PubMed] [Google Scholar]
- 16.Yao L et al. A selective gut bacterial bile salt hydrolase alters host metabolism. eLife 7, 675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q et al. Developing Irreversible Inhibitors of the Protein Kinase Cysteinome. Chemistry & Biology 20, 146–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson AJ, Kerns JK, Callahan JF & Moody CJ Keap calm, and carry on covalently. Journal of Medicinal Chemistry 56, 7463–7476 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Serafimova IM et al. Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles. Nature Chemical Biology 8, 471–476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henise JC & Taunton J Irreversible Nek2 kinase inhibitors with cellular activity. Journal of Medicinal Chemistry 54, 4133–4146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie T et al. Pharmacological targeting of the pseudokinase Her3. Nature Chemical Biology 10, 1006–1012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quintás-Cardama A, Kantarjian H, Cortes J & Verstovsek S Janus kinase inhibitors for the treatment of myeloproliferative neoplasias and beyond. Nature Reviews Drug Discovery 10, 127–140 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Cohen MS, Zhang C, Shokat KM & Taunton J Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science 308, 1318–1321 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angliker H, Wikstrom P, Rauber P & Shaw E The synthesis of lysylfluoromethanes and their properties as inhibitors of trypsin, plasmin and cathepsin B. Biochem. J 241, 871–875 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller RM & Taunton J Targeting protein kinases with selective and semipromiscuous covalent inhibitors. Meth. Enzymol 548, 93–116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coleman JP & Hudson LL Cloning and characterization of a conjugated bile acid hydrolase gene from Clostridium perfringens. Appl. Environ. Microbiol 61, 2514–2520 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka H, Hashiba H, Kok J & Mierau I Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl. Environ. Microbiol 66, 2502–2512 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z et al. Identification and Characterization of a Bile Salt Hydrolase from Lactobacillus salivarius for Development of Novel Alternatives to Antibiotic Growth Promoters. Appl. Environ. Microbiol 78, 8795–8802 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stellwag EJ & Hylemon PB Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochimica et Biophysica Acta (BBA) - Enzymology 452, 165–176 (1976). [DOI] [PubMed] [Google Scholar]
- 30.Smith K, Zeng X & Lin J Discovery of bile salt hydrolase inhibitors using an efficient high-throughput screening system. PLOS ONE 9, e85344 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayin SI et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Li F et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun 4, 2384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofmann AF The function of bile salts in fat absorption. The solvent properties of dilute micellar solutions of conjugated bile acids. Biochem. J 89, 57–68 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferruzza S, Rossi C, Scarino ML & Sambuy Y A protocol for differentiation of human intestinal Caco-2 cells in asymmetric serum-containing medium. Toxicol In Vitro 26, 1252–1255 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Lanning BR et al. A road map to evaluate the proteome-wide selectivity of covalent kinase inhibitors. Nature Chemical Biology 10, 760–767 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parasar B et al. Chemoproteomic Profiling of Gut Microbiota-Associated Bile Salt Hydrolase Activity. ACS Cent Sci 5, 867–873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alnouti Y Bile Acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol. Sci. 108, 225–246 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Padmanabhan P, Grosse J, Asad ABMA, Radda GK & Golay X Gastrointestinal transit measurements in mice with 99mTc-DTPA-labeled activated charcoal using NanoSPECT-CT. EJNMMI Res 3, 60–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh J, Petter RC, Baillie TA & Whitty A The resurgence of covalent drugs. Nature Reviews Drug Discovery 10, 307–317 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Turk B Targeting proteases: successes, failures and future prospects. Nature Reviews Drug Discovery 5, 785–799 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Johnson DS, Weerapana E & Cravatt BF Strategies for discovering and derisking covalent, irreversible enzyme inhibitors. Future Med Chem 2, 949–964 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joyce SA et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. U.S.A. 111, 7421–7426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma C et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, eaan5931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallace BD et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie C et al. An Intestinal Farnesoid X Receptor-Ceramide Signaling Axis Modulates Hepatic Gluconeogenesis in Mice. Diabetes 66, 613–626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morin A et al. Collaboration gets the most out of software. eLife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ficarro SB, Alexander WM & Marto JA mzStudio: A Dynamic Digital Canvas for User-Driven Interrogation of Mass Spectrometry Data. Proteomes 5, 20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhoeckx K et al. Caco-2 Cell Line. The Impact of Food Bioactives on Health 175, 103–111 (2015). [Google Scholar]
- 52.Weerapana E, Speers AE & Cravatt BF Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc 2, 1414–1425 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Ficarro SB et al. Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Anal. Chem. 81, 3440–3447 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Alexander WM, Ficarro SB, Adelmant G & Marto JA multiplierz v2.0: A Python-based ecosystem for shared access and analysis of native mass spectrometry data. Proteomics 17, 1700091 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Zybailov B et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res 5, 2339–2347 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Callahan BJ et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caporaso JG et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schloss PD et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weber N et al. Nephele: a cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics 34, 1411–1413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wrzosek L et al. Transplantation of human microbiota into conventional mice durably reshapes the gut microbiota. Sci Rep 8, 6854–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rDNA datasets analyzed in the manuscript are available throughs NCBI under accession number PRJNA574158. The coordinates for both the apo and covalently inhibited forms of the BSH are deposited in the PDB and have PDB ID codes 6UFY and 6UH4, respectively. Raw mass spectrometry data were deposited at MassIVE (massive.ucsd.edu). Native mass spectrometry data files are available for download from the MassIVE archive at UCSD (ftp://massive.ucsd.edu/MSV000084491/). All other data generated or analyzed during this study are included in this article and its supplementary information files.