Björn Usadel and colleagues report the genome sequence of the wild tomato species Solanum pennellii. The authors identify genes important for stress tolerance, metabolism and fruit maturation and suggest that transposable elements have had an important role in the evolution of the S. penellii stress response.
Supplementary information
The online version of this article (doi:10.1038/ng.3046) contains supplementary material, which is available to authorized users.
Subject terms: Plant molecular biology, Plant genetics, Genomics
Abstract
Solanum pennellii is a wild tomato species endemic to Andean regions in South America, where it has evolved to thrive in arid habitats. Because of its extreme stress tolerance and unusual morphology, it is an important donor of germplasm for the cultivated tomato Solanum lycopersicum1. Introgression lines (ILs) in which large genomic regions of S. lycopersicum are replaced with the corresponding segments from S. pennellii can show remarkably superior agronomic performance2. Here we describe a high-quality genome assembly of the parents of the IL population. By anchoring the S. pennellii genome to the genetic map, we define candidate genes for stress tolerance and provide evidence that transposable elements had a role in the evolution of these traits. Our work paves a path toward further tomato improvement and for deciphering the mechanisms underlying the myriad other agronomic traits that can be improved with S. pennellii germplasm.
Supplementary information
The online version of this article (doi:10.1038/ng.3046) contains supplementary material, which is available to authorized users.
Main
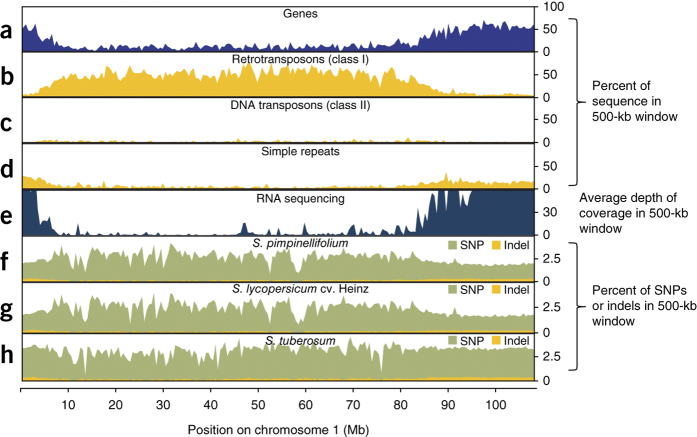
Crosses between distantly related plants can lead to substantial improvements in performance. Notably, S. pennellii × S. lycopersicum ILs have been used to define numerous quantitative trait loci (QTLs) for superior yield, chemical composition, morphology, abiotic stress tolerance and extreme heterosis3,4. Although genetic studies have proven informative, few genes underlying specific QTLs have been cloned, largely because of the lack of a S. pennellii genome sequence. To support QTL analyses, we sequenced the genome of S. pennellii using Illumina sequencing with ∼190-fold coverage (Fig. 1 and Supplementary Tables 1–5). The initial assembly size was 942 Mb, with a scaffold N50 value of 1.7 Mb and N90 value of 0.43 Mb (Table 1 and Supplementary Tables 6 and 7). We estimated the total genome size to be about 1.2 Gb using a k-mer–based analysis (Supplementary Fig. 1 and Supplementary Table 8), in accordance with previous estimations3,4. We anchored 97.1% of the genome assembly to chromosomes using genetic maps and restriction site–associated DNA sequencing (RAD-seq)-based markers from the IL population5 (Supplementary Note). Comparison of the assembly to publicly available BAC sequences indicated an accuracy of >99.9%, and a satisfactory accuracy of gap-filled regions was shown by realigning reads (Supplementary Fig. 2 and Supplementary Table 9). Of the 307,350 S. lycopersicum and 7,812 S. pennellii publicly available ESTs, 93% and >96% could be aligned to the genome, respectively (Supplementary Table 10), indicating comprehensive coverage of the gene-rich regions. We predicted 32,273 high-confidence genes and a potential set of 44,966 protein-coding genes and checked these for codon usage (Supplementary Table 11). We found that 20,076 (ref. 6) protein-coding genes were functionally annotated and compared to other species (Supplementary Figs. 3–6, Supplementary Tables 12 and 13, and Supplementary Data Sets 1–6). The average gene had 5.7 exons and coded for a 518-amino-acid protein. Multiple statistical analyses indicated a complex interplay between Copia transposable elements and stress-related genes.
Figure 1. Genomic landscape of S. pennellii chromosome 1.
(a–d) Densities of genes (a), retrotransposons (b), DNA transposons (c) and simple repeats (d) are shown for a 500-kb window. (e) Average RNA sequencing coverage in a 500-kb window. (f–h) Percentages of variants relative to S. pimpinellifolium (f), S. lycopersicum (g) and S. tuberosum (h).
Table 1.
S. pennellii genome assembly statistics
| Stage | N50 | N90 | Contigs/scaffolds | n > N50 | Assembled size (bp) | Unknown nucleotides | Anchored |
|---|---|---|---|---|---|---|---|
| Contigs | 2,176 | 68 | 4,315,954 | 81,824 | 1,117,562,721 | 0 | |
| Scaffolding | 1,603,317 | 98,078 | 407,506 | 177 | 1,021,472,455 | 125,806,430 | |
| Gap filling | 1,590,935 | 95,443 | 407,506 | 177 | 1,012,612,203 | 67,624,937 | |
| Final | 1,741,129 | 437,042 | 4,579 | 156 | 942,595,034 | 67,190,021 | 97.1% |
n > N50, number of sequences longer than the N50 length.
In addition, we sequenced the S. lycopersicum cv. M82 genome—the recurrent parent of the IL population—at approximately 44-fold coverage. We compared this sequence to the S. lycopersicum cv. Heinz genome7, identifying 1,338,510 variants, of which 1,188,524 were SNPs. There was a clear enrichment for variants on chromosomes 4, 5 and 11, likely as a result of putative Solanum pimpinellifolium introgressions into the M82 cultivar, as evidenced by pairwise SNP analyses (Supplementary Figs. 7–9). Previous analyses7 suggested introgression of S. pimpinellifolium into the Heinz cultivar genome, and our genome-wide data seem to suggest an additional introgression on chromosome 4 as well.
Almost 82% of the nongapped S. pennellii assembly consisted of repeats (Supplementary Fig. 10). Long terminal repeat (LTR)-retrotransposons (LTR-RT) elements were by far the most abundant repeats, comprising ∼45% of the S. pennellii genome assembly. As in S. lycopersicum, there were many more Gypsy-type (349 Mb) than Copia-type (79 Mb) LTR-RTs in the S. pennellii genome (Supplementary Fig. 11). LTR-RTs have a substantial role in genome size variation, representing 355 Mb of the 781-Mb S. lycopersicum genome assembly and 428 Mb of 942 Mb in S. pennellii. We assessed the recent activity of LTR-RTs in these two tomato genomes and the potato (Solanum tuberosum) genome using a sequence alignment of full-length LTR-RTs and transformed distances into ages. This analysis showed that S. pennellii has a higher abundance of young LTR-RTs (∼15% showed close to zero divergence; Supplementary Figs. 12 and 13) than S. lycopersicum (less than 5% showed very low divergence), which is especially pronounced for Copia elements. These results point toward different genome dynamics in these two species since their separation from a common ancestor.
We next compared orthologous gene pairs from S. pennellii and S. lycopersicum to find genes with evidence for positive selection (Supplementary Fig. 14, Supplementary Tables 14–17 and Supplementary Data Set 7), as indicated by a Ka/Ks ratio (ratio of nonsynonymous to synonymous nucleotide changes) of >1.0. In the resulting gene list, we found an enrichment of genes encoding acyl-carrier proteins, which are known to be involved in lipid biosynthesis. This enrichment might reflect an adaptation of the S. pennellii hydrophobic cuticle to minimize transpiration water loss and thus promote survival in desert habitats.
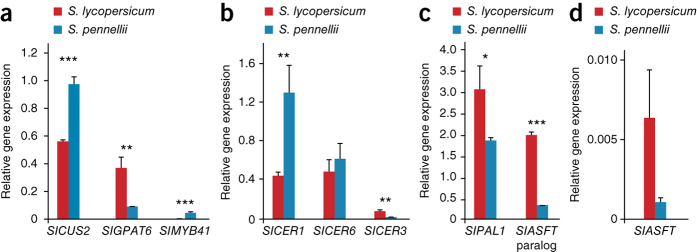
Previous studies on the fruit cuticles of wild tomato species documented considerable diversity in waxes and cutin8. We therefore analyzed the leaf cuticular waxes of both S. pennellii and S. lycopersicum and observed a threefold greater abundance of waxes, primarily very-long-chain alkanes, in S. pennellii (Supplementary Fig. 15). The alkane abundance in S. pennellii and the higher ratio of alkanes to triterpenoids have previously been suggested as mechanisms for increasing resistance to water flux across the cuticle9,10. Additionally, the phenylpropanoid component of S. pennellii cutin was reduced to ∼20% of that found in S. lycopersicum. We thus compared the expression of orthologs of known cuticle biosynthesis–related genes in the two species (Fig. 2, Supplementary Fig. 16 and Supplementary Tables 18 and 19). The expression of CER1, an ortholog of a key regulator of alkane concentrations in Arabidopsis thaliana11, was substantially higher in S. pennellii, consistent with the greater abundance of alkanes. In contrast, a consistent differential expression pattern was not evident for genes associated with the biosynthesis of aliphatic cutin components, in agreement with the absence of major differences in cutin biochemical data. Lastly, the abundance of cutin phenylpropanoids in S. lycopersicum correlated with the much higher expression of two feruloyltransferase homologs and PAL1, which encodes the phenylpropanoid pathway gateway enzyme. This comparative analysis of the genome sequences of S. pennellii and S. lycopersicum, together with gene expression studies, demonstrates their potential for elucidating the ecological constraints and adaptations underlying traits of agronomic importance.
Figure 2. Expression of cuticle biosynthesis–related genes.
(a–d) The expression of genes related to cutin biosynthesis (a) and wax biosynthesis (b) and of genes putatively (c) or known to be (d) associated with the formation of cuticular aromatic components was analyzed using quantitative PCR to validate data from RNA sequencing experiments and biochemical analysis. Statistical analysis was performed using a two-tailed t test. *P < 0.05, **P < 0.01, ***P < 0.001. Gene expression shown is relative to that for actin (Solyc05g054480). Error bars, s.e.m.; four biological replicates, each with three technical replicates.
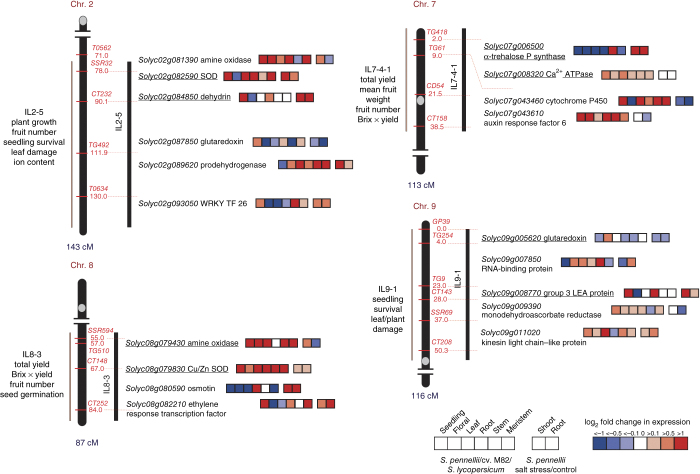
The high stress tolerance of S. pennellii, which has also been shown in some ILs (Supplementary Tables 20 and 21), led us to investigate its underlying mechanism. Stress tolerance has frequently been associated with allelic polymorphisms12, copy number variation13 and constitutive or inducible differential expression of numerous genes involved in regulatory or metabolic pathways14. To specifically target stress-related mechanisms, we identified 389 potential stress-related genes by mining the literature (Supplementary Table 22). These genes were filtered against drought- or salt-related QTLs detected in the ILs14,15,16,17,18,19, which resulted in the identification of 100 candidate genes (Fig. 3 and Supplementary Note). Alignment of the M82 and S. pennellii protein sequences indicated only minor differences, with no outliers in Ka/Ks ratios (Supplementary Data Set 7). Most nonsynonymous changes, if any, occurred in the less-conserved portions of the protein sequences (Supplementary Note), with a few instances occurring in highly conserved portions of the protein sequences, as was the case for dehydration-responsive element–binding protein 1 (DREB1, Solyc06g050520)20.
Figure 3. Chromosome mapping of stress-related candidate genes.
Candidate genes related to salt stress and drought are visualized on their respective IL QTLs for selected ILs 2-5, 7-4-1, 8-3 and 9-1. Colored squares next to each gene represent the magnitude of differential expression (log2) across a range of different tissues and conditions. Red indicates higher expression in S. pennellii, and blue indicates higher expression in S. lycopersicum. Underlined genes are those characterized by large differences in expression between S. lycopersicum cv. M82 and S. pennellii at the promoter and/or coding sequence level. Large differences are characterized as large (>30-bp) indels in the promoter region and/or at least one significant amino acid change in the coding sequence as determined by the P value predicted by SIFT Blink45. Brix × yield, total agronomic yield.
Further investigation showed that around half of the candidate stress-related genes had extensive polymorphisms in the regions upstream of their initiation codons. These sequence polymorphisms correlated with the magnitude of differential expression detected between M82 and S. pennellii21 (Supplementary Tables 23 and 24). One S. pennellii gene, a member of the drought tolerance–promoting dehydrin family (Solyc02g084850), contained two upstream insertions. A search for cis-acting elements in these two insertions showed the presence of several overlapping motifs, including some cis elements specifically responsive to dehydration (for example, MYCATRD22). This gene has been reported to show higher expression in the more drought-tolerant S. pennellii seedlings in comparison to M82 (ref. 21).
We further used the stress-related gene set to survey the colocalization of these genes with retrotransposons in the two species (Supplementary Note and Supplementary Data Sets 8–12). Stress-related genes (Supplementary Table 25) were found to be enriched in a 5-kb window around Copia-like retrotransposons in S. pennellii and S. lycopersicum. However, this enrichment was much more pronounced in S. pennellii (P < 0.0001 versus P = 0.001), and the enrichment around Gypsy elements was significant (P = 0.0021) in S. pennellii only (Supplementary Fig. 17 and Supplementary Note). Unsurprisingly, Copia elements in the proximal promoters (500 bp in length) of S. pennellii genes led to lower expression of these genes than for orthologous genes in S. lycopersicum that lacked such a Copia element, under standard conditions (Supplementary Note). We then investigated the correlation between the distribution of Copia-like elements and gene expression data from a published data set22 studying drought stress in leaves from S. pennellii and S. lycopersicum. We found a significant enrichment of S. pennellii–specific Copia elements within 5 kb of genes that were more stress responsive in S. pennellii than their S. lycopersicum orthologs (66 of 293 upregulated genes, P < 0.038 and 69 of 399 downregulated genes, P < 0.022). Genes that were more stress responsive in S. lycopersicum than their orthologs in S. pennellii were not significantly associated with S. lycopersicum–specific Copia-like elements (Supplementary Note).
This differential distribution of Gypsy and Copia-like elements, along with their association with stress responsiveness, suggests that, at least in some instances, LTR-RTs may exert a potential role in the regulation of gene expression. Such a role was proposed decades ago23, and some evidence of a role for transposable elements in the modulation of proximal gene expression has accumulated24,25,26. Our S. pennellii data set thus provide grounds, in future investigations, to assess the hypothesis that, in specific gene neighborhoods, Copia-like elements might represent 'conditional' regulatory sequences co-opted by the host genome upon exposure to different environmental stresses.
S. pennellii has additionally been widely documented to show phenotypes divergent from S. lycopersicum in relation to fruit development, maturation and metabolism27,28,29,30,31,32,33 (Supplementary Note and Supplementary Data Set 13). In terms of fruit maturation, several differences in the sequence and expression of some key regulatory genes were apparent. Most notable were differences in positive regulators, such as the FUL1 MADS-box gene34 that might be masked by the redundant FUL2 gene, and reduced expression and predicted activity of AP2A35, a negative regulator of ethylene synthesis that might compensate for the reduced ethylene production of maturing S. pennellii fruit36.
Analyses of differential gene expression in mature fruit were dominated by photosynthesis-related genes, which showed considerably higher expression in S. pennellii, consistent with the lack of a chloroplast-to-chromoplast transition in this species. Elevated expression of GLK2 (refs. 37,38) in S. pennellii fruit is consistent with these phenotypes.
Primary metabolism–associated genes did not show any unexpected changes in behavior, with core metabolic pathways having essentially conserved gene structures and similar expression patterns in the two species. Secondary metabolism in S. pennellii is considerably more complex, as evidenced by its glycoalkaloid, acyl sugar, terpene, carotenoid and volatile content. The additional glycoalkaloids in S. pennellii can be toxic, and their abundance is reflected in the relative expression of decorative enzymes of glycoalkaloid biosynthesis in S. pennellii and S. lycopersicum39 (Supplementary Note), with similar patterns also observed for acyl sugars and terpenes29,33. Variants seen in several carotenoid pathway genes, including PSY1 and lyc-B genes, are consistent with the lack of lycopene accumulation in mature S. pennellii fruit (Supplementary Note). Domestication has selected against anti-nutrients and bitter-tasting flavors in S. lycopersicum but has also inadvertently led to poorer tasting tomatoes, which is in part a consequence of reduced volatile production40,41. A detailed analysis showed divergence in both the sequence and expression of different volatile content–associated genes (Supplementary Note). A more complete understanding of these candidate genes, alongside those involved in the flavonoid and vitamin biosynthetic pathways, will likely greatly facilitate the breeding of better tasting and more nutritious fruit.
In conclusion, we have generated a valuable resource for the characterization of QTLs in the S. pennellii × S. lycopersicum IL population and have provided examples of how this genomics resource can be used to understand changes in response to water deficit, fruit maturation and metabolism. These investigations demonstrate the power of sequencing the parental lines of immortalized genetic populations for which a broad spectrum of phenotypic data has been collected42. One of the next challenges is to combine these phenotypic data with data gleaned from the genomes to explain the myriad of QTLs that have already been identified. For a comprehensive understanding of the evolutionary mechanism at play in the diverse Solanum genus, access to even more genomes is crucial.
Notably, this study suggests that the distinctive morphological characteristics of S. pennellii involved in differential stress resistance might be underpinned both by alterations in cuticle composition and nonrandom association of specific gene sets with transposable elements. Further investigations are needed to shed light on the role of candidate transposable elements in transcriptional rewiring and genome evolution43,44 within the tomato clade.
Methods
The genome of S. pennellii LA0716 was sequenced using the whole-genome shotgun (WGS) approach on the Illumina platform. The three libraries of paired-end sequencing, with insert sizes of 205 bp, 275 bp and 515 bp, provided a total of 229.4 Gb of sequence data in 2.1 billion reads and were supplemented by 13 libraries of mate-pair sequencing, with insert sizes ranging from 3 kb to 40 kb, yielding an additional 348 million read pairs containing longer-distance structural information. Additionally, a BAC library was prepared and end sequenced using the Sanger approach.
Next-generation sequencing data were filtered to remove adaptors and low-quality base calls. Genome size estimation was performed using the k-mer counting approach (Supplementary Fig. 1). Filtered data were corrected, assembled, scaffolded and then gap filled. Contaminant scaffolds were detected by alignment of the assembly against the NCBI non-redundant nucleotide database, and scaffolds that were found to be closest to a reference sequence of non-plant origin were removed. Finally, the paired-end libraries were remapped to the assembly, and the resulting SNPs were used to correct the assembly.
Scaffolds were anchored to chromosome regions using a combination of traditional markers, BAC-end sequences and 'de novo' markers extracted from an existing RAD-seq data set of the S. pennellii IL population. Some scaffolds that were likely to have been misassembled were detected and split during this process. Nine existing BAC sequences were used to independently validate the genome.
The chloroplast was separately assembled by selecting for reads from very-high-coverage regions and assembling these, followed by manual arrangement to best align with the existing S. lycopersicum chloroplast sequence.
Gene annotation was performed using parameters trained for S. lycopersicum, supplemented by evidence from EST and RNA sequencing data. In addition to the publicly available EST and RNA sequencing data, 70 new RNA sequencing samples were sequenced covering a variety of conditions and tissues. The resulting gene transcripts were filtered on the basis of orthology against known related protein sequences and RNA support, at two different thresholds, resulting in a primary gene set and a high-confidence subset. These gene models were validated against the proteomes of Arabidopsis, S. lycopersicum and S. tuberosum.
Functional gene annotation and assignment to MapMan classes was performed using the Mercator pipeline. To enable cross-species comparison,the same pipeline version was used to annotate the S. lycopersicum and S. tuberosum gene models. MapMan classes that showed significant differences in their proportion in S. pennellii in comparison to the other species were further investigated manually. Orthology analysis was performed to identify simple orthologous pairs between S. pennellii and S. lycopersicum and also to identify orthologous gene families between S. lycopersicum, S. tuberosum, Arabidopsis and Oryza sativa (Supplementary Fig. 5).
Protein domains of S. pennellii, S. lycopersicum and S. tuberosum were analyzed using Interproscan and Pfam, identifying some notable differences. Pectin esterase and P450 proteins (Supplementary Figs. 18–22) were aligned against the appropriate hidden Markov model (HMM), and phylogenetic trees were created. Evolutionary analysis was performed on orthologous gene pairs by first aligning their protein sequence and then calculating their Ka/Ks ratio.
The coding sequence, promoter sequence and expression of genes known to be involved in primary metabolism, secondary metabolism, fruit development and ripening were further investigated in S. pennellii and S. lycopersicum.
Expression patterns were compared by aligning RNA reads from six tissue types against the S. pennellii transcriptome (Supplementary Figs. 23 and 24, Supplementary Tables 26–29 and Supplementary Data Sets 14–16). The RPKM (reads per kilobase (of transcript) per million reads mapped) values from this analysis were hierarchically clustered with those available from S. lycopersicum and S. pimpinellifolium, using the pairwise orthologs described above.
Repeat analysis was performed by de novo identification of repeats followed by whole-genome annotation. Repeats were classified using the REPET classification utility followed by semi-manual curation. LTR insertion times were estimated by finding full-length LTR transposons, and sequence alignment and evolutionary distances were calculated and converted to ages using a previously established rate.
Various stress-related gene sets were tested for proximity to transposable elements. The initial investigation used a set of 389 genes manually identified from the literature, which were expanded using the gene family clusters to form a more statistically powerful gene set. This gene set was tested for enrichment around both Gypsy and Copia elements, using the Fisher's exact test. Further similar analyses were performed using salt stress–responsive genes in S. pennellii, as well as a cross-species comparison of drought-responsive genes between S. pennellii and S. lycopersicum.
The S. lycopersicum cv. M82 genome was resequenced using the existing S. lycopersicum cv. Heinz genome as a reference. One paired-end library was created with an insert size of 190 bp and sequenced to provide 574 million reads totaling 47.4 billion bases. These data were initially aligned to the Heinz reference genome, and this alignment was further refined. The detected variants were then applied to the genome, and a further round of alignment was performed, where additional SNPs were detected and applied to create the final M82 genome.
Waxes and cutin from S. lycopersicum cv. Heinz and S. pennellii leaves were extracted, derivatized and then analyzed by gas chromatography–mass spectrometry. Metabolic differences were cross-referenced with quantitative PCR–derived expression data for genes known to be involved in wax and cutin synthesis.
Detailed methods and their associated references can be found in the Supplementary Note.
Accession codes.
The S. pennellii (LA716) raw data have been deposited with the European Bioinformatics Institute (EBI) under project number PRJEB5809. The genome sequence is available from the European Nucleotide Archive (ENA) under accessions HG975439–HG975452. The S. lycopersicum cv. M82 raw data have been deposited at the EBI under project number PRJEB6302. The genome sequence is available from the ENA under accessions HG975512–HG975525.
Supplementary information
Supplementary Figures 1–24, Supplementary Tables 1–7, 9–12, 14, 18–21, 23, 28 and 29, and Supplementary Note. (PDF 8130 kb)
Supplementary Tables 8, 13, 15–17, 22 and 24–27. (XLSX 1177 kb)
Supplementary Data Sets 1–16. (XLSX 74137 kb)
Acknowledgements
We gratefully acknowledge the support of the Max Planck Society in the form of an exceptional grant to cover the costs of sequencing the tomato species. In addition, work in the laboratories of D.Z. and A.R.F. was funded in the framework of Deutsche Israeli Project FE 552/12-1, which is administered by the Deutsche Forschungsgemeinschaft. B.U. and M.E.B. acknowledge the support of the Bundesministerium f¨r Bildung und Forschung (BMBF)-funded primary database FKZ 0315961. B.A. thanks the Deutsche Forschungsgemeinschaft International Research Training Groups (IRTG) 1525. F.S. acknowledges the support of the CRA Young Investigator Program. Work in the laboratory of F.C. was funded by INTA, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Agencia Nacional de Promoción Científica y Tecnológica (ANPCYPT) grants. Work in the laboratories of N.S. and J.N.M. was funded by NSF grant IOS-0820854. J.R. was supported by the US National Science Foundation (Plant Genome Program; DBI-0606595 and DBI-1313887) and by Agriculture and Food Research Initiative competitive grant 2011-04197 from the USDA National Institute of Food and Agriculture. Work in the laboratories of J.G. and Z.F. was additionally supported by US National Science Foundation Plant Genome Program grants DBI-0820612 and IOS-0923312. A.B. would like to thank Y. Brotman for preparing the Pseudomonas syringae–infected S. pennellii samples.
Author Contributions
A.B., B.U. and A.R.F. managed the project. A.B., F.S., M.E.B., F.M., T.T., K.S., R.S., I.O., J.N.M., N.S., Z.F., J.K.C.R., D.Z., F.C., J.J.G., D.W., B.U. and A.R.F. designed the analysis. F.S., M.E.B., H.K., C.L., M.C., Y.X., S.A.A., M.R., B.U. and A.V. conducted DNA and RNA preparation and sequencing. C.L., J.M.J.-G., S.K., J.V., E.S., R.K., D.K., L.R.H., R.C.H.J.v.H., R.K.L., R.P., D.W. contributed new reagents and analytical tools. A.B., F.S., G.L., F.M., R.S., B.U. and A.R.F. conducted the data analyses. H.Q., S.A., I.S., E.A.F., S.O., L.M. and B.A. identified and evaluated candidate genes. B.U., A.B., M.E.B., F.S., D.W., F.M., J.K.C.R. and A.R.F. wrote the manuscript with help from all authors.
Accession codes
Primary accessions
European Nucleotide Archive
Competing interests
D.Z. is a cofounder of the company Phenom Networks, which develops phenotype bioinformatics tools.
References
- 1.Rick C, Tanksley S. Genetic variation in Solanum pennellii: comparisons with two other sympatric tomato species. Plant Syst. Evol. 1981;139:11–45. [Google Scholar]
- 2.Gur A, Zamir D. Unused natural variation can lift yield barriers in plant breeding. PLoS Biol. 2004;2:e245. doi: 10.1371/journal.pbio.0020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 1991;9:208–218. [Google Scholar]
- 4.Kamenetzky L, et al. Genomic analysis of wild tomato introgressions determining metabolism- and yield-associated traits. Plant Physiol. 2010;152:1772–1786. doi: 10.1104/pp.109.150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chitwood DH, et al. A quantitative genetic basis for leaf morphology in a set of precisely defined tomato introgression lines. Plant Cell. 2013;25:2465–2481. doi: 10.1105/tpc.113.112391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohse M, et al. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014;37:1250–1258. doi: 10.1111/pce.12231. [DOI] [PubMed] [Google Scholar]
- 7.Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature485, 635–641 (2012). [DOI] [PMC free article] [PubMed]
- 8.Yeats TH, et al. The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J. 2012;69:655–666. doi: 10.1111/j.1365-313X.2011.04820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsons EP, et al. Fruit cuticle lipid composition and fruit post-harvest water loss in an advanced backcross generation of pepper (Capsicum sp.) Physiol. Plant. 2012;146:15–25. doi: 10.1111/j.1399-3054.2012.01592.x. [DOI] [PubMed] [Google Scholar]
- 10.Vogg G, et al. Tomato fruit cuticular waxes and their effects on transpiration barrier properties: functional characterization of a mutant deficient in a very-long-chain fatty acid beta-ketoacyl-CoA synthase. J. Exp. Bot. 2004;55:1401–1410. doi: 10.1093/jxb/erh149. [DOI] [PubMed] [Google Scholar]
- 11.Bourdenx B, et al. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 2011;156:29–45. doi: 10.1104/pp.111.172320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asins MJ, et al. Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+ /K+ homeostasis. Plant Cell Environ. 2013;36:1171–1191. doi: 10.1111/pce.12051. [DOI] [PubMed] [Google Scholar]
- 13.Maron LG, et al. Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc. Natl. Acad. Sci. USA. 2013;110:5241–5246. doi: 10.1073/pnas.1220766110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong P, et al. Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J. Exp. Bot. 2010;61:3563–3575. doi: 10.1093/jxb/erq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eshed Y, Abu-Abied M, Saranga Y, Zamir D. Lycopersicon esculentum lines containing small overlapping introgressions from L. pennellii. Theor. Appl. Genet. 1992;83:1027–1034. doi: 10.1007/BF00232968. [DOI] [PubMed] [Google Scholar]
- 16.Frary A, et al. Salt tolerance in Solanum pennellii: antioxidant response and related QTL. BMC Plant Biol. 2010;10:58. doi: 10.1186/1471-2229-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frary A, Keles D, Pinar H, Gol D, Doganlar S. NaCl tolerance in Lycopersicon pennellii introgression lines: QTL related to physiological responses. Biol. Plant. 2011;55:461–468. [Google Scholar]
- 18.Gur A, et al. Yield quantitative trait loci from wild tomato are predominately expressed by the shoot. Theor. Appl. Genet. 2011;122:405–420. doi: 10.1007/s00122-010-1456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uozumi A, et al. Tolerance to salt stress and blossom-end rot in an introgression line, IL8–3, of tomato. Sci. Hortic. (Amsterdam) 2012;138:1–6. [Google Scholar]
- 20.Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011;62:4731–4748. doi: 10.1093/jxb/err210. [DOI] [PubMed] [Google Scholar]
- 21.Koenig D, et al. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. USA. 2013;110:E2655–E2662. doi: 10.1073/pnas.1309606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filippis I, Lopez-Cobollo R, Abbott J, Butcher S, Bishop GJ. Using a periclinal chimera to unravel layer-specific gene expression in plants. Plant J. 2013;75:1039–1049. doi: 10.1111/tpj.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClintock B. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Butelli E, et al. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 2012;24:1242–1255. doi: 10.1105/tpc.111.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 26.Quadrana L, et al. Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 2014;5:4027. doi: 10.1038/ncomms5027. [DOI] [PubMed] [Google Scholar]
- 27.Schauer N, et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 2006;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- 28.Schauer N, et al. Mode of inheritance of primary metabolic traits in tomato. Plant Cell. 2008;20:509–523. doi: 10.1105/tpc.107.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schilmiller AL, et al. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl. Acad. Sci. USA. 2009;106:10865–10870. doi: 10.1073/pnas.0904113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tieman D, et al. Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J. 2010;62:113–123. doi: 10.1111/j.1365-313X.2010.04128.x. [DOI] [PubMed] [Google Scholar]
- 31.Kochevenko A, Fernie AR. The genetic architecture of branched-chain amino acid accumulation in tomato fruits. J. Exp. Bot. 2011;62:3895–3906. doi: 10.1093/jxb/err091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mageroy MH, Tieman DM, Floystad A, Taylor MG, Klee HJ. A Solanum lycopersicum catechol-O-methyltransferase involved in synthesis of the flavor molecule guaiacol. Plant J. 2012;69:1043–1051. doi: 10.1111/j.1365-313X.2011.04854.x. [DOI] [PubMed] [Google Scholar]
- 33.Schilmiller AL, Charbonneau AL, Last RL. Identification of a BAHD acetyltransferase that produces protective acyl sugars in tomato trichomes. Proc. Natl. Acad. Sci. USA. 2012;109:16377–16382. doi: 10.1073/pnas.1207906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bemer M, et al. The Tomato FRUITFULL Homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell. 2012;24:4437–4451. doi: 10.1105/tpc.112.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung MY, et al. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 2010;64:936–947. doi: 10.1111/j.1365-313X.2010.04384.x. [DOI] [PubMed] [Google Scholar]
- 36.Grumet R, Fobes JF, Herner RC. Ripening behavior of wild tomato species. Plant Physiol. 1981;68:1428–1432. doi: 10.1104/pp.68.6.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen CV, et al. Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell. 2014;26:585–601. doi: 10.1105/tpc.113.118794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powell AL, et al. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science. 2012;336:1711–1715. doi: 10.1126/science.1222218. [DOI] [PubMed] [Google Scholar]
- 39.Iijima Y, et al. Steroidal glycoalkaloid profiling and structures of glycoalkaloids in wild tomato fruit. Phytochemistry. 2013;95:145–157. doi: 10.1016/j.phytochem.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 40.Tieman D, et al. Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. USA. 2006;103:8287–8292. doi: 10.1073/pnas.0602469103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klee HJ. Purple tomatoes: longer lasting, less disease, and better for you. Curr. Biol. 2013;23:R520–R521. doi: 10.1016/j.cub.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 42.Zamir D. Where have all the crop phenotypes gone? PLoS Biol. 2013;11:e1001595. doi: 10.1371/journal.pbio.1001595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bui, Q. & Grandbastien, M.-A. in Plant Transposable Elements Vol. 24 (eds. Grandbastien, M.-A. & Casacuberta, J.M.) 273–296 (Springer, Berlin, 2012).
- 44.Lisch D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013;14:49–61. doi: 10.1038/nrg3374. [DOI] [PubMed] [Google Scholar]
- 45.Kumar P, et al. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–24, Supplementary Tables 1–7, 9–12, 14, 18–21, 23, 28 and 29, and Supplementary Note. (PDF 8130 kb)
Supplementary Tables 8, 13, 15–17, 22 and 24–27. (XLSX 1177 kb)
Supplementary Data Sets 1–16. (XLSX 74137 kb)