Mingsheng Chen, Klaus Mayer, Steve Rounsley, Rod Wing and colleagues report the genome sequence of African rice (Oryza glaberrima), a different species than Asian rice. The authors resequenced 20 O. glaberrima accessions and 94 Oryza barthii accessions (the putative progenitor species of O. glaberrima), and their analyses support the hypothesis that O. glaberrima was domesticated in a single region along the upper Niger river.
Supplementary information
The online version of this article (doi:10.1038/ng.3044) contains supplementary material, which is available to authorized users.
Subject terms: Genomics, Plant sciences
Abstract
The cultivation of rice in Africa dates back more than 3,000 years. Interestingly, African rice is not of the same origin as Asian rice (Oryza sativa L.) but rather is an entirely different species (i.e., Oryza glaberrima Steud.). Here we present a high-quality assembly and annotation of the O. glaberrima genome and detailed analyses of its evolutionary history of domestication and selection. Population genomics analyses of 20 O. glaberrima and 94 Oryza barthii accessions support the hypothesis that O. glaberrima was domesticated in a single region along the Niger river as opposed to noncentric domestication events across Africa. We detected evidence for artificial selection at a genome-wide scale, as well as with a set of O. glaberrima genes orthologous to O. sativa genes that are known to be associated with domestication, thus indicating convergent yet independent selection of a common set of genes during two geographically and culturally distinct domestication processes.
Supplementary information
The online version of this article (doi:10.1038/ng.3044) contains supplementary material, which is available to authorized users.
Main
O. glaberrima Steud. is an African species of rice that was independently domesticated from the wild progenitor O. barthii ∼3,000 years ago1, 6,000–7,000 years after the domestication of Asian rice (O. sativa)2,3. Rice cultivation had a central role in building strong, knowledgeable and vibrant agrarian cultures in west Africa. It was this know-how, i.e., cultivation in lowland, upland and mangrove environments, harvesting and milling, that brought rice cultivation to the New World through slavery4,5. O. glaberrima, likely the first rice cultivated in the New World, was probably introduced aboard Portuguese slave ships, but higher yielding Asian rice varieties (O. sativa) subsequently supplanted it6.
O. glaberrima is well adapted for cultivation in west Africa and possesses traits for increased tolerance to biotic and abiotic stresses, including drought, soil acidity, iron and aluminum toxicity, as well as weed competitiveness7,8. The O. glaberrima accession CG14, sequenced here, is one of the parents used in the generation of the 'New Rice for Africa' (NERICA) cultivars that revolutionized rice cultivation in west Africa by combining the high-yielding traits of Asian rice with the adaptive traits of west African rice9.
Here we present a high-quality assembly and annotation of the O. glaberrima genome and detailed population genomics analyses of its domestication history and selection.
Results
Genome sequencing and assembly
We sequenced the O. glaberrima genome (International Rice Germplasm Collection (IRGC) accession #96717, var. CG14) using a minimum tiling path (MTP) of 3,485 BAC clones selected from a BAC-based physical map aligned to the O. sativa ssp. japonica reference genome (RefSeq)10,11 with (i) a hybrid BAC pool (3,319 BACs) and whole genome shotgun approach using Roche/454 GS-FLX Titanium sequencing technology for 11.5 chromosomes12 and (ii) a BAC-by-BAC (166 BACs) Sanger method for the short arm of chromosome 3 (Chr3S). The overall run statistics are summarized in Supplementary Table 1. The genome was assembled as outlined in Supplementary Figure 1 and is composed of 5,309 scaffolds (scaffold N50, 217 kb) assembled into 12 pseudomolecules, resulting in a total assembly size of 316 Mb (Supplementary Table 2). About 90% of the genome assembly (scaffold N50, 231 kb) could be ordered and oriented unambiguously based on the O. sativa ssp. japonica RefSeq11 (Supplementary Table 3).
We evaluated the final assembly for accuracy and completeness using four previously Sanger-sequenced and finished BACs located on chromosomes 1 (1 BAC), 5 (1 BAC) and 6 (2 BACs). Overall, more than 98% of the query BAC sequences were detected and localized in the correct chromosomal locations (549,598 bp query/557,270 bp subject) with a sequence accuracy of 99.6%. The 7.7 kb of missing sequence was located across 23 sequence gaps in the pseudomolecules (Supplementary Table 4).
Protein coding and tRNA predictions
The identification of protein coding gene models was based on consensus predictions derived from several types of evidence: ab initio gene finders, protein homology from finished plant genome projects and optimal spliced alignments of ESTs and tentative consensus transcripts. In addition to consensus models, we also searched for potentially missing candidate models using a Markov model13 that provides the likelihood for the coding potential of a given sequence. tRNA genes were identified by tRNAscan-SE14 using default parameters.
In total, we derived 33,164 gene models, including potential gene models and 701 tRNA genes (Fig. 1). Table 1 shows a comparison of the predicted protein coding and tRNA gene content of four sequenced Oryza species, including O. glaberrima. The gene number in O. glaberrima falls between that of Oryza brachyantha15 (30,952 genes) and that of the gold standard, O. sativa RefSeq11 (41,620 genes).
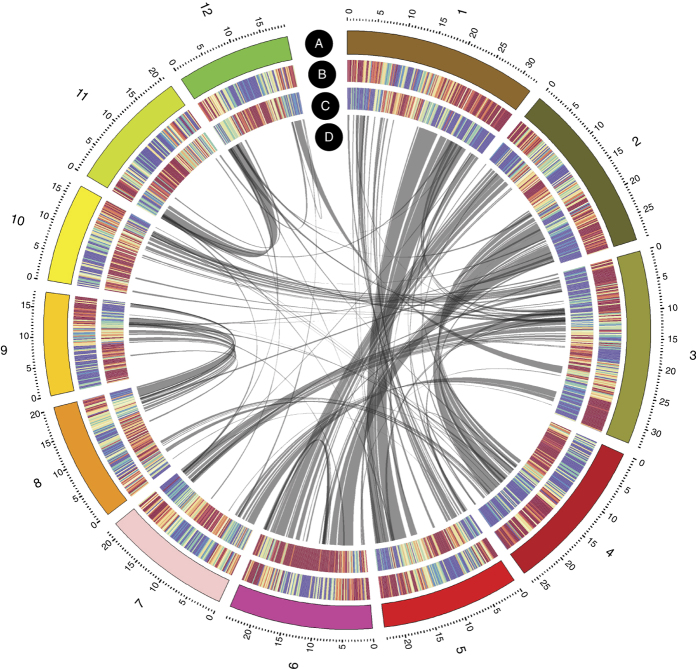
Figure 1. The O. glaberrima genome (CG14 v1).
Concentric circles show structural, functional and evolutionary aspects of the genome: A, chromosome number; B, heat map view of genes; C, repeat (RNA and DNA TEs without MITEs) density in 200-kb windows (red, average +1 s.d.; blue, average −1 s.d.; yellow, gene and repeat density between red and blue); and D paralogous relationships between O. glaberrima chromosomes.
Table 1.
Gene feature statistics of four Oryza genomes
| Feature | O. glaberrima | O. sativa ssp. japonica (MSU V6.1) | O. sativa ssp. indica | O. brachyantha |
|---|---|---|---|---|
| Gene count | 33,164 | 41,620 | 40,745 | 30,952 |
| Exon count | 142,095 | 171,163 | 170,496 | 141,987 |
| Intron count | 108,931 | 129,543 | 129,751 | 111,035 |
| Gene size (bp) | 2,393 | 2,306 | 2,439 | 2,677 |
| CDS size (bp) | 1,094 | 1,044 | 1,117 | 1,044 |
| Exon size (bp) | 255 | 254 | 267 | 228 |
| Intron size (bp) | 396 | 407 | 416 | 456 |
| Exons per gene | 4.28 | 4.11 | 4.18 | 4.59 |
| tRNA | 701 | 722 | 707 | 516 |
Shown are the gene, exon and intron counts, the sizes of genes, coding regions, exons and introns, the number of exons per gene (shown as mean values) and the number of tRNAs identified in the O. glaberrima genome assembly. For O. sativa ssp. japonica, only non–transposon related genes were used. On average, genes in O. brachyantha are larger than those in the other three species, a result that is driven by larger introns. CDS, coding DNA sequence.
To determine the extent of conservation of functional genes in O. glaberrima, we selected a set of O. sativa genes from three important pathways associated with flowering time, light response and stress resistance (WRKY genes) and searched for orthologous genes in the O. glaberrima genome. We found that 95.5% (170/178) of the O. sativa genes tested were intact and syntenic within the O. glaberrima genome (Supplementary Tables 5, 6, 7, 8, 9).
To determine whether the eight missing genes were indeed not present in the O. glaberrima genome or were absent because of missing data or problems with the assembly, we scanned our baseline O. glaberrima RNA sequencing (RNA-Seq) data set for evidence of transcription of these genes. The results (Supplementary Fig. 2) demonstrate that seven of the eight genes are actually present and transcribed in the O. glaberrima genome but are located within or in close proximity to gaps in the genome assembly (Supplementary Table 10). The last gene, Hd1 (heading date 1), was shown previously to be deleted in O. glaberrima16. Our assembly and RNA-Seq data support this finding (Supplementary Fig. 3).
Repeat annotation and dynamics
We found that transposable elements (TEs) represented 104 Mb (i.e., 34.25%) in the O. glaberrima genome assembly (Fig. 1). The largest classes of TEs are the long terminal repeat retrotransposons (LTR-RTs) (16.65% total; Copia, 2.70%; Gypsy, 10.41%; unclassified elements, 3.55%), followed by DNA TEs (13.48% total, including miniature inverted-repeat TEs (MITEs)) (Supplementary Table 11).
MITE- and LTR-RT–related sequences represent 44.4% and 26.2% of the total number of TE sequences, respectively. The average distance between a TE-related sequence and the closest gene was 1,002 bp. Of the 28,249 predicted gene models located closer to TE sequences than to scaffold ends, ∼63% of the TE-related sequences were MITEs and ∼16% were LTR-RTs, so MITEs were enriched in their proximity to genes, whereas LTR-RTs were depleted. A similar trend holds true for the 16,136 predicted genes that include TE fragments: 9,685 insertions (60.02%) were MITEs and 2,703 (16.75%) were LTR-RT fragments (Supplementary Table 12).
The estimated divergence time between O. sativa and O. glaberrima lineages, i.e., ∼600,000 years17, provides a unique opportunity to study the dynamics of TE-driven structural changes through a genome-wide comparative approach. TEs constitute 104 Mb of the O. glaberrima genome, and they represent 156 Mb of the O. sativa genome. The 52-Mb difference is consistent with the overall size difference between the two genomes, which shows that this difference could be accounted for solely by transposable elements. Moreover, given both the high synteny level and sequence identity between the two genomes, we conducted a thorough comparative survey of all TE insertions, thus distinguishing between insertions that occurred before (common in both genomes) and those that occurred after divergence from their common ancestor (either O. sativa– or O. glaberrima–specific insertions). When we considered Helitrons, short interspersed elements (SINEs) and LTR-RTs (both complete elements and solo LTRs), we found that 54.2% of the insertions occurred before their divergence, whereas 11.5% occurred in O. glaberrima and 34.3% occurred in O. sativa after their divergence (Supplementary Table 13). Specifically, this finding confirms that recent transpositional activity has been greater in the O. sativa lineage. A closer examination of LTR-RT insertions showed that distinct families were active in each genome. For example, the Hopi family, amplified recently in the O. sativa genome18, has over 500 complete copies representing more than 5 Mb of sequence in O. sativa, whereas we found only one truncated copy in the O. glaberrima genome.
Our data demonstrate that TE-driven genome differentiation occurred on a short time scale between O. glaberrima and O. sativa through the activation of distinct families in each lineage.
Domestication history of O. glaberrima
The geographical origins of crop domestication in Africa have been seen through two lenses—centric and noncentric19. Portères (1962)20 first proposed that O. glaberrima was domesticated in the inland delta of the upper Niger river and diffused to two secondary centers of diversification, one along the Senegambian coast and the other in the interior Guinea highlands. Harlan19 proposed a noncentric model of domestication for most African plants (such as sorghum) but by 1976 (ref. 21) agreed with Portères' hypothesis, treating O. glaberrima as an exception to this general pattern. Recent genetic analyses of 14 genes have supported Portères' hypothesis22.
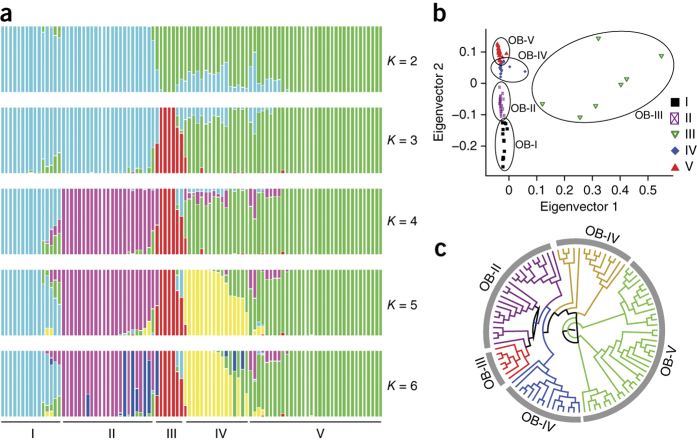
Because O. barthii can be found in situ across most of the African continent, it is important to establish the population structure, if any, of a diverse collection of O. barthii accessions and to associate one or more of these populations with a collection of domesticated O. glaberrima varieties. To perform these experiments, we first resequenced 94 O. barthii accessions (threefold coverage each) from 12 African nations (Supplementary Table 14) and determined their population structure and genetic distance with respect to the CG14 O. glaberrima genome. A total of 8,174,678 O. barthii SNPs were identified and input into the maximum-likelihood clustering program ADMIXTURE23 with K values ranging from two to six. ADMIXTURE partitioned the O. barthii accessions into four subgroups (OB-I, OB-II, OB-III and OB-V), as well as one apparent admixture subgroup (OB-IV) (Fig. 2a, Supplementary Fig. 4 and Supplementary Table 15). This subgrouping was further supported by principal component analysis (PCA) (Fig. 2b) and both neighbor-joining (NJ) (Fig. 2c and Supplementary Fig. 5) and maximum likelihood trees (Supplementary Fig. 6) using the same SNP data set.
Figure 2. Population structure analysis of 94 O. barthii accessions.
(a) Population structure of 94 O. barthii accessions inferred using ADMIXTURE23. Each color represents one population. The length of each segment in each vertical bar represents the proportion contributed by ancestral populations. The O. barthii population is partitioned into four subgroups (OB-I, OB-II, OB-III and OB-V), as well as an admixed group (OB-IV). (b) PCA of 94 O. barthii accessions using all identified SNPs as markers. The O. barthii accessions from the same subgroup are clustered together. (c) NJ phylogenetic tree of 94 O. barthii accessions.
It should be noted that O. barthii group III (OB-III) appears to be genetically distinct from the rest of the O. barthii accessions tested. Although the molecular and functional nature of this distinction has yet to be investigated, we do know that the majority of the accessions in OB-III were collected from Nigeria, and thus this pattern could be associated with local adaptation.
Next we resequenced 20 diverse O. glaberrima accessions (Supplementary Table 16) to threefold coverage, called SNPs relative to the CG14 genome assembly and constructed a NJ tree using the combined O. glaberrima and O. barthii SNP data. The NJ tree showed that that all but one O. glaberrima accession clustered with O. barthii subgroup OB-V (Fig. 3a and Supplementary Fig. 7). To measure population differences and similarities, we calculated the fixation index values (FST)24 between each O. barthii subgroup and O. glaberrima. The pairwise FST value between OB-V and O. glaberrima (0.009) was smaller than the pairwise FST values between other subgroups and O. glaberrima (range, 0.055–0.369) (Supplementary Table 17). Thus, our data suggest that O. glaberrima was domesticated directly from O. barthii subgroup OB-V.
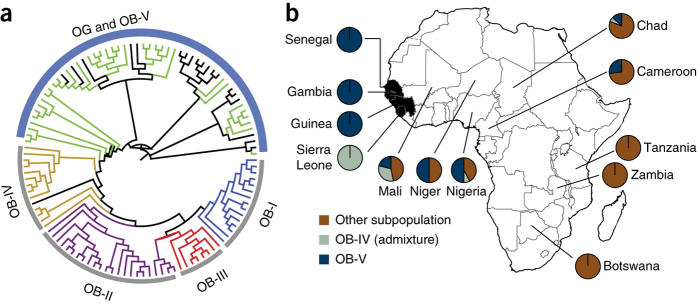
Figure 3. Identification of the domestication center of O. glaberrima.
(a) NJ phylogenetic tree of 20 O. glaberrima and 94 O. barthii accessions. All but one of the O. glaberrima accessions (black) are clustered with O. barthii accessions from group OB-V (green). (b) The proportion of each group of O. barthii accessions originating from different countries in Africa. All O. barthii accessions collected from the countries in the proposed domestication center (highlighted in black) are from the OB-V and OB-IV admixture groups. The proportion of O. barthii from the OB-V and OB-IV admixture groups found in each country decreased with distance from the domestication center, whereas the O. barthii accessions from other subgroups showed the opposite trend.
The combined population genetic analyses described above provide evidence that O. glaberrima was domesticated directly from the OB-V O. barthii subgroup and allowed us to estimate the approximate domestication center of O. glaberrima by calculating the proportion of each O. barthii subgroup originating from different geographical regions in Africa (Supplementary Table 18). All of the O. barthii accessions collected from the countries in the proposed domestication center (i.e., Gambia, Guinea, Senegal and Sierra Leone) are from the OB-V and OB-IV admixture groups, whereas the other sampled regions possess fewer and fewer OB-V accessions relative to their distance from the proposed domestication center (Fig. 3b). These results support the domestication theory proposed by Portères and are consistent with those reported by Li et al. (2011)22.
Evidence of artificial selection in O. glaberrima
Results from the analyses described above indicate that O. glaberrima was domesticated from O. barthii in a single domestication center in west Africa. Studies of domestication in many plant species have demonstrated a reduction in genetic diversity in domesticated crops relative to their wild progenitors25,26,27. To compare the genetic diversity present within African rice and its wild progenitor and identify signals of artificial selection during domestication, we first resequenced 20 O. glaberrima and 19 O. barthii accessions (Supplementary Tables 16 and 19) to a deeper coverage (10–30×) than that described in the previous section. A total of 4,447,424 and 8,735,082 SNPs were identified across the 20 O. glaberrima accessions and 19 O. barthii accessions, respectively, and were used to measure genome-wide levels of nucleotide diversity (π), the minor allele frequency (MAF) spectrum, the decay of linkage disequilibrium (LD) and Tajima's D values across all annotated genes and intergenic regions. The levels of nucleotide variability in O. glaberrima (π per kb, 2.281) were significantly lower than those in O. barthii (π per kb, 5.324) (P < 1 × 10−5) (Supplementary Figs. 8 and 9). The levels of nucleotide variability of all the annotated genes in O. glaberrima (π per kb, 4.457) were significantly lower than those in O. barthii (π per kb, 6.835) (P < 1 × 10−5) (Supplementary Fig. 9). The levels of nucleotide variability of intergenic regions in O. glaberrima (π per kb, 4.799) were also significantly lower than those in O. barthii (π per kb, 8.506) (P < 1 × 10−5) (Supplementary Fig. 9). The MAF spectrum of O. glaberrima and O. barthii detected an excess of low-frequency alleles in O. glaberrima compared with O. barthii (Supplementary Fig. 10). Tajima's D values in all annotated genes were significantly more negative in O. glaberrima than in O. barthii (P < 1 × 10−5). Furthermore, LD decay analysis revealed that LD decay blocks were four times larger in O. glaberrima (251 kb) than in O. barthii (57 kb) (Supplementary Fig. 11). These combined results are strongly suggestive of a significant reduction in genetic diversity in O. glaberrima as a consequence of domestication.
To identify genomic regions under artificial selection in O. glaberrima, we calculated the nucleotide diversity ratio (πw/πc) between O. barthii subgroup OB-V (wild rice) and O. glaberrima (cultivated rice) using 100-kb windows25,28. A total of 73 regions were identified in the top 2.5 percentile of genetic diversity cutoffs (πw/πc > 5.8) (Supplementary Fig. 12 and Supplementary Table 20), one of which contains an ortholog of the O. sativa domestication-related gene badh2 (ref. 29), which is associated with fragrance in Asian rice.
To detect evidence of recent selective sweeps in the O. glaberrima genome, we used the SNP data set described above to search for site frequency spectrum (SFS) deviations relative to genome-wide patterns using the composite likelihood ratio statistic30 (Fig. 4). We conducted the same analyses in the O. barthii subgroup OB-V to identify regions that only showed evidence of selective sweeps in O. glaberrima (Supplementary Fig. 13). Scanning the genome in 3-kb windows, we identified the top 0.5% outlier regions and first searched for regions close to or encompassing homologs of known and hypothesized O. sativa domestication genes. Homologs of seven agronomically important O. sativa genes with diverse functions (Sd1 (ref. 31), OsNAC6 (ref. 32), EP3 (ref. 33), Sh4 (ref. 34), Ep2 (ref. 35), Sub1 (ref. 36) and Xa26 (ref. 37)) are close—within the window of LD decay—to regions with strong evidence of recent selective sweeps in O. glaberrima (Supplementary Table 21). In addition, we conducted a genome scan of O. glaberrima for regions of high haplotype homozygosity using the integrated haplotype score (iHS) statistic38. These analyses identified several regions with unusually long haplotypes relative to the whole genome, suggesting recent incomplete selective sweeps (Supplementary Figs. 14 and 15). We examined the top 500 SNPs with the largest |iHS| (|iHS| > 4, corresponding to 0.028% of the analyzed SNPs genome wide) and identified 24 candidate regions of multiple contiguous SNPs with |iHS| > 4. Three of those regions were close (8–190 kb)—within the window of LD decay—to genes previously identified to be under domestication in Asian rice (Supplementary Table 22).
Figure 4. Identification of candidate regions of artificial selection during O. glaberrima domestication.
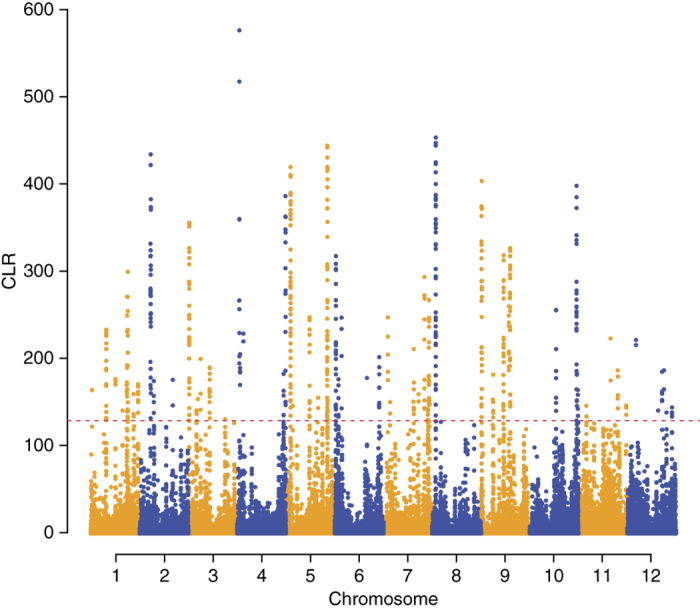
Plot of the composite likelihood ratio (CLR) across O. glaberrima genome. The red dashed line indicates the cutoff value for the 0.5% outlier regions with significant deviations from neutrality, indicating evidence of recent selective sweeps.
The regions identified with signals of artificial selection from all the tests described above can be considered candidate regions for future genetic and functional analyses.
Evidence for independent domestication of O. glaberrima
Although the domestication process of Asian rice has been widely investigated25,39, it remains unclear whether the domestication of O. glaberrima underwent similar or independent paths. That is, did ancient Asian and African farmers artificially select, in parallel, the same or similar traits and genes during the domestication process, did they select a completely different set of traits and genes, or did they use a combination of both processes? Two recent studies have suggested that both scenarios could be in play. First, Gross et al.40 showed that Rc, the red pericarp gene, has signatures of selection in both Asian and African rice but that the genetic diversity profile of Rc is different and independent for each species, which is suggestive of convergent but independent domestication. As described above, Hd1, a key gene in Asian rice that controls synchronized flowering, is completely deleted in O. glaberrima16. However, this gene is intact and expressed in O. barthii (IRGC accession #105608; Supplementary Fig. 3), further supporting the hypothesis of independent domestication of a common trait but by the selection of a different set of gene(s) and/or quantitative trait loci.
To make the comparative investigation of domestication in Asian and African rice even more complex, there is evidence that the domestication of African rice in more recent times may have been influenced by the introduction of Asian rice into west Africa and subsequent intercrossing, as suggested by the microsatellite study of Semon et al.41 that inferred substantial admixture between some accessions of both species. To address this latter concern and look for evidence of introgression of Asian rice alleles into African rice, we first probed the O. glaberrima genome assembly with resequencing reads of 14 O. sativa (7 O. sativa spp. japonica and 7 O. sativa ssp. indica) (Supplementary Table 23) and 20 O. glaberrima accessions. We then used a newly developed haplotype-based summary statistic that compares the minimum number of nucleotide differences between sequences from two different species to the average number of between-species differences to identify regions of the genome that have experienced recent introgression42,43. Our genome-wide analyses were unable to detect any evidence of recent introgression between the sequenced accessions of the two species (Supplementary Fig. 16), thus indicating that O. glaberrima and O. sativa were domesticated independently.
As we detected no evidence of recent introgression of O. sativa into O. glaberrima, we next investigated whether a known set of O. sativa domestication genes was also artificially selected during the domestication of African rice. We used 19 O. sativa domestication genes (Supplementary Table 24) to probe the O. glaberrima genome, resulting in the identification of 16 clear orthologous loci. Three of these 16 genes (qSh1 (ref. 44), Sd1 (ref. 31) and Dep1 (ref. 45)) yielded substantial deviations from neutral expectations in Tajima's D tests (Supplementary Table 25). These results were further supported by comparing the nucleotide diversity of these genes in O. glaberrima and O. barthii to the genome-wide nucleotide diversity of intergenic regions in both species (Supplementary Tables 26, 27, 28). In addition, the nucleotide diversity of these genes was greatly reduced in O. glaberrima relative to O. barthii (Supplementary Figs. 17, 18, 19). These data suggest that a minimum of four genes, including Rc40,46, that are associated with domestication in O. sativa may have been under recent selection in O. glaberrima.
The selection of non-shattering panicles has been associated with crop domestication for a wide variety of crops47,48. Because orthologs of a gene that controls shattering in O. sativa (qSh1) were shown to possess signals of artificial selection in O. glaberrima, we investigated the molecular structure and transcriptional nature of this gene and two additional shattering gene orthologs from O. sativa, Shattering 1 and Shattering 4, to search for evidence of independent domestication.
The O. sativa Shattering 1 gene (OsSh1: LOC_Os03g44710) encodes a YABBY transcription factor whose gene has an ∼4-kb insertion in its third intron, thus leading to reduced transcription and a shattering-resistant phenotype in O. sativa compared to Oryza rufipogon49. Functional orthologs of this gene have also been identified in sorghum and maize, confirming the independent domestication of shattering genes in cereals in general. Sequence analysis of the orthologous region in O. glaberrima revealed the presence of a 45-kb deletion that resulted in the complete ablation of the O. glaberrima OsSh1 ortholog and three additional genes (Fig. 5a and Supplementary Fig. 20). We confirmed the presence of this gene in the wild progenitor, and its absence in the domesticated species, by sequence analysis of BACs encompassing this orthologous region from O. barthii (IRGC accession #105608) and O. glaberrima (CG14) and RNA-Seq data (Supplementary Fig. 21).
Figure 5. Sequence comparisons of OsSh1 and Sh4.
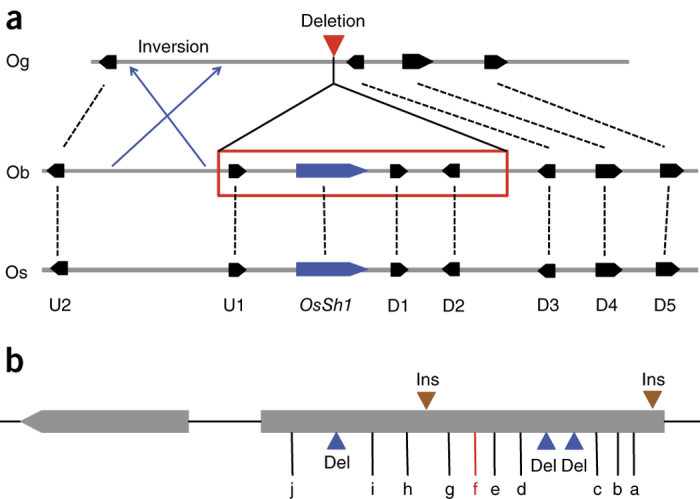
(a) Orthologous gene relationship of the OsSh1 region of O. sativa ssp. japonica (Os) with those of O. barthii (Ob) and O. glaberrima (Og). The 45-kb deletion (red triangle) resulted in the complete removal of the OsSh1 ortholog and three additional genes in O. glaberrima (red rectangle). Inversion is indicated with blue arrows. U and D represent upstream and downstream genes relative to OsSh1, respectively (U1, LOC_Os03g44680; U2, LOC_Os03g44690; OsSh1, LOC_Os03g44710; D1, LOC_Os03g44720; D2, LOC_Os03g44740; D3, LOC_Os03g44750; D4, LOC_Os03g44760; D5, LOC_Os03g44780). (b) Sequence comparison of Sh4 of O. sativa ssp. japonica and O. glaberrima. Two insertions (Ins) and three deletions (Del) of O. glaberrima compared to O. sativa ssp. japonica are shown as brown triangles and blue triangles, respectively. Ten SNPs are labeled a–j. The causative mutation (f) of the non-shattering phenotype in O. sativa is highlighted in red. This mutation did not exist in O. glaberrima.
The O. sativa Shattering 4 gene (Sh4: LOC_Os04g57530) encodes a Myb3 transcription factor in which a single-nucleotide substitution leads to reduced transcription and a shattering-resistant phenotype in O. sativa compared to O. nivara34. Sequence analysis of the orthologous genic region in O. glaberrima revealed a completely different set of sequence variation relative to the O. sativa gene that includes the presence of ten SNPs and five small insertion/deletions. Most notably, the causative mutation revealing the non-shattering phenotype in O. sativa was not found in O. glaberrima (Fig. 5b). Analysis of O. glaberrima and O. barthii RNA-Seq expression data from mixed-stage panicles from both O. glaberrima and O. barthii showed a differential expression pattern with no transcription detected in O. glaberrima, whereas transcription was detected in O. barthii (Supplementary Fig. 22). As the expression level of OgSh4 is absent or reduced in O. glaberrima and this gene is located close to a region under a selective sweep, it is possible that the causative mutation may reside in the upstream promoter region. Thus, we searched the promoter regions of OgSh4 in O. glaberrima and O. barthii for signatures of selection and found that the promoter region of O. glaberrima shows substantial deviation from neutral expectations using Tajima's D test (Supplementary Table 25). The result was further supported by comparing the nucleotide diversity of the promoter region in O. glaberrima and O. barthii to the genome-wide nucleotide diversity of intergenic regions in O. glaberrima and O. barthii (Supplementary Table 29). In addition, the nucleotide diversity of the promoter region was greatly reduced in O. glaberrima relative to O. barthii (Supplementary Fig. 23). This result suggests that the promoter region of OgSh4 may have been selected during domestication, leading to reduction of the expression of OgSh4.
The O. sativa qSh1 (LOC_Os01g62920) locus encodes a transcription factor that has a single SNP located 12 kb upstream of the transcription start site. This mutation prevents transcription in the abscission layer that results in a shattering-resistant phenotype in O. sativa ssp. indica compared with O. sativa ssp. japonica44. Sequence comparison of the O. sativa qSh1 locus and its orthologous locus in O. glaberrima showed a high level of sequence identity but did not reveal the presence of the causal O. sativa SNP that leads to the shattering phenotype in O. sativa. RNA-Seq analysis of the qSh1 ortholog showed that the gene was transcribed in both O. glaberrima and O. barthii panicles and presumably functions normally in comparison to the gene expression pattern observed in Asian rice subspecies (Supplementary Fig. 24).
The comparative analysis described above provides persuasive evidence that ancient African and Asian farmers unknowingly selected for mutations in all three orthologous genes investigated that prevented or reduced panicle shattering. We also demonstrate that the mutation profiles of each orthologous pair of these genes (OsSh1, Sh4 and qSh1) are completely different. Thus, we conclude that the control of panicle shattering in African and Asian rice is the result of independent domestication. Combined with the results of genome-wide introgression analysis of O. glaberrima and O. sativa, we provide strong evidence that O. glaberrima was domesticated independently from O. barthii.
Discussion
As the world population is projected to increase from 7.1 billion to over 9 billion by 2050, plant biologists must forge a second green revolution with the creation of crops that have two to three times the current yield with reduced inputs (i.e., less water, fertilizers and pesticides and the ability to grow on marginal soils)50. Rice will have a key role in helping to solve the problem of how to feed 9 billion people51.
The release of the O. glaberrima genome, its annotation and comparative population genomics data sets enables an unprecedented opportunity for the identification and utilization of adaptive traits that are important for rice agriculture, especially in west Africa, a region whose population is expected to grow rapidly over the next 50 years.
Methods
O. glaberrima genome sequencing.
A total of 3,485 MTP BAC clones were selected from the O. glaberrima BAC-based physical map. Of those, 166 BACs representing Chr3S were individually Sanger sequenced, and the remaining BACs were divided into 115 chromosome-specific BAC pools (28–30 chromosomally ordered BACs per pool). All MTP BACs were re–end sequenced and compared to the O. glaberrima BAC-end data set to validate the clone identity. About 3–5 μg of pooled BAC DNA was used to construct each Roche/454 Titanium library with multiplex identifiers (MID) using the manufacturer's protocol, followed by Bioanalyzer and fluorometry analyses to assess library quality and quantity, respectively. Clonal amplification of individual libraries was performed by emulsion-based clonal amplification (emPCR amplification), and a group of five to six MID-tagged DNA beads was pooled and loaded onto one picotiter plate (PTP) and sequenced on a Roche/454 GS FLX instrument using Titanium chemistry following the manufacturer's protocols. Chromosome-specific libraries (3-kb paired ends (PEs)) were constructed from the BAC pools and sequenced on one half PTP per chromosome. Additionally, about 5× genome–equivalent Titanium reads were generated using a whole genome shotgun (WGS) method. Overall sequencing statistics are summarized in Supplementary Table 1.
O. glaberrima genome assembly.
The O. glaberrima genome was assembled using ∼12.4 Gb of high-quality bases with the Newbler V2.3 assembler following a six-step protocol (Supplementary Fig. 1). (i) Generating pool-based assemblies: the Roche/454 Titanium reads were independently assembled for each pool to obtain pool-wise assembled contigs. PE reads corresponding to each chromosome were mapped to the contigs of the respective chromosomes and PE reads belonging to each chromosome pool were extracted. A second round of assembly with both Titanium and PE reads corresponding to each pool was performed to obtain pool-level scaffolds. (ii) Incorporation of WGS reads into pool-wise assemblies: to identify WGS reads corresponding to each chromosome pool, the WGS reads were separately assembled in combination with PE reads from all 12 chromosomes, and the WGS scaffolds corresponding to each pool were identified by comparing the scaffolds to the pool-wise scaffolds obtained in the previous step using BLASTN52. WGS reads corresponding to each pool were then extracted, and a third round of assembly was performed with the Titanium, PE and WGS reads for each pool. (iii) Identification of misassembled scaffolds: each scaffold was aligned to the corresponding chromosome of the O. sativa ssp. japonica reference sequence (RefSeq)11 using Nucmer53, and potential misassembled scaffolds and misassembly breakpoints were identified. Putative misassemblies in scaffolds were evaluated by inspecting PE depth and Titanium depth across each breakpoint. Any scaffolds that had a PE depth ≤1 and coverage of ≤4× were confirmed as misassemblies, and each scaffold was split at the breakpoints into multiple scaffolds. (iv) Merging of overlapping regions between pools: to remove any redundant sequences between adjacent pools, scaffolds from neighboring pools were aligned using Nucmer, and redundant regions were identified. Reads from overlapping scaffolds were extracted and reassembled to generate a non-redundant set of scaffolds. This process was repeated for all neighboring pools for all chromosomes to obtain a non-redundant set of scaffolds for each chromosome. (v) Ordering and orienting of scaffolds: scaffolds for each chromosome were aligned with the corresponding chromosome of the O. sativa ssp. japonica reference genome using Nucmer, and the scaffolds were grouped into either the mapped or unmapped category. All mapped scaffolds were ordered and oriented as per the O. sativa ssp. japonica chromosomes. (vi) Incorporation of Sanger sequences: as a final step, the existing Sanger BAC sequences from the short arm of chromosome 3 and from chromosomes 6, 8, 11 and 12 were spliced into the existing 454 assembly. The 454 scaffolds corresponding to Sanger sequences were identified by BLASTN, and the 454 scaffolds were replaced by the Sanger sequences.
Protein-coding and tRNA gene predictions.
Gene models were determined using a combination approach of a consensus prediction from ab initio gene predictions (Fgenesh++54, ProtoMap55 and GeneID13) and an optimal spliced alignment from protein and transcript homology searches using GenomeThreader56 with a splice site model for rice. Homologous protein evidence mappings were derived from annotations of the following finished plant genome projects: O. sativa (version TIGR6.1 (ref. 57) and RAP2 (ref. 58)), Sorghum bicolor (high confidence set of version 1.4)59, Arabidopsis thaliana (version TAIR9)60 and Brachypodium (version 1.2)61. Optimal spliced alignments of TIGR transcript assemblies62, comprising several monocotyledonous species (Zea mays, Saccharum officinale, O. sativa, Hordeum vulgare, Triticum aestivum and Brachypodium distachyon) as well as assembled RNA-Seq reads of O. glaberrima, were used for gene predictions based on homology and/or experimental evidence. Additionally, Markov models and position-specific scoring matrices were applied to estimate the coding potential for the models without protein homology and to evaluate splice site and start and stop codons, respectively. We used coding sequences for the Markov models that had a minimum protein similarity and alignment coverage of ≥95% to known O. sativa models, a minimum mean expression of three RNA-Seq reads and coverage ≥98% by RNA-Seq reads. The gene models without homology, i.e., E value greater than 10−10, were selected for a minimum size of at least 60 amino acids, minimum coding potential >0, maximal overlap with repeats of 20% and mean RNA-Seq expression >0.5. tRNA genes were identified with tRNAscan-SE14 using default parameters.
The flowering time, light response and WRKY genes were selected from the O. sativa ssp. japonica RefSeq. The orthologous genes in O. glaberrima were determined using MegaBLAST with an identity of 90% for flowering time and light response genes. The orthologous WRKY genes in O. glaberrima and O. brachyantha were determined by tBLASTN using the conserved domain and predicted proteins from the O. sativa ssp. japonica genome annotation. Translated proteins with identity and coverage greater than 80% were considered in the analysis. We further checked the syntenic region to confirm the orthologous status of these genes.
RNA sequencing and analysis.
The O. glaberrima, O. barthii, O. sativa ssp. japonica, O. rufipogon and O. nivara accessions used for RNA-Seq analysis were obtained from the International Rice Research Institute (IRRI) Genetic Resource Center (GRC). Leaf tissue for library construction was collected from plants at the four-leaf stage. Root tissue for library construction was collected from plants growing in hydroponic conditions. The panicles were collected at different stages of flower development for the construction of sequencing libraries. Strand-specific RNA-Seq libraries of the different species were constructed using the protocol described previously63 and sequenced on one lane of an Illumina HiSeq 2000 instrument. Raw pair-end reads were de novo assembled using Trinity r2012-05-18 (ref. 64). The assembled sequences were aligned to O. sativa ssp. japonica RefSeq using Bowtie V2.1.0 (ref. 65). The results of the alignment were visualized and annotated using the Integrative Genomics Viewer (IGV) V2.3 (ref. 66).
Repeat annotation.
Known repeats were annotated using RepeatMasker with Rebase TE67, TIGR/MSU repeat57 and in-house Oryza TE databases. De novo repeats were identified with RepeatScout68. RepeatMasker and LTR-FINDER69 were used to identify complete LTR-RTs by examination of target site duplications (TSDs), primer binding sites, polypurinic tracts and element boundaries. Putative solo LTRs were identified by comparing the RepeatMasker match length with the LTR query length. Solo LTR candidates were then manually inspected for evidence of TSDs.
The 5′ and 3′ ends (40 bp per end) of an O. sativa complete Helitron data set were mapped to identify putative Helitrons in O. glaberrima using RepeatMasker with the following rules: (i) the 5′ and 3′ ends should be aligned within a distance of 1–20 kb on concordant strands with the expected orientation, (ii) allow less than four mismatches for the 5′ end and one mismatch in the 3′ CTAG end and (iii) inspect for insertion at the dinucleotide AT.
SINEs were identified using the strategy outlined in Baucom et al.70. MITE sequences were identified by searching for inverted repeats that were flanked by TSDs (2 bp for Stowaway and 3 bp for Tourist). For Stowaway, we searched specifically for the canonical terminal inverted repeat motif CTCCTCCC and a TA target site duplication. Terminal inverted repeats of at least 8 bp that started with a G or C were considered for Tourist elements. Approximately 400 candidate sequences were used for a multiple alignment with CLUSTALW71 to identify groups, and redundancy (for example, forward and reverse sequences of the same family) was removed to yield the initial set of families. Consensus sequences were mapped to the genome by BLASTN to identify all copies for each family.
Identified repetitive elements were further characterized and classified using the criteria described by Wicker et al.72, with the exception of LTR-RTs. An LTR-RT was assigned to a family based on the best BLAST hit against RepBase. Insertion times of complete LTR-RT elements were estimated using a molecular paleontology approach described by SanMiguel et al.73, with a mutation rate of 1.3 × 10−8 substitutions per site per year, as proposed by Ma and Bennetzen74.
Sample preparation and sequencing for evolutionary analysis.
The O. glaberrima and O. barthii accessions (Supplementary Tables 12 and 14) were obtained from the African Rice Center (ARC) and IRRI-GRC. The O. sativa ssp. japonica accessions were obtained from IRRI-GRC. Leaf tissue was collected for each accession at the five-leaf stage, and genomic DNAs were extracted for sequencing using Qiagen DNeasy Plant Mini kits (Qiagen, Maryland, USA) following the manufacturer's protocol. GAIIx libraries were constructed using Illumina standard adaptors and sequenced on an Illumina GAIIx instrument with 2 × 75-bp sequencing chemistry. HiSeq libraries were constructed on a SPRIworks System I (Beckman Coulter, USA) with custom MID adaptors as described in Huang et al. (2009)75 and sequenced on an Illumina HiSeq 2000 instrument using 2 × 100-bp cycle sequencing chemistry. For O. barthii accessions sequenced with 5× coverage, 20 individuals were sequenced in one lane of the HiSeq 2000 instrument. For other accessions, one individual was sequenced in one lane of the GAIIx instrument, or five individuals were sequenced in one lane of the HiSeq 2000 instrument.
SNP detection.
Sequence reads from O. glaberrima and O. barthii accessions were separated according to the 3-bp barcode added in the adaptor using custom Perl scripts. Burrows-Wheeler Aligner (BWA) V0.5.9 (ref. 76) was used to map raw pair-end reads onto the O. glaberrima CG14 genome assembly. All reads with a mapping quality score of less than 20 were discarded. The mean coverage of each accession was evaluated using BEDtools V2.13.3 (ref. 77). Alignment files were then input into Genome Analysis Toolkit (GATK) V1.2 (ref. 78) to identify SNPs. Multiple SNP calling was performed using the UnifiedGenotyper from GATK. SNPs were filtered for quality score >20, coverage greater than two and less than twice the mean coverage of all accessions. A SNP was retained if more than one accession had the SNP called at the identical position.
History of O. glaberrima domestication.
Several methods were applied to identify the population structure of O. barthii. First, the SNPs of the O. barthii population were loaded into the program ADMIXTURE23 to perform maximum likelihood clustering analysis. The result from ADMIXTURE was plotted using an R script. Second, a NJ tree of the O. barthii population was constructed for all SNP positions using the program Phylip with 1,000 bootstrap steps. Third, PCA was performed to assess the population structure of O. barthii using SMARTPCA79. The result of PCA was plotted using an R script. Finally, 10,000 SNP positions were randomly selected from the SNP set using a PERL script. The SNPs were input into the program PhyML 3.0 (ref. 80) to construct a maximum likelihood tree using the HKY85 model with 100 bootstrap steps.
In total, 8,374,445 SNPs were used to investigate the genetic relationship of O. glaberrima and O. barthii. Genetic differentiation (FST) between O. glaberrima and each of the O. barthii subpopulations was calculated using VCFtools V0.1.12 (ref. 81). The NJ tree of the O. glaberrima and O. barthii accessions was constructed for all SNP positions using the program Phylip with 1,000 bootstrap steps.
The country of origin for each O. barthii accession was obtained from ARC and IRRI-GRC. The proportions of O. barthii accessions from different subgroups were calculated for each country. The proportions of O. barthii accessions of different types were also calculated for each country.
Genetic variation analysis of O. glaberrima and O. barthii.
Genome-wide genetic diversity (π) was calculated using VCFtools V0.1.12 and POPBAM V0.3 (ref. 43) for nonoverlapping 100-kb windows across the genome. The genetic diversity (π) of all annotated genes and intergenic regions was calculated using POPBAM V0.3. The Wilcoxon test for comparing genome-wide π values of O. glaberrima and O. barthii was performed using an R script. The distribution and boxplot of π values were generated using an R script. In total, 3,373,099 SNPs were input into VCFtools V0.1.12 to identify the MAF for O. glaberrima and O. barthii. The MAF spectrum was plotted using an R script. The correlation coefficient (r2) of alleles was calculated to measure LD level in O. glaberrima and O. barthii using plink V1.07 (ref. 82). The distance that the LD decays to half of the highest value was calculated, and then plotted, using an R script.
Identification of regions under artificial selection.
The reduction of genetic diversity (πw/πc) was calculated for nonoverlapping 100-kb windows across the genome using VCFtools V0.1.12. The windows that were in the 2.5% right tail of the distribution (πw/πc > 5.8) were selected as candidate regions. The distribution of πw/πc along the chromosome was plotted using an R script.
SweeD83 was used for detecting complete selective sweeps in the O. glaberrima genome with default settings. A total of 4,436,289 SNPs were input into the SweeD program after filtering some SNPs according to the program's requirements. This program implements the composite likelihood ratio statistic30, which identifies regions with significant deviations from the neutral SFS and performs better with genome-wide SNP data than SweepFinder30 does. The O. punctata genome sequence (AVCL00000000.1) was used to determine whether alleles were ancestral or derived in order to use the unfolded version of the SFS. Haplotypes were inferred using HapMap tools, and derived or ancestral alleles were identified using the genome sequence of O. punctata. A total of 1,746,642 SNPs were analyzed. We used the integrated haplotype score (iHS) statistic38 to identify regions with unusually long haplotypes relative to the whole genome. Large negative values of iHS indicated high haplotype homozygosity because of an increase in frequency of the derived allele, and positive values reflected high frequency of the ancestral allele. Analyses were conducted using the R package rehh84.
Evidence of independent domestication of O. glaberrima.
To investigate the introgression of Asian rice into African rice, resequencing reads from 14 O. sativa and 20 O. glaberrima accessions (10–30× coverage) were mapped to the O. glaberrima RefSeq data using BWA V0.5.9. All reads with a mapping quality score of less than 20 were discarded. The alignment files of the O. sativa and O. glaberrima accessions were then input into POPBAM V0.3 to detect evidence of introgression from O. sativa into O. glaberrima using the recently developed Gmin statistic42.
O. sativa domestication genes were selected according to criteria described in Meyer and Purugganan48. Orthologous genes of the O. sativa domestication genes in O. glaberrima were identified by nucleotide and amino acid BLAST searches. The Tajima's D value of annotated genes and intergenic regions was calculated using POPBAM V0.3. The Tajima's D value of O. glaberrima orthologs of domestication genes was calculated using POPBAM V0.3. The reduction of diversity (πw/πc) of genes that deviated significantly from neutrality was calculated for 300-bp sliding windows with 200-bp overlapping using the SNPs generated from 20 O. glaberrima and 19 O. barthii accessions using VCFtools V0.1.12. Distributions of the reduction of diversity of genes were plotted using an R script.
To identify the OsSh1 orthologous region, five upstream and downstream genes from OsSh1 (LOC_Os03g44710) were mapped to the O. glaberrima genome. Two BAC clones encompassing the OsSh1 region (OG_BBa0035M03 for O. glaberrima and OB_ABa0085D15 for O. barthii) were selected by aligning BAC-end sequences to the O. sativa ssp. japonica and O. glaberrima genomes. Two different insert libraries (500 bp and 10 kb) were constructed, sequenced and assembled using a Pacific Biosciences RS II instrument and HGAP software according to the manufacturer's recommendations. Both BACs were assembled into single contigs (139,241 bp for OG_BBa0035M03 and 176,785 bp for OB_ABa0085D15) without further finishing efforts. Sequence comparisons were performed using Dotter and Nucmer, and orthologs were identified using Nucmer and BLAST. To identify the Sh4 orthologous region, Sh4 gene sequences from O. sativa ssp. japonica were mapped to the O. glaberrima genome sequences. One BAC clone encompassing Sh4 in O. glaberrima (OG_BBa0063K01) was selected. The BAC was assembled into a single contig (175,355 bp) without further finishing efforts using the same method described above. Sequence comparisons were performed using BLAST. The orthologous gene of qSh1 was identified and compared using BLAST and manual inspection.
URLs.
RepeatMasker, http://www.repeatmasker.org/; SNPs used for O. barthii population structure analysis and for investigating the genetic relationship of O. glaberrima and O. barthii can be downloaded at http://de.iplantcollaborative.org/dl/d/11EA9662-D292-4C64-8862-69466C087936/Oglaberrima_Obarthii_population_variation.vcf; SNPs used for the study of genetic variation of O. glaberrima and O. barthii can be downloaded at http://ensembl.gramene.org/Oryza_glaberrima/Info/Index.
Accession codes.
The assembly and annotation of the O. glaberrima genome are available at http://ensembl.gramene.org/Oryza_glaberrima/Info/Index and have been deposited under GenBank accession number ADWL00000000.1. The assembly of the Oryza punctata genome has been deposited under GenBank accession number AVCL00000000.1. The BAC sequence assemblies have been deposited under GenBank accession numbers KF284071, KF284072 and KJ650506. The resequencing reads of O. glaberrima, O. barthii, O. sativa ssp. japonica and O. sativa ssp. indica have been deposited under GenBank accession numbers SRX502298–SRX502317, SRX502162–SRX502255, SRX621487–SRX621493 and SRX621496–SRX621502, respectively. The RNA-Seq reads of O. sativa ssp. japonica leaf, panicle and root have been deposited under GenBank accession numbers SRX477950, SRX477951 and SRX477952, respectively. The RNA-Seq reads of O. glaberrima leaf, panicle and root have been deposited under GenBank accession numbers SRX474528, SRX474529 and SRX474530, respectively. The RNA-Seq reads of leaf and panicle of O. barthii, O. rufipogon and O. nivara have been deposited under GenBank accession numbers SRX471823, SRX472434, SRX512340, SRX512341, SRX472708 and SRX472710 respectively.
Supplementary information
Supplementary Figures 1–24 and Supplementary Tables 1–4, 7, 8, 10–21 and 23–29 (PDF 26645 kb)
Homologues of flowering time genes in O. glaberrima (XLSX 41 kb)
Homologues of light response genes in O. glaberrima (XLSX 35 kb)
Homologues of WRKY genes in the O. glaberrima genome (XLSX 49 kb)
List of the top 500 SNPs with the largest |iHS| in O. glaberrima (XLSX 54 kb)
Acknowledgements
This material is based on work supported by the US National Science Foundation under grants #0321678, #0638541, #0822284 and #1026200 to Y.Y., C.A.M., S.R. and R.A.W. and the Bud Antle Endowed Chair of Excellence in Agriculture and Life Sciences to R.A.W. Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the US National Science Foundation. We thank coauthor J. Carney, author of Black Rice, for her inspiration and encouragement in our attempt to convey the deep-rooted and far-reaching history of African rice farming through the O. glaberrima genome sequence. We thank Z. Lu from Arizona Statistical Consulting Laboratory for statistical advice and J. Ross-Ibarra for comments.
Author Contributions
M. Wang performed all population sequencing and light response gene analyses. M. Wang and C.A.M. performed the population genomic analysis. Y.Y. directed all genome sequencing efforts. G.H. performed all genome-wide gene and tRNA annotations and the analysis of tandemly duplicated genes. P.R.M. assembled the O. glaberrima genome. C.F. performed all flowering time genes analyses. C.F. and M. Wang performed domestication-genes artificial selection analysis. J.L.G. edited the O. glaberrima physical map and selected the MTP. A.Z., R.M.C., O.P. and T.W. performed all repeat annotation and analyses. X.S., M. Wang, Y.Y. and D.K. performed analysis of the shattering genes. D.K. and J.S.S.A. performed overall project management. C.M. performed all WRKY gene family analyses. S.L., N.S., K.d.B., W.G., M. Wissotski and W.K. performed all hands-on activities related to O. glaberrima genome sequencing. P.S. grew, validated and maintained all Oryza accessions used in the project. M.-N.N. and K.S. provided all O. barthii accessions used for population genomics analysis. M.L. provided advice on the analysis of molecular evolution and population genomic data. J. Carney provided advice and consultation on the geographical and historical aspects of west African rice and slavery. M.C. and J. Chen provided early access to the O. brachyantha genome sequence and general consultation throughout the project. Y.Y., S.R., K.F.X.M., M.C. and R.A.W. conceived and guided the entire project and wrote the paper with contributions and input from all project members.
Accession codes
Primary accessions
NCBI Reference Sequence
Sequence Read Archive
Competing interests
The authors declare no competing financial interests.
Footnotes
Muhua Wang, Yeisoo Yu, Georg Haberer and Pradeep Reddy Marri: These authors contributed equally to this work.
Contributor Information
Mingsheng Chen, Email: mschen@genetics.ac.cn.
Klaus F X Mayer, Email: k.mayer@helmholtz-muenchen.de.
Steve Rounsley, Email: steve.rounsley@gmail.com.
Rod A Wing, Email: rwing@mail.arizona.edu.
References
- 1.Sweeney M, McCouch S. The complex history of the domestication of rice. Ann. Bot. 2007;100:951–957. doi: 10.1093/aob/mcm128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Portères, R. in Origins of African Plant Domestication (ed. Harlan, J.R.) 409–452 (Mouton Publishers, 1976).
- 3.Vaughan DA, Lu BR, Tomooka N. The evolving story of rice evolution. Plant Sci. 2008;174:394–408. [Google Scholar]
- 4.Littlefield, D.C. Rice and Slaves: Ethnicity and the Slave Trade in Colonial South Carolina (University of Illinois Press, 1991).
- 5.Portères, R. in Papers in African Prehistory (eds. Fage, J.D. & Oliver, R.A.) 43–58 (Cambridge University Press, 1970).
- 6.Carney, J.A. Black Rice: the African Origins of Rice Cultivation in the Americas (Harvard University Press, 2001).
- 7.Brar DS, Khush GS. Alien introgression in rice. Plant Mol. Biol. 1997;35:35–47. [PubMed] [Google Scholar]
- 8.Sanchez, P.L., Wing, R.A. & Brar, D.S. in Genetics and Genomics of Rice (eds. Zhang, Q. & Wing, R.A.) 9–25 (Springer, New York, 2013).
- 9.Jones MP, Dingkuhn M, Aluko GK, Semon M. Interspecific Oryza sativa L. X O. glaberrima Steud. progenies in upland rice improvement. Euphytica. 1997;94:237–246. [Google Scholar]
- 10.Kim H, et al. Construction, alignment and analysis of twelve framework physical maps that represent the ten genome types of the genus Oryza. Genome Biol. 2008;9:R45. doi: 10.1186/gb-2008-9-2-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki Takuji. The map-based sequence of the rice genome. Nature. 2005;436(7052):793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 12.Rounsley S, et al. De novo next generation sequencing of plant genomes. Rice. 2009;2:35–43. [Google Scholar]
- 13.Parra G, Blanco E, Guigo R. GeneID in Drosophila. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe TM. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, et al. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat. Commun. 2013;4:1595. doi: 10.1038/ncomms2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanyal A, et al. Orthologous comparisons of the Hd1 region across genera reveal Hd1 gene lability within diploid Oryza species and disruptions to microsynteny in Sorghum. Mol. Biol. Evol. 2010;27:2487–2506. doi: 10.1093/molbev/msq133. [DOI] [PubMed] [Google Scholar]
- 17.Ammiraju JSS, et al. Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell. 2008;20:3191–3209. doi: 10.1105/tpc.108.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vitte C, Panaud O, Quesneville H. LTR retrotransposons in rice (Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genomics. 2007;8:218. doi: 10.1186/1471-2164-8-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harlan JR. Agricultural origins: centers and noncenters. Science. 1971;174:468–474. doi: 10.1126/science.174.4008.468. [DOI] [PubMed] [Google Scholar]
- 20.Portéres R. Berceaux agricoles primaires sur le continent Africain. J. Afr. Hist. 1962;3:195–210. [Google Scholar]
- 21.Harlan, J., De Wet, J. & Stemler, A. in Origins of African Plant Domestication (ed. Harlan, J.R.) 3–20 (Mouton Publishers, 1976).
- 22.Li ZM, Zheng XM, Ge S. Genetic diversity and domestication history of African rice (Oryza glaberrima) as inferred from multiple gene sequences. Theor. Appl. Genet. 2011;123:21–31. doi: 10.1007/s00122-011-1563-2. [DOI] [PubMed] [Google Scholar]
- 23.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat. Rev. Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam HM, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 27.Wright SI, et al. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- 28.Xu X, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012;30:105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 29.Kovach MJ, Calingacion MN, Fitzgerald MA, McCouch SR. The origin and evolution of fragrance in rice (Oryza sativa L.) Proc. Natl. Acad. Sci. USA. 2009;106:14444–14449. doi: 10.1073/pnas.0904077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen R, et al. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA. 2002;99:9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakashima K, et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- 33.Piao R, et al. Map-based cloning of the ERECT PANICLE 3 gene in rice. Theor. Appl. Genet. 2009;119:1497–1506. doi: 10.1007/s00122-009-1151-x. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Zhou A, Sang T. Rice domestication by reducing shattering. Science. 2006;311:1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 35.Zhu K, et al. ERECT PANICLE2 encodes a novel protein that regulates panicle erectness in indica rice. Genetics. 2010;184:343–350. doi: 10.1534/genetics.109.112045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu K, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- 37.Sun X, et al. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313x.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- 38.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molina J, et al. Molecular evidence for a single evolutionary origin of domesticated rice. Proc. Natl. Acad. Sci. USA. 2011;108:8351–8356. doi: 10.1073/pnas.1104686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross BL, Steffen FT, Olsen KM. The molecular basis of white pericarps in African domesticated rice: novel mutations at the Rc gene. J. Evol. Biol. 2010;23:2747–2753. doi: 10.1111/j.1420-9101.2010.02125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semon M, Nielsen R, Jones MP, McCouch SR. The population structure of African cultivated rice Oryza glaberrima (Steud.): evidence for elevated levels of linkage disequilibrium caused by admixture with O. sativa and ecological adaptation. Genetics. 2005;169:1639–1647. doi: 10.1534/genetics.104.033175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geneva, A.J., Lovato, L.M., Kingan, S.B. & Garrigan, D. A new method to scan genomes for introgression in a secondary contact model. http://arxiv.org/abs/1403.1552 (2014). [DOI] [PMC free article] [PubMed]
- 43.Garrigan D. POPBAM: tools for evolutionary analysis of short read sequence alignments. Evol. Bioinform. Online. 2013;9:343–353. doi: 10.4137/EBO.S12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konishi S, et al. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 45.Huang X, et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009;41:494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- 46.Sweeney MT, Thomson MJ, Pfeil BE, McCouch S. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell. 2006;18:283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 48.Meyer RS, Purugganan MD. Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 2013;14:840–852. doi: 10.1038/nrg3605. [DOI] [PubMed] [Google Scholar]
- 49.Lin Z, et al. Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 2012;44:720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q. Strategies for developing Green Super Rice. Proc. Natl. Acad. Sci. USA. 2007;104:16402–16409. doi: 10.1073/pnas.0708013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Godfray HC, et al. Food security: the challenge of feeding 9 billion people. Science. 2010;327:812–818. doi: 10.1126/science.1185383. [DOI] [PubMed] [Google Scholar]
- 52.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 53.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yona G, Linial N, Linial M. ProtoMap: automatic classification of protein sequences and hierarchy of protein families. Nucleic Acids Res. 2000;28:49–55. doi: 10.1093/nar/28.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gremme G, Brendel V, Sparks ME, Kurtz S. Engineering a software tool for gene structure prediction in higher organisms. Inf. Softw. Technol. 2005;47:965–978. [Google Scholar]
- 57.Ouyang S, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rice Annotation Project. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res.36, D1028–D1033 (2008). [DOI] [PMC free article] [PubMed]
- 59.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 60.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature463, 763–768 (2010). [DOI] [PubMed]
- 62.Childs KL, et al. The TIGR Plant Transcript Assemblies database. Nucleic Acids Res. 2007;35:D846–D851. doi: 10.1093/nar/gkl785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, et al. A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RNA-seq. PLoS ONE. 2011;6:e26426. doi: 10.1371/journal.pone.0026426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 68.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(suppl. 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 69.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baucom RS, et al. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 2009;5:e1000732. doi: 10.1371/journal.pgen.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson JD, Higgins DG, Gibson TJ. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 73.SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat. Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 74.Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA. 2004;101:12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang X, et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009;19:1068–1076. doi: 10.1101/gr.089516.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 81.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pavlidis P, Zivkovic D, Stamatakis A, Alachiotis N. SweeD: likelihood-based detection of selective sweeps in thousands of genomes. Mol. Biol. Evol. 2013;30:2224–2234. doi: 10.1093/molbev/mst112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28:1176–1177. doi: 10.1093/bioinformatics/bts115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–24 and Supplementary Tables 1–4, 7, 8, 10–21 and 23–29 (PDF 26645 kb)
Homologues of flowering time genes in O. glaberrima (XLSX 41 kb)
Homologues of light response genes in O. glaberrima (XLSX 35 kb)
Homologues of WRKY genes in the O. glaberrima genome (XLSX 49 kb)
List of the top 500 SNPs with the largest |iHS| in O. glaberrima (XLSX 54 kb)