Summary
Bacteria are continually challenged by foreign invaders including bacteriophages, and have evolved a variety of defenses against these invaders. Here, we describe the structural and biochemical mechanisms of a bacteriophage immunity pathway found in a broad array of bacteria, including E. coli and Pseudomonas aeruginosa. This pathway employs eukaryotic-like HORMA domain proteins that recognize specific peptides, then bind and activate a cGAS/DncV-like nucleotidyltransferase (CD-NTase) to generate a cyclic triadenylate (cAAA) second messenger; cAAA in turn activates an endonuclease effector, NucC. Signaling is attenuated by a homolog of the AAA+ ATPase Pch2/TRIP13, which binds and disassembles the active HORMA-CD-NTase complex. When expressed in non-pathogenic E. coli, this pathway confers immunity against bacteriophage λ through an abortive infection mechanism. Our findings reveal the molecular mechanisms of a bacterial defense pathway integrating a cGAS-like nucleotidyltransferase with HORMA domain proteins for threat sensing through protein detection, and negative regulation by a Trip13 ATPase.
Keywords: Second messenger signaling, bacteriophage immunity, abortive infection, CD-NTase, HORMA domain, AAA+ ATPase remodeler
Introduction
All cellular life depends on signaling pathways that can rapidly sense and respond to changes in both a cell’s internal state and its external environment. Many such signaling pathways rely on diffusible second messenger molecules, which are synthesized by sensor proteins and activate effector proteins to drive a physiological response. In bacteria, the cyclic dinucleotide second messengers cyclic di-AMP, cyclic di-GMP, and cyclic GMP-AMP (cGAMP) play key roles in cellular homeostasis and pathogenesis (Corrigan and Gründling, 2013; Davies et al., 2012; Hengge, 2009). In mammals, cGAMP and linear oligoadenylate are important innate-immune signals, synthesized by cGAS (cyclic GMP-AMP synthase) and OAS (oligoadenylate synthase) family proteins in response to cytoplasmic DNA or double-stranded RNA (Chen et al., 2016; Hornung et al., 2014). Nucleotide-based second messengers also mediate cross-kingdom signaling, with mammalian innate immune receptors able to recognize and respond to a variety of bacterially-generated second messengers (Burdette et al., 2011; McFarland et al., 2017; Whiteley et al., 2019).
A major family of second messenger biosynthetic enzymes found across kingdoms is distantly related to DNA polymerase β and includes bacterial DncV (Dinucleotide cyclase in Vibrio), which synthesizes cGAMP (Davies et al., 2012; Zhu et al., 2014), and mammalian cGAS and OAS. Comparative genomics studies in bacteria have identified over 6000 distinct cGAS/DncV-like nucleotidyltransferases (CD-NTases) in environmental and human patient-derived bacterial strains, with examples found in over 10% of sequenced bacterial genomes (Burroughs et al., 2015; Cohen et al., 2019; Whiteley et al., 2019). These enzymes synthesize a marked variety of second messengers, including cyclic dinucleotides with both purine and pyrimidine bases, cyclic trinucleotides, and other as-yet uncharacterized molecules (Whiteley et al., 2019). The majority of bacterial CD-NTases are found within “defense islands” alongside known anti-phage defense systems (Cohen et al., 2019), and are encoded in operons that also encode putative effector proteins including predicted nucleases, proteases, and phospholipases (Burroughs et al., 2015; Whiteley et al., 2019). Two CD-NTase containing operons, one from V. cholerae encoding a phospholipase effector, and another from Bacillus cereus encoding a predicted transmembrane protein effector, have been shown to confer immunity against a range of bacteriophages (Cohen et al., 2019). In both cases, immunity is mediated by abortive infection, in which infected cells die prior to completion of phage replication and thereby prevent additional rounds of infection (Cohen et al., 2019). This new class of anti-bacteriophage defense systems has been termed CBASS (Cyclic oligonucleotide-Based Anti-phage Signaling System) (Cohen et al., 2019).
Biochemical analysis of 66 purified bacterial CD-NTases from a variety of CBASS systems showed that the majority of these enzymes are inactive in vitro, suggesting that their activity is tightly regulated in cells (Whiteley et al., 2019). Intriguingly, about 10% of CBASS systems encode one or two proteins with weak homology to eukaryotic HORMA domain proteins, along with an ortholog of the AAA+ ATPase regulator of HORMA domain proteins, Pch2/TRIP13 (Burroughs et al., 2015). In eukaryotes, HORMA domains (named for the first three proteins shown to possess this domain: Hop1, Rev7, and Mad2) bind short regions within their interaction partners called “closure motifs,” and assemble into signaling complexes that control inter-homolog recombination in meiosis (Hop1), DNA repair (Rev7), the mitotic spindle assembly checkpoint (Mad2), and autophagy (Atg13, Atg101) (Aravind and Koonin, 1998; Rosenberg and Corbett, 2015). The HORMA domain possesses two distinct folded states: an “open” state unable to bind closure motifs, and a “closed” state in which the HORMA domain C-terminus wraps entirely around a bound closure motif (Luo and Yu, 2008; Rosenberg and Corbett, 2015). The Pch2/TRIP13 ATPase disassembles HORMA:closure motif complexes by partially unfolding the N-terminus of the HORMA domain, dissociating the complex and in some cases converting the HORMA domain from the “closed” to the “open” state (Ma and Poon, 2016; Ye et al., 2015, 2017).
While HORMA domains play key signaling roles in eukaryotes, these proteins have not been previously described in bacteria. The presence of putative HORMA domain proteins in a subset of CBASS systems suggests that these proteins may mediate phage sensing, perhaps utilizing the HORMA domain’s peptide binding and conformational conversion abilities to control second messenger synthesis by their cognate CD-NTases. Here, we show that this is indeed the case: bacterial HORMA domain proteins adopt the canonical HORMA domain fold, undergo conformational conversion between open and closed states, and bind specific closure motif peptides. Binding of a “closed” HORMA domain protein to its cognate CD-NTase activates synthesis of a cyclic tri-AMP (cAAA) second messenger, which in turn activates an effector endonuclease also encoded in the operon. Bacterial Trip13-like ATPases function as negative regulators of signaling, binding and likely disassembling the active CD-NTase:HORMA complex. Finally, we show that a HORMA+Trip13 CBASS system from a patient-derived E. coli strain confers immunity against bacteriophage λ infection via abortive infection. Together, these results define the molecular mechanisms of a new bacterial defense system that employs HORMA domain proteins as a regulator of second messenger signaling.
Results
Bacterial HORMA domain proteins bind and activate second messenger synthesis by their associated CD-NTases
CBASS systems encoding putative HORMA domain proteins and an ortholog of the AAA+ ATPase Pch2/TRIP13 (hereon termed Trip13) fall into two major classes, one of which encodes a single HORMA domain protein (here termed HORMA1) and the second of which encodes two such proteins (HORMA2 and HORMA3) (Figure 1A and Figure S1A) (Burroughs et al., 2015). The HORMA1 and HORMA2/3 operons also encode diverged Trip13 ATPases, suggesting that the HORMA and Trip13 genes have co-evolved within each family of operons (Figure S1A–C) (Tromer et al., 2019). Bacterial CD-NTases have been grouped into different clades based on their sequence similarity (Whiteley et al., 2019), and we find that HORMA1 operons mainly encode clade C01 CD-NTases (hereon termed CdnC), while HORMA2/3 operons encode clade D05 CD-NTases (termed CdnD). To address the function of these operons, we chose a CdnC+HORMA1+Trip13 operon from E. coli strain MS115-1, and a CdnD+HORMA2/3+Trip13 operon from Pseudomonas aeruginosa strain ATCC27853, for further study (Figure 1A). The E. coli operon is found in several patient-derived E. coli strains, and the P. aeruginosa operon is found in over a quarter of patient-derived P. aeruginosa strains. Both operons also encode related putative endonuclease effectors, termed REase by Burroughs et al. (Burroughs et al., 2015) and here termed NucC (Nuclease, CD-NTase associated).
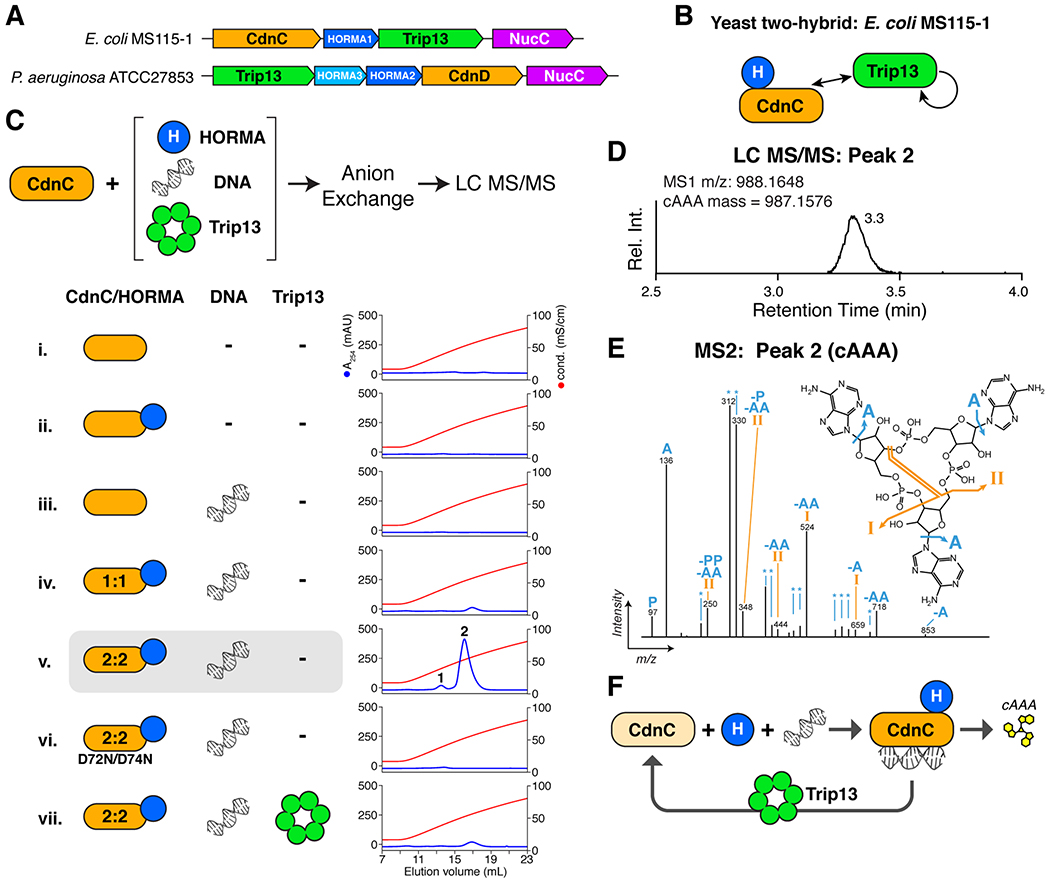
Figure 1. HORMA+Trip13-associated CD-NTases synthesize cAAA.
(A) Schematic of CD-NTase+HORMA+Trip13 operons from E. coli MS115-1 (top) and P. aeruginosa ATCC27853 (bottom). See Figure S1 for phylogenetic analysis of Trip13 and HORMA proteins. See Table S4 for protein sequences. (B) Summary of yeast two-hybrid and three-hybrid assays with the E. coli MS115-1 CdnC, HORMA, and Trip13. See Figure S2A for yeast two-hybrid and three-hybrid results, Figure S2B for purification and stoichiometry of a CdnC:HORMA complex, and Figure S2C for purification of a CdnC:HORMA:Trip13EQ complex. See Figure S2D–G for equivalent assays with the P. aeruginosa ATCC27853 operon. (C) Top: Schematic of second messenger synthesis assays. Ec CdnC was incubated with potential regulators (HORMA, plasmid DNA, and Trip13) plus ATP, then the products were separated by anion-exchange chromatography and analyzed by liquid chromatography with tandem mass spectrometry (LC-MS/MS). Bottom: Anion exchange elution profiles from second messenger synthesis assays using Ec CdnC, HORMA, DNA, and Trip13. Blue lines show absorbance at 254 nm, and red lines show conductivity for the 0.2-2.0 M ammonium acetate gradient elution. CdnC D72N/D74N contains aspartate-to-asparagine mutations in the putative active-site residues 72 and 74. See Figure S3A for assays with Ec CdnC and different DNAs, and Figure S3B for equivalent assays with Pa CdnD. (D) Liquid chromatography elution profile of the major product of Ec CdnC (peak 2 from panel C, sample v), with measured MS1 m/z and theoretical mass of cAAA. (E) MS2 fragmentation spectrum of the major product of Ec CdnC, annotated according to expected fragments of cAAA. The m/z of the extracted ion (corresponding to the [M+H] adduct of cAAA) was 988.1648, consistent with cAAA (monoisotopic mass = 987.1576 amu; molecular weight = 987.6263 amu). The minor product of Ec CdnC (peak 1 from panel C, sample v) was also analyzed by LC MS/MS and confirmed to be cAA (not shown). (F) Model for CdnC activation by HORMA and DNA binding, with negative regulation by Trip13.
As a first step, we performed yeast two-hybrid analysis with the E. coli MS115-1 (Ec) proteins, and found that Ec CdnC and Ec HORMA bind one another (Figure 1B and Figure S2A). When co-expressed in E. coli, Ec CdnC and Ec HORMA form two complexes with both 1:1 and 2:2 stoichiometry (Figure S2B). In a yeast three-hybrid experiment, a stabilized hexamer mutant of Ec Trip13 (Walker B motif residue Glu159 to Gln; Trip13EQ) binds to an Ec CdnC:HORMA complex (Figure 1B and Figure S2A), and in vitro reconstitution reveals that this ternary complex has a stoichiometry of 6 TRIP13EQ:1 CdnC:1 HORMA (Figure S2C). Similar yeast two-hybrid analysis with the P. aeruginosa ATCC27853 (Pa) proteins showed that Pa HORMA2 and HORMA3 interact with one another, and that both HORMA proteins interact with Pa CdnD (Figure S2E). In vitro reconstitutions reveal that these proteins form a CdnD:HORMA2:HORMA3 complex with 1:1:1 stoichiometry (Figure S2F–H). Finally, both Ec NucC and Pa NucC self-interact in yeast two-hybrid assays, suggesting that NucC forms homodimers or larger oligomers (Figure S2A and Figure S2E).
Both Ec CdnC and Pa CdnD were previously found to be inactive in vitro (Whiteley et al., 2019), leading us to test if their activity depends on their cognate HORMA and Trip13 proteins. For this purpose, we tested purified proteins in an in vitro second messenger synthesis assay based on prior work with a related CD-NTase, E. cloacae CdnD02 (Figure 1C) (Whiteley et al., 2019). We first tested for activity of Ec CdnC and the Ec CdnC:HORMA complex (both 1:1 and 2:2 complexes). As the related cGAS and OAS enzymes both require nucleic acid binding for activation (Hornung et al., 2014), we also included double-stranded DNA as a potential activator. We find that while CdnC is inactive on its own (Figure 1C, sample i), the 2:2 complex of Ec CdnC and HORMA synthesizes two phosphatase-resistant products in the presence of DNA (Figure 1C, sample v). Curiously, the 1:1 CdnC:HORMA complex is inactive (Figure 1C, sample iv). CdnC+HORMA is activated by long double-stranded DNAs, but not by single-stranded DNA or short (40 bp) double-stranded DNA (Figure S3A). CdnC activity is disrupted by mutation of the putative active site residues Asp72 and Asp74, demonstrating that CdnC uses a similar biosynthetic mechanism as related CD-NTases (Figure 1C, sample vi). Finally, we found that addition of sub-stoichiometric amounts of Ec Trip13 (1:60 molar ratio of active Trip13 hexamers to CdnC+HORMA) strongly reduced second messenger synthesis (Figure 1C, sample vii). Together, these data support a model in which Ec CdnC is activated by binding its cognate HORMA protein and DNA, and inactivated by Trip13, potentially through direct disassembly of the CdnC:HORMA complex (Figure 1F).
We next used liquid chromatography coupled to tandem mass spectrometry (LC MS/MS) to analyze the major and minor products of Ec CdnC. We identified the major product as cyclic tri-AMP (cAAA; Figure 1D–E), and the minor product as cyclic di-AMP (cAA; not shown). Ec CdnC synthesizes only cAAA/cAA even in the presence of both GTP and ATP, indicating that the enzyme uses only ATP as a substrate (not shown). Thus, Ec CdnC synthesizes a cyclic trinucleotide second messenger upon activation by its cognate HORMA domain protein and double-stranded DNA.
We next tested the activity of Pa CdnD in the presence of Pa HORMA2, Pa HORMA3, and Pa Trip13. While Pa CdnD shows lower activity than Ec CdnC in vitro and requires Mn2+ instead of Mg2+ as a cofactor, we find that CdnD also primarily synthesizes cAAA (Figure S3B). Pa CdnD requires HORMA2 for activity, but is not activated by HORMA3 (Figure S3B), perhaps reflecting functional specialization of the two HORMA domain proteins in this operon. As with the E. coli proteins, addition of Pa Trip13 to a reaction with Pa CdnD and HORMA2 strongly inhibits second messenger synthesis (Figure S3B). Finally, we find that in contrast to Ec CdnC, DNA is not required for Pa CdnD activity, nor does addition of DNA further stimulate cAAA production (Figure S3B). Thus, while Ec CdnC and Pa CdnD are regulated differently, the two enzymes share a common dependence on HORMA domain protein binding for activation, and generate the same cyclic trinucleotide second messenger.
Structures of HORMA-associated CD-NTases
To address the structural and biochemical mechanisms by which Ec CdnC and Pa CdnD synthesize cAAA in a regulated manner, we first purified and determined high-resolution crystal structures of both enzymes (Figure 2A–B). Both Ec CdnC and Pa CdnD adopt a polymerase β-like nucleotidyltransferase fold, with two lobes sandwiching a large active site. Ec CdnC and Pa CdnD adopt similar overall structures, with a 3.95 Å Cα r.m.s.d. (over 250 residues), and are more structurally related to one another than to either DncV (CD-NTase clade A) or CdnE, both of which synthesize cyclic dinucleotide second messengers (Figure 2C).
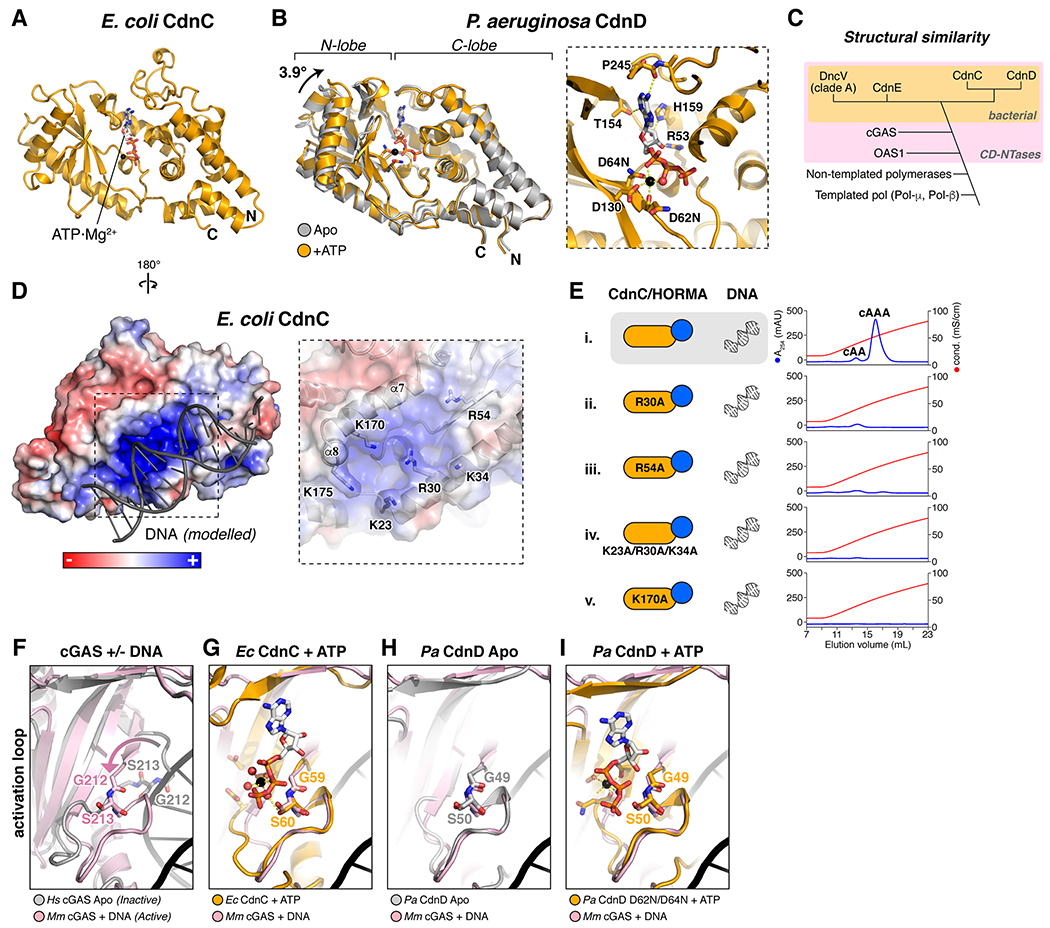
Figure 2. Structures of HORMA+Trip13-associated CD-NTases.
(A) Structure of Ec CdnC, with bound ATP·Mg2+ shown as sticks. (B) Overlay of Pa CdnD in the Apo state (gray) and bound to ATP (orange), with bound ATP·Mg2+ shown as sticks. Right: Closeup view of ATP·Mg2+ binding to Pa CdnD. See Figure S4A for ITC assays measuring nucleotide binding to Pa CdnD. (C) Schematic of structural similarity in Pol-β type nucleotidyltransferases. CD-NTases are shaded in pink, and bacterial CD-NTases are shaded in orange. (D) Reverse view of Ec CdnC, showing surface charge and DNA (gray) modelled from a superposition with M. musculus cGAS bound to DNA (PDB 4K9B) (Gao et al., 2013). For surface conservation of Ec CdnC, see Figure S4B. For surface conservation and charge of Pa CdnD, see Figure S4C–D. Closeup view (right) shows positively-charged residues, with helices α7 and α8 (equivalent to mammalian cGAS DNA-binding surface) labeled. (E) Anion exchange elution profiles from second messenger synthesis assays with wild-type Ec CdnC (sample i) and mutants to the putative DNA-binding surface (samples ii-v). (F) Overlay of inactive H. sapiens cGAS (gray; PDB ID 4O69 (Zhang et al., 2014)) with active, DNA-bound M. musculus cGAS (pink; PDB ID 4O6A (Zhang et al., 2014)) showing motion of the cGAS activation loop upon DNA binding. (G) Overlay of ATP-bound Ec CdnC (orange) with active cGAS (pink). (H) Overlay of Apo Pa CdnD (gray) with active cGAS (pink). (I) Overlay of ATP-bound Pa CdnD (orange) with active cGAS (pink).
In Ec CdnC, the active site is occupied by one molecule of ATP that was co-purified with the protein (Figure 2A); we were unable to remove this tightly-bound nucleotide. We could purify Pa CdnD without a bound nucleotide, and found by isothermal titration calorimetry that the enzyme binds strongly to ATP, but not to GTP (Figure S4A). These data agree with our findings that both Ec CdnC and Pa CdnD primarily synthesize cAAA. We determined structures of Pa CdnD in both Apo and ATP-bound states, and found that the two states differ by a 3.9° rotation of the enzyme’s N- and C-lobes with respect one another, with the ATP-bound state more “closed” than the Apo state (Figure 2B).
Our biochemical data show that Ec CdnC requires dsDNA for activation, similar to mammalian cGAS (Figure 1C). We modelled DNA binding to CdnC by overlaying our structure with a cGAS:DNA complex (Gao et al., 2013). In the resulting model, DNA contacts a surface of Ec CdnC that is strongly positively charged and highly conserved (Figure 2D and Figure S4B). To test the idea that this surface is responsible for DNA binding, we mutated several positively-charged residues on this surface of CdnC and measured both DNA binding and DNA-stimulated cAAA synthesis. The four mutants we tested had variable effects on DNA binding (Figure S4C), but all four eliminated cAAA synthesis by CdnC in the presence of HORMA+DNA (Figure 2E). These data support a role for this surface in DNA binding and activation of CdnC.
In mammalian cGAS, DNA binding to α-helices α7 and α8 restructures an “activation loop” in the enzyme’s active site to promote substrate binding and second messenger synthesis (Figure 2F) (Civril et al., 2013; Kato et al., 2013; Li et al., 2013; Zhang et al., 2014). In our structures of Ec CdnC and Pa CdnD, the conformation of the activation loop is equivalent to the activated state of cGAS (Figure 2G–I). Thus, while DNA binding is required for activation of Ec CdnC, the structural mechanism of activation is likely distinct from that of mammalian cGAS.
Pa CdnD does not require DNA for cAAA synthesis in vitro. Nonetheless, Pa CdnD shows a similar, albeit less pronounced, pattern of high conservation and positive charge on the surface equivalent to the cGAS DNA-binding surface (Figure S4D–E). We therefore tested DNA binding directly, but did not detect binding (Figure S4C). These data recall earlier findings with V. cholerae DncV, which possesses an equivalent conserved, positively charged surface, but whose synthesis of cGAMP is not stimulated by DNA (Kranzusch et al., 2014).
Bacterial HORMA domain proteins adopt canonical open and closed states
We next sought to determine whether the putative bacterial HORMA domain proteins share the canonical HORMA domain fold and mechanism of closure motif binding (Figure 3A). We first determined a structure of Pa HORMA2, and found that the protein adopts a canonical “closed” HORMA domain structure, with a central three-stranded β-sheet (β-strands 4, 5, and 6) backed by three α-helices (αA, B, and C) (Figure 3B). Characteristic of closed HORMA domains, the N-terminus of Pa HORMA2 is disordered, and the C-terminus wraps around the protein and forms a β-hairpin (β-strands 8′ and 8′′) against strand β5. Strikingly, the C-terminal safety belt of Pa HORMA2 embraces a peptide from the disordered N-terminus of a crystallographic symmetry-related molecule in the canonical closure motif binding site (Figure 3B). Thus, Pa HORMA2 represents a bona fide HORMA domain with a closure-motif binding C-terminal safety belt.
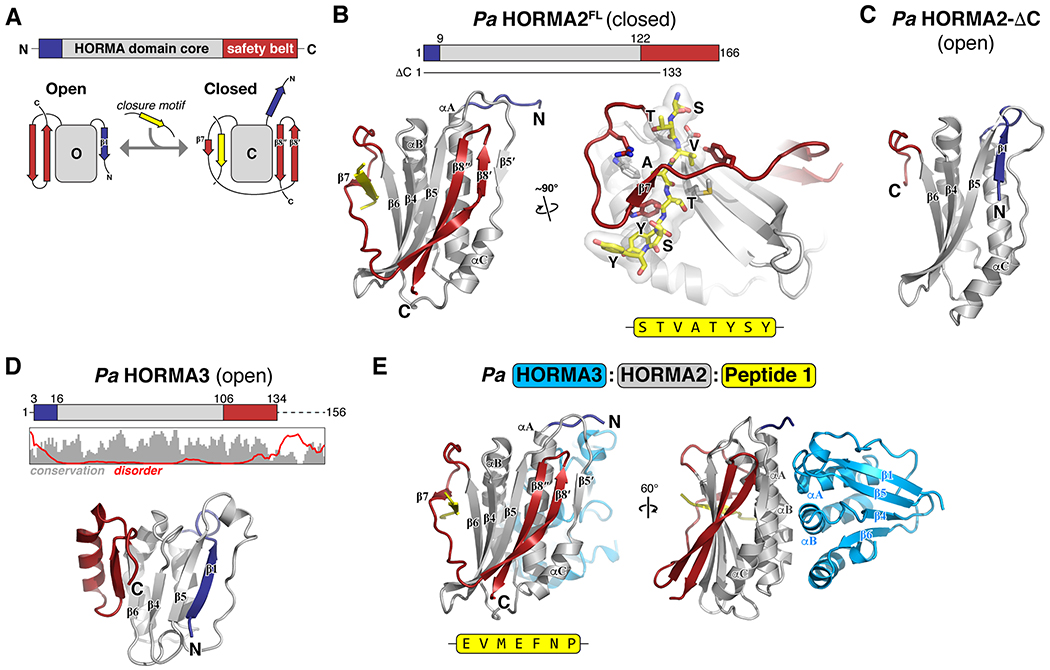
Figure 3. Structure and closure motif binding of bacterial HORMA proteins.
(A) Top: Schematic of HORMA domain primary structure, with the HORMA domain core gray, N-terminus blue, and C-terminal safety belt red. Bottom: Schematic of the open and closed states of the canonical HORMA domain protein MAD2. The transition from open to closed involves movement of both the N-terminus (blue) and C-terminal safety belt (red), and binding of a closure motif peptide (yellow) by the safety belt. (B) Left: Structure of Pa HORMA2 in the closed state, colored as in panel (A). Secondary-structure elements are labeled as in MAD2, except for strand β5′, which is not observed in other HORMA domain proteins. Right: Closeup view of Pa HORMA2 binding the extended N-terminus of a symmetry-related Pa HORMA2 molecule, which mimics a closure motif (yellow). (C) Structure of Pa HORMA2-ΔC (lacking residues 134–166) in the open state. (D) Top: Schematic of Pa HORMA3, with plot showing the Jalview alignment conservation score (3-point smoothed; gray) (Livingstone and Barton, 1993) and DISOPRED3 disorder propensity (red) for aligned bacterial HORMA3 proteins (Jones and Cozzetto, 2015). Bottom: Structure of Pa HORMA3 in the open state. (E) Structure of the Pa HORMA3:HORMA2:Peptide 1 complex. See Figure S5 for identification of closure motif sequences for Pa HORMA2 and Ec HORMA by phage display.
To identify preferred closure motif sequences for bacterial HORMA proteins, we performed phage display with purified Ec HORMA, Pa HORMA2, and Pa HORMA3, and a library of 7-mer peptides. We identified consensus binding motifs enriched in hydrophobic amino acids for both Pa HORMA2 and Ec HORMA (Figure S5A–B), and confirmed binding by coexpression and purification of peptide-HORMA complexes (Figure S5C–D). Thus, both Ec HORMA and Pa HORMA2 can likely adopt the closed conformation and bind specific closure motif peptides.
To determine whether Pa HORMA2 can also adopt an “open” HORMA domain state, we generated a truncated construct lacking the C-terminal safety belt (Pa HORMA2-ΔC). The crystal structure of Pa HORMA2-ΔC shows that in contrast to full-length Pa HORMA2, the N-terminus of Pa HORMA2-ΔC is well-ordered and packs as a β-strand (β1) against strand β5 (Figure 3C). This transition of the N-terminus from disordered to an ordered β1 strand is characteristic of open HORMA domains (Rosenberg and Corbett, 2015). Thus, bacterial HORMA domain proteins can adopt both the open and closed states, and bind closure motif peptides equivalently to their eukaryotic relatives.
We next determined a structure of Pa HORMA3, and found that this protein also adopts an open state with an ordered N-terminal β1 strand (Figure 3D). Based on the sequence of the Pa HORMA3 N-terminus and the predicted disorder of its poorly-conserved C-terminus (Figure 3D), we propose that this protein may be “locked” in the open state similarly to the eukaryotic HORMA domain protein Atg101 (Qi et al., 2015; Suzuki et al., 2015). Consistent with this idea, we were unable to identify Pa HORMA3-binding peptide sequences by phage display (not shown). Our data therefore suggests that in HORMA2/3 containing CBASS systems, HORMA2 can adopt the open and closed states and bind closure motif peptides, while HORMA3 adopts an open state and does not bind peptides.
We next determined a structure of a Pa HORMA3:HORMA2:peptide complex (Peptide 1: EVMEFNP). Strikingly, the Pa HORMA3:HORMA2 dimer bears no resemblance to dimers of eukaryotic HORMA domains, which assemble through their αC helices (Mapelli et al., 2007; Rosenberg and Corbett, 2015). Rather, Pa HORMA3 and Pa HORMA2 interact through both proteins’ αA and αB helices, plus the short α-helix in the C-terminal region of Pa HORMA3 (Figure 3E). The interface does not involve any elements of Pa HORMA2 that change conformation significantly between the open and closed states, and yeast two-hybrid analysis shows that the open Pa HORMA2-ΔC construct can also bind Pa HORMA3 (Figure S5E–G). Thus, while Pa HORMA3 apparently does not bind closure motif peptides or undergo conformational conversion, it can likely scaffold larger complexes by binding Pa HORMA2 and CdnD.
HORMA proteins bind their cognate CD-NTases specifically in the closed state
We next sought to understand how bacterial HORMA domain proteins regulate their cognate CD-NTases. We first purified and determined the structure of a complex containing Pa CdnD, HORMA2, and Peptide 1, and found that Pa HORMA2 binds the C-lobe of CdnD (Figure 4A). Compared to its Apo state, HORMA2-bound Pa CdnD shows a 6.6° rotation of the N-lobe toward the C-lobe, consistent with the idea that binding of HORMA2 may aid activation of CdnD. HORMA2 interacts with CdnD through its β8′ strand and αC helix (Figure 4B); as the β8′ strand does not form in the open state, this binding mode is likely dependent on HORMA2 adopting a closed conformation. Indeed, we found by yeast two-hybrid analysis that Pa HORMA2-ΔC, which adopts an open conformation, does not interact with Pa CdnD (Figure 4C). Consistent with our ability to reconstitute a CdnD:HORMA2:HORMA3 complex, we could model a structure of Pa HORMA2 bound to both CdnD and HORMA3 with no apparent clashes (Figure S6A).
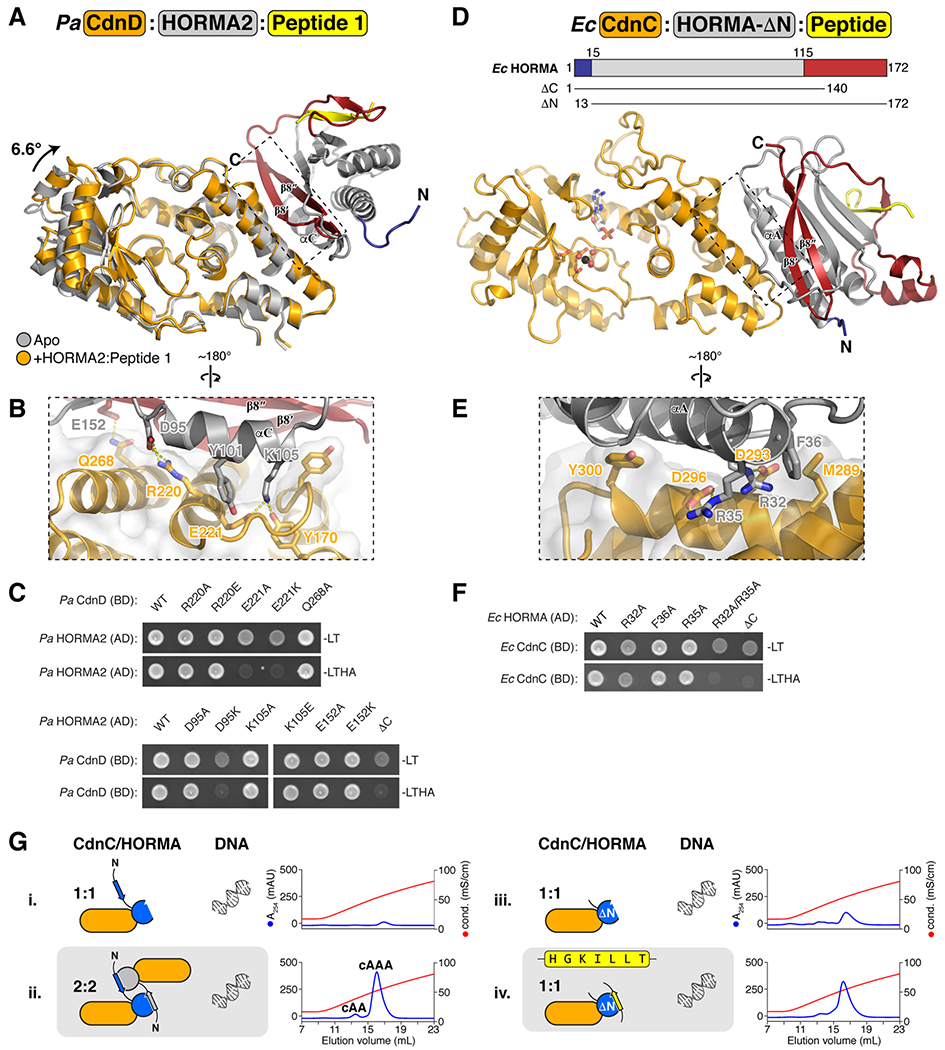
Figure 4. Structures of CD-NTase:HORMA complexes.
(A) Overall structure of the Pa CdnD:HORMA2:Peptide 1 complex, with CdnD orange, HORMA2 gray with safety-belt red and N-terminus blue, and Peptide 1 yellow. CdnD is overlaid with Apo CdnD (gray), showing the 6.6° N-lobe rotation in the HORMA2-bound state. (B) Closeup view of the Pa CdnD:HORMA2 interface. (C) Yeast two-hybrid analysis of the Pa CdnD-HORMA2 interaction. BD: fusion to the Gal4 DNA-binding domain; AD: fusion to the Gal4 activation domain. -LT: media lacking leucine and tryptophan (non-selective); -LTHA: media lacking leucine, tryptophan, histidine, and adenine (stringent selection). (D) Overall structure of the Ec CdnC:HORMAΔN complex, with CdnC orange, HORMA gray with safety-belt red and N-terminus blue, and bound peptide yellow (see Figure S6B). (E) Closeup view of the Ec CdnC:HORMA interface. (F) Yeast two-hybrid analysis of the Ec CdnC:HORMA interaction. (G) Anion exchange elution profiles from second messenger synthesis assays with separately-purified 1:1 (sample i) and 2:2 (sample ii) samples of the full-length Ec CdnC:HORMA complex (with intact His6-tag on HORMA, same as Figure 1C, samples iv and v.) and Ec CdnC:HORMA-ΔN with His6-tag removed, without addition of peptide (sample iii) or with addition of consensus HORMA-binding closure motif peptide HGKILLT (sample iv). See Figure S6D for size exclusion elution profiles of Ec CdnC:HORMA-ΔN. Residual activity in sample iii is likely due to incomplete dissociation of the cleaved His6-tag from HORMA-ΔN.
We next purified and determined the structure of an Ec CdnC:HORMA-ΔN complex, which lacks the disordered N-terminal 11 residues of Ec HORMA. In this complex, the structure of Ec HORMA closely resembles Pa HORMA2 in the closed state, with a well-ordered C-terminal safety belt forming a β-hairpin (β8′-8″) against strand β5 (Figure 4D). Notably, in this structure we find that Ec HORMA-ΔN is bound to a short peptide from the N-terminal His6-tag of a symmetry-related HORMA-ΔN molecule, resulting in a CdnC:HORMA-ΔN complex with 2:2 stoichiometry (Figure S6B). Unlike our structure of Ec CdnC alone, in which the protein is bound to a single ATP molecule co-purified from E. coli, the two copies of Ec CdnC in the CdnC:HORMA-ΔN structure contain either no ligand or AMP in the active site (Figure 4D). This suggests that in comparison to isolated Ec CdnC, the CdnC:HORMA complex may be more conformationally flexible and enzymatically active.
The interaction of Ec HORMA with Ec CdnC closely resembles that of Pa HORMA2 with Pa CdnD (Figure 4D). Ec HORMA binds the CdnC C-lobe, adjacent to CdnC’s putative DNA-binding surface. Ec HORMA binds CdnC through a similar interface as the one used by Pa HORMA2, binding CdnC largely through its αA helix (Figure 4D–E). Ec HORMA’s β8′ strand does not contact CdnC, yet truncation of the protein’s C-terminus (Ec HORMA-ΔC, lacking residues 141-172) disrupts CdnC binding, suggesting that the Ec HORMA-CdnC interaction likely also depends on the closed HORMA domain state (Figure 4F).
In our structure of CdnC:HORMA-ΔN, Ec HORMA binds a symmetry-related HORMA N-terminus as a short β-strand, but with an N-to-C polarity opposite that observed in our structure of Pa HORMA2 bound to Peptide 1 (and indeed opposite that of all known eukaryotic HORMA:closure motif complexes). We interpret this switched polarity of closure motif binding as an artifact of a non-biological complex assembly, rather than an indication that this HORMA domain binds closure motifs in an unique manner. We modelled binding of a consensus closure motif sequence identified by phage display (HGKILLT) with the canonical N-to-C polarity, revealing that the hydrophobic residues in the Ec HORMA-binding consensus would all fit into hydrophobic pockets on Ec HORMA (Figure S6C). This supports the idea that our phage display experiment identified sequences that bind Ec HORMA as a closure motif.
The observation of a 2:2 complex in the structure of Ec CdnC:HORMA-ΔN suggests that the 2:2 complex we observe when purifying Ec CdnC:HORMA may be assembled through reciprocal binding of HORMA N-termini to opposite HORMA protomers. To test this idea, we purified an Ec CdnC:HORMA-ΔN complex in which the N-terminal His6-tag was removed, and found that this complex forms only 1:1 complexes and is only minimally active (Figure 4G and Figure S6D). When we co-purified the same complex with a consensus closure motif peptide (HGKILLT), we found that it too forms a 1:1 CdnC:HORMA complex, but is now active in vitro (Figure 4G and Figure S6D). These data explain why in our earlier assays, only 2:2 CdnC:HORMA complexes were active for cAAA synthesis: these complexes’ HORMA proteins are bound to closure motif-mimicking peptides, while the HORMA proteins in 1:1 complexes are unbound. Thus, these data support the idea that activation of Ec CdnC requires a closure motif-bound HORMA protein.
Structure of a bacterial Trip13 engaged with its CD-NTase:HORMA substrate
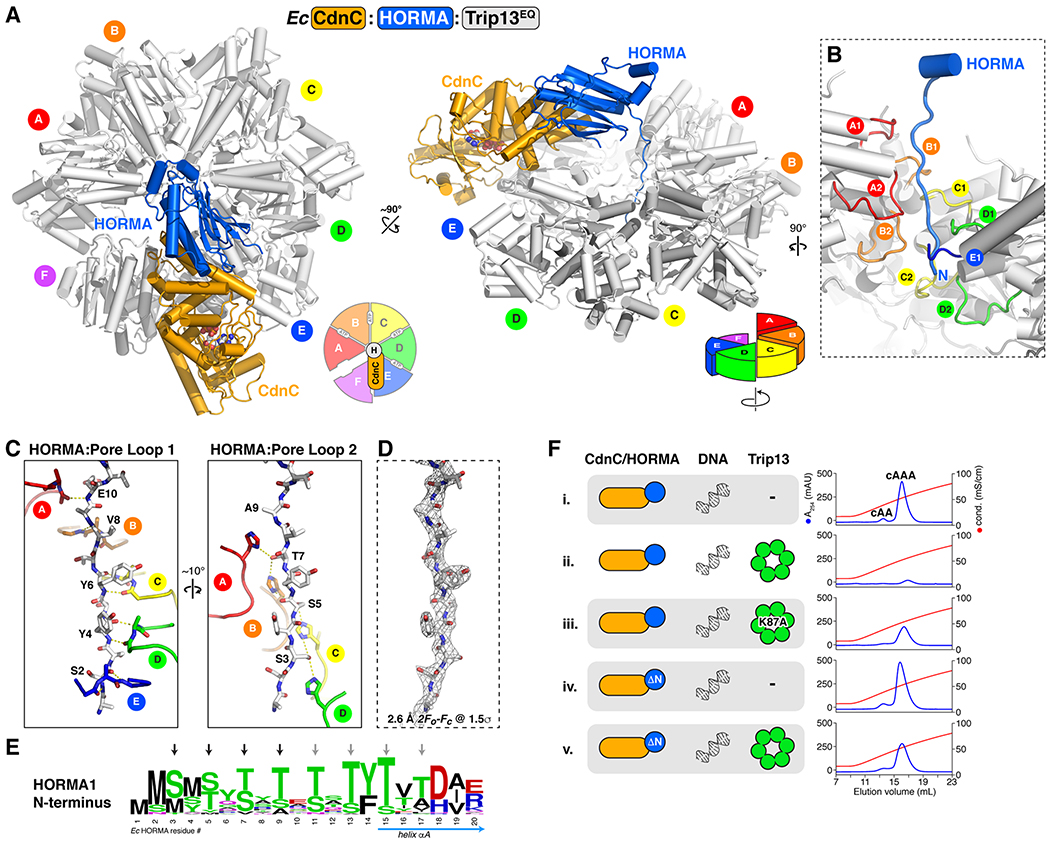
The Trip13-like ATPase encoded by bacterial HORMA-containing operons negatively regulates the production of cAAA by the HORMA-CD-NTase complex (Figure 1C). To address the structural mechanism of this activity, and to compare this mechanism to that of eukaryotic Pch2/TRIP3 in control of the eukaryotic HORMA domain protein Mad2, we determined the structures of both an isolated bacterial Trip13 hexamer (Figure S7A–D) and of an Ec CdnC:HORMA:Trip13EQ complex (Figure 5A; the Trip13 E159Q mutation stabilizes hexamer assembly and substrate interactions by disallowing ATP hydrolysis (Ye et al., 2015, 2017)). The crystal structure of Ec CdnC:HORMA:Trip13EQ reveals a mode of substrate binding and engagement for bacterial Trip13 that is similar to eukaryotic Pch2/TRIP13, with the high resolution of 2.6 Å providing unprecedented insight into substrate engagement and unfolding by AAA+ ATPases.
Figure 5. Structure of the Ec CdnC:HORMA:Trip13EQ complex.
(A) Top and side views of the Ec CdnC:HORMA:Trip13EQ complex, with CdnC orange, HORMA blue, and Trip13EQ white. See Figure S8 for structural analysis of Trip13EQ. Trip13 forms a spiral conformation, with its A subunit at top and F subunit at bottom (see schematics). CdnC binds the top surface of Trip13 subunits E and F (see Figure S8B for buried surface area). See Figure S6E–G for yeast two-hybrid analysis of the CdnC-Trip13 and HORMA-TRIP13 interfaces. (B) Closeup view of the Ec HORMA N-terminus (blue) engaged by Trip13 pore loops 1 (upper) and 2 (lower) from chains A-E. (C) Detail views of interactions between the Ec HORMA N-terminus (white sticks) with Trip13 pore loop 1 (left) and pore loop 2 (right). HORMA residues Ser4, Tyr4, Tyr6, Val8, and Glu10 form an extended backbone hydrogen-bonding network with Trip13 pore loop 1 residues Gly119 (main-chain carbonyl) and Val121 (main-chain amine) from monomers A-E (Trip13 Arg120 side-chains are not shown for clarity). HORMA residues Ser3, Ser5, Thr7, and Ala9 form an extended hydrogen-bond network with Trip13 pore loop 2 residue His173 from monomers A-D. (D) View equivalent to panel (C) showing 2Fo-Fc electron density for the HORMA N-terminus at 1.5 σ (2.6 Å resolution). (E) Sequence logo showing conservation of the N-termini of HORMA1 proteins. See Figure S7E–F for analysis of Trip13 pore loop 1 and 2 conservation, and equivalent logos of Trip13 and HORMA from two-HORMA operons. (F) Anion exchange elution profiles from second messenger synthesis assays with wild-type proteins (samples i-ii), Trip13 Walker A mutant K87A (sample iii), and HORMA-ΔN (missing N-terminal residues 1–12; samples iv-v). See Figure S3B for equivalent assays with Pa CdnD and Trip13 mutants.
A number of recent studies have reported moderate-resolution (3.2-5.0 Å) cryo-electron microscopy structures of AAA+ ATPase remodelers engaged with their substrates, including mammalian TRIP13 bound to a p31comet:Mad2 complex (Alfieri et al., 2018; Deville et al., 2017; Gates et al., 2017; Han et al., 2017; Puchades et al., 2017; Ripstein et al., 2017; White et al., 2018; Yu et al., 2018). These structures have revealed that active AAA+ remodelers adopt a right-handed spiral or “lock-washer” conformation, with 4-5 nucleotide-bound subunits forming a tight helical spiral and engaging an extended substrate peptide with their “pore loops”. In the case of TRIP13, the structures revealed that the “adapter” protein p31comet binds the top surface of the TRIP13 hexamer, thereby positioning the substrate Mad2 such that its extended N-terminus drapes into the hexamer pore for engagement and remodeling (Alfieri et al., 2018; Ye et al., 2017). In our structure of Ec CdnC:HORMA:Trip13EQ, the Ec Trip13EQ hexamer adopts a right-handed spiral conformation, with four subunits (monomers A-D) bound to ATP, and two subunit (monomers E and F) unbound (Figure S8A–B). The four ATP-bound subunits overlay closely, while monomers E and F show rotation of the large and small AAA subdomains with respect to one another (Figure S8C). Monomer F is positioned at the “bottom” of the spiral, and as such shows the most significant conformational differences compared to the other five subunits. Overall, the structure of the Ec Trip13EQ hexamer closely resembles that of other substrate-engaged AAA+ ATPases, including mammalian TRIP13 (Alfieri et al., 2018).
As noted above, eukaryotic TRIP13 recognizes its HORMA domain substrate Mad2 through the adapter protein p31comet (Alfieri et al., 2018; Ye et al., 2017). In the Ec CdnC:HORMA:Trip13EQ complex, CdnC serves as the adapter, binding the top face of the Trip13 hexamer at the interface of monomers E and F (Figure 5A). CdnC-Trip13 binding positions Ec HORMA over the Trip13 hexamer pore, with its extended N-terminus draping into the pore and interacting with the pore loops from Trip13 monomers A-E (Figure 5B). These pore loops form a tight spiral and engage the extended HORMA N-terminus with two-residue periodicity: Trip13 pore loop 1 residues Gly119 and Val121 in monomers A-E engage even-numbered residues from HORMA (Ser2, Tyr4, Tyr6, Val8, and Glu10) through main-chain hydrogen bonding (Figure 5C–D). At the same time, conserved histidine residues (His173) in pore loop 2 of Trip13 monomers A-D form a “ladder” with each histidine side-chain forming two hydrogen bonds with the side-chains of serine/threonine residues at odd-numbered positions in the HORMA N-terminus (Ser3, Ser5, and Thr7) (Figure 5C–D). In keeping with these highly specific interactions, we observe a striking pattern of conservation in the N-termini of HORMA1 proteins (which includes Ec HORMA), with conserved serine/threonine residues in every second position (Figure 5E). This conservation pattern extends to Ec HORMA residue 17, strongly suggesting that Ec Trip13 can undergo at least 3-4 cycles of ATP hydrolysis and translocation before disengaging, sufficient to partially unfold helix αA and disrupt both HORMA-CdnC binding (which primarily involves helix αA) and HORMA-closure motif binding (by destabilizing the C-terminal safety belt). HORMA2 proteins do not possess this pattern of conserved serine/threonine residues in their N-termini, and Trip13 proteins in HORMA2/3 operons lack a conserved pore loop 2 histidine residue (Figure S7E–F), suggesting a different mode of HORMA-Trip13 recognition in these operons. Thus, our structure of the Ec CdnC:HORMA:Trip13EQ complex supports a model in which Trip13 disassembles a CD-NTase:HORMA complex by remodeling the HORMA domain protein, and provides the highest-resolution view to date of a AAA+ ATPase hexamer engaged with its substrate.
Our biochemical data shows that addition of Trip13 inactivates second messenger synthesis by Ec CdnC+HORMA+DNA, with Trip13 effectively inhibiting second messenger synthesis at a 1:60 ratio of Trip13 hexamer to CdnC+HORMA (Figure 1C, sample vi). This data is consistent with the idea that Trip13 catalytically disassembles and inactivates the CdnC:HORMA complex, likely through engagement and unfolding of the HORMA N-terminus. In agreement with this idea, we found that Trip13 is unable to effectively inhibit second messenger synthesis when ATP binding is disrupted by a mutation to the Walker A motif (K87A; Figure 5F, sample iii). Further, Trip13 is unable to inhibit second messenger synthesis by an Ec CdnC:HORMA-ΔN complex, which lacks the extended N-terminus of Ec HORMA and thus cannot be engaged by Trip13 (Figure 5F, samples iv-v). These data support a model in which bacterial Trip13 enzymes attenuate signaling by their cognate CD-NTases by disassembling an active CD-NTase:HORMA complex.
NucC is a cAAA-activated DNA endonuclease
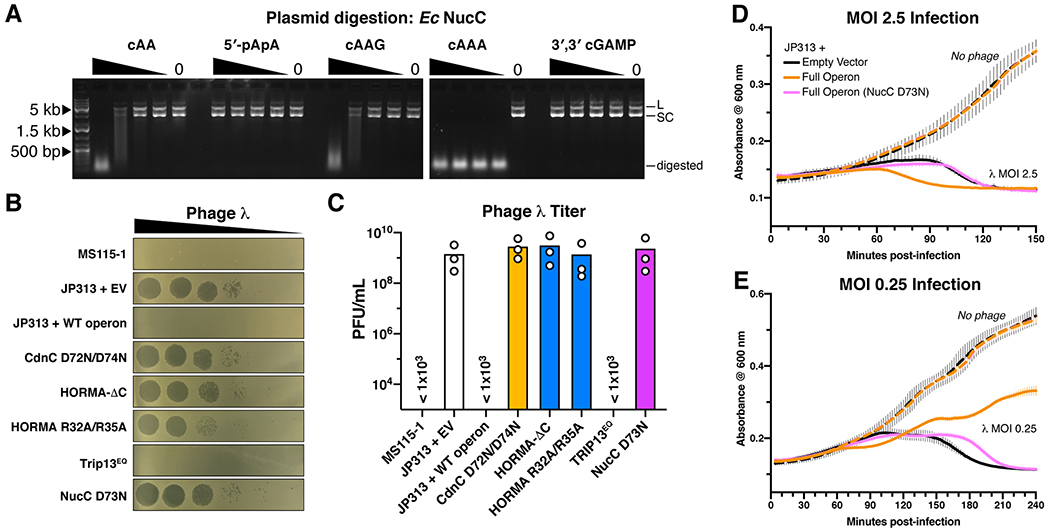
Bacterial CBASS systems encode one of several putative effector proteins, which include proteases, phospholipases, and at least three families of putative endo- and exonucleases (Burroughs et al., 2015). Both the E. coli MS115-1 and P. aeruginosa ATCC27853 systems encode a restriction endonuclease-related protein we term NucC (Nuclease, CD-NTase associated). We purified E. coli MS115-1 NucC and tested its ability to degrade purified plasmid DNA in the presence of second messengers including cyclic and linear di-AMP, and the cyclic trinucleotides cAAA and cAAG. We found that NucC degrades DNA to ~50-100 bp fragments in the presence of low-nanomolar concentrations of cAAA, and is also activated to a lesser extent by cyclic di-AMP and cAAG (Figure 6A). NucC is apparently insensitive to DNA methylation: the enzyme is equally active on unmethylated PCR product, plasmid purified from a K-12 based E. coli strain (NovaBlue), and plasmid purified from strain MS115-1, which harbors the CBASS system and several predicted DNA methylases not found in K-12 strains (Figure S9A). Thus, NucC is DNA endonuclease activated by cAAA, the second messenger product of its cognate CD-NTases.
Figure 6. The E. coli MS115-1 CBASS system confers bacteriophage immunity through activation of a DNA endonuclease.
(A) Plasmid digestion assay with Ec NucC (10 nM) and the indicated second messenger molecules at 400/100/25/6.25/0 nM. L: linear plasmid, SC: supercoiled plasmid. (B) Dilution of phage λ on E. coli MS115-1 (top) and JP313 (wild-type laboratory strain) with the plasmid-encoded E. coli MS115-1 CBASS system with wild-type proteins (WT) or the indicated mutations. EV: empty vector. Six 10-fold phage dilutions are shown. (C) Quantitation of phage λ infectivity. Shown is the average +/− standard deviation of three trials at a single bacteriophage dilution. The three strains marked “< 1x103“ showed no plaques with the highest-tested bacteriophage concentration. Bars represent mean of three individual measurements (white circles). (D) Growth curves for E. coli JP313 transformed with the indicated plasmids and infected at 0 minutes with bacteriophage λ at an MOI of 2.45 (solid lines) or without phage addition (dashed lines). Lines represent the mean of four replicates, and error bars indicate standard deviation. (E) Growth curves of cell cultures infected at an MOI of 0.245.
The E. coli MS115-1 CBASS system confers bacteriophage immunity through an abortive infection mechanism
Recent work has demonstrated that two CBASS systems with different architectures provide immunity against a range of bacteriophages, acting through an abortive infection mechanism to kill cells prior to the completion of phage replication (Cohen et al., 2019). Our structural and biochemical data suggest that in HORMA+Trip13 containing CBASS systems, HORMA domain proteins recognize specific peptides from a bacteriophage, then bind and activate their cognate CD-NTase to produce cAAA, which in turn activates a defensive response by the effector nuclease NucC. We tested the resistance of E. coli MS115-1 to bacteriophage λcI- (lacking the lysogeny gene cI), and found that in contrast to a laboratory strain of E. coli (JP313; (Economou et al., 1995)), E. coli MS115-1 is immune to bacteriophage λcI- infection as judged by plaque formation (Figure 6B–C). We next inserted the entire E. coli MS115-1 operon (including CdnC, HORMA, Trip13, and NucC) into E. coli JP313 on a plasmid vector, and found that the operon confers robust immunity to bacteriophage λcI- (Figure 6B–C). Immunity was compromised when we disrupted second messenger synthesis by CdnC (CdnC D72N/D74N mutant), HORMA-CdnC binding (HORMA-ΔC or R32A/R35A mutants), or NucC’s DNA cleavage activity (NucC D73N mutant; based on sequence alignments with type II restriction endonucleases), demonstrating that immunity depends on both the sensor and effector modules in this pathway (Figure 6B–C). Disruption of Trip13’s ATP hydrolysis activity (Trip13EQ mutant) did not compromise immunity, consistent with the idea that Trip13 is a negative regulator of this pathway (Figure 6B–C).
To determine whether the E. coli MS115-1 CBASS system functions by abortive infection, we tracked the growth of bacterial cultures infected at high multiplicity of infection (MOI) with bacteriophage λcI-. In cells lacking the CBASS operon infected at an MOI of 2.5, we observed complete culture collapse beginning ~90 minutes post-infection due to phage-induced cell lysis (Figure 6D). In contrast, cells harboring the CBASS operon showed culture collapse beginning ~60 minutes post-infection, fully 30 minutes prior to phage-induced cell lysis. At an MOI of 0.25, we observed a sharp drop in growth rate at ~60 minutes post-infection in CBASS-harboring cells, followed by recovery of growth, while cells lacking CBASS showed culture collapse beginning at ~90 minutes post-infection (Figure 6E). Introduction of the NucC D73N active-site mutation eliminated these effects (Figure 6D–E). Together, these data support the idea that the E. coli MS115-1 CBASS system functions by abortive infection, causing the death of infected cells prior to the completion of phage replication. The requirement for NucC further suggests that cell death is caused by NucC-mediated host cell genome destruction.
Discussion
The recent discovery and classification of a diverse family of cGAS/DncV-like nucleotidyltransferases (CD-NTases) that are sparsely distributed among important environmental and pathogenic bacteria raised many important questions about these proteins’ molecular mechanisms and the biological functions of their associated operons (Burroughs et al., 2015; Cohen et al., 2019; Whiteley et al., 2019). Two such operons have been shown to confer immunity against bacteriophage infection, acting through an abortive infection mechanism in which infected cells die prior to completion of the bacteriophage replication cycle, and are now termed CBASS (Cyclic oligonucleotide-Based Anti-phage Signaling System) (Cohen et al., 2019). Here, we address the molecular mechanisms of CBASS defense systems and in particular define the mechanisms of a CBASS family encoding eukaryotic-like HORMA domain proteins and Trip13-like ATPase regulators. We demonstrate CD-NTase activation by binding of a HORMA-closure motif complex, identify the second messenger product of these enzymes, and show that second messenger production activates the effector nuclease NucC. Together, these proteins constitute a pathway that confers robust bacteriophage immunity onto its bacterial host through an abortive infection mechanism.
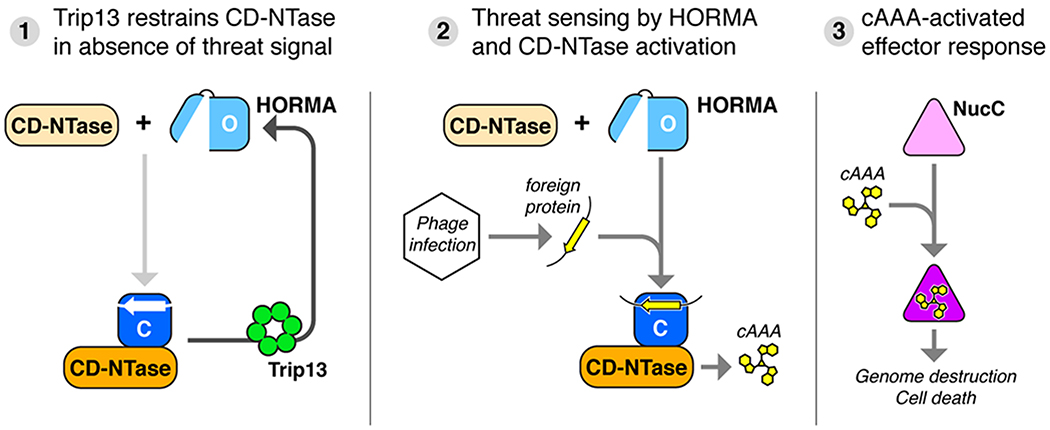
Our data support a model for HORMA/Trip13 regulation of CD-NTase activity in bacteria that is remarkably reminiscent of the roles of MAD2 and Pch2/TRIP13 in the eukaryotic spindle assembly checkpoint. In this checkpoint pathway, the HORMA domain protein MAD2 is kept in its inactive open state through constant conformational conversion by TRIP13 (Kim et al., 2018; Ma and Poon, 2016, 2018). Upon entrance into mitosis, large-scale assembly of MAD2 into a “mitotic checkpoint complex” with the closure motif-bearing protein CDC20 overwhelms the disassembly capacity of TRIP13, resulting in checkpoint activation. After successful kinetochore-microtubule attachment and cessation of mitotic checkpoint complex assembly, the checkpoint is inactivated as existing complexes are disassembled by TRIP13 and an alternative pathway (Kim et al., 2018). In a similar manner, we propose that in the absence of a foreign threat, bacterial HORMA domain proteins are maintained in the inactive “open” state by continual activity of Trip13 (Figure 7, step 1), thereby inhibiting CD-NTase activation and second messenger synthesis. Maintenance of the HORMA proteins in the open state by Trip13 primes them for binding to specific bacteriophage proteins upon infection or lysogenic induction. Following binding and conversion to the closed state, the HORMA domain protein binds and activates its cognate CD-NTase to synthesize cAAA (Figure 7, step 2). cAAA in turn activates the effector nuclease NucC, which indiscriminately destroys both the invading phage genome and the bacterial host genome, resulting in cell death and abortive infection (Figure 7, step 3). In related work, we demonstrate how NucC is allosterically activated by cAAA binding, and confirm that NucC destroys cellular DNA in infected cells to cause cell death (Lau et al., 2019).
Figure 7. Model for bacteriophage sensing and immunity by CD-NTase+HORMA+Trip13 operons.
(1) In the absence of a bacteriophage threat, Trip13 maintains HORMA proteins in the open state (light blue) by disassembling complexes of closed-HORMA (dark blue) + CdnC, thereby restraining CD-NTase activation. (2) Upon infection, open HORMA proteins recognize closure motif sequences in foreign proteins (yellow), convert to the closed state, then bind their cognate CD-NTase (orange). The CD-NTase:HORMA complex (which further requires bound DNA in some cases; not shown) synthesizes a nucleotide-based second messenger (cAAA). (3) Second messengers bind and activate effector proteins including NucC (Lau et al., 2019), causing cell death and abortive infection.
While we demonstrate the structural mechanisms of bacterial HORMA domain protein-mediated CD-NTase activation, important questions remain regarding these proteins’ evolution and function. First, it will be important to identify the closure motif-containing proteins recognized by bacterial HORMA domain proteins. The consensus closure motif sequences we identified by phage display do not allow clear identification of likely targets by sequence searches, so more direct tests will be required to identify these targets. The operon’s utility to bacterial survival would be maximized if the HORMA proteins were to recognize a widely-conserved bacteriophage protein, rather than a protein that is only found in a few bacteriophages. It would also be beneficial to recognize phage infection as early as possible, potentially by recognizing initial invasion through tail spike detection, or by recognizing a protein that is transcribed and translated early in the phage infection cycle.
A key question is the evolutionary origin of bacterial HORMA and Trip13 proteins, and their relationship with eukaryotic HORMA domain proteins and Pch2/TRIP13 ATPases. A recent bioinformatics analysis of bacterial and eukaryotic HORMA proteins showed that the bacterial proteins fall into two major families (HORMA1 and HORMA2), and that these two families co-occur with diverged Trip13 ATPases (Tromer et al., 2019). This study also showed that eukaryotic HORMA domain proteins – including Mad2, p31comet, Rev7, Atg13/101, and the meiotic HORMAD proteins – form a monophyletic group within the bacterial HORMA2 family, and that eukaryotic Pch2/TRIP13 ATPases form a monophyletic group within HORMA2-associated bacterial Trip13 proteins (Figure S8G) (Tromer et al., 2019). These findings indicate that HORMA domain proteins and Trip13 ATPases originated in bacteria, and their presence in CBASS systems indicates an ancestral function in protein sensing in bacteriophage defense.
Our structural data have enabled us to identify a third family of bacterial HORMA protein, which we term HORMA3, that is found in CBASS systems alongside HORMA2 proteins. We find that these proteins are likely “locked” in an open state, and form heterodimeric complexes with their cognate HORMA2 proteins. As HORMA2 binds closure motif peptides and can adopt both open and closed conformations, this protein likely functions as the peptide sensor in HORMA2/3 operons. The biological role of HORMA3 remains unknown, but it may function either to stabilize HORMA2 or scaffold larger signaling complexes.
Well-characterized CD-NTases including bacterial DncV and mammalian cGAS catalyze cyclic dinucleotide synthesis by binding two nucleotides, and catalyzing nucleophilic attack of each nucleotide’s 2′ or 3′ ribose oxygen by the other nucleotide’s α-phosphate. OAS proteins, on the other hand, synthesize linear 2′−5′ linked oligoadenylates through a similar mechanism that likely involves a single nucleophilic attack, followed by translocation and binding/attack by another ATP (Lohöfener et al., 2015). In this context, a key unanswered question is how bacterial CD-NTases in clades C and D – including Ec CdnC, Pa CdnD, and E. cloacae CdnD02 (Whiteley et al., 2019) – catalyze specific synthesis of cyclic trinucleotide second messengers. The enzymes’ active sites likely cannot accommodate three nucleotides, necessitating translocation of a linear intermediate during synthesis prior to the final cyclization reaction. Both Ec CdnC and Pa CdnD synthesize cAA as a minor product, suggesting that the enzymes can cyclize both 2-base or 3-base linear intermediates. Additional studies will be required to understand the mechanism and specificity of this reaction.
Here, we have defined the molecular mechanisms and biological role of a class of bacterial CBASS systems with associated HORMA and Trip13-like regulators and the nuclease effector NucC. Bacterial CBASS systems are extraordinarily diverse, however, with multiple regulatory/sensor systems including the HORMA/Trip13 systems described here and a putative ubiquitin-conjugation system, and a variety of putative effectors including nucleases, proteases, and phospholipases (Burroughs et al., 2015). While we have demonstrated that HORMA+TRIP13-containing CBASS systems likely sense phage proteins through their HORMA domain proteins, how other CBASS systems sense a bacteriophage infection, and whether all CBASS systems act via abortive infection remain key open questions. Finally, more work will be required to determine whether all CBASS systems function in bacteriophage defense or whether some have evolved other functions, protecting bacteria against other threats, contributing to homeostasis of bacterial communities, or even contributing to pathogenicity in unanticipated ways.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kevin D. Corbett (kcorbett@ucsd.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
All proteins were produced in E. coli strain Rosetta2 pLysS (EMD Millipore). Cells were grown in standard media (2XYT broth) with appropriate antibiotics, and standard temperatures (37°C for growth, 20°C for protein expression induction). For cellular assays with E. coli MS115-1 (BEI Resources #HM-344) and JP313 (Economou et al., 1995), cells were grown in LB broth at 30°C with appropriate antibiotics.
METHOD DETAILS
Yeast Two-hybrid
Coding sequences for genes from operon 300414846 from E. coli MS 115-1 (CD-NTase018 from (Whiteley et al., 2019)) were synthesized (GeneArt) and inserted into vectors pBridge and pGADT7 (Clontech) by isothermal assembly. Coding sequence for genes from P. aeruginosa strain ATCC27853 (CD-NTase023 from (Whiteley et al., 2019)) were amplified from genomic DNA (American Type Culture Collection) and inserted into vectors pBridge and pGADT7 by isothermal assembly. See Table S4 for all protein sequences. pBridge vectors were transformed into S. cerevisiae strain AH109 and selected on SC media lacking tryptophan (-TRP). pGADT7 AD vectors were transformed into S. cerevisiae strain Y187 and selected on SC media lacking leucine (-LEU). Haploid strains were mated and diploids selected on SC -TRP/-LEU. Diploid cells were diluted in water and replated onto SC -TRP/-LEU (control), -TRP/-LEU/-HIS (histidine) (low stringency), and -TRP/-LEU/-HIS/-ADE (adenine) (high stringency), incubated for 2-3 days at 30°C, then examined for growth.
For yeast three-hybrid assays, pBridge vectors containing one protein in MCS I were further modified by NotI cleavage at the MCS II site followed by isothermal assembly-mediated insertion of a second gene, resulting in a single vector encoding two genes, with the Gal4-BD tag fused to the N-terminus of the gene in MCS I. These vectors were transformed into AH109 and mated with pGADT7 AD vectors encoding other proteins.
Protein Expression, Purification, and Characterization
All proteins were cloned into UC Berkeley Macrolab vectors 2AT (for untagged expression) or 2BT (encoding an N-terminal TEV protease-cleavable His6-tag). Co-expression cassettes were assembled by amplifying genes from these vectors by PCR, and re-inserting into vector 2BT so that one protein is tagged. For coexpression of HORMA proteins with closure motif peptides, the peptide (Peptide 1: EVMEFNP; Peptide 2: HGKILLT) was cloned into UC Berkeley Macrolab vector 2CT (encoding an N-terminal TEV protease-cleavable His6-maltose binding protein tag), then assembled into co-expression vectors with Ec HORMA or Pa HORMA2 by PCR. All point-mutants were generated by PCR-mediated mutagenesis.
Proteins were expressed in E. coli strain Rosetta 2 (DE3) pLysS (EMD Millipore, Billerica MA). Cultures were grown at 37°C to A600=0.5, then induced with 0.25 mM IPTG and shifted to 20°C for 15 hours. Cells were harvested by centrifugation and resuspended in buffer A (20 mM Tris pH 7.5 (Tris pH 8.5 for CdnC/CdnD), 10% glycerol) plus 400 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol. Proteins were purified by Ni2+-affinity (Ni-NTA agarose, Qiagen) then passed over an anion-exchange column (Hitrap Q HP, GE Life Sciences, Piscataway NJ) in Buffer A plus 5 mM β-mercaptoethanol and 0.1-1 M NaCl, collecting flow-through or peak fractions. Tags were cleaved with TEV protease (Tropea et al., 2009), and cleaved protein was passed over another Ni2+ column (collecting flow-through fractions) to remove uncleaved protein, cleaved tags, and tagged TEV protease. The protein was passed over a size exclusion column (Superdex 200, GE Life Sciences) in buffer GF (buffer A plus 300 mM NaCl and 1 mM dithiothreitol (DTT)), then concentrated by ultrafiltration (Amicon Ultra, EMD Millipore) to 10-20 mg/ml and stored at 4°C. For selenomethionine derivatization, protein expression was carried out in M9 minimal media supplemented with amino acids plus selenomethionine prior to IPTG induction (Van Duyne et al., 1993), and proteins were exchanged into buffer containing 1 mM tris(2-carboxyethyl)phosphine (TCEP) after purification to maintain the selenomethionine residues in the reduced state.
For characterization of oligomeric state by size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS), 100 μL of purified protein/complex at 2–5 mg/mL was injected onto a Superdex 200 Increase 10/300 GL or Superose 6 Increase 10/300 GL column (GE Life Sciences) in a buffer containing 20 mM HEPES pH 7.5, 300 mM NaCl, 5% glycerol, and 1 mM DTT. Light scattering and refractive index profiles were collected by miniDAWN TREOS and Optilab T-rEX detectors (Wyatt Technology), respectively, and molecular weight was calculated using ASTRA v. 6 software (Wyatt Technology).
Crystallization and structure determination
E. coli CdnC
We obtained crystals of Ec CdnC using two different methods. First, small-scale dialysis of Ec CdnC at 10 mg/mL from buffer GF to a buffer containing 0% glycerol and 200 mM NaCl resulted in the formation of large prism crystals. Second, mixing CdnC (10 mg/mL in buffer GF) 1:1 with a well solution containing 20 mM HEPES pH 8.1, 10-12% PEG 4000, 100 mM NaCl, 10 mM MgCl2 (plus 1 mM TCEP for selenomethionine-derivatized protein) in hanging-drop format resulted in smaller prism crystals. All crystals were cryoprotected by the addition of 2-Methyl-2,4-pentanediol (MPD; 30% for dialysis crystals, 15% for hanging-drop), and flash-frozen in liquid nitrogen. We collected diffraction data at the Advanced Photon Source NE-CAT beamline 24ID-E (support statement below), and all datasets were processed with the RAPD data-processing pipeline, which uses XDS (Kabsch, 2010) for data indexing and reduction, AIMLESS (Evans and Murshudov, 2013) for scaling, and TRUNCATE (Winn et al., 2011) for conversion to structure factors. We determined the structure by single-wavelength anomalous diffraction methods using a 1.92 Å dataset from selenomethionine-derivatized protein, in the PHENIX Autosol wizard (Terwilliger et al., 2009). We manually rebuilt the initial model in COOT (Emsley et al., 2010), and refined against a 1.44 Å native dataset in phenix.refine (Afonine et al., 2012) using positional and individual anisotropic B-factor refinement (Table S3). Since ATP was not added to Ec CdnC purification or crystallization buffers, we conclude that ATP observed in the Ec CdnC active site was co-purified with the enzyme.
P. aeruginosa CdnD
Crystals of wild-type Pa CdnD (Apo state) were obtained by mixing protein (20 mg/mL) in crystallization buffer (25 mM Tris-HCl pH 7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM TCEP) 1:1 with well solution containing 0.1 M HEPES pH 7.5 (or imidazole pH 8.0) and 1.8-2.0 M Ammonium sulfate in hanging-drop format. Crystals were cryoprotected by the addition of 30% glycerol, and flash-frozen in liquid nitrogen. Diffraction data were collected at the Advanced Photon Source beamline 24ID-E and processed with the RAPD data-processing pipeline. Crystals of Pa CdnD D62N/D64N (ATP-bound) were obtained by mixing protein (20 mg/mL) in crystallization buffer plus 10 mM MgCl2 and 5 mM ATP 1:1 with well solution containing 0.1 M Tris pH 8.5, 0.2 M sodium formate, and 20-22% PEG 6000 in hanging drop format. Crystals were cryoprotected by the addition of 10% glycerol and flash-frozen in liquid nitrogen. Diffraction data were collected at the Advanced Photon Source beamline 24ID-E and processed with the RAPD data-processing pipeline.
We determined the structure of Apo Pa CdnD using a 2.05 Å-resolution single-wavelength dataset from a crystal of selenomethionine-derivatized protein, using the PHENIX Autosol wizard. We manually rebuilt the initial model in COOT, followed by refinement in phenix.refine using positional and individual B-factor refinement. The structure of ATP-bound Pa CdnD D62N/D64N was determined by molecular replacement in PHASER (McCoy et al., 2007).
P. aeruginosa HORMA3
Crystals of Pa HORMA3 were obtained by mixing protein (10 mg/mL) in crystallization buffer 1:1 with well solution containing 100 mM imidazole pH 8.0, 200 mM CaCl2, and 32% PEG 3350 in hanging-drop format. Crystals were flash-frozen in liquid nitrogen directly from the crystallization drop. We collected diffraction data at the Advanced Photon Source NE-CAT beamline 24ID-C, and all datasets were processed with the RAPD pipeline. In order to obtain phases by selenomethionine derivatization, we designed several mutants with conserved hydrophobic residues mutated to methionine. One mutant, V102M/L146M, crystallized more robustly than wild-type protein and was used for all later crystallographic analysis. We determined the structure of Pa HORMA3 V102M/L146M from a 1.86 Å resolution dataset from crystals derivatized with NaBr (1 minute soak in cryoprotectant solution containing 1 M NaBr). Data was processed with the RAPD pipeline, Br sites were identified with hkl2map (Pape et al., 2004; Sheldrick, 2010), and the structure was determined using the phenix AUTOSOL wizard. The initial models was manually rebuilt with COOT and refined against a 1.64 Å resolution native dataset using phenix.refine.
P. aeruginosa HORMA2
We obtained crystals of Pa HORMA2 by mixing protein (10 mg/mL) in crystallization buffer 1:1 in hanging drop format with well solution containing 100 mM Imidazole pH 8.0, 0.8 M NaH2PO4, and 0.55-0.8 M KH2PO4. Crystals were cryoprotected with an additional 30% glycerol and flash-frozen in liquid nitrogen. Native data (to 2.1 Å resolution) were collected at APS beamline 24ID-C, and a single-wavelength anomalous diffraction dataset (to 2.5 Å resolution) on a crystal grown from selenomethionine-derivatized protein was collected at SSRL beamline 9-2. Data were processed by the RAPD pipeline (native) or the autoxds pipeline (selenomethionine). The structure was determined by SAD phasing using the PHENIX AutoSol wizard, models were manually rebuilt with COOT, and refined against the native dataset using phenix.refine.
P. aeruginosa HORMA2-ΔC
We purified Pa HORMA2-ΔC (residues 2-133) as above (replacing Tris-HCl with HEPES pH 8.0 in all buffers), then dimethylated surface lysine residues by mixing protein at 1 mg/mL with 50 mM freshly-prepared dimethylamine borane complex and 100 mM formaldehyde, incubating at 4°C for one hour, then quenching by addition of 25 mM glycine. Dimethylated Pa HORMA2-ΔC was then re-concentrated to 17 mg/mL and buffer-exchanged into GF buffer to remove residual reaction components.
We obtained crystals of dimethylated Pa HORMA2-ΔC by mixing mixed protein (17 mg/mL) in crystallization buffer 1:1 in hanging drop format with well solution containing 100 mM Tris-HCl pH 8.5, 10 mM NiCl2, and 20% PEG 2000 MME. Crystals were cryoprotected with an additional 10% glycerol or 10% PEG 400, then looped and frozen in liquid nitrogen. Diffraction data were collected at APS beamline 24ID-C and processed by the RAPD pipeline. We determined the structure by molecular replacement using the structure of Pa HORMA2 as a search model (the successful model lacked the C-terminal safety belt region and the N-terminus, which both undergo conformational changes between the open and closed states. Initial models were manually rebuilt with COOT and refined using phenix.refine.
P. aeruginosa HORMA3:HORMA2:Peptide 1
A complex of Pa HORMA3 (residues 1-133), HORMA2, and Peptide 1 (EVMEFNP) was obtained by co-expressing and purifying His6-MBP-tagged Peptide 1 with untagged HORMA3 and HORMA2, with the tag cleaved prior to crystallization. Crystals were obtained by mixing protein (10 mg/mL) in crystallization buffer 1:1 in hanging drop format with well solution containing 100 mM HEPES pH 7.5, 0.5 M sodium acetate, 31% PEG 3350, and 1 mM ATP. Crystals were flash-frozen in liquid nitrogen directly from the crystallization drop. We collected a 2.0 Å diffraction dataset on SSRL beamline 9-2, processed the data using the autoxds pipeline, and determined the structure by molecular replacement with PHASER, using the structures of Pa HORMA3 and Pa HORMA2. Initial models were manually rebuilt with COOT and refined using phenix.refine.
Rhizobiales sp. Trip13
Rh Trip13 was purified as above, except all purification buffers contained 5 mM MgCl2. We obtained crystals of selenomethionine-derivatized Rh Trip13 by mixing protein (24 mg/mL) in crystallization buffer 1:1 in hanging drop format with well solution containing 100 mM sodium citrate pH 6.0, 1.6 M NH2SO4, and 0.2 M sodium/potassium tartrate. Crystals were cryoprotected with 30% glycerol, and flash-frozen in liquid nitrogen. We collected a 2.05 Å-resolution single-wavelength anomalous diffraction dataset on APS beamline 24ID-E, processed the data with the RAPD pipeline, and determined the structure using the PHENIX Autosol wizard. Initial models were manually rebuilt with COOT and refined using phenix.refine. The structure of Rh Trip13 is shown in Figure S7.
P. aeruginosa HORMA2:Peptide 1:CdnD
We obtained crystals of Pa HORMA2:Peptide 1:CdnD by mixing purified complex (15 mg/mL) in crystallization buffer 1:1 in hanging drop format with well solution containing 100 mM Bis-Tris pH 5.5, 0.2 M ammonium acetate, and 30% PEG 3350. Crystals were flash-frozen in liquid nitrogen directly from the crystallization drop. Diffraction data were collected at APS beamline 24ID-C, and processed by the RAPD pipeline. The structure was determined by molecular replacement in PHASER using the structures of Pa HORMA2 and CdnD as search models. Initial models were manually rebuilt with COOT, and refined using phenix.refine.
E. coli HORMA:CdnC:Trip13EQ
To assemble a HORMA:CdnC:Trip13EQ complex, we separately purified Ec HORMA:CdnC (coexpressed, with an N-terminal His6-tag on HORMA) and Ec Trip13EQ (E159Q), then mixed at a ratio of one 1:1 HORMA:CdnC complex per Trip13EQ hexamer in buffer GF plus 1 mM ATP and 5 mM MgCl2 at 4°C. After a one-hour incubation, the mixture was passed over a Superdex 200 size exclusion column (GE Life Sciences), and assembled complexes were collected and concentrated by ultrafiltration. Crystals were obtained by mixing protein (10 mg/mL) 1:1 with well solution containing 0.1 M MOPS pH 7.0, 0.2 M MgCl2, and 14% PEG 3350 in hanging-drop format. We collected a 2.64 Å resolution dataset at APS beamline 24ID-E, and processed the data using the RAPD pipeline. Crystals adopt space group P21, with one 1 HORMA, 1 CdnC, and 6 Trip13EQ monomers per asymmetric unit. We determined the structure by molecular replacement in PHASER, using the structure of Ec CdnC and a Trip13 hexamer model built by separately overlaying the large and small AAA subdomains of six Rh Trip13 monomers onto a recent cryo-EM structure of human TRIP13 bound to p31comet:MAD2 (Alfieri et al., 2018). A model of Pa HORMA2 was used as a guide to manually build Ec HORMA into difference density obtained from initial refinement. Initial models were manually rebuilt with COOT and refined using phenix.refine.
E. coli CdnC:HORMA-ΔN
We obtained crystals of Ec CdnC:HORMA-ΔN by mixing protein (12 mg/mL) in crystallization buffer 1:1 in hanging drop format with well solution containing 0.1 M CHES pH 9.5, 0.1 M magnesium acetate, and 17% PEG 3350. Crystals were cryoprotected by the addition of 20% glycerol and flash-frozen in liquid nitrogen. Diffraction data were collected at APS beamline 24ID-C, and processed by the RAPD pipeline. The structure was determined by molecular replacement in PHASER using the structures of Ec HORMA and Ec CdnC as search models. Initial models were manually rebuilt with COOT, and refined using phenix.refine.
APS NE-CAT Support Statement
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Eiger 16M detector on 24-ID-E beam line is funded by a NIH-ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
SSRL Support Statement
Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Isothermal Titration Calorimetry
Isothermal titration calorimetry was performed at the Sanford Burnham Prebys Medical Discovery Institute Protein Analysis Core. Measurements were performed on a MicroCal iTC200 in buffer GF, by injecting ATP or GTP (1 mM) into a solution containing 0.1 mM Pa CdnD (wild-type or D62N/D64N mutant). Two injections were performed per protein/nucleotide combination.
DNA Binding
For DNA binding assays, a 40 bp double-stranded DNA was produced by annealing 5′-FAM Oligo #1 and Oligo #2 (see Key Resources Table). Binding reactions (30 μL) contained 50 nM dsDNA probe and 0.39-50 μM protein in binding buffer (25 mM Tris-HCl pH 8.5, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 5% glycerol), and were incubated 30 minutes at room temperature for binding. Pre-treated nitrocellulose (top; GE Healthcare) and nylon (GE Healthcare Hybond-N; bottom) membranes were assembled into a Bio-Rad bio-dot 96-well vacuum filtration apparatus. Samples were passed through the filtration apparatus, washed three times with 200 μL binding buffer, then imaged using a Bio-Rad Chemi-Doc gel imager. For Kd value calculation, images were quantified with ImageJ (Schneider et al., 2012), then binding curves were fit in Prism v. 8 (GraphPad Software).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli MS115-1 | BEI Resources | HM-344 |
| E. coli Rosetta2(DE3) pLysS | EMD Millipore | #71403 |
| E. coli JP313 | Joe Pogliano, UCSD (Economou et al., 1995) | |
| Bacteriophage lambda cI- | Joe Pogliano, UCSD | |
| Biological Samples | ||
| Genomic DNA for P. aeruginosa ATCC27853 | American Type Culture Collection (ATCC) | #27853 |
| Sheared salmon sperm DNA | Invitrogen | AM9680 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 5′-pApA | Invivogen | #tlrl-napapa |
| 3′,3′ cyclic di-AMP | Invivogen | #tlrl-nacda |
| 2′,3′ cyclic di-AMP | Invivogen | #tlrl-nacda23 |
| 3′,3′ cyclic GMP-AMP (cGAMP) | Invivogen | #tlrl-nacga |
| 5′pGpG | Invivogen | #tlrl-napapa |
| 3′,3′ cyclic di-GMP | Invivogen | #tlrl-nacdg |
| 3′,3′,3′ cyclic tri-AMP (cAAA) | (Lau et al., 2019; Whiteley et al., 2019) | |
| 3′,3′,3′ cyclic AMP-AMP-GMP (cAAG) | (Lau et al., 2019; Whiteley et al., 2019) | |
| Recombinant TEV protease (homemade) | (Tropea et al., 2009) | |
| Enterobacter cloacae CdnD (homemade) | (Whiteley et al., 2019) | |
| Critical Commercial Assays | ||
| Ph.D.-7 Phage Display Peptide Library Kit | New England Biolabs | E8100S |
| Deposited Data | ||
| Structure of E. coli CdnC | This work | Protein Data Bank (http://www.rcsb.org) 6P80 DOI: 10.2210/pdb6P80/pdb |
| Diffraction data for E. coli CdnC (Native) | This work | SBGrid Data Bank (https://data.sbgrid.org) 668 DOI: 10.15785/SBGRID/668 |
| Diffraction data for E. coli CdnC (SeMet) | This work | SBGrid-DB 669 DOI: 10.15785/SBGRID/669 |
| Structure of P. aeruginosa CdnD | This work | PDB 6P82 DOI: 10.2210/pdb6P82/pdb |
| Diffraction data for P. aeruginosa CdnD | This work | SBGrid-DB 670 DOI: 10.15785/SBGRID/670 |
| Structure of P. aeruginosa CdnD D62N/D64N+ATP | This work | PDB 6P8J DOI: 10.2210/pdb6P8J/pdb |
| Diffraction data for P. aeruginosa CdnD D62N/D64N+ATP | This work | SBGrid-DB 671 DOI: 10.15785/SBGRID/671 |
| Structure of P. aeruginosa HORMA1 | This work | PDB 6P8P DOI: 10.2210/pdb6P8P/pdb |
| Diffraction data for P. aeruginosa HORMA1 (Native) | This work | SBGrid-DB 673 DOI: 10.15785/SBGRID/673 |
| Diffraction data for P. aeruginosa HORMA1 (NaBr) | This work | SBGrid-DB 674 DOI: 10.15785/SBGRID/674 |
| Structure of P. aeruginosa HORMA2 | This work | PDB 6P8R DOI: 10.2210/pdb6P8R/pdb |
| Diffraction data for P. aeruginosa HORMA2 | This work | SBGrid-DB 676 DOI: 10.15785/SBGRID/676 |
| Diffraction data for P. aeruginosa HORMA2 (SeMet) | This work | SBGrid-DB 677 DOI: 10.15785/SBGRID/677 |
| Structure of P. aeruginosa HORMA2 -ΔC | This work | PDB 6P8O DOI: 10.2210/pdb6P8O/pdb |
| Diffraction data for P. aeruginosa HORMA2-Δ | This work | SBGrid-DB 672 DOI: 10.15785/SBGRID/672 |
| Structure of P. aeruginosa HORMA1:HORMA2:Pep1 | This work | PDB 6P8S DOI: 10.2210/pdb6P8S/pdb |
| Diffraction data for P. aeruginosa HORMA1:HORMA2:Pep1 | This work | SBGrid-DB 678 DOI: 10.15785/SBGRID/678 |
| Structure of P. aeruginosa CdnD:HORMA2:Pep1 | This work | PDB 6P8U DOI: 10.2210/pdb6P8U/pdb |
| Diffraction data for P. aeruginosa CdnD:HORMA2:Pep1 | This work | SBGrid-DB 679 DOI: 10.15785/SBGRID/679 |
| Structure of Rhizobiales Pch2 | This work | PDB 6PB3 DOI: 10.2210/pdb6PB3/pdb |
| Diffraction data for Rhizobiales Pch2 | This work | SBGrid-DB 681 DOI: 10.15785/SBGRID/681 |
| Structure of E. coli HORMA:CdnC:Pch2 | This work | PDB 6P8V DOI: 10.2210/pdb6P8V/pdb |
| Diffraction data for E. coli HORMA:CdnC:Pch2 | This work | SBGrid-DB 680 DOI: 10.15785/SBGRID/680 |
| Structure of E. coli HORMA-ΔN:CdnC | This work | PDB 6U7B DOI: 10.2210/pdb6U7B/pdb |
| Diffraction data for E. coli HORMA-ΔN:CdnC | This work | SBGrid-DB 711 DOI: 10.15785/SBGRID/711 |
| Experimental Models: Cell Lines | ||
| S. cerevisiae strain Y187 | Clontech | #630457 |
| S. cerevisiae strain AH109 | Clontech | (discontinued) |
| Oligonucleotides | ||
| CCTTTCGAAGACTCATCACGGAGAGCAGACATCATCTGGC | IDT | Oligo #1 |
| 5’FAM-CCTTTCGAAGACTCATCACGGAGAGCAGACATCATCTGGC | IDT | 5’-FAM Oligo #1 |
| GCCAGATGATGTCTGCTCTCCGTGATGAGTCTTCGAAAGG | IDT | Oligo #2 |
| Recombinant DNA | ||
| pGADT7 vector | Clontech | #630442 |
| pBridge vector | Clontech | #630404 |
| UC Berkeley Macrolab Vector 2AT | Addgene | #29665 |
| UC Berkeley Macrolab Vector 2BT | Addgene | #29666 |
| pLAC22 vector | (Warren et al., 2000) | |
| Software and Algorithms | ||
| RAPD Data-processing pipeline | https://github.com/RAPD/RAPD | |
| autoxds data-processing pipeline | In-house script (SSRL) | |
| XDS | (Kabsch, 2010) | |
| AIMLESS | (Evans and Murshudov, 2013) | |
| TRUNCATE | (Winn et al., 2011) | |
| hkl2map | (Pape et al., 2004) | |
| PHENIX AutoSol | (Terwilliger et al., 2009) | |
| PHASER | (McCoy et al., 2007) | |
| COOT | (Emsley et al., 2010) | |
| phenix.refine | (Afonine et al., 2012) | |
| PyMOL v. 2.3 | Schrödinger | |
| Prism v.8 | GraphPad Software | |
| Other | ||
Phage Display
For phage display, we used a commercial assay kit (Ph.D.-7 Phage Display Peptide Library Kit, New England Biolabs) and followed the recommended protocol for “solution phase panning with affinity bead capture” with the following modifications. We used His6-tagged Ec HORMA, Pa HORMA2, and Pa HORMA3 as bait proteins. For affinity purification, we diluted 2 μL of bait protein at 0.2 mM and 10 μL phage library into 200 μL volume in TBST (50mM Tris pH7.5, 150mM NaCl, plus 0.1% Tween (first round) or 0.5% Tween (subsequent rounds)). After a 20 minute incubation at room temperature, 50 μL of magnetic Ni-NTA beads (Qiagen; pre-blocked with blocking buffer (0.1M NaHCO3, pH 8.6, 5mg/mL BSA, 0.02% NaN3) and washed 3x with TBST) were added, and the mixture was incubated a further 15 minutes at room temperature. Beads were washed 10x with wash buffer (50 mM Tris-HCl pH 7.5, 300 mM NaCl, plus 0.1% Tween (first round) or 0.5% Tween (subsequent rounds)). For elution, 1 mL elution buffer (0.2M Glycine-HCl, pH 2.2, 1mg/mL BSA) was added, incubated for 10 minutes at room temperature, then neutralized the eluate by adding 150 μL Tris-HCl pH 9.1. Eluted phage were titered and amplified according to standard protocols, and the selection was repeated, Following four rounds of selection, phages were isolated and the variable peptide region sequenced for at least 20 individual clones.
Second messenger synthesis
Cyclic trinucleotide synthesis was performed essentially as described (Whiteley et al., 2019). Briefly, 2 mL synthesis reactions contained 1 μM (for Ec CdnC) or 3 μM (for Pa CdnD) CD-NTase or CD-NTase:HORMA complex and 0.25 mM ATP (plus 0.25 mM GTP for Pa CdnD) in reaction buffer with 12.5 mM NaCl, 20 mM MgCl2, 1 mM DTT, and 10 mM Tris-HCl pH 8.5 (or HEPES-NaOH pH 7.5 for reactions with Pa Trip13). For reactions with DNA, plasmid DNA (pUC18) was used at 0.15 μg/mL, 4 kb linear DNA was used at 0.1 μg/mL, single-stranded DNA (sheared salmon sperm DNA; Invitrogen #AM9680) was used at 1 μg/mL, and 40 bp dsDNA (annealed Oligos #1 and #2; see Key Resources Table) was used at 13 μg/mL. For reactions with Trip13, Ec Trip13 was used at 100 nM monomer/16.67 nM hexamer and Pa Trip13 was used at 300 nM monomer/50 nM hexamer; this represents a 60-fold lower molar concentration of active Trip13 hexamers compared to CdnC/HORMA in the reactions. Reactions were incubated at 37°C for 16 hours, then 2.5 units/mL reaction volume (5 units for 2 mL reaction) calf intestinal phosphatase was added and the reaction incubated a further 2 hours at 37°C. The reaction was heated to 65°C for 30 minutes, and centrifuged 10 minutes at 15,000 RPM to remove precipitated protein. Reaction products were separated by ion-exchange chromatography (1 mL Hitrap Q HP, GE Life Sciences) using a gradient from 0.2 to 2 M ammonium acetate. Product peaks were pooled, evaporated using a speedvac, and analyzed by LC-MS/MS.
Mass spectrometry
To characterize the products of CD-NTase reactions, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS). LC-MS/MS analysis was performed using a Thermo Scientific Vanquish UHPLC coupled to a Thermo Scientific Q Exactive™ HF Hybrid Quadrupole-Orbitrap™ Mass Spectrometer, utilizing a ZIC-pHILIC polymeric column (100 mm x 2.1 mm, 5 μm) (EMD Millipore) maintained at 45°C and flowing at 0.4 mL/min. Separation of cyclic trinucleotide isolates was achieved by injecting 2 μL of prepared sample onto the column and eluting using the following linear gradient: (A) 20 mM ammonium bicarbonate in water, pH 9.6, and (B) acetonitrile; 90% B for 0.25 minutes, followed by a linear gradient to 55% B at 4 minutes, sustained until 6 minutes. The column was re-equilibrated for 2.50 minutes at 90% B.
Detection was performed in positive ionization mode using a heated electrospray ionization source (HESI) under the following parameters: spray voltage of 3.5 kV; sheath and auxiliary gas flow rate of 40 and 20 arbitrary units, respectively; sweep gas flow rate of 2 arbitrary units; capillary temperature of 275°C; auxiliary gas heater temperature of 350 °C. Profile MS1 spectra were acquired with the following settings; mass resolution of 35,000, AGC volume of 1×106, maximum IT of 75 ms, with a scan range of 475 to 1050 m/z to include z=1 and z=2 ions of cyclic trinucleotides. Data dependent MS/MS spectra acquisition was performed using collision-induced dissociation (CID) with the following settings: mass resolution of 17,500; AGC volume of 1×105; maximum IT of 50 ms; a loop count of 5; isolation window of 1.0 m/z; normalized collision energy of 25 eV; dynamic exclusion was not used. Data reported is for the z=1 acquisition for each indicated cyclic trinucleotide.
Nuclease Assays
For all nuclease assays, UC Berkeley Macrolab plasmid 2AT (Addgene #29665; 4,731 bp) was used. Ec NucC (10 nM unless otherwise indicated) and second messenger molecules were mixed with 1 μg plasmid DNA or PCR product in a buffer containing 10 mM Tris-HCl (pH 7.5), 25 mM NaCl, 10 mM MgCl2, and 1 mM DTT (50 μL reaction volume), incubated 10 min at 37°C, then separated on a 1.2% agarose gel. Gels were stained with ethidium bromide and imaged by UV illumination. Second messenger molecules were either purchased from Invivogen or synthesized enzymatically by Enterobacter cloacae CdnD02 and purified as previously described (Lau et al., 2019; Whiteley et al., 2019).
Plaque Assays
To measure bacteriophage immunity, the four-protein CBASS operon from E. coli MS115-1 (See Methods S1 for sequence) was PCR-amplified and inserted into vector pLAC22 for IPTG-inducible expression, then modified by PCR-based mutagenesis to generate mutant/tagged protein variants. Vectors were transformed into E. coli strain JP313 (Economou et al., 1995) for plaque assays.
For bacteriophage infection plaque assays, a single bacterial colony was picked from a fresh LB agar plate and grown in LB broth containing proper selection antibiotic at 37°C to an OD600 of 0.1-0.2. CdnC operon expression was induced through the addition of 0.2 mM IPTG, followed by further growth for one hour to an OD600 of 0.6-0.7. For phage dilution assays, 500 μL of cells were mixed with 4.5 mL of of 0.35% LB top agar was added and entire sample was poured on LB plates containing appropriate selection and/or IPTG (0.1 mM). Plates were spotted with 5 μL of bacteriophage λcI- (cI gene deleted to inhibit lysogeny and promote clear plaque formation) diluted in phage buffer (10 mM Tris, pH 7.5, 10 mM MgCl2, 68 mM NaCl) plus 1 mM CaCl2 at six dilutions: ~2.4x106 PFU/mL and five 10-fold dilutions thereof. Plates were incubated at 37°C for 18 hours, then imaged. For plaque numbers, 500 μL of cells at OD600 0.6-0.7 were mixed with 10 μL of bacteriophage λcI (stock concentration ~2.4x107 PFU/mL as measured by infection of JP313, diluted 1000-fold prior to mixing) diluted in phage buffer (10 mM Tris, pH 7.5, 10 mM MgCl2, 68 mM NaCl) plus 1 mM CaCl2, and incubated at room temperature for 30 min. 4.5 mL of 0.35% LB top agar was added and entire sample was poured on LB plates containing appropriate selection and IPTG (0.1 mM) and incubated at 37°C. Plaques were counted after 18 hours of growth.
Bacterial Growth Assays
pLAC22 vectors were constructed encoding either the full CBASS operon from E. coli strain MS115-1 (see Methods S1) or a variant encoding the NucC D73N mutant under the control of an IPTG-inducible promoter. E. coli strain JP313 (a derivative of E. coli MC4100) (Economou et al., 1995) was transformed with these vectors and incubated in 5 mL LB broth with 0.1 mg/ml ampicillin in culture tubes at 30°C with rolling. OD at 600nm was measured after 3 hours and cells were diluted to an OD600 of 0.1 in a reservoir containing LB with 100ug/ml ampicillin and 0.2mM IPTG. 100 μl of cells were loaded into a 96-well plate (four replicates per strain) and 5 μl of bacteriophage λcI- diluted in phage buffer with 1mM CaCl2 was added (final multiplicity of infection either 0.25 or 2.5). The 96-well plate was shaken at 30°C between measurements of OD600, as an average of 25 flashes every 2 minutes, by a Tecan Infinite M200 plate reader.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis of primary diffraction datasets was performed with standard measures including Rsym, Rmeas, and CC1/2 to determine data quality and resolution cutoffs. Statistical analysis of refined crystal structures used standard measures including R and Rfree factors, and geometry analysis by the MolProbity server (Chen et al., 2010).
DATA AND CODE AVAILABILITY
All primary diffraction datasets are available at the SBGrid Data Bank (https://data.sbgrid.org) under the accession numbers noted in Table S3 and the Key Resources Table. All reduced diffraction datasets and refined molecular coordinates are available at the RCSB Protein Data Bank (https://www.rcsb.org) under the accession numbers noted in Table S3 and the Key Resources Table. The original imaging data for Figure 1 and Figure S1 is available at Mendeley Data (https://data.mendeley.com) under DOI 10.17632/dr8mjtwh2d.1.
Supplementary Material
Table S1. (Related to Figure S1) Bacterial HORMA domain protein sequences
(Provided as a separate Microsoft Excel file)
Table S2. (Related to Figure S1) Bacterial Trip13 protein sequences
(Provided as a separate Microsoft Excel file)
Table S3. (Related to Figure 2, Figure 3, Figure 4, and Figure 5) Crystallographic data collection and refinement
(Provided as a separate Microsoft Excel file)
Acknowledgements
The authors thank the staffs of the Stanford Synchrotron Radiation Lightsource and the Advanced Photon Source NE-CAT beamlines for assistance with crystallographic data collection; A. Bobkov (Sanford Burnham Prebys Medical Discovery Institute, Protein Analysis Core) for assistance with isothermal titration calorimetry; M. Raffatellu for valuable advice on bacterial genetics and experimental design; E. Tromer for input on HORMA/Trip13 sequence analysis; and A. Desai, M. Daugherty, and P. Kranzusch for critical reading and helpful suggestions. RKL was supported by the UC San Diego Quantitative and Integrative Biology training grant (NIH T32 GM127235). ITM was supported by NIH T32 HL007444, T32 GM007752 and F31 CA236405. JDW was supported by NIH K01 DK116917 and P30 DK063491. MJ was supported by NIH S10 OD020025 and R01 ES027595. JP was supported by NIH R01 GM129245. KDC was supported by the Ludwig Institute for Cancer Research and the University of California, San Diego.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD, et al. (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D, Biol. Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfieri C, Chang L, and Barford D (2018). Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13. Nature 559, 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, and Koonin EV (1998). The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci 23, 284–286. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, and Vance RE (2011). STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Zhang D, Schäffer DE, Iyer LM, and Aravind L (2015). Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43, gkv1267–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun L, and Chen ZJ (2016). Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol 17, 1142–1149. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D, Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, and Hopfner K-P (2013). Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Melamed S, Millman A, Shulman G, Oppenheimer-Shaanan Y, Kacen A, Doron S, Amitai G, and Sorek R (2019). Cyclic GMP-AMP signaling protects bacterial against viral infection. Nature 574, 691–695. [DOI] [PubMed] [Google Scholar]
- Corrigan RM, and Gründling A (2013). Cyclic di-AMP: another second messenger enters the fray. Nat. Rev. Microbiol 11, 513–524. [DOI] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, and Mekalanos JJ (2012). Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deville C, Carroni M, Franke KB, Topf M, Bukau B, Mogk A, and Saibil HR (2017). Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci. Adv 3, e1701726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, and Clardy J (1993). Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Economou A, Pogliano JA, Beckwith J, Oliver DB, and Wickner W (1995). SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83, 1171–1181. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. Sect. D, Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, and Murshudov GN (2013). How good are my data and what is the resolution? Acta Crystallogr. Sect. D, Biol. Crystallogr 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. (2013). Cyclic [G(2’,5’)pA(3’,5’)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates SN, Yokom AL, Lin J, Jackrel ME, Rizo AN, Kendsersky NM, Buell CE, Sweeny EA, Mack KL, Chuang E, et al. (2017). Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science (80-. ). 357, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Monroe N, Sundquist WI, Shen PS, and Hill CP (2017). The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. Elife 6, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol 7, 263–273. [DOI] [PubMed] [Google Scholar]
- Hornung V, Hartmann R, Ablasser A, and Hopfner K-P (2014). OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol 14, 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, and Cozzetto D (2015). DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta Crystallogr. Sect. D, Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Ishii R, Goto E, Ishitani R, Tokunaga F, and Nureki O (2013). Structural and functional analyses of DNA-sensing and immune activation by human cGAS. PLoS One 8, e76983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Han JS, Ly P, Ye Q, McMahon MA, Myung K, Corbett KD, and Cleveland DW (2018). TRIP13 and APC15 drive mitotic exit by turnover of interphase- and unattached kinetochore-produced MCC. Nat. Commun 9, 4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee ASY, Wilson SC, Solovykh MS, Vance RE, Berger JM, and Doudna JA (2014). Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell 158, 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau RK, Ye Q, Berg KR, Mathews IT, Watrous JD, Ego KM, Whiteley AT, Lowey B, Mekalanos JJ, Kranzusch PJ, et al. (2019). Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Submitted. [DOI] [PMC free article] [PubMed]
- Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, and Li P (2013). Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39, 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone CD, and Barton GJ (1993). Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci 9, 745–756. [DOI] [PubMed] [Google Scholar]
- Lohöfener J, Steinke N, Kay-Fedorov P, Baruch P, Nikulin A, Tishchenko S, Manstein DJ, and Fedorov R (2015). The Activation Mechanism of 2’-5’-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity. Structure 23, 851–862. [DOI] [PubMed] [Google Scholar]
- Luo X, and Yu H (2008). Protein metamorphosis: the two-state behavior of Mad2. Structure 16, 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HT, and Poon RYC (2016). TRIP13 Regulates Both the Activation and Inactivation of the Spindle-Assembly Checkpoint. Cell Rep. 14, 1086–1099. [DOI] [PubMed] [Google Scholar]
- Ma HT, and Poon RYC (2018). TRIP13 Functions in the Establishment of the Spindle Assembly Checkpoint by Replenishing O-MAD2. Cell Rep. 22, 1439–1450. [DOI] [PubMed] [Google Scholar]
- Mapelli M, Massimiliano L, Santaguida S, and Musacchio A (2007). The Mad2 Conformational Dimer: Structure and Implications for the Spindle Assembly Checkpoint. Cell 131, 730–743. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland AP, Luo S, Ahmed-Qadri F, Zuck M, Thayer EF, Goo YA, Hybiske K, Tong L, and Woodward JJ (2017). Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-κB Activation and Shapes a Proinflammatory Antibacterial State. Immunity 46, 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Schneider TR, and IUCr (2004). HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr 37, 843–844. [Google Scholar]
- Puchades C, Rampello AJ, Shin M, Giuliano CJ, Wiseman RL, Glynn SE, and Lander GC (2017). Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science (80-. ). 358, eaao0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Kim DJ, Stjepanovic G, and Hurley JH (2015). Structure of the Human Atg13-Atg101 HORMA Heterodimer: an Interaction Hub within the ULK1 Complex. Structure 23, 1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripstein ZA, Huang R, Augustyniak R, Kay LE, and Rubinstein JL (2017). Structure of a AAA+ unfoldase in the process of unfolding substrate. Elife 6, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SC, and Corbett KD (2015). The multifaceted roles of the HORMA domain in cellular signaling. J. Cell Biol 211, 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM (2010). Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. Sect. D, Biol. Crystallogr 66, 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kaizuka T, Mizushima N, and Noda NN (2015). Structure of the Atg101–Atg13 complex reveals essential roles of Atg101 in autophagy initiation. Nat. Struct. Mol. Biol 22, 572–580. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, and Hung L-W (2009). Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. Sect. D, Biol. Crystallogr 65, 582–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromer EC, van Hooff JJE, Kops GJPL, and Snel B (2019). Mosaic origin of the eukaryotic kinetochore. Proc. Natl. Acad. Sci. U. S. A 116, 12873–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropea JE, Cherry S, and Waugh DS (2009). Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol. Biol. (Clifton, NJ) 498, 297–307. [DOI] [PubMed] [Google Scholar]
- White KI, Zhao M, Choi UB, Pfuetzner RA, and Brunger AT (2018). Structural principles of SNARE complex recognition by the AAA+ protein NSF. Elife 7, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AT, Eaglesham JB, de Oliveira Mann CC, Morehouse BR, Lowey B, Nieminen EA, Danilchanka O, King DS, Lee ASY, Mekalanos JJ, et al. (2019). Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D, Biol. Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Rosenberg SC, Moeller A, Speir JA, Su TY, and Corbett KD (2015). TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. Elife 2015, 1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Kim DH, Dereli I, Rosenberg SC, Hagemann G, Herzog F, Tóth A, Cleveland DW, and Corbett KD (2017). The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding. EMBO J. 36, 2419–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lupoli TJ, Kovach A, Meng X, Zhao G, Nathan CF, and Li H (2018). ATP hydrolysis-coupled peptide translocation mechanism of Mycobacterium tuberculosis ClpB. Proc. Natl. Acad. Sci 115, E9560–E9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wu J, Du F, Xu H, Sun L, Chen Z, Brautigam CA, Zhang X, and Chen ZJ (2014). The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 6, 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Wang L, Shang G, Liu X, Zhu J, Lu D, Wang L, Kan B, Zhang J-R, and Xiang Y (2014). Structural biochemistry of a Vibrio cholerae dinucleotide cyclase reveals cyclase activity regulation by folates. Mol. Cell 55, 931–937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. (Related to Figure S1) Bacterial HORMA domain protein sequences
(Provided as a separate Microsoft Excel file)
Table S2. (Related to Figure S1) Bacterial Trip13 protein sequences
(Provided as a separate Microsoft Excel file)
Table S3. (Related to Figure 2, Figure 3, Figure 4, and Figure 5) Crystallographic data collection and refinement
(Provided as a separate Microsoft Excel file)
Data Availability Statement
All primary diffraction datasets are available at the SBGrid Data Bank (https://data.sbgrid.org) under the accession numbers noted in Table S3 and the Key Resources Table. All reduced diffraction datasets and refined molecular coordinates are available at the RCSB Protein Data Bank (https://www.rcsb.org) under the accession numbers noted in Table S3 and the Key Resources Table. The original imaging data for Figure 1 and Figure S1 is available at Mendeley Data (https://data.mendeley.com) under DOI 10.17632/dr8mjtwh2d.1.