Abstract
Background:
Uncontrolled lung inflammation is one of the prominent features in the pathogenesis of lung infection associated acute lung injury (ALI). Microvesicles (MVs) are extracellular nanovesicles that are generated via direct membrane budding.
Methods:
Bronchoalveolar lavage fluids (BALF) samples were collected from mice with or without intratracheal lipopolysaccharide (LPS) instillation. BALF MVs were characterized and MV-containing miRNA profiles were assessed and confirmed. Secretion and function of MV-containing miR-223/142 (MV-miR-223/142) were analyzed in vivo.
Results:
In BALF, MVs are mainly derived from macrophages in response to LPS. After intratracheal instillation (i.t.) of LPS or Klebsiella pneumoniae (K.pneu), MV-containing miR-223/142 are dramatically induced in both BALF and serum. Mechanistically, miRNA 3’ end uridylation mediates the packing of miR-223/142 into MVs. To investigate the functional role of MV-miR-223/142, we loaded miR-223/142 mimics into unstimulated MVs and delivered them into the murine lungs via i.t.. The miR-223/142 mimics-enriched MVs selectively targeted lung macrophages and suppressed the inflammatory lung responses that were triggered by LPS or K.pneu. Mechanistically, miR-223 and 142 synergistically suppress Nlrp3 inflammasome activation in macrophages via inhibition of Nlrp3 and Asc, respectively.
Conclusion:
In the pathogenesis of lung macrophage-mediated inflammatory responses, MV-miR-223/142 secretion is robustly enhanced and detectable in BALF and serum. Furthermore, restoration of intracellular miR-223/142 via vesicle-mediated delivery suppresses macrophage activation and lung inflammation via inhibition of Nlrp3 inflammasome activation.
Keywords: microvesicle, exosome, microRNA, bacterial infection, LPS, macrophage, lung injury
INTRODUCTION
Inflammatory responses are a characteristic feature of the host immune response to bacterial infections. As a double-edged sword, inflammatory responses play an essential role in host defense and immune responses against bacterial infections.1 On the other hand, the aims and regulation of inflammatory responses are imprecise and often become a run-away cascade, which can cause collateral damage in tissues.2 Developing a specific biomarker that both reflects the stages of inflammatory responses and can be detected in the circulating blood stream or body fluid is very important. However, currently known biomarkers such as erythrocyte sedimentation rate and C-reactive protein are often non-specific.
MicroRNAs (miRNAs) are a class of small non-coding RNAs that are conserved across species.3 MiRNAs can enter body fluids through multiple pathways including leakage from dying cells, active secretion from live cells via microvesicles (MVs), and secretion with RNA-binding proteins.4 Since 2007, accumulating evidence has shown that circulating miRNAs serve as biomarkers for cancer development, metastasis, and prognosis.5 Emerging data further supports that circulating miRNAs may be used as novel diagnostic and therapeutic targets for a variety of non-cancer diseases such as metabolic abnormalities and cardiovascular disorders.6
Extracellular vesicles (EVs) are released by all cells and are ubiquitous in all body fluids, thus carrying a great potential to serve as diagnostic biomarkers and therapeutic targets.7 EVs are now classified into exosomes (Exos), microvesicles (MVs) and apoptotic bodies (ABs) based on the size, components, and mechanisms of generation.8 In our previous studies, we have demonstrated that EVs transfer miRNAs from the “mother” cells to recipient cells, and that this transfer of their molecular/genetic cargo re-programs the function of the recipient cell.8,9 Therefore, EV-containing cargos not only serve as potential diagnostic markers for disease progression but also as novel targets to develop therapeutic agents. Accumulating evidence suggests that miRNAs are selectively incorporated into EVs rather than randomly wrapped in.10 Therefore, the miRNA profiles in EVs potentially reflect the biological status of the parent cells from which the secreted EVs are derived.
Lung infections place a major burden on public health worldwide and are the leading cause of death in the United States.11 Infections caused by gram-negative (G-) bacteria have features that are of particular concern, such as being highly efficient at acquiring antibiotic resistance. The high mortality and morbidity after bacterial infection often result from an imbalance in host defenses between bactericidal and an excessive inflammatory response that leads to tissue damage.12 In this report, we analyzed the macrophage-secreted MVs in response to G- bacteria or LPS. In these macrophage-derived MVs, significantly altered miRNA profiles were explored. We attempted to identify the diagnostic and therapeutic targets for profound lung inflammation by analyzing the MV-shuttling miRNAs derived from G- bacteria or LPS-activated macrophages.
MATERIALS AND METHODS
Reagents
LPS from Escherichia coli O111:B4, miR-223–3p/miR-142–3p mimics, inhibitors, and respective controls were purchased from Sigma-Aldrich. Phosphate-buffered saline (PBS), fetal bovine serum (FBS), RPMI-1640 and DMEM were purchased from Gibco. EV-depleted FBS (System Biosciences). The canonical 3’ end adenylated hsa-miR-223–3p (5’ UGUCAGUUUGUCAAAUACCCCAAAA 3’) and 3’ end uridylated miR-223 (5’ UGUCAGUUUGUCAAAUACCCCAUUU 3’) were synthesized by Integrated DNA Technologies (IDT). IDT also synthesized all the adapters and primers used in this study, as described below. Lipofectamine® 3000 reagent, gentamicin, and protease inhibitor cocktail were purchased from Thermo Fisher Scientific. NLRP3 and CD68 antibodies were purchased from Abcam. CD11c, F4/80, Ly-6G/Ly-6C, and CD45 antibodies were ordered from BD Biosciences. E-cadherin, ASC, β-actin and CD40L antibodies were ordered from Cell Signaling Technology. K. pneu (strain from ATCC#43186) was provided by Dr. Dela Cruz at Yale University School of Medicine.
Bacteria culture
Cultures of K.pneu were grown overnight in Luria-Bertani medium at 37°C in a rotator at 250 rpm. They were then sub-cultured into fresh medium and grown to mid-log phase. After culturing, bacteria were pelleted and resuspended in PBS. Bacterial concentrations were assessed by serial dilutions. Bacteria count was estimated by OD600 and was diluted to final colony forming unit (CFU) concentrations as needed for each experiment.
Bone marrow-derived macrophages (BMDM) isolation and cell culture
BMDM were isolated as previously described13 and cultured with 30% L929 cells conditioned medium in DMEM complete medium for 7 days before any further experimental procedure. Human THP-1 monocytes were obtained from American Type Culture Collection (ATCC) and maintained in RPMI-1640 with 10% fetal bovine serum (FBS). THP-1 cells were differentiated into macrophage-like cells for all the experiments by 150 nM phorbol myristate acetate (PMA) treatment for 24 hour. HEK293T cells were purchased from ATCC and cultured in DMEM with 10% FBS. To reduce the interference of MVs derived from bovine serum, EV-depleted FBS was used instead. All the cells were maintained at 37°C in a humidified atmosphere of 5% CO2 - 95% air.
In vitro proinflammatory stimulation
For LPS treatment, cultured BMDM were incubated with LPS at a final concentration of 100 ng/mL for the indicated time. For bacterial stimulation, 107 CFU of K. pneu were added to 105 BMDM for 1 h. After the incubation, bacteria were removed by washing, and 50 μg/mL of gentamicin was added to kill any remaining extracellular bacteria, as described before.14 Supernatants and cells from treated cultures were harvested following an additional incubation as indicated.
Cloning, transfection and luciferase assay
To identify which target gene might be responsible for miR-223/142–mediated anti-inflammatory effects, we used TargetScan (Release 7.1) to predict both highly and poorly conserved binding targets. The fragment of the Asc and Nlrp3 3’UTRs were amplified from murine cDNA by PCR and inserted into the pRL-TK vector containing a cDNA encoding Renilla luciferase and HSV-TK promoter (Promega). QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) was purchased and used for 3’UTRs mutation (mut). The primer sequences used for cloning and mutation are shown in table 1. The plasmid pGL-3 (Promega) coding firefly luciferase was co-transfected and firefly luciferase activity was used as normalization control. Transfection of miRNA mimics, inhibitors and plasmids were performed using Lipofectamine 3000 according to the manufacturer’s instructions. At 48 h after transfection, a luciferase assay was performed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. Firefly and Renilla luciferase activities were measured.
Table 1.
Sequences of DNA oligos for cloning, mutation and 3’-Dumbbell-PCR
| Asc 3’UTR forward | TGCTCTAGAGCCAGTGTCCCTGCTCAGAGTA | |
| Asc 3’UTR reverse | GAAAAAAGCGGCCGCCTTTGCACTAGAATGGAGAAGC | |
| Nlrp3 3’UTR forward | TGCTCTAGAGAAGCAGGACCACCAGGTGCCT | |
| Nlrp3 3’UTR reverse | GAAAAAAGCGGCCGCAACAATGTAGCTGAGAGGCTGC | |
| Asc 3’UTR-mut-F | ATGTCCTTTGTGATGTGCTTTGTTCCATCC | |
| Asc 3’UTR-mut-R | CTTGTCTTGGCTGGTGGTCTCTGCACGA | |
| Nlrp3 3’UTR-mut-F | GTTTTGACTGCATCCCGCATAAGGAGCTGC | |
| Nlrp3 3’UTR-mut-R | AGGAAGACAAGACTGAAGGTATGGGCAA | |
| adenylated (3’-AAA) miR-223 | Adapter | Phos/CTCAGTGCAGGGTCCGAGGTATTCGCACTGAGTTTTGG |
| Primer-F | GCGCTGTCAGTTTGTCAAATACCC | |
| Primer-R | CGAATACCTCGGACC | |
| Probe | 6-FAM/CAAAACTCA/ZEN/GTG/IABkFQ | |
| uridylated (3’-UUU) miR-223 | Adapter | Phos/CTCAGTGCAGGGTCCGAGGTATTCGCACTGAGAAATGG |
| Primer-F | GCGCTGTCAGTTTGTCAAATACCC | |
| Primer-R | CGAATACCTCGGACC | |
| Probe | 6-FAM/CATTTCTCA/ZEN/GTG/IABkFQ | |
| adenylated (3’-AAA) miR-142 | Adapter | Phos/CTCAGTGCAGGGTCCGAGGTATTCGCACTGAGTTTTCC |
| Primer-F | GCGCTGTAGTGTTTCCTACTTTATG | |
| Primer-R | CGAATACCTCGGACC | |
| Probe | 6-FAM/GAAAACTCA/ZEN/GTG/IABkFQ | |
| uridylated (3’-UUU) miR-142 | Adapter | Phos/CTCAGTGCAGGGTCCGAGGTATTCGCACTGAGAAATCC |
| Primer-F | GCGCTGTAGTGTTTCCTACTTTATG | |
| Primer-R | CGAATACCTCGGACC | |
| Probe | 6-FAM/GATTTCTCA/ZEN/GTG/IABkFQ | |
The table shows the sequences of primers that were used for Asc and Nlrp3 3’UTR cloning/site mutation, as well as 3’-Dumbbell-PCR to detect 3’ adenylation and uridylation of miR-223/142 in the RNA samples as indicated.
Animal study
WT C57BL/6 and Mir223 knockout mice of both genders (8 weeks of age) were obtained from The Jackson Laboratory. To induce lung inflammation, mice were anesthetized and intubated intratracheally using a blunt-ended feeding needle. 1 μg LPS or 104 CFU of K. pneu in 50 μL PBS was given through the needle. Mice were euthanized by an aerosolized isoflurane overdose at the indicated time point after instillation. The number of mice used per group was 5 to 8. For hyperoxia-induced acute lung injury model, C57BL/6 mice of both genders (8 – 10 weeks of age) were placed in a Plexiglas chamber maintained at 100% O2 (hyperoxia group) or in a chamber open to room air (room air group) for the indicated time. All the protocols involving animals in this study were approved by the institutional animal care and use committee (IACUC) of Boston University.
Preparation and characterization of extracellular vesicles
MVs were prepared by using sequential centrifugation protocols described previously.15 Murine BALF or conditioned media was collected and centrifuged at 300 ×g for 10 min to remove floating cells. The supernatant was further centrifuged at 2000 ×g for 20 min to pellet apoptotic bodies (ABs). To isolate MVs, the AB-depleted supernatant was passed through a 0.8-μm-pore filter followed by centrifugation at 16,000 ×g for 40 min. To track the MVs distribution in the lung, PKH26 Fluorescent Cell Linker Kits for General Cell Membrane Labeling (Sigma-Aldrich) was purchased and used according to the manufacturer’s protocol. For TEM, a commercial kit for transmission electron microscopy imaging was obtained from 101Bio Corporation. The TEM images were taken using a Philips CM120 EM at the Experimental Pathology Laboratory Service Core (Boston University School of Medicine). Nanoparticle tracking analysis (NTA) data were obtained using Nanosight NS500 at the Nanomedicines Characterization Core Facility (The University of North Carolina at Chapel Hill). To measure the change of MVs secretion in BALF, flow cytometric analysis was performed as described previously.8 Purified MVs were coupled to 10 mL of aldehyde/sulfate latex beads (Thermo Fisher Scientific) for 2 h. After blocking, the bead-bound MVs were fixed and incubated with antibodies against different biomarkers as indicated.
Preparation and delivery of small RNA loaded-MVs
Electroporation was used to introduce small RNAs, including miR-223/142 mimics and miR-223/142 inhibitors and their respective controls, into MVs as described before.16 Briefly, 100 pmol miRNA mimics or inhibitors or their respective controls were mixed with 100 μg BALF MVs (quantified by protein content). The final volume was adjusted to 100 μL using sterile PBS. The mixture was loaded into the 100 μL Neon Tip and electroporated at 0.5 kV using 10-ms pulse five times using the Neon Transfection System (Thermo Fisher Scientific). To wash the MVs, 900 μL cold PBS was added after electroporation. MVs were pelleted by centrifugation at 16,000 ×g for 40 min and resuspended in 50 μL PBS. Then, small RNA-loaded MVs were intratracheally instilled into the lung.
Lung inflammation and caspase-1 activity
For cytospin preparations, the cell suspension was cytocentrifuged at 300 ×g for 5 min using a Shandon Cytospin 4 (Thermo Fisher Scientific). Total inflammatory cell counts in the BALF were determined using a hemocytometer as previously described.17 BALF cells and lung sections were air-dried and stained with PROTOCOL Hema 3 fixative and solutions (Fisher Scientific). The caspase-1 activity in the murine lung tissue was measured using the Caspase-Glo® 1 Inflammasome Assay Kit purchased from Promega Corporation. The total protein was used as normalization control.
Immunofluorescence staining
Immunofluorescence staining was performed as previously described.16 Briefly, lung sections were incubated overnight at 4°C with an antibody against mouse CD68 (macrophage marker) or a Ly-6G/Ly-6C antibody (granulocyte marker). After washing, Alexa 488-conjugated secondary antibodies (Thermo Fisher Scientific) were applied. Nuclei were counterstained with DAPI. Images of the stained lung sections were visualized and captured using a fluorescence microscope (Axioplan-2, Zeiss), a high speed 5 megapixel microscope camera (AxioCam, Zeiss), and the software package (AxioVision, Zeiss) with an N-Achroplan 20×/0.45 (Zeiss) objective lens.
RNA preparation, reverse transcription, qRT-PCR, PCR array and 3’-Dumbbell-PCR
miRNeasy Mini Kit (QIAGEN) was used for purification of total RNA. For miR-223 and miR-142 detection, qRT-PCR was performed using TaqMan® PCR kit (Thermo Fisher Scientific) as previously described.18 Hprt1 was used as a normalization control. To detect the canonical 3’ end adenylated (3’-AAA) or uridylated (3’-UUU) miR-223 and miR-142, 3’-Dumbbell-PCR was used as described before.19 The sequences of adapters and primers used for 3’-Dumbbell-PCR are shown in table 1.
miRNA Quantification by the FirePlex Assay
Pooled RNA samples from MVs (three groups including PBS, 6 hours and 24 hours after LPS treatment) were provided to Abcam (Cambridge, MA) and processed to analyze using the FirePlex miRNA Panel - Immunology (Abcam). Results were background subtracted and normalized. A geNorm algorithm has been applied to generate the normalized data. Data are shown as the signals reported in Arbitrary Units (AU) of fluorescence per miRNA target. The signals are proportional to the average amount of fluorescent target bound to the particles. The heat maps were generated using the Firefly™ Analysis Workbench software.
Western blot analysis and ELISA
Western blot analysis was performed as described previously.20 In brief, cells or MVs were homogenized in RIPA lysis buffer supplemented with protease inhibitor cocktail. Protein lysates were resolved on SDS-PAGE gels before being transferred to the PVDF membrane. DuoSet ELISA Kits for mouse IL-1 beta and IL-18 were purchased from R&D Systems to determine the levels of cytokines in BALF according to the manufacturer’s protocol.
Statistical analysis
Data were analyzed with SigmaPlot (Systat Software, San Jose, CA). Differences between two groups were analyzed using the two-tailed unpaired Student’s t-test for variables with a normal distribution or the Mann-Whitney U-test for variables without a normal distribution. Differences between three or more groups were analyzed using a one-way ANOVA with a Tukey HSD test for variables with normal distribution or the Kruskal-Wallis One-Way Analysis followed by pair-wise comparisons using Mann-Whitney U-tests for those without a normal distribution. Parametric data are presented as mean with SD. Non-parametric data are presented as box-plots showing medians and 25th and 75th percentiles and whisker showing 10th and 90th percentiles. p ≤ 0.05 was considered to be statistically significant.
RESULTS
BALF MVs are altered in response to LPS
BALF MVs obtained from control or LPS-treated mice were purified and examined using Transmission Electron Microscopy (TEM). The captured MVs in both groups have a typical cup shape (figure 1A). The count and size of BALF MVs were determined using Nanoparticle Tracking Analysis (NTA) (figure 1B, C). A variety of cell markers were analyzed on the isolated MVs, including markers of leukocytes (CD45), macrophages (CD68), epithelial cells (E-cadherin), and MVs (CD40L). Notably, the enhanced expressions of CD45 and CD68 were observed in MVs after LPS stimulation (figure 1D). To confirm this observation, we further analyzed the BALF MVS using nanoFACS (figure 1E). LPS stimulation increases the percentage of F4/80+/CD11c+ beads from to 32.54% to 61.34% (95% CI 17.17 to 40.43%) (p = 0.0004) (figure 1F). We confirmed that LPS induced the MVs that were secreted from lung macrophages.
Figure 1. LPS stimulation alters the profile of MVs derived from BALF.
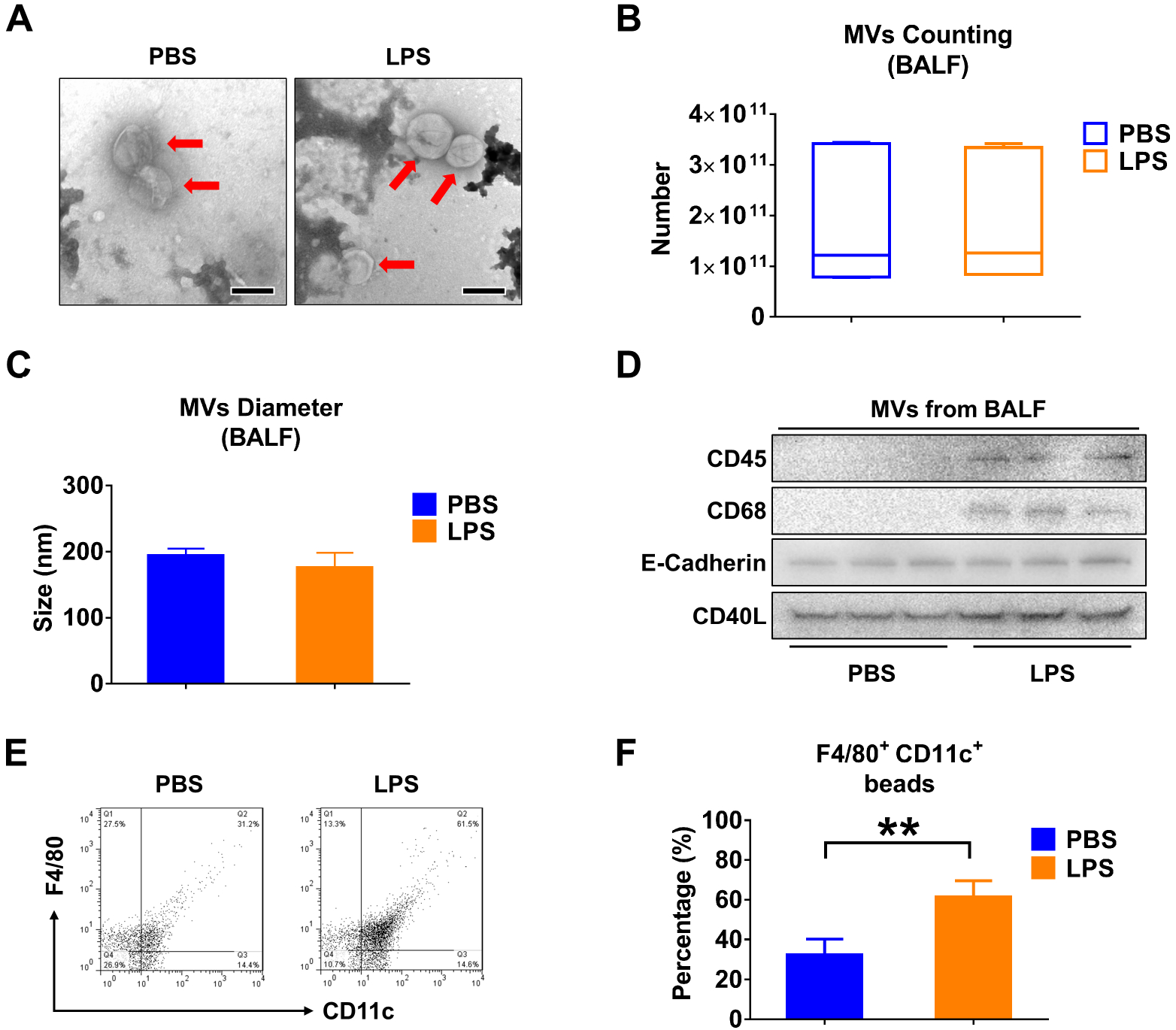
Mice received 50 μL PBS or 50 μL PBS containing 1 μg LPS via i.t.. After 24 hours, MVs were isolated from BALF. (A) TEM images of BALF-derived MVs were shown, scale bar = 200 nm. (B) The number of BALF MVs isolated from mice was counted. The boxes in the box-plots show the medians with 25th and 75th percentiles, and the whiskers show the 10th and 90th percentiles; n = 7 mice per group. Data were analyzed using a Kruskal-Wallis One-Way ANOVA followed by pair-wise testing with Mann-Whitney U tests. (C) The diameter of BALF MVs isolated from mice was measured using NTA. Data are mean + SD; n = 6 mice per group. Data were analyzed using a One-Way ANOVA followed by pair-wise testing with Student’s t-tests. (D) CD45 (leucocyte marker), CD68 (macrophage marker), E-Cadherin (epithelial cell marker) and CD40L (MVs marker) were detected in 100 μg MVs protein using western blot. (E and F) MVs derived from BALF were to flow cytometry analysis (E). The percentage of macrophage-derived MVs (F4/80+CD11c+) in response to LPS stimulation are shown (F). Data are mean + SD; n = 5 mice per group. Data were analyzed using a One-Way ANOVA followed by pair-wise testing with Student’s t-tests. **, p < 0.01 versus the PBS group.
Infectious stimuli increase miR-223/142 levels in MVs
We analyzed the miRNA cargo in the isolated BALF MVs using the FirePlex® miRNA Immunology Panel (figure 2A). 24h after LPS exposure, miR-223 and miR-142 exhibited the greatest induction of expression in MVs, reaching 26.41-fold and 24.21-fold increases compared to controls, respectively (figure 2B). We next validated the enhanced expression of miR-223/142 in MVs using qPCR. In figure 2C, miR-223 in BALF MVs was strikingly up-regulated by LPS stimulation with a 47.77-fold increase in expression (p ≤ 0.001). Similarly, miR-142 had a 3.49-fold increase (p = 0.002) (figure 2D). Furthermore, the levels of MV-miR-223/142 were detected after LPS treatment in vitro. As shown in figure 2E, F, LPS treatment increases MV-miR-233 derived from both BMDM (19.34-fold, p = 0.029) and THP-1 (9.63-fold, p = 0.000153), as well as MV-miR-142 secreted from BMDM (14.28-fold, p = 0.029) and THP-1 (3.99-fold, p = 0.0374). Furthermore, we confirmed the above observations using mice which were exposed to Klebsiella pneumoniae (K.pneu). Consistently, MV-miR-223/142 levels were remarkably elevated both in vivo and in vitro after K.pneu (figure 2G–J).
Figure 2. Infectious stimuli elevate miR-223/142 in BALF MVs.
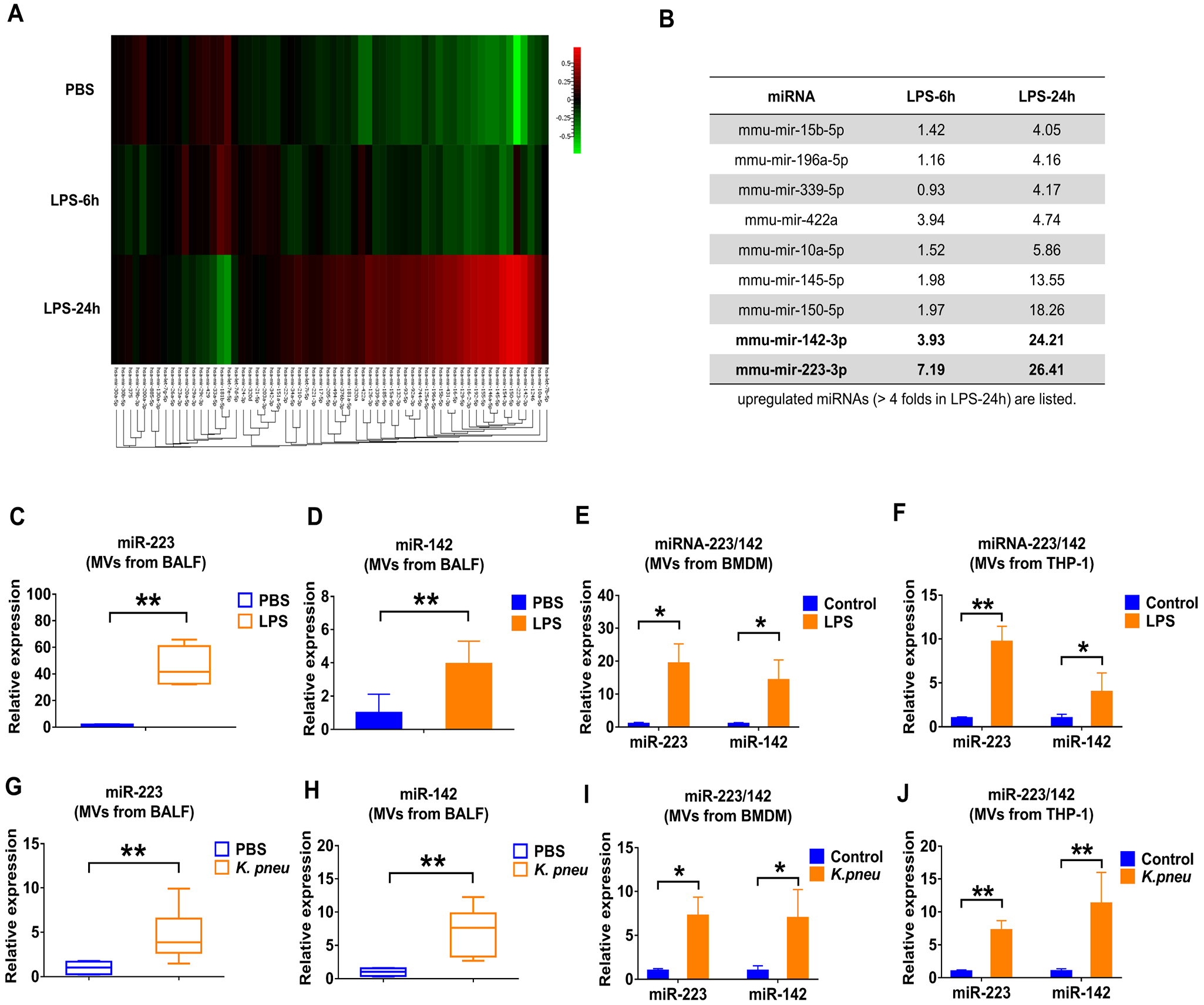
(A and B) WT mice (n = 7 mice per group) received 50 μL PBS or 50 μL PBS containing 1 μg LPS via i.t.. MVs were isolated from BALF at 6 h or 24 h after treatment. Pooled RNA from BALF MVs was profiled using the FirePlex® miRNA Immunology Panel. The differential expression of miRNAs in MVs from BALF following LPS challenge was shown in the heat map (A). Red and green indicate high expression level and low expression level, respectively. Table showing the upregulated miRNAs (B) (fold of increase > 4 in MVs derived from BALF, 24 h after LPS instillation). (C and D) WT mice (n = 7 mice per group) received 50 μL PBS or 50 μL PBS containing 1 μg LPS via i.t.. After 24 h, the relative expression of miR-223 (C) and miR-142 (D) in MVs was detected using qRT-PCR. (E and F) MVs were isolated from BMDM (E) and THP-1 (F) culture media with or without LPS treatment (100 ng/ml) for 24 h. The expressions of miR-223/142 in MVs were detected using qRT-PCR. Results are from 3 independent experiments. (G and H) WT mice (n= 6 mice per group) received 50 μL PBS or K. pneu (104 CFU in 50 μL PBS) via i.t.. After 24 h, the relative expression of miR-223 (G) and miR-142 (H) in MVs were compared using qRT-PCR. (I and J) MVs were isolated from BMDM (I) and THP-1 (J) culture media with or without K. pneu treatment (107 CFU bacteria/105 macrophages, 1-hour incubation). After 24 h, the expression of miR-223/142 in MVs was detected using qRT-PCR. Results are from 3 independent experiments. In panel C, G and H, the boxes in the box-plots show the medians with 25th and 75th percentiles, and the whiskers show the 10th and 90th percentiles. Data were analyzed using a Kruskal-Wallis One-Way ANOVA followed by pair-wise testing with Mann-Whitney U tests. In panel D, E, F, I and J, data are presented as mean + SD and were analyzed using a One-Way ANOVA followed by pair-wise testing with Student’s t-tests. *, p < 0.05; **, p < 0.01 versus the PBS or control group, respectively.
3’ end uridylation mediates the package of miR-223/142 into MVs
Small RNA compositions in the secreted MVs are different from that in host cells.21 3’ end nucleotide additions have been reported to mediate the selection of miRNAs.21,22 We measured the canonical 3’ end adenylated (3’-AAA) or uridylated (3’-UUU) miR-223/142 in BMDM and BMDM-derived MVs, respectively. We show that 3’ end adenylated miR-223 was relatively enriched in cells (adenylated : uridylated = 2.17 : 1, p = 0.00824), while 3’ end uridylated miR-223/142 were highly elevated in MVs (adenylated : uridylated = 1.43 : 2.66, p = 0.0253) (figure 3A, B). Additionally, we observed that adenylated or uridylated miR-142 has a similar distribution in macrophages and MVs, suggesting that 3’ end nucleotides play a role in the packaging of miR-223/142 into MVs. To further support this hypothesis, we transfected miR-223-depleted BMDMs with synthesized adenylated (3’-AAA) and uridylated (3’-UUU) miR-223 (figure 3C). 24 hours after transfection, we found that uridylated miR-223 is significantly more prevalent in secreted MVs than in adenylated miR-223 (adenylated : uridylated = 16.60 : 40.68, p = 0.0103) (figure 3D). As previously reported, Tut4 (Zcchc11) and Tut7 (Zcchc6) regulate mature miRNA uridylation22. Interestingly, we found that LPS dramatically induced Tut4 (4.85-fold, p = 0.0010 at 6 hours; 4.74-fold, p = 0.002 at 12 hours; 3.16-fold, p = 0.039 at 24 hours) and Tut7 (4.91-fold, p < 0.001 at 6 hours; 6.49-fold, p < 0.001 at 12 hours; 3.12-fold, p = 0.024 at 24 hours) in BMDM (figure 3E). In addition, we knocked down Tut4 and Tut7 in BMDM in the presence of LPS using siRNAs. As shown in figure 3F, miR-223 was significantly decreased in MVs released from BMDM transfected with Tut4 (0.50-fold, p = 0.009) and Tut7 (0.60-fold, p = 0.025) siRNAs. Similarly, MV-miR-142 was decreased after knockdown of Tut4 (0.34-fold, p = 0.002) and Tut7 (0.65-fold, p = 0.034) (figure 3F).
Figure 3. 3’ end uridylation mediates the packaging of miR-223/142 into MVs.
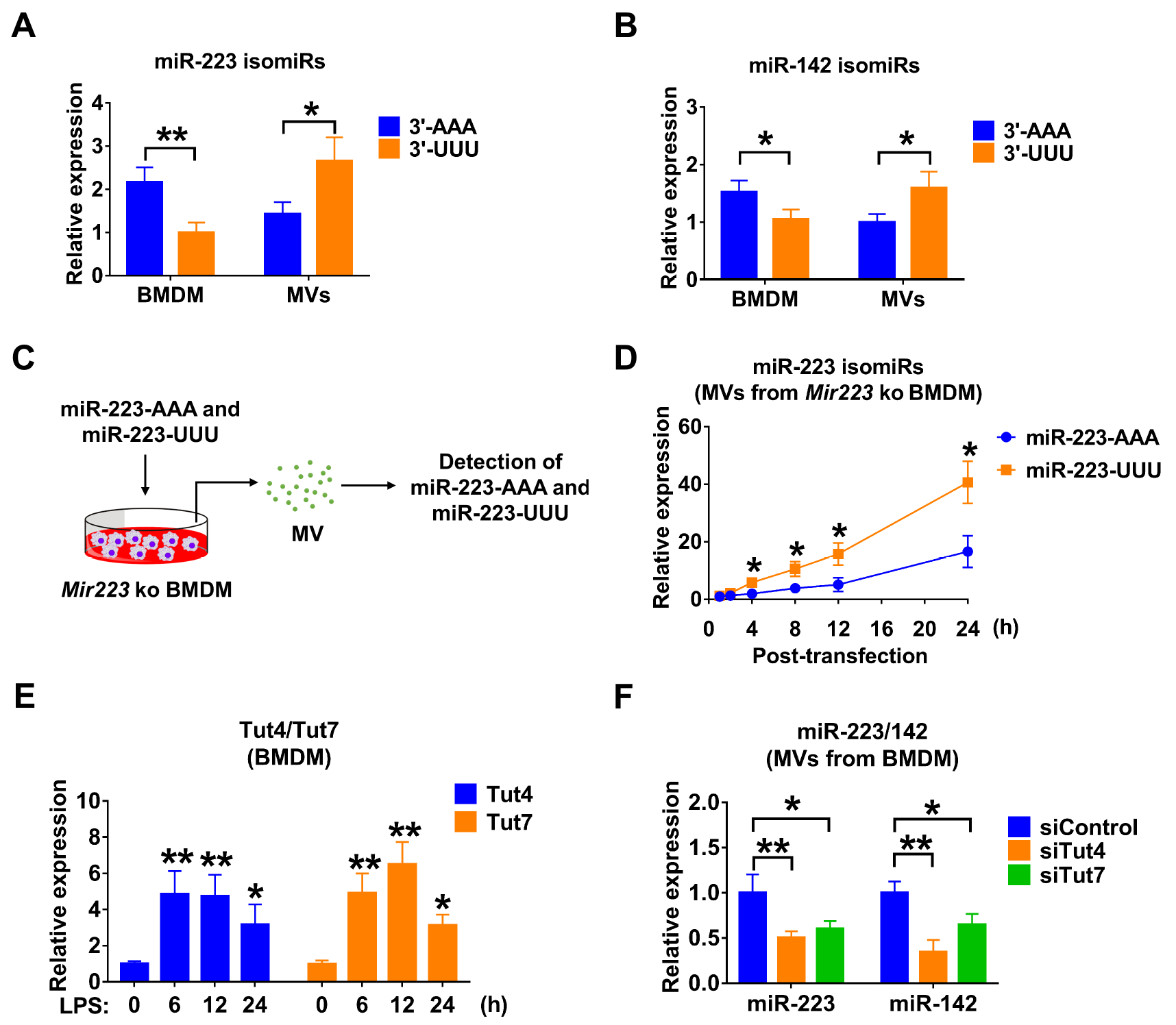
(A and B) Detection of canonical 3’ end adenylated (3’-AAA) or uridylated (3’-UUU) miR-223 (A) and miR-142 (B) in BMDM and BMDM-derived MVs using 3’-Dumbbell-PCR. (C and D) The experimental design for testing the role of 3’ end adenylation/uridylation-mediated miR-223 packaging into MVs (C). Mir223 depleted BMDM were transfected with synthesized 3’ end adenylated (3’-AAA) or uridylated (3’-UUU) miR-223. After transfection, MVs were purified from culture media at the indicated time points. The level of adenylated or uridylated miR-223 in MVs was detected using 3’-Dumbbell-PCR (D). (E) BMDM were stimulated with LPS (100 ng/mL) for the indicated time points. The mRNA expressions of Tut4 and Tut7 were detected using qRT-PCR. (F) BMDM were transfected with siRNA control, Tut4 siRNA or Tut7 siRNA. After 24 hours, BMDM were treated with LPS (100 ng/mL) for an additional 24 hours. MVs were purified from culture media and subjected to RNA isolation. The expression of miR-223 and miR-142 in MVs were detected using qRT-PCR. All of these results are presented as mean + SD and are from 3 independent experiments. In panel A and B, data were analyzed using a One-Way ANOVA followed by pair-wise testing with Student’s t-tests. In panel D, E and F, data were analyzed using a One-Way ANOVA followed by Tukey’s HSD. *, p < 0.05; **, p < 0.01 versus their corresponding control.
Circulating MV- miR-223/142 is a potential marker for lung inflammation
Since BALF MV- miR-223/142 were dramatically elevated in response to infectious stimulation (figure 2), we tested whether circulating MV- miR-223/142 potentially reflect inflammatory lung responses. Interestingly, serum MV-miR-223 were significantly increased in response to LPS (255.34-fold of increase, p = 0.007) and K.pneu (320.22-fold of increase, p = 0.001) (figure 4A). Meanwhile, serum derived MV-miR-142 was also upregulated after LPS (68.67-fold of increase, p = 0.022) and K.pneu (158.31-fold of increase, p = 0.001) treatment. Additionally, the expression of free miR-223/142 was detected in total serum. However, no significant differences were observed between the control group and infectious stimuli-treated groups (LPS and K.pneu) as shown in figure 4C and D. Furthermore, we examined miRNA profiles in the BALF EVs (figure 4E) and the serum EVs (figure 4F) which were isolated from hyperoxia-treated mice (a noninfectious ALI model), using the FirePlex miRNA Immunology Panel The miRNA profiles obtained from hyperoxia-treated mice (figure 4E and 4F) were markedly different comparing with those obtained using infectious models (figure 2B).
Figure 4. Elevated miR-223/142 in serum-derived MVs during lung inflammation.
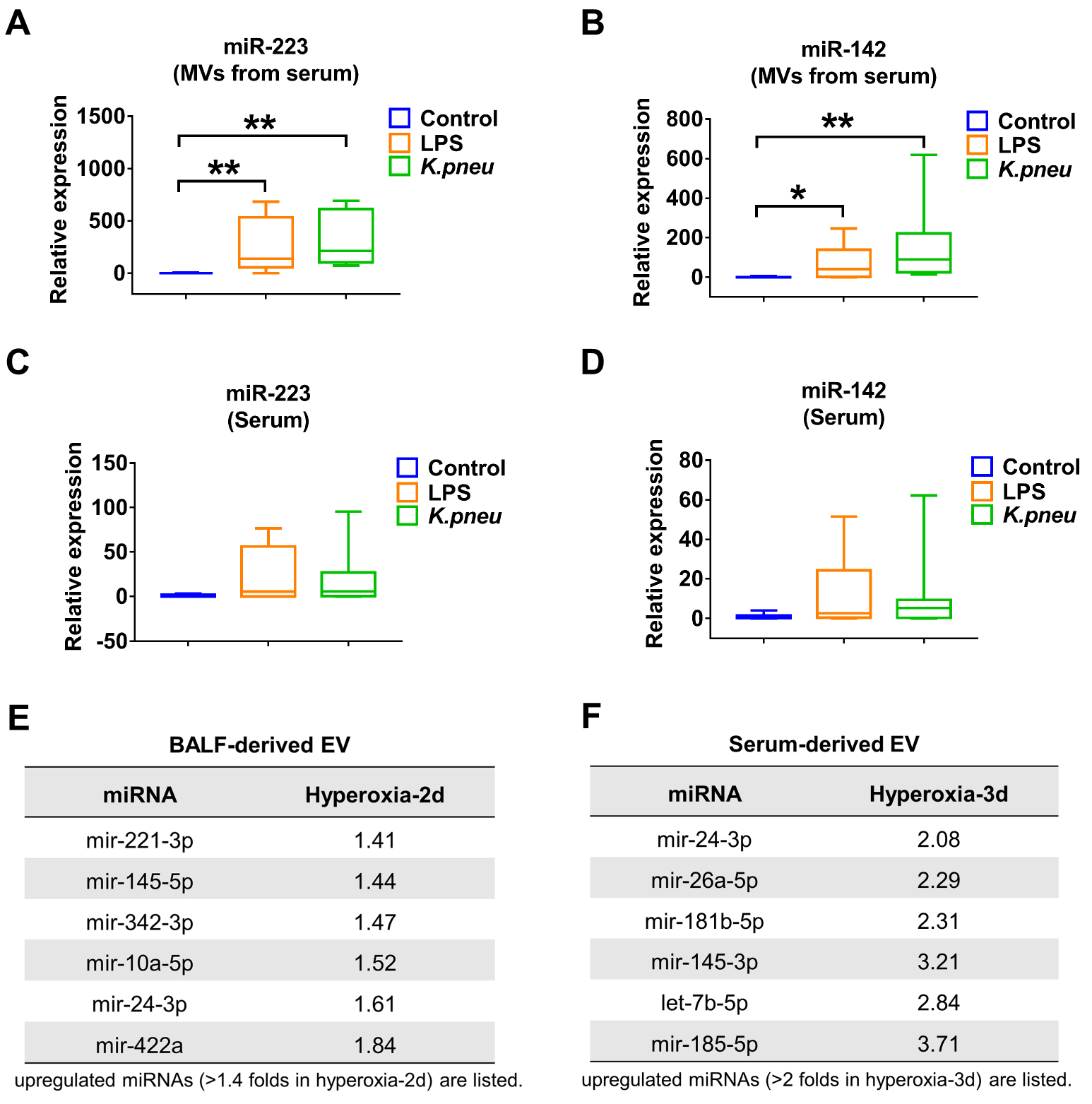
(A and B) Relative levels of miR-223 (A) and miR-142 (B) in serum-derived MVs were detected in control, LPS- and K.pneu-treated mice using qRT-PCR. (C and D) Relative levels of miR-223 (C) and miR-142 (D) in total serum were detected in control, LPS- and K.pneu-treated mice using qRT-PCR. The boxes in the box-plots show the medians with 25th and 75th percentiles, and the whiskers show the 10th and 90th percentiles; n = 8 mice per group. Data were analyzed using a Kruskal-Wallis One-Way ANOVA followed by pair-wise testing with Mann-Whitney U tests. *, p < 0.05; **, p < 0.01 versus the control group. (E and F) C57BL/6J mice (n = 6 mice per group) were exposed to room air or hyperoxia (100%). EVs were isolated from BALF or serum after 2 or 3 days as indicated. Pooled RNA from BALF EVs (E) or serum EVs (F) was profiled using the FirePlex® miRNA Immunology Panel. The upregulated miRNAs in BALF EVs (E) or serum EVs (F) after hyperoxia exposure were shown in tables.
MiR-223/142 regulate Nlrp3 inflammasome components in macrophages
MiR-223 and miR-142 have been identified as anti-inflammatory miRNAs in several studies.23–26 After LPS or K.pneu stimulation, the expression of miR-223/142 in BMDM and THP-1 was decreased while the macrophages were activated (figure 5A–D), indicating that miR-223/142 play an important role in macrophage classical activation in response to infectious stimuli. Furthermore, we identified that NLRP3 and ASC were potential targets of miR-223 and miR-142, respectively (figure 5E). Next, after transfection of miR-223/142 inhibitors or mimics, Nlrp3 (miR-223 mimics: 0.48-fold, p = 0.021; miR-223 inhibitors: 1.66-fold, p = 0.017) and Asc (miR-142 mimics: 0.44-fold, p = 0.047; miR-142 inhibitors: 1.55-fold, p = 0.018) protein levels were directly altered, as detected by western blot analysis (figure 5F, G). Dual luciferase assay demonstrated that miR-223/142 mimics were able to repress the 3’ UTR of Nlrp3 (0.29-fold, p < 0.001) and Asc (0.43-fold, p < 0.001), respectively (figure 5H), and the inhibitory effects were rescued when the predicted miRNA bind site was mutated (figure 5E, H). It is known that NLRP3 and ASC are key components of the NLRP3 inflammasome. Therefore, we then measured the caspase-1 activity in primed BMDMs that were transfected with miR-223/142 mimics. We found a significant inhibition of caspase-1 activity in miR-223/142 over-expressed BMDMs (mimics control: 1054931.625 ± 296326.596 relative luminometer units (RLU) vs miR-223 mimics: 473999.250 ± 244124.986 RLU (p = 0.02); vs miR-142 mimics: 512505.250 ± 253929.356 RLU (p = 0.029); vs miR-223/142 mimics: 238909.025 ± 84816.262 RLU (p = 0.002)) (figure 5I).
Figure 5. miR-223/142 control Nlrp3 inflammasome activity in macrophages.
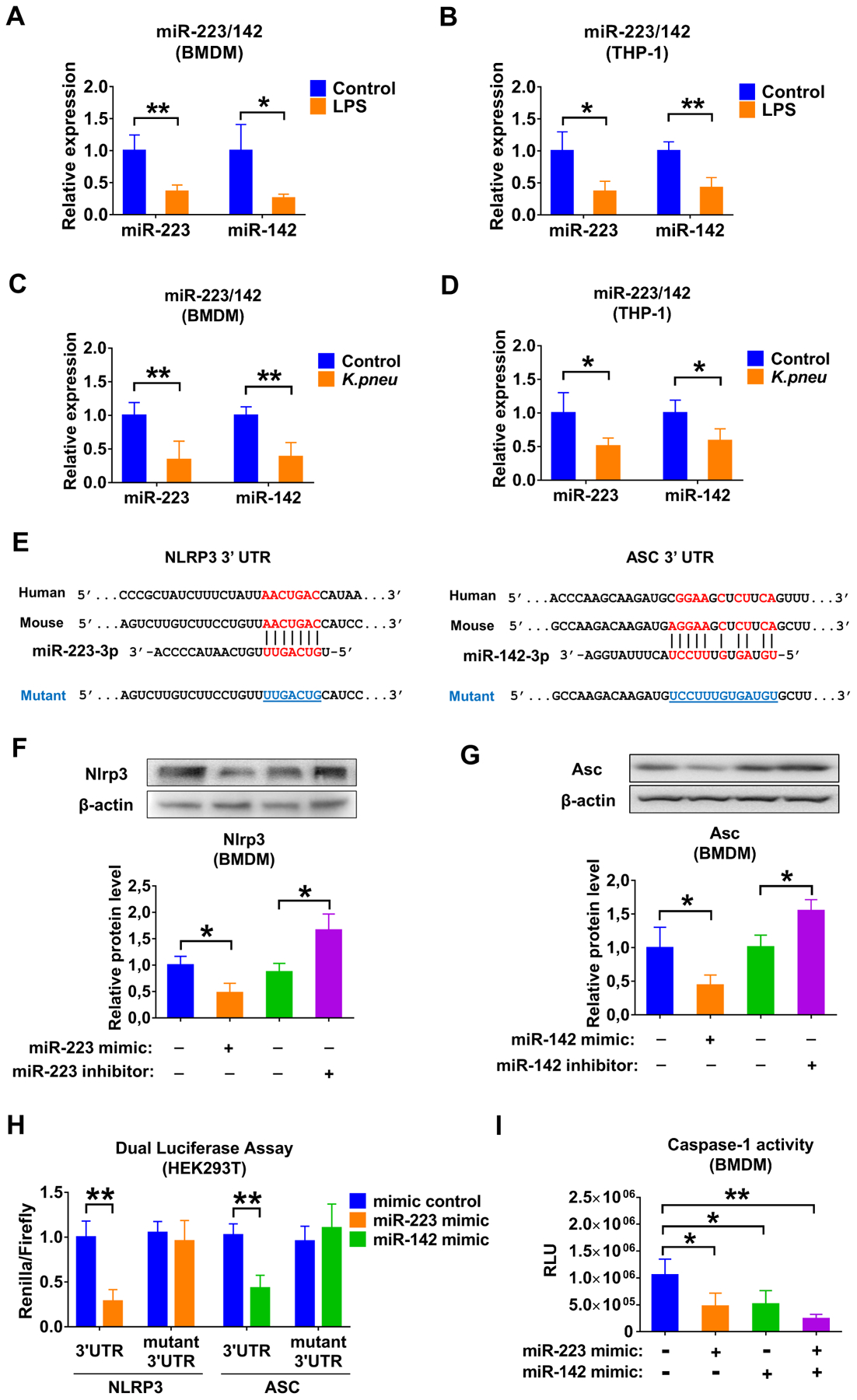
(A and B) Macrophages were cultured with or without LPS (100 ng/mL) for 24 h. Total RNA was purified from BMDM (A) or THP-1 (B), the expression of miR-223 and miR-142 was detected using qRT-PCR. (C and D) Macrophages were cultured with or without K. pneu treatment (107 CFU bacteria/105 macrophages, 1 h incubation). After 24 h, total RNA was purified from BMDM (C) or THP-1 (D), and the expression of miR-223 and miR-142 was detected using qRT-PCR. (E) Schematic of the miR-223 binding site to the 3’UTR of the NLRP3 and also the miR-142 binding site to the 3′ UTR of the ASC. Mutations (underlined) were introduced into NLRP3 and ASC 3’ UTR to disrupt base-pairing with miR-223 and miR-142, respectively. (F and G) Overexpression and inhibition of miR-223 or miR-142 were performed using transfection. 24 h later, Nlrp3 (F) and Asc (G) in BMDM were detected using western blot analysis. The relative protein level was quantified and normalized to β-actin. (F) Renilla luciferase reporter containing wild-type or mutant NLRP3 3′ UTR was co-transfected into HEK293T cells with miR-223 mimic or mimic control, respectively. Renilla luciferase reporter containing wild-type or mutant ASC 3′ UTR was co-transfected into HEK293T cells with miR-142 mimic or mimic control, respectively. 48 h after transfection, renilla luciferase activity was measured and normalized to firefly luciferase activity. (G) Mimic control, miR-223 and/or miR-142 mimic was transfected into BMDM. After transfection, BMDM were primed with LPS (1 μg/mL) for 5 min followed by addition of ATP (5 mM) for 30 min without removal of LPS. Caspase-1 activity in BMDM was measured 24 h after transfection. All these results presented as mean + SD are from 3 or 4 independent experiments. In panel A to D, data were analyzed using a One-Way ANOVA followed by pair-wise testing with Student’s t-tests. In panel F to I, data were analyzed using a One-Way ANOVA followed by Tukey’s HSD. *, p < 0.05; **, p < 0.01 versus their corresponding control.
MiR-223/142 delivery via MVs attenuates the K.pneu-induced lung inflammation.
Previously, we developed a protocol using serum-derived Exos as vehicles to deliver designated small RNA molecules into lung macrophages in vivo.16 Here, we slightly modified the method by using BALF MVs. First, we tracked the recipient cells in K.pneu-pretreated mice that took up instilled MVs (figure 6A). Similarly to our previous observation, the majority of PKH26-positive cells were CD68 (green, a marker of monocyte lineage) positive, but Ly-6G/Ly-6C (a marker of polymorphonuclear neutrophils (PMNs)) negative (figure 6B), suggesting that MVs were selectively taken by lung macrophages rather than PMNs.
Figure 6. Delivery of miR-223/142 via MVs attenuates K.pneu-induced lung inflammation.
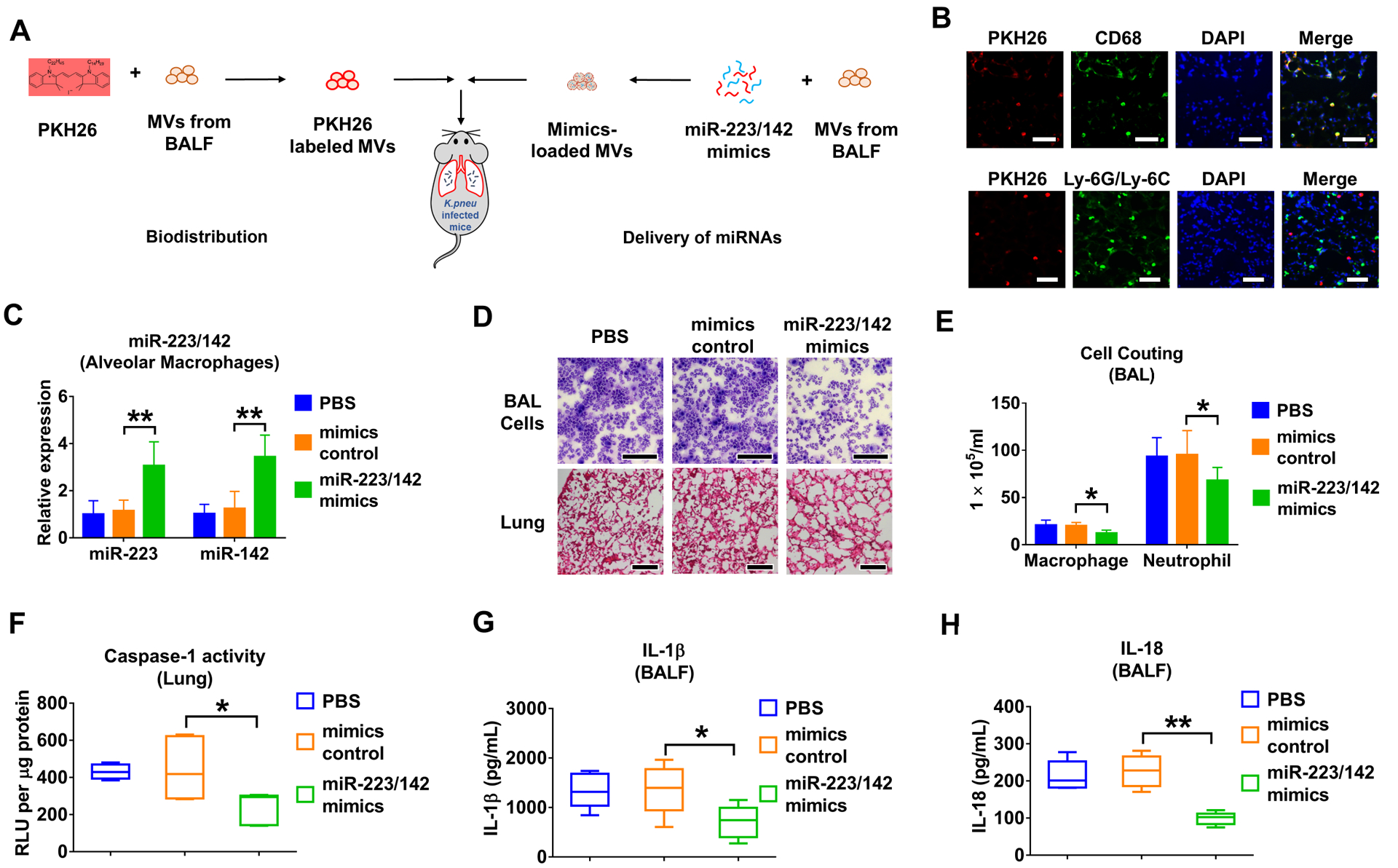
(A) Schematic illustration of tracking PKH26-labeled MVs after i.t. or delivery of miR-223/142 mimics into K.pneu-pretreated mice via MVs. (B) WT mice received K. pneu (104 CFU in 50 μL PBS) via i.t. 3 hours before MVs administration. 100 μg PKH26-labeled MVs in 50 μL PBS were given to K.pneu-pretreated mice intratracheally. Mice were then sacrificed 24 h after MVs administration (n = 5 mice per group). Immunofluorescence staining was performed in lung sections using antibodies against CD68 (macrophage marker) or Ly-6G (neutrophil marker) to track MVs uptake. Scale bars = 100 μm. (C-H) 100 pmol mimics control or miR-223/142 mimics were electroporated into 100 μg BALF MVs. 50 μL PBS (as a negative control) or small RNA-loaded MVs (in 50 μL PBS) were administrated via i.t.. Mice were sacrificed 24 h after MVs administration. The levels of miR-223 and miR-142 were detected in macrophages isolated from BALF (n = 5 mice per group) (C). H&E staining was performed using BALF cells and lung sections. Scale bars = 100 μm (D) (n = 5 mice per group). The number of macrophages or neutrophils collected from BALF was counted (E) (n = 6 mice per group). The activity of caspase-1 in murine lung tissue was measured and normalized to total protein (F) (n = 6 mice per group). The secretion of IL-1β (G) and IL-18 (H) in BALF was detected using ELISA (n = 6 mice per group). In panel C and E, results presented as mean + SD were analyzed using a One-Way ANOVA followed by Tukey’s HSD. In panel F, G and H, the boxes in the box-plots show the medians with 25th and 75th percentiles, and the whiskers show the 10th and 90th percentiles. Data were analyzed using a Kruskal-Wallis One-Way ANOVA followed by pair-wise testing with Mann-Whitney U tests. *, p < 0.05; **, p < 0.01 versus mimics control group.
Next, to evaluate whether miR-223/142-loaded MVs potentially modulate inflammatory lung responses, WT mice were pre-infected with K.pneu. After 3 h, the infected mice received MV-mediated delivery of miRNA mimics control or MV-miR-223/142 mimics intratracheally (figure 6A). The increased levels of miR-223 (3.06-fold of increase, p = 0.004) and miR-142 (3.44-fold of increase, p = 0.001) were confirmed in macrophages obtained from BALF using qRT-PCR (figure 6C). Significantly less cellular infiltration was observed in lung tissue obtained from the miR-223/142 mimics-treated mice than in the mimics control group (figure 6D). Consistently, total cell counts of macrophages (mimics control: 20.100 ± 3.423 × 105 mL vs miR-223/142 mimics: 12.150 ± 3.313 × 105 mL, p = 0.014) and neutrophils (mimics control: 95.283 ± 25.674 × 105 mL vs miR-223/142 mimics: 67.917 ± 13.956 × 105 mL, p = 0.045) were significantly reduced in miR-223/142 mimics-treated mice (figure 6E). Moreover, MV-mediated delivery of miR-223/142 mimics suppressed the inflammasome activity (mimics control: 443.146 ± 153.129 RLU per μg protein vs miR-223/142 mimics: 246.326 ± 81.203 per μg protein, p = 0.012) (figure 6F), and secretion of IL-1β (mimics control: 1354.765 ± 479.887 pg/mL vs miR-223/142 mimics: 716.014 ± 337.261 pg/mL , p = 0.032) and IL-18 (mimics control: 226.671 ± 41.678 pg/mL vs miR-223/142 mimics: 99.509 ± 16.392, p < 0.001) (figure 6G, H) in the lung. Consistently, we observed miR-223/142 inhibitor delivered via MVs promoted LPS induced lung inflammation (data not shown). These data suggested that the delivery of miR-223/142 mimics via MVs promoted anti-inflammatory effect and attenuated the K.pneu-induced lung inflammation.
DISCUSSION
In the past decade, miRNAs have become attractive candidates as novel biomarkers to evaluate human disease development, particularly in the cancer field.27 Compared to protein markers, miRNA levels have unique merits.5 For example, the miRNA level can be measured rapidly and accurately owing to high-throughput sequencing technology.28 Furthermore, the combination of multiple miRNAs which are differentially expressed in different pathways would provide more information than protein markers.29 This is particularly important when developing novel biomarkers using body fluids. Moreover, compared to protein-based markers, detecting miRNAs using polymerase chain reaction (PCR) and modern sequencing technologies requires much less labor and time than developing a specific antibody against a particular protein. The high stability of miRNAs in body fluids, probably due to their relatively shorter sequences compared with other long non-coding RNAs, represents additional advantages to serve as a biomarker.5,29 Approximately a decade ago, miRNAs were first detectable in circulation within lipid-encapsulated microvesicles.30 More recently, these lipid-encapsulated microvesicles have become named extracellular vesicles (EVs). EV-encapsulating miRNAs draw intense interest due to their potential of being developed into noninvasive biomarkers and drug-delivery agents.7,31 The EV cargo is composed of mostly nucleotides and proteins. EVs protect these encapsulated components from degradation.32 Furthermore, EVs carry the same surface protein markers as their originated (parent) cells. This feature can be better used as a biomarker to reflect the pathogenesis of the cell-of-origin.33 Additionally, EVs can serve as a cell-specific carrier to facilitate the delivery of miRNAs as we reported previously.16
It is known that miR-223 and miR-142 belong to hematopoietic tissue-specific miRNAs.34 MiR-223 and miR-142 were both reported previously to regulate inflammation in tissue and organs other than lungs.23–26,34 The first merit of our studies is that miR-223/142 synergistically suppressed NLRP3 inflammasome activation and bacterial infection-associated lung inflammation in vivo. We chose MVs in our study because miR-223/142 are enriched in MVs when compared to exosomes (data not shown). Consistently, previous publications from other groups also show that exosomes contain fewer miRNAs.35 More importantly, we found that both miR-223 and miR-142 were selectively secreted from macrophages in an MV-mediated manner. MiRNA 3’ end uridylation was essential in the miR-223/142 encapsulation into MVs. Interestingly, in eukaryotes, uridylation has been reported to alter the half-life of RNA molecules.36 Our data suggest that uridylation of miR-223/142 facilitate the reduction of intracellular levels of miR-223/142, probably via MV-mediated secretion. We added one novel role of uridylation in regulating intracellular levels of miRNAs in response to noxious stimuli.
The significance of the above findings is that MV-miR-223/142 detected in body fluids, including both BALF and serum, are potentially practical biomarkers that reflect the activation of macrophages towards pro-inflammatory directions. Compared with the detection of non-EV-containing miR-223/142, MV-containing miR-223/142 potentially suggest the specific organs/locations in which macrophages are being activated. MV surface markers are often the same as the surface proteins of their “parent” cells,32 given that MVs are directly generated via surface blebs.37 Furthermore, we do not observe elevated MV-miR-223/142 derived from BALF or serum in hyperoxia-induced ALI mice (figure 4E, F), which is a noninfectious condition. Taken together, our work potentially suggests a novel method by which circulating MV-miR-223/142 can be used to predict lung inflammation and its dynamics post bacterial infections.
Another insight of this current report is that the recipient cells of exogenously delivered MVs are mainly lung macrophages. Based on this feature, we delivered the miR-223/142 mimics in vivo using MVs. Our work confirmed exogenously delivered miR-223/142 prohibited the activation of the NLRP3 inflammasome and provided an anti-inflammation effect in the lung during K.pneu-induced infection. Therefore, MV-mediated miRNA molecules potentially provide a novel therapeutic strategy in a cell-specific manner. Using MVs as a vehicle to deliver exogenous nucleotides has several potential advantages compared with other nanoparticle-mediated delivery methods. First, MVs can be obtained from the blood of the host.38 Therefore, MV-mediated delivery potentially triggers less immune response and may act like the autograft transplantation.39 Next, as stated above, the macrophage-specific MV-mediated delivery illustrated in our report potentially adds more efficacy and less off-target effects when using miRNA molecules as therapeutics.
In our current studies, we compared the levels between MV-miR-223/142 and secreted non-MV-miR-223/142 (figure 4C, D). MiRNAs are released in multiple ways, including, but not limited to, a MV-mediated manner. Our data showed that non-MV-shuttling miR-223/142 are not altered in serum. These observation suggested that the detection of the MV-encapsulating miR-223/142 potentially indicates the cellular origin of these released miRNAs. Moreover, we used a systemic inflammation mouse model induced by cecal ligation and puncture (CLP). Total miR-223 level in the serum was detected in both CLP and sham surgery groups. We observed that systemic inflammation induced by CLP increases total miR-233 level in mouse serum, which is different than from lung inflammation (data not shown). Therefore, by detecting the circulating EV-miR-223/142, it might provide a way to distinguish lung inflammation from systemic inflammation.
Currently, we did not address the kinetics of MV-miR-223/142 in our manuscript. MVs are constantly generated and taken up by surrounding cells.40 After noxious stimuli, the secretion of MVs from cells is much more significant compared to their uptake. One of our future directions will focus on addressing the dynamic turnover of MVs and MV-miR-223/142 in our disease models.
As summarized in figure 7, our work demonstrates that miR-223/142 are actively secreted from macrophages via MVs at the time when macrophages are activated towards a proinflammatory direction. Mechanistically, 3’ uridylation plays a role in the miR-223/142 encapsulation into MVs and their secretion into extracellular spaces. MV-miR-223/142 detected in circulation may serve as novel biomarkers to indicate lung macrophage activation and/or lung inflammation. Delivery of MV-containing miR-223/142 mimics in vivo effectively modifies the intracellular level of miR-223/142 and subsequently suppresses macrophage activation via inhibiting NLRP3 inflammasome activation and inflammatory lung responses.
Figure 7. Schematic review of the MVs-mediated secretion and function of miR-223/142 in macrophages during lung inflammation.
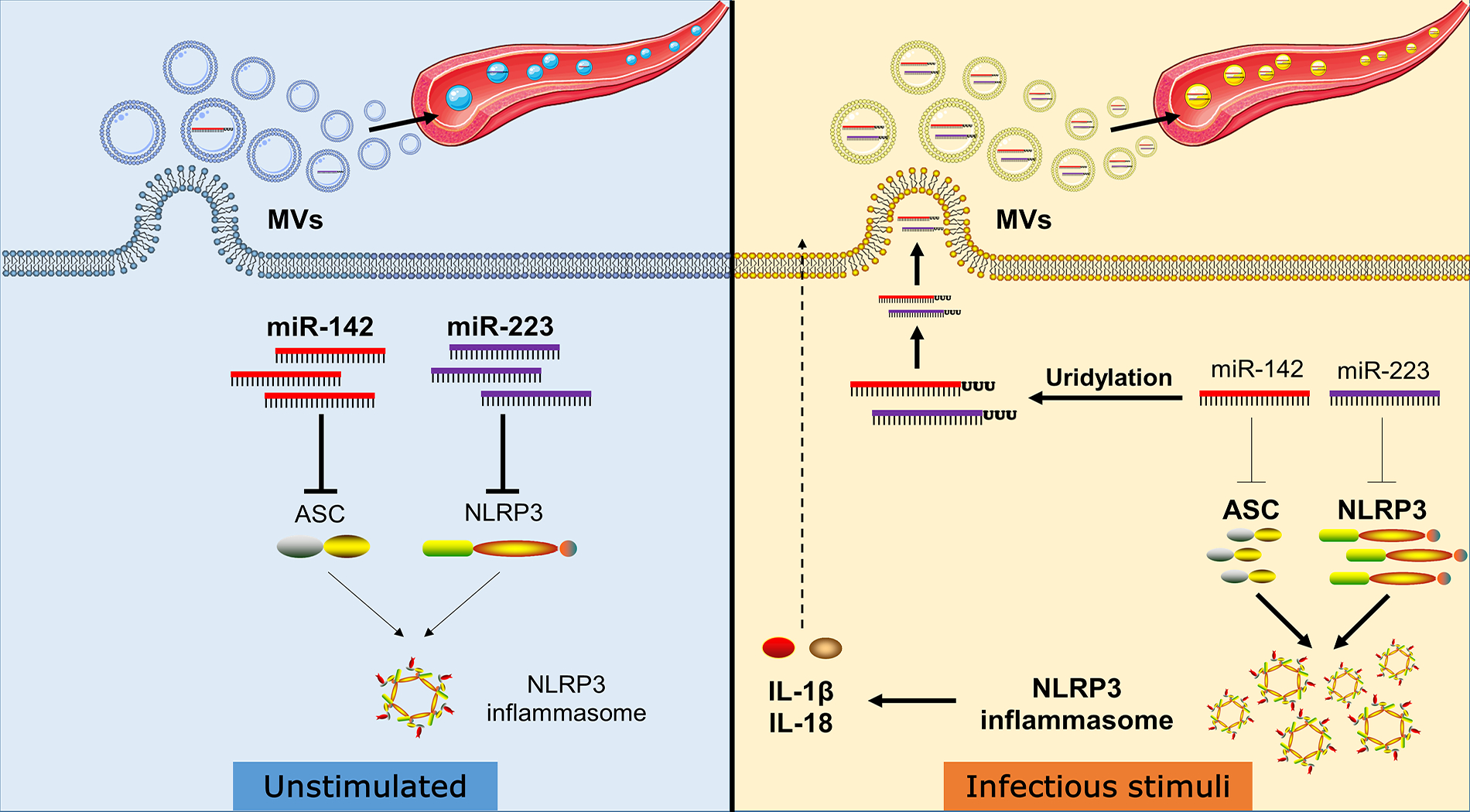
Infectious stimuli increase in non-templated 3’ uridylation of miR-223/142, which enhances their packaging into MVs. The reduced levels of miR-223/142 via MVs-mediated release in macrophages lead to increased expression of NLRP3 and ASC protein, two key components of the NLRP3 inflammasome. Accumulated NLRP3 and ASC proteins facilitate the NLRP3 inflammasome activity and promote the maturation and secretion of pro-inflammatory cytokines IL-1β and IL-18. Besides, miR-223/142 enriched MVs are released into the circulation and might serves as a diagnostic biomarker for lung inflammation.
What is the key question?
The primary question in this manuscript is to delineate whether BALF MV and MV-miRNA cargo are altered after infectious stimuli both in vivo and in vitro. We further investigated whether BALF MV and MV-miRNA cargo can serve as a novel target to develop diagnostic and therapeutic methods.
What is the bottom line?
Intracellular miR-223/142 suppress NLRP3 inflammasome activation in lung macrophages without stimulation. Infectious stimuli lead to 3’ uridylation of miR-223/142 and subsequently a robust release of miR-223/142 via MVs, which occurs along with the NLRP3 inflammasome activation.
Why read on?
Our study suggests circulating MV-containing miR-223/142 potentially serve as a novel diagnostic marker for LPS/ G- bacterial-induced lung inflammation. Moreover, delivery of miR-223/142 via MVs selectively target lung macrophages and provides a novel anti-inflammatory strategy during G- bacterial infection.
Funding
This work was supported by National Institutes of Health (NIH) grants R21 AI121644, R33 AI121644, R01 GM111313, R01 GM127596, Wing Tat Lee award (all to YJ); by NIH grant K99HL141685 (to DZ).
Footnotes
Competing interests None declared.
REFERENCES
- 1.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 2009;22(2):240–73, Table of Contents. doi: 10.1128/CMR.00046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ulloa L, Brunner M, Ramos L, et al. Scientific and clinical challenges in sepsis. Curr Pharm Des 2009;15(16):1918–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felekkis K, Touvana E, Stefanou C, et al. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia 2010;14(4):236–40. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu H, Fan GC. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am J Cardiovasc Dis 2011;1(2):138–49. [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Peng R, Wang J, et al. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenetics 2018;10:59. doi: 10.1186/s13148-018-0492-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang YC, Li Y, Wang XY, et al. Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. Diabetologia 2013;56(10):2275–85. doi: 10.1007/s00125-013-2996-8 [DOI] [PubMed] [Google Scholar]
- 7.Sadovska L, Eglitis J, Line A. Extracellular Vesicles as Biomarkers and Therapeutic Targets in Breast Cancer. Anticancer Res 2015;35(12):6379–90. [PubMed] [Google Scholar]
- 8.Lee H, Zhang D, Laskin DL, et al. Functional Evidence of Pulmonary Extracellular Vesicles in Infectious and Noninfectious Lung Inflammation. J Immunol 2018;201(5):1500–09. doi: 10.4049/jimmunol.1800264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Z, Zhang D, Lee H, et al. Macrophage-derived apoptotic bodies promote the proliferation of the recipient cells via shuttling microRNA-221/222. J Leukoc Biol 2017;101(6):1349–59. doi: 10.1189/jlb.3A1116-483R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med 2006;3(11):e442. doi: 10.1371/journal.pmed.0030442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Si-Tahar M, Touqui L, Chignard M. Innate immunity and inflammation--two facets of the same anti-infectious reaction. Clin Exp Immunol 2009;156(2):194–8. doi: 10.1111/j.1365-2249.2009.03893.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H, Zhang D, Zhu Z, et al. Epithelial cell-derived microvesicles activate macrophages and promote inflammation via microvesicle-containing microRNAs. Sci Rep 2016;6:35250. doi: 10.1038/srep35250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Happel KI, Zheng M, Young E, et al. Cutting edge: roles of Toll-like receptor 4 and IL-23 in IL-17 expression in response to Klebsiella pneumoniae infection. J Immunol 2003;170(9):4432–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H, Zhang D, Wu J, et al. Lung Epithelial Cell-Derived Microvesicles Regulate Macrophage Migration via MicroRNA-17/221-Induced Integrin beta1 Recycling. J Immunol 2017;199(4):1453–64. doi: 10.4049/jimmunol.1700165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, Lee H, Wang X, et al. Exosome-Mediated Small RNA Delivery: A Novel Therapeutic Approach for Inflammatory Lung Responses. Mol Ther 2018;26(9):2119–30. doi: 10.1016/j.ymthe.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Polverino F, Rojas-Quintero J, et al. A Disintegrin and A Metalloproteinase-9 (ADAM9): A Novel Proteinase Culprit with Multifarious Contributions to COPD. Am J Respir Crit Care Med 2018. doi: 10.1164/rccm.201711-2300OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D, Lee H, Cao Y, et al. miR-185 mediates lung epithelial cell death after oxidative stress. Am J Physiol Lung Cell Mol Physiol 2016;310(7):L700–10. doi: 10.1152/ajplung.00392.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honda S, Kirino Y. Dumbbell-PCR: a method to quantify specific small RNA variants with a single nucleotide resolution at terminal sequences. Nucleic Acids Res 2015;43(12):e77. doi: 10.1093/nar/gkv218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang D, Lee H, Haspel JA, et al. Long noncoding RNA FOXD3-AS1 regulates oxidative stress-induced apoptosis via sponging microRNA-150. FASEB J 2017;31(10):4472–81. doi: 10.1096/fj.201700091R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsatsaronis JA, Franch-Arroyo S, Resch U, et al. Extracellular Vesicle RNA: A Universal Mediator of Microbial Communication? Trends Microbiol 2018;26(5):401–10. doi: 10.1016/j.tim.2018.02.009 [DOI] [PubMed] [Google Scholar]
- 22.Thornton JE, Du P, Jing L, et al. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res 2014;42(18):11777–91. doi: 10.1093/nar/gku805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhen J, Chen W. MiR-142 inhibits cecal ligation and puncture (CLP)-induced inflammation via inhibiting PD-L1 expression in macrophages and improves survival in septic mice. Biomed Pharmacother 2018;97:1479–85. doi: 10.1016/j.biopha.2017.11.058 [DOI] [PubMed] [Google Scholar]
- 24.Sun Y, Varambally S, Maher CA, et al. Targeting of microRNA-142–3p in dendritic cells regulates endotoxin-induced mortality. Blood 2011;117(23):6172–83. doi: 10.1182/blood-2010-12-325647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang G, Meng C, Guo X, et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 2012;125(23):2892–903. doi: 10.1161/CIRCULATIONAHA.111.087817 [DOI] [PubMed] [Google Scholar]
- 26.Neudecker V, Haneklaus M, Jensen O, et al. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med 2017;214(6):1737–52. doi: 10.1084/jem.20160462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stokowy T, Eszlinger M, Swierniak M, et al. Analysis options for high-throughput sequencing in miRNA expression profiling. BMC Res Notes 2014;7:144. doi: 10.1186/1756-0500-7-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2012;4(3):143–59. doi: 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter MP, Ismail N, Zhang X, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 2008;3(11):e3694. doi: 10.1371/journal.pone.0003694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H, Zhang D, Rai A, et al. The Obstacles to Current Extracellular Vesicle-Mediated Drug Delivery Research. J Pharm Pharm 2017;4(2):156–58. doi: 10.15436/2377-1313.17.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whiteside TL. Extracellular vesicles isolation and their biomarker potential: are we ready for testing? Ann Transl Med 2017;5(3):54. doi: 10.21037/atm.2017.01.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol 2014;14(3):195–208. doi: 10.1038/nri3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramkissoon SH, Mainwaring LA, Ogasawara Y, et al. Hematopoietic-specific microRNA expression in human cells. Leuk Res 2006;30(5):643–7. doi: 10.1016/j.leukres.2005.09.001 [DOI] [PubMed] [Google Scholar]
- 35.Chevillet JR, Kang Q, Ruf IK, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A 2014;111(41):14888–93. doi: 10.1073/pnas.1408301111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim J, Ha M, Chang H, et al. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 2014;159(6):1365–76. doi: 10.1016/j.cell.2014.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200(4):373–83. doi: 10.1083/jcb.201211138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavallari C, Ranghino A, Tapparo M, et al. Serum-derived extracellular vesicles (EVs) impact on vascular remodeling and prevent muscle damage in acute hind limb ischemia. Sci Rep 2017;7(1):8180. doi: 10.1038/s41598-017-08250-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeung JC, Keshavjee S. Overview of clinical lung transplantation. Cold Spring Harb Perspect Med 2014;4(1):a015628. doi: 10.1101/cshperspect.a015628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaborowski MP, Balaj L, Breakefield XO, et al. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015;65(8):783–97. doi: 10.1093/biosci/biv084 [DOI] [PMC free article] [PubMed] [Google Scholar]


