Abstract
Background
Rodents constitute an important part of the diet of many carnivore species. This predator-prey food chain is exploited by helminth parasites, such as cestodes, whose larval stages develop in rodents and then mature to the adult stage in predators. The main aim of our study was to use molecular techniques for identification of cestode species recovered from both intermediate and definitive hosts, with a particular focus on the genus Mesocestoides.
Methods
Larval cestodes were obtained during our long-term studies on rodent helminth communities in the Mazury Lake District in the north-east Poland in 2000–2018. Cestode larvae/cysts were collected from body cavities or internal organs (e.g. liver) during autopsies. Adult tapeworms were derived from nine red foxes, three Eurasian badgers and one Eurasian lynx. PCR amplification, sequencing and phylogenetic analyses were conducted employing three genetic markers: 18S rDNA, mitochondrial (mt) 12S rDNA and the mt cytochrome c oxydase subunit 1 (cox1) gene fragment.
Results
Altogether 19 Mesocestoides samples were analyzed, including 13 adult tapeworms from definitive hosts and six larval samples from 4 bank voles and 2 yellow-necked mice. Phylogenetic analyses revealed three well-supported trees of similar topology. In each case the Mesocestoides samples formed two separate clades. All isolates from foxes, the lynx isolate and two isolates from rodents grouped with Mesocestoides litteratus. Four isolates from rodents and all three isolates from Eurasian badgers were resolved in a separate clade, most similar to North American M. vogae (syn. M. corti). Examination of fixed, stained adult specimens from Eurasian badgers revealed consistency with the morphology of Mesocestoides melesi. Therefore, this clade is likely to represent M. melesi, a species first described in 1985 from the Eurasian badger Meles meles. Molecular analysis allowed also the identification of Taenia crassiceps, Hydatigera kamiyai and Cladotaenia globifera among larvae derived from rodents.
Conclusions
Molecular and phylogenetic analyses support the recognition of M. melesi as a valid species. Our data represent the first record of the larvae of this species in rodents. This is the first report on the occurrence of H. kamiyai in rodents from Poland.
Keywords: Mesocestoides, Hydatigera, Taenia crassiceps, Rodents, Fox, Badger, Lynx
Background
Rodents constitute an important part of the diet of many carnivorous species. This predator-prey food chain is exploited by helminth parasites, such as cestodes, whose larval stages develop in rodents and then mature to the adult stage in predators (both carnivorous mammals and birds of prey). The role of rodents as obligatory intermediate or paratenic hosts of tapeworms exploiting this route of transmission (families Mesocestoididae, Taeniidae and Paruterinidae) is therefore indispensable in enabling the completion of their life-cycles.
In our previous studies on parasite communities of rodents from north-east Poland, we investigated the larval cestodes present in different body cavities and in the liver [1–5]. The larval stages of several cestode species were recognized in bank voles (Myodes glareolus) by morphological features, including Mesocestoides sp., Cladotaenia globifera, Taenia martis, Taenia mustelae and Hydatigera taeniaeformis (syn. Taenia taeniaeformis). However, in recent years molecular studies have revealed that some of these species actually comprise complexes that include cryptic species which could not be distinguished earlier by conventional morphological examination. Hence re-description of these species has been necessary and driven primarily by their genetic signatures, i.e. H. taeniaeformis parasitizing voles has been re-described as Hydatigera kamiyai and T. mustelae as Versteria mustelae [6, 7]. To the best of our knowledge, no such molecular studies, reporting the presence of newly raised species, have been carried out to date on cestode isolates from rodents in Poland.
Tapeworms of the genus Mesocestoides (Cyclophyllidea, Mesocestoididae) have been reported to parasitize a range of wild and domestic carnivores and even birds of prey as definitive hosts [8–10]. The systematics of Mesocestoides spp. is still not fully resolved [11, 12] and the unarmed scolex and pleomorphic metacestodes/larvae (tetrathyridia) found in rodents and other intermediate hosts (insectivore mammals, birds, reptiles, etc.), do not provide sufficient characteristic features to enable unambiguous differentiation between species. To date, 4–7 Mesocestoides species have been reported from Europe [13–17]. The two most commonly reported species are M. litteratus found in red foxes (originally described as from a ‘fox’), rodents, grey wolves, dogs and cats among others; and M. lineatus that has been reported from domestic/wild cats (originally described from wild cats, Felis sylvestris) and dogs, jackals and other carnivores [18]. In Poland, only one molecular study has been completed on Mesocestoides larvae from rodent hosts, and this identified M. litteratus in striped field mice Apodemus agrarius and M. glareolus from the Wrocław area (western Poland) [19]. Red foxes (Vulpes vulpes) are considered to be the principal hosts of adult Mesocestoides spp. in Poland [20]. In recent years we have carried out extensive studies on different parasites of the red fox from different regions of Poland and we have confirmed the high overall prevalence of Mesocestoides in foxes, with a prevalence of 88% in all of the sampled populations [21], as in Karamon et al. [20].
The main aim of our current study was to use molecular techniques for identification of, and comparison between, cestode species recovered from both intermediate and definitive hosts: sylvatic rodents, red foxes and other definitive hosts, with a particular focus on Mesocestoides spp.
Methods
Larval cestodes were obtained during our long-term studies on rodent helminths in the Mazury Lake District in north-east Poland in 2000–2018 [1–5]. In addition, one Mesocestoides sample was obtained from a yellow-necked mouse (Apodemus flavicollis) from the Białowieża Forest region, north-east Poland. Altogether, ten infected rodents were examined, including five bank voles M. glareolus, two yellow-necked mice A. flavicollis, two common voles Microtus arvalis and one striped field mouse A. agrarius (Table 1). Cestode larvae from body cavities, identified preliminarily as Mesocestoides spp., were obtained from seven rodents, including one sample identified later by molecular typing as an undeveloped Hydatigera larva. In one sample, cysts found in the body cavity were morphologically identified as T. crassiceps. Two larval samples were derived from rodent livers: one mature strobilocercus of Hydatigera sp. and numerous C. globifera larvae. The host species for each specimen are recorded in Table 1.
Table 1.
Origin (host species, region and site) and results of genotyping for larval and adult cestodes involved in the study
| Host group | Host ID | Host species | Region, site, year | Developmental stage, localization | Cestode species (morphological) | Cestode species (molecular) | GenBank ID | ||
|---|---|---|---|---|---|---|---|---|---|
| 18S rDNA | 12S rDNA | cox1 | |||||||
| Rodents | 0005 | M. glareolus | Masuria, U, 2018 | Larvae, PC | Mesocestoides sp. | M. melesi | MN512706 | MN505192 | MN514024 |
| 0029 | M. glareolus | Masuria, U, 2018 | Larvae, PC | Mesocestoides sp. | M. melesi | MN512707 | MN505193 | MN514025 | |
| 0066 | M. glareolus | Masuria, U, 2018 | Larvae, PC | Mesocestoides sp. | M. melesi | MN401347 | MN505194 | MN514026 | |
| 0130 | M. glareolus | Masuria, T, 2018 | Larvae, PC, Liv | Mesocestoides sp. | M. litteratus | MN401340 | MN505195 | MN514027 | |
| 00M3 | A. flavicollis | Masuria, U, 2001 | Larvae, PC | Mesocestoides sp. | M. melesi | MN401345 | MN505196 | MN514028 | |
| 0177 | A. agrarius | Masuria, T, 2018 | Larvae, Liv | C. globifera | C. globifera | nd | MN505197 | MN514029 | |
| 0D45 | M. arvalis | Masuria, U, 2000 | Larvae, Liv | H. taeniaeformis | H. kamiyai | nd | MN505198 | MN514030 | |
| 0D53 | M. arvalis | Masuria, U, 2000 | Larvae, PC | T. crassiceps | T. crassiceps | nd | nd | MN514031 | |
| D172 | M. glareolus | Masuria, U, 2000 | Larvae, PC | Mesocestoides sp. | H. kamiyai | nd | nd | MN514032 | |
| 0IS4 | A. flavicollis | Podlaskie, B, 2016 | Larvae, PC | Mesocestoides sp. | M. litteratus | MN401344 | MN505199 | MN514033 | |
| Canids | 0079 | V. vulpes | Mazovia, J, 2017 | Adult, SI | M. litteratus | M. litteratus | MN401342 | MN505200 | MN514034 |
| 0125 | V. vulpes | Kujawsko-Pomorskie, K, 2017 | Adult, SI | M. litteratus | M. litteratus | MN512708 | MN505201 | MN514035 | |
| 0138 | V. vulpes | Kujawsko-Pomorskie, K, 2017 | Adult, SI | M. litteratus | M. litteratus | MN512709 | nd | MN514036 | |
| 0143 | V. vulpes | Kujawsko-Pomorskie, K, 2017 | Adult, SI | M. litteratus | M. litteratus | MN401343 | MN505202 | MN514037 | |
| 0145 | V. vulpes | Kujawsko-Pomorskie, K, 2017 | Adult, SI | M. litteratus | M. litteratus | MN512710 | nd | MN514038 | |
| 0146 | V. vulpes | Kujawsko-Pomorskie, K, 2017 | Adult, SI | M. litteratus | M. litteratus | MN512711 | MN505203 | MN514039 | |
| 0321 | V. vulpes | Łódzkie, Wo, 2018 | Adult, SI | M. litteratus | M. litteratus | MN401341 | MN505204 | MN514040 | |
| 0322 | V. vulpes | Łódzkie, M, 2018 | Adult, SI | M. litteratus | M. litteratus | MN512712 | MN505205 | MN514041 | |
| 0280 | V. vulpes | Mazovia, W, 2018 | Adult, SI | T. crassiceps | T. crassiceps | MN512713 | MN505206 | MN514042 | |
| Mustelids | 0366 | M. meles | Masuria, Je, 2018 | Adult, SI | Mesocestoides sp. | M. melesi | MN512714 | MN505207 | nd |
| 0367 | M. meles | Masuria, Je, 2018 | Adult, SI | Mesocestoides sp. | M. melesi | MN401346 | MN505208 | MN514043 | |
| 0368 | M. meles | Masuria, Je, 2018 | Adult, SI | Mesocestoides sp. | M. melesi | nd | MN505209 | MN514044 | |
| Felids | 0IS1 | L. lynx | Podkarpackie, L, 2013 | Adult, SI | Mesocestoides sp. | M. litteratus | nd | MN505210 | nd |
Abbreviations: U, Urwitałt; T, Tałty; W, Warsaw; J, Jagodne; K, Kłóbka; Wo, Wolbórz, My, Myślenice, Je, Jedwabno, B, Białowieża, L, Lubaczów; PC, peritoneal cavity; Liv, liver; SI, small intestine; nd, not done
Adult Mesocestoides tapeworms were selected from eight red foxes (V. vulpes) originating from three administrative regions of Poland: the Mazowieckie, Łódzkie and Kujawsko-Pomorskie Voivodeships (Table 1). One adult T. crassiceps from a red fox was also included in the study for comparison with rodent samples. Additionally, adult Mesocestoides specimens from one Eurasian lynx (Lynx lynx) [22] and from three Eurasian badgers (Meles meles), from Podkarpackie Voivodeship, south-east Poland and the Mazury Lake District, north-east Poland, respectively, were also included (Table 1).
Morphological examination of Mesocestoides spp.
Larval Mesocestoides from rodents and adult Mesocestoides from badgers were flattened and fixed in AFA solution (100 ml 40% formaldehyde, 250 ml 95% ethanol, 100 ml glycerine, 50 ml glacial acetic acid, 500 ml distilled water) and stained using borax carmine, dehydrated in an ethanol series and mounted in Canada balsam for microscopical examination. Slides were examined and selected measurements were recorded using a NIKON Eclipse E-600 microscope with differential interference contrast, equipped with the NIS Elements Br 3.1 software (Nikon Instruments Co., Tokyo, Japan) for image processing and recording. Photographs were taken using a NIKON DX-1200 digital camera connected to the microscope.
DNA extraction and amplification
Genomic DNA was extracted from specimens fixed in ethanol (about 20 mg of tissue) using the DNAeasy Blood & Tissue kit (Qiagen, Hilden, Germany) and stored at a temperature of − 20 °C.
Molecular typing of tapeworms was performed by amplification and sequencing of three markers: (i) a fragment of c.1100 bp of 18S rDNA was amplified using the primers Worm A (5′-GCG AAT GGC TCA TTA AAT AG-3′) and 1270R (5′-CCG TCA ATT CCT TTA AGT TT-3′) [23]; (ii) a fragment of c.350 bp of mitochondrial (mt) 12S rDNA was amplified using the primers P60 for (5′-TTA AGA TAT ATG TGG TAC AGG ATT AGA TAC CC-3′) and P375 rev (5′-AAC CGA GGG TGA CGG GCG GTG TGT ACC-3′) [24]; (iii) a fragment of c.400 bp of the cytochrome c oxidase subunit 1 (cox1) was amplified using the primers JB3 (5′-TTT TTT GGG CAT CCT GAG GTT TAT-3′) and JB45 (5′-TAA AGA AAG AAC ATA ATG AAA ATG-3′) [25]. The PCR reactions were performed in a volume of 20 μl, including 1× PCR Dream Taq Green buffer (Thermo Fisher Scientific, Waltham, Massachusetts, USA), 1U Dream Taq polymerase (Thermo Fisher Scientific), 0.33 mM dNTPs, 1 μM of each primer and 2 μl of the extracted DNA sample. Negative controls were performed with nuclease-free distilled water, in the absence of template DNA.
All PCR reactions were carried out in identical cycling conditions: primary denaturation at 94 °C for 3 min, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 1 min, and elongation at 72 °C for 1 min, followed by a final elongation step at 72 °C for 7 min and a hold step at 4 °C.
PCR products were subjected to electrophoresis on a 1.5% agarose gel, stained with Midori Green stain (Nippon Genetics, GmbH). PCR products were directly sequenced in both directions by Genomed S.A. (Warsaw, Poland) with the primers used for DNA amplification. Sequences were aligned and visually inspected using Clustal W in MEGA v.7.0 [25]. Consensus sequences were compared with sequences deposited in the GenBank database.
Phylogenetic analyses were conducted separately for each molecular marker (Table 2). Sequences were aligned using E-INS-i algorithm implemented in Mafft version 7.271 [26, 27]. Maximum likelihood trees were obtained in RAxML version 8.2.4 [28] assuming a GTR + G model for the nucleotide substitution process. The topology and branch lengths were optimized starting the analysis 200 times with distinct randomized maximum parsimony trees. Branch support values were obtained during 1000 rapid bootstrap replicates. Bayesian phylogenetic inference was conducted in MrBayes parallel version 3.2.6 [29] with selection of the model of nucleotide substitution (for 12S rDNA: GTR + G; for 18S rDNA: K80 + G; for cox1: GTR + G) by using the BIC implemented in Partition Finder2 [30, 31]. The Bayesian analysis was run for 10 million generations with two independent runs sampled every 1000 generations. The results were combined after discarding 25% of trees considered as ‘burn-in’ phase. The remaining 30,000 trees were summarized as a 50% majority rule consensus tree. Convergence of independent runs and the effective sample size of sampled parameters were inspected in Tracer version 1.6.
Table 2.
Characteristics of the nucleotide datasets used in phylogenetic analyses
| 12S rDNA | 18S rDNA | cox1 | |
|---|---|---|---|
| Number of sequences | 51 | 33 | 63 |
| Sequence length variation (bp) | 222–337 | 593–1133 | 366–388 |
| Number of aligned positions | |||
| Total | 350 | 1206 | 388 |
| Constant | 193 | 1010 | 235 |
| Autapomorphic | 28 | 86 | 9 |
| Parsimony informative | 129 | 110 | 144 |
| Containing gaps | 171 | 707 | 29 |
| Percentage of gaps/missing data | 10.35 | 11.42 | 0.93 |
Results
Molecular identification of Mesocestoides spp.
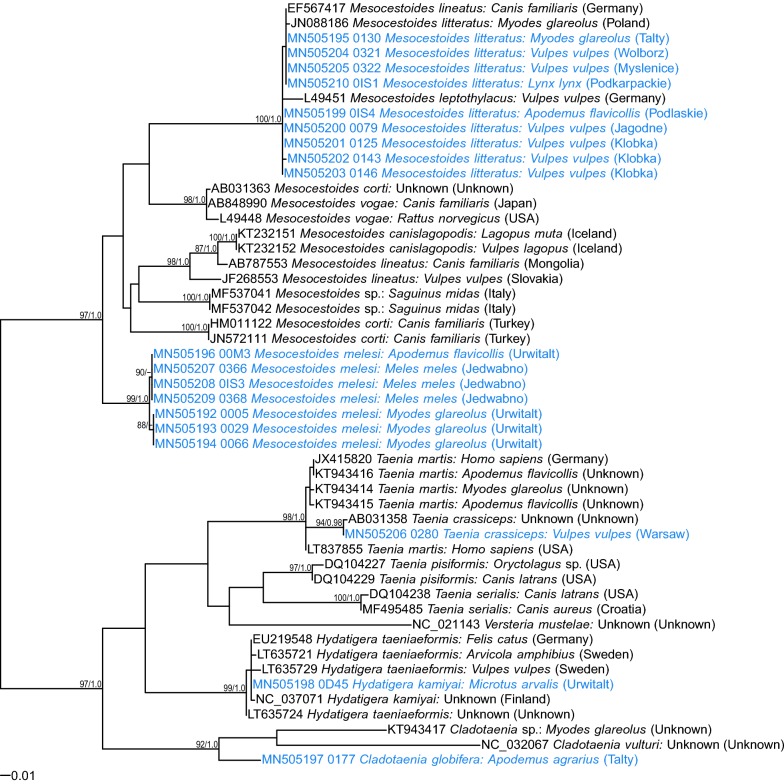
All eight adult Mesocestoides specimens from red foxes and one adult Mesocestoides from the Eurasian lynx were identified as M. litteratus based on 98–100% identity of the three markers with M. litteratus sequences deposited in GenBank (Additional file 1: Tables S1–S3). All three applied genetic markers were successful in amplifying Mesocestoides spp. DNA from foxes; however, only 12S rDNA could be amplified from the lynx sample. All the sequences obtained in the present study grouped with sequences of M. litteratus from carnivores from a range of European countries (Figs. 1, 2, 3).
Fig. 1.
Maximum likelihood tree for Mesocestoides and relatives based on 18S rDNA (GTR + G model). Numbers along branches are bootstrap support (BS) and posterior probability (PP) values if corresponding bipartition was found in Bayesian 50% majority rule consensus tree. Only values of BS higher than 75% and PP higher than 0.95 are shown. The scale-bar indicates the expected number of nucleotide substitutions per site
Fig. 2.
Maximum likelihood tree for Mesocestoides and relatives based on mt 12S rDNA (GTR + G model). Numbers along branches are bootstrap support (BS) and posterior probability (PP) values if corresponding bipartition was found in Bayesian 50% majority rule consensus tree. Only values of BS higher than 75% and PP higher than 0.95 are shown. The scale-bar indicates the expected number of nucleotide substitutions per site
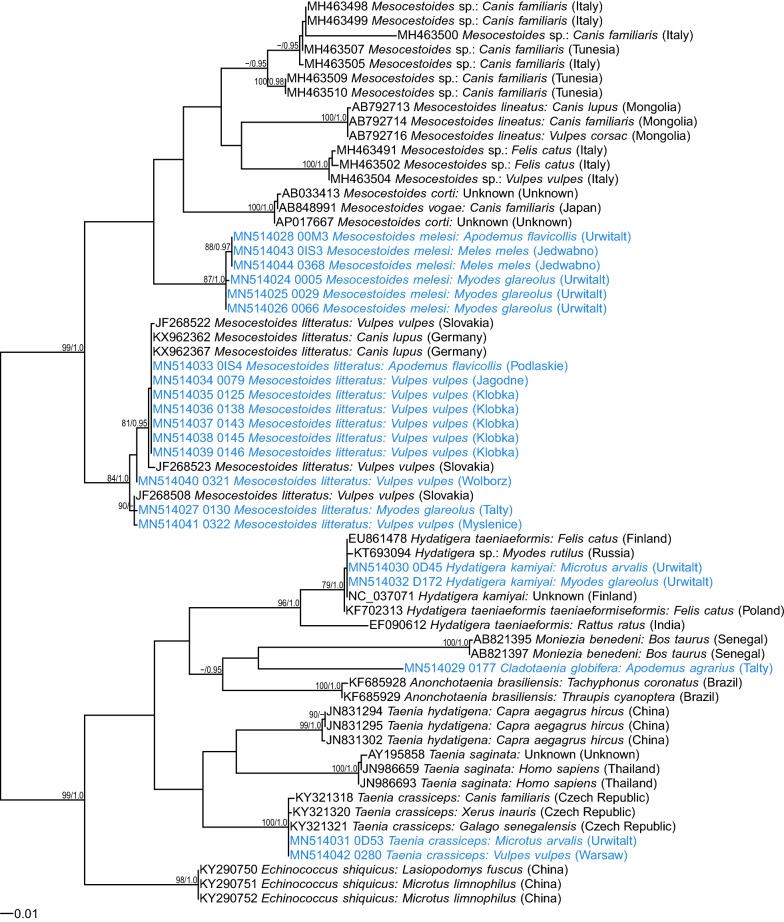
Fig. 3.
Maximum likelihood tree for Mesocestoides and relatives based on cox1 gene fragment (GTR + G model). Numbers along branches are bootstrap support (BS) and posterior probability (PP) values if corresponding bipartition was found in Bayesian 50% majority rule consensus tree. Only values of BS higher than 75% and PP higher than 0.95 are shown. The scale-bar indicates the expected number of nucleotide substitutions per site
Among six larval Mesocestoides isolates from rodents, only two (one from a bank vole from Masuria and one from a yellow-necked mouse from Białowieża) were identified as M. litteratus, based on 98–100% identity of the three markers used for analysis with M. litteratus sequences deposited in GenBank (Additional file 1: Tables S1–S3). A group of four sequences could not be identified due to the lack of identical sequences of 18S rDNA, mt 12S rDNA and cox1 in the GenBank database. These sequences, one derived from A. flavicollis and three from M. glareolus, both from the Mazury Lake District, displayed the highest similarity (97.4–99.4%) to M. vogae (syn. M. corti) based on 18S rDNA (Additional file 1: Table S2). Based on mt 12S rDNA and cox1 sequences, percent similarity was markedly lower (90.2–90.5% in 12S rDNA and 88–89% in cox1; Additional file 1: Tables S1, S3), suggesting the presence of a distinct species.
In phylogenetic analyses, these four isolates grouped separately (Figs. 1, 2, 3), distant from M. litteratus, M. lineatus or M. canislagopodis, but displaying closer similarity with North American M. vogae (syn. M. corti) (Figs.1, 3). Maximum likelihood and Bayesian trees had very similar topology and therefore we show only ML trees with posterior probability for corresponding bipartitions (Figs. 1, 2, 3).
Interestingly, all three Mesocestoides sequences derived from adult worms from Eurasian badgers were very similar (Additional file 1: Tables S1–S3) to these four isolates from rodents. In all phylogenetic trees, the four sequences from rodents and all available sequences from badgers formed one phylogenetic group, distant from M. litteratus, other species and a range of recently identified Mesocestoides genotypes from Italy and Tunisia [32, 33]. This group of sequences displayed the highest similarity to M. vogae (syn. M. corti) based on 18S rDNA and cox1 markers (Figs. 1, 3). Some minor diversity (1–3 SNPs) among this group of sequences was also observed (Figs. 1, 2, 3; Additional file 1: Tables S1–S3). There were also some differences between different M. litteratus sequences/isolates (Figs. 2, 3; 12S and cox1).
Morphological examination of Mesocestoides spp.
The larvae of putative M. melesi were half the size of M. litteratus larvae (Additional file 2: Figure S1) and additional morphological evaluation of slides with stained adult tapeworms from Eurasian badgers (Additional file 3: Figure S2) revealed no obvious differences between the present cestodes and these described as M. melesi. Although mean sucker length and width of the adult tapeworms from badgers were slightly larger than the means reported by Yanchev and Petrov [34] (Additional file 4: Table S4), they were well within the range described for M. melesi. Interestingly, the dimensions of the larval suckers of M. melesi identified in this study were half the size of the reported dimensions of suckers in adult worms. Fixed, stained preparations of these worms were compared also with other Mesocestoides spp. in the collection of the Natural History Museum, London (R. A. Bray and P. Olson, personal communication) and it was concluded that M. melesi could not be eliminated as the identity of these worms and with the additional genetic evidence provided in this paper, it was concluded that they were most likely to be M. melesi. A slide with adult tapeworms has been deposited in the Natural History Museum, London, UK, under the accession number NHMUK 2019.9.23.1.
Molecular identification of other larval and adult cestodes
Two isolates were identified as Taenia crassiceps based on 100% identity of the newly generated cox1 sequences with a sequence from the GenBank database (KY321321). One isolate was derived from an adult tapeworm from a red fox from the Mazowieckie Voivodeship and the second was a larva from the common vole, trapped in Masuria in 2000 (Table 1). Unfortunately, we were able to amplify only the cox1 gene fragment from the latter isolate. These two isolates grouped with other T. crassiceps in one clade of the phylogenetic tree based on cox1 sequences (Fig. 3).
The two Hydatigera larvae were identified as H. kamiyai based on 100% similarity of our cox1 sequences with sequences from the GenBank database (NC037071). Again, for these larval isolates from bank voles and common voles sampled in 2000, only cox1 and cox1 and 12S rDNA sequences, respectively, were amplified successfully. These two cox1 sequences localized in one clade with the H. kamiyai reference sequences from voles [6].
We were able to obtain cox1 and 12S rDNA sequences for C. globifera larvae from A. agrarius. However, we found no match with any available sequences deposited in GenBank for both markers, so the sequences were deposited as C. globifera based on morphological identification (number and dimensions of larval hooks).
Discussion
In the present study, three genetic markers were used for identification of cestode species recovered from both intermediate (rodents) and definitive hosts (red fox, Eurasian lynx and Eurasian badger) with a particular focus on Mesocestoides spp. We demonstrated that M. litteratus is a dominant species, occurring in red foxes in Poland and also in the Eurasian lynx from Podkarpackie, south-east Poland and in rodents. However, four isolates from rodents from the Mazury Lake District and all three isolates from Eurasian badgers from the same region created a separate clade, distant from all known species or genotypes available in the GenBank database, but most similar to North American M. vogae (syn. M. corti) or recently described M. canislagopodis [16]. Although genetic divergence for 18S rDNA between our unique isolates and these Mesocestoides spp. was only about 1–3%, much higher divergence was noted for the mitochondrial markers, 9–10% for 12S rDNA and 11–12% for cox1, which is enough to consider that these isolates must be a distinct tapeworm species with a novel genetic signature [6, 11, 35, 36]. On balance, taking into account both our morphological observations on adult worms and the genetic analysis, the samples in this clade are most likely to represent M. melesi. Our larval and adult cestodes of putative M. melesi revealed no obvious differences with the description of M. melesi, a species that was first described in 1985 from the Eurasian badger M. meles [34]. This first robust description of M. melesi was based on a significant number of tapeworms from 42 Eurasian badgers from Bulgaria and detailed several morphological features enabling differentiation of these worms as a new species distinct from M. lineatus and M. erschovi. The authors did not suggest any intermediate hosts for the new species at that time.
Moreover, although our four M. melesi samples from rodents displayed the highest genetic similarity to M. vogae (syn. M. corti), it is unlikely that they could represent a variant of M. vogae. Phylogenetic analyses clearly separated our sequences from M. vogae. Besides, M. corti was described in the USA by Hoeppli [37] based on about 100 tapeworms (adults, 8 cm long) recovered from the intestines of Mus musculus in Colorado in 1909 and recorded in the collection of Professor W.W. Cort. Later, others found only tetrathyridia in mice and rodents and small adults in cats, dogs and skunks [38, 39]. The original description by Hoeppli [37] was eventually questioned [38], especially as the original description was based solely on one archival field sample and rodents are now known not to serve as definitive hosts of Mesocestoides spp. These serious concerns led to the description of a new species by Etges [39], M. vogae, based on metacestodes from the body cavities and livers of fence lizards (Sceloporus occidentalis biseriatus) from California [40]. This description was approved and M. corti was synonymized with M. vogae. However, no data on definitive hosts was presented in the description of this new species. Then in 2004, Padgett and Boyce [8] provided detailed molecular data on the definitive hosts of M. vogae, including coyotes (Canis latrans) and domestic dogs, and proposed rodents (deer mice Peromyscus maniculatus) as intermediate hosts of this cestode. This biological data support differentiation of M. vogae (syn. M. corti) from M. melesi, with its life-cycle based on Eurasian badgers and European rodents (Myodes spp., Apodemus spp.).
To the best of our knowledge, our study is one of the first presenting the molecular characteristics of tapeworms derived from both intermediate and definitive hosts. Our analyses have demonstrated clearly that larval and adult Mesocestoides derived from rodents and Eurasian badgers, respectively, are closely related and genetically very similar, distant from other Mesocestoides species/genotypes, representing a badger-specific species. Thus, taking into account the previous description of Mesocestoides from Eurasian badgers as a new species by Yanchev and Petrov [34], we provide evidence for recognition of M. melesi as a valid species.
Our study supports the dominant occurrence of M. litteratus in rodents and carnivores from central Europe, in accordance with previous studies [14, 15, 41]. This species appears to be a generalist, occurring in a wide range of carnivores (but not in Eurasian badgers); in our study it was found in red foxes from different regions of Poland and in a Eurasian lynx from south-east Poland (Podkarpackie Voivodeship). In a recent molecular study of tapeworms, only this Mesocestoides species was found in dogs and cats in south-east Poland [42]. A few years ago, tetrathyridia of M. litteratus were identified molecularly in M. glareolus and A. agrarius from the Wrocław area, south-west Poland [19]. Both rodent species, in which we identified M. litteratus larvae, M. glareolus and A. flavicollis, are known intermediate hosts of this species. Interestingly, phylogenetic analyses of M. litteratus mitochondrial sequences obtained in this study from carnivores and rodents revealed some degree of diversity, suggesting the existence of several genotypes within the species.
The molecular characteristics of tapeworms derived from both intermediate and final hosts allowed us to conclude that the same genotype of T. crassiceps was present in rodents (M. arvalis) and red foxes, the definitive hosts of this species.
In our previous studies, cysts containing strobilocercus larvae, morphologically identified as T. taeniaeformis, were found in the livers of M. glareolus [3–5] and Arvicola terrestris (Bajer, unpublished) from the same region of Poland. However, following a recent reappraisal of H. taeniaeformis and the description of H. kamiyai (previously Taenia taeniaeformis complex; [6, 7]), here we were able to confirm the occurrence of H. kamiyai in voles as intermediate hosts. Moreover, we have now added a third species of Microtus, the common vole M. arvalis, and the bank vole Myodes glareolus to the published list of intermediate hosts for this cestode [6]. To the best of our knowledge, the present study is also the first to report the molecular detection of H. kamiyai in Poland, in addition to the recent identification of H. taeniaeformis in cats [42].
Conclusions
Molecular and phylogenetic analyses support the recognition of M. melesi as a valid species. To the best of our knowledge, our data represent the first record of the larvae of this species in rodents and the first report of the occurrence of H. kamiyai in rodents from Poland.
Supplementary information
Additional file 1: Table S1. Similarity (%) between selected 12S rDNA sequences of Mesocestoides generated in the present study and sequences of Mesocestoides spp. from the GenBank database. Table S2. Similarity (%) between selected 18S rDNA sequences of Mesocestoides generated in the present study and sequences of Mesocestoides spp. from the GenBank database. Table S3. Similarity (%) between selected cox1 sequences of Mesocestoides generated in the present study and sequences of Mesocestoides spp. from the GenBank database.
Additional file 2: Figure S1. Images of the larvae of M. melesi. Larvae from bank vole no. 029: free larva from peritoneal cavity (a-c) and liver cyst (d).
Additional file 3: Figure S2. Images of the adult M. melesi from the Eurasian badger no. 367. a Scolex with suckers. b Uterine proglottid, cirrus pouch visible. c Gravid proglottids with paruterine organ and cirrus pouch. d Scolex and gravid proglottid.
Additional file 4: Table S4. Comparison of the measurements of larval and adult M. melesi with M. litteratus and data from Yanchev and Petrov [34].
Acknowledgements
We are grateful to Drs Rod Bray and Peter Olson, the Natural History Museum, London, UK, for examining stained preparations of the adult stages of M. melesi from the Eurasian badgers in this study. Eurasian badger carcasses were obtained during realization of the bird conservation project: “Active protection of the black grouse on the grounds managed by the State Forests in Poland in 2017–2022”. We would like also to thank the employees of the Mammal Research Institute, Polish Academy of Sciences, Krzysztof Schmidt, Jan Boratyński, Ewelina Hapunik, and Dariusz Chilecki, for their help in mammal autopsies and collection of parasites.
Abbreviations
- PCR
polymerase chain reaction
- SNP
single nucleotide polymorphism
- mt
mitochondrial
Authors’ contributions
The study was designed and performed by AB. MA performed molecular and phylogenetic analyses. ŁB participated in phylogenetic analyses. DD, EJM, MKS, JBB, MG, KT, NK, ŁS, PO, MKP, GG, MA and JMB participated in collection of the material from rodents, badgers, lynx, and foxes. AB and JMB drafted the manuscript. All authors read and approved the final manuscript.
Funding
The study was partially supported by the National Science Centre (NCN) Sonata Bis grant no. 2014/14/E/NZ7/00153. Collection of the material was partially supported by NCN grant no. 2016/21/B/NZ8/02429. The publication is co-finansed within the framework of Ministry of Science and Higher Education programme as “Regional Initiative Excellence” in years 2019-2022, project number 005/RID/2018/19. None of the funding sources were involved in study design, data collection, data analysis, data interpretation or in writing the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files. Representative sequences are submitted to the GenBank database (accession numbers are provided in Table 1). Tapeworms from three Eurasian badgers, one Eurasian lynx, and one yellow-necked mouse were deposited in the scientific collection of the MRI PAS in Białowieża, Poland. A slide with adult tapeworms M. melesi has been deposited in the Natural History Museum, London, UK, under the accession number NHMUK 2019.9.23.1.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Anna Bajer and Mohammed Alsarraf contributed equally to this work
Contributor Information
Anna Bajer, Email: anabena@biol.uw.edu.pl.
Mohammed Alsarraf, Email: muha@biol.uw.edu.pl.
Dorota Dwużnik, Email: dorota.dwuznik@biol.uw.edu.pl.
Ewa J. Mierzejewska, Email: ewajuliamierzejewska@gmail.com
Marta Kołodziej-Sobocińska, Email: mksobocinska@ibs.bialowieza.pl.
Jolanta Behnke-Borowczyk, Email: jolanta.behnke@up.poznan.pl.
Łukasz Banasiak, Email: banasiak@biol.uw.edu.pl.
Maciej Grzybek, Email: maciej.grzybek@gumed.edu.pl.
Katarzyna Tołkacz, Email: k.h.tolkacz@biol.uw.edu.pl.
Natalia Kartawik, Email: nataliakartawik@gmail.com.
Łukasz Stańczak, Email: lukasz.stanczak@up.poznan.pl.
Patrycja Opalińska, Email: patrycja.opalinska@up.poznan.pl.
Małgorzata Krokowska-Paluszak, Email: malgorzata.krokowska-paluszak@up.poznan.pl.
Grzegorz Górecki, Email: gorecki@up.poznan.pl.
Mustafa Alsarraf, Email: elmusto0@gmail.com.
Jerzy M. Behnke, Email: jerzy.behnke@nottingham.ac.uk
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-020-3961-2.
References
- 1.Bajer A, Behnke JM, Pawelczyk A, Kuliś K, Sereda MJ, Siński E. Medium-term temporal stability of the helminth component community structure in bank voles (Clethrionomys glareolus) from the Mazury Lake District region of Poland. Parasitology. 2005;130:213–228. doi: 10.1017/S0031182004006389. [DOI] [PubMed] [Google Scholar]
- 2.Behnke JM, Barnard CJ, Bajer A, Bray D, Dinmore J, Frake K, et al. Variation in the helminth community structure in bank voles (Clethrionomys glareolus) from three comparable localities in the Mazury Lake District region of Poland. Parasitology. 2001;123:401–414. doi: 10.1017/S0031182001008605. [DOI] [PubMed] [Google Scholar]
- 3.Behnke JM, Bajer A, Harris PD, Newington L, Pidgeon E, Rowlands G, et al. Temporal and between-site variation in helminth communities of bank voles (Myodes glareolus) from NE Poland. 1. Regional fauna and component community levels. Parasitology. 2008;135:985–997. doi: 10.1017/S0031182008004393. [DOI] [PubMed] [Google Scholar]
- 4.Behnke JM, Bajer A, Harris PD, Newington L, Pidgeon E, Rowlands G, et al. Temporal and between-site variation in helminth communities of bank voles (Myodes glareolus) from NE Poland. 2. The infracommunity level. Parasitology. 2008;135:999–1018. doi: 10.1017/S0031182008004484. [DOI] [PubMed] [Google Scholar]
- 5.Grzybek M, Bajer A, Bednarska M, Alsarraf M, Behnke-Borowczyk J, Harris P, et al. Long-term spatiotemporal stability and dynamic changes in helminth infracommunities of bank voles (Myodes glareolus) in NE Poland. Parasitology. 2015;142:1722–1743. doi: 10.1017/S0031182015001225. [DOI] [PubMed] [Google Scholar]
- 6.Lavikainen A, Iwaki T, Haukisalmi V, Konyaev SV, Casiraghi M, Dokuchaev NE, et al. Reappraisal of Hydatigera taeniaeformis (Batsch, 1786) (Cestoda: Taeniidae) sensu lato with description of Hydatigera kamiyai n. sp. Int J Parasitol. 2016;46:36174. doi: 10.1016/j.ijpara.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Nakao M, Lavikainen A, Iwaki T, Haukisalmi V, Konyaev S, Oku Y, et al. Molecular phylogeny of the genus Taenia (Cestoda: Taeniidae): proposals for the resurrection of Hydatigera Lamarck, 1816 and the creation of a new genus Versteria. Int J Parasitol. 2013;43:427–437. doi: 10.1016/j.ijpara.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 8.Padgett KA, Boyce WM. Life-history studies on two molecular strains of Mesocestoides (Cestoda: Mesocestoididae): identification of sylvatic hosts and infectivity of immature life stages. J Parasitol. 2004;90:108–113. doi: 10.1645/GE-100R1. [DOI] [PubMed] [Google Scholar]
- 9.Padgett KA, Nadler SA, Munson L, Sacks B, Boyce WM. Systematics of Mesocestoides (Cestoda: Mesocestoididae): evaluation of molecular and morphological variation among isolates. J Parasitol. 2005;91:1435–1443. doi: 10.1645/GE-3461.1. [DOI] [PubMed] [Google Scholar]
- 10.Padgett KA, Crosbie PR, Boyce WM. Mesocestoides. In: Liu D, editor. Molecular detection of human pathogens. Boca Raton: CRC Press, Taylor and Francis Group; 2013. pp. 277–285. [Google Scholar]
- 11.Kubecka BW, Traub NJ, Tkach VV, Shirley TN, Rollins D, Fedynich A. Mesocestoides sp. in wild northern bobwhite (Colinus virginianus)and scaled quail (Callipepla squamata) J Wild Dis. 2018;54:612–616. doi: 10.7589/2017-11-275. [DOI] [PubMed] [Google Scholar]
- 12.McAllister CT, Tkach VV, Conn DB. Morphological and molecular characterization of post-larval pre-tetrathyridia of Mesocestoides sp. (Cestoda: Cyclophyllidea) from ground skink, Scincella lateralis (Sauria: Scincidae), from southeastern Oklahoma. J Parasitol. 2018;104:246–253. doi: 10.1645/17-178. [DOI] [PubMed] [Google Scholar]
- 13.Chertkova AN, Kosupko GA. Pododrjad Mesocestoidata Skrjabin, 1940. Osnovy cestodologii, 9. Moskva: Nauka; 1978. pp. 118–229. [Google Scholar]
- 14.Literák I, Olson PD, Georgiev BB, Spakulová M. First record of metacestodes of Mesocestoides sp. in the common starling (Sturnus vulgaris) in Europe, with an 18S rDNA characterisation of the isolate. Folia Parasitol (Praha). 2004;51:45–49. doi: 10.14411/fp.2004.006. [DOI] [PubMed] [Google Scholar]
- 15.Literák I, Tenora F, Letkova V, Goldova M, Torres J, Olson PD. Mesocestoides litteratus (Batsch, 1786) (Cestoda: Cyclophyllidea: Mesocestoididae) from the red fox: morphological and 18S rDNA characterization of European isolates. Helminthologia. 2006;43:191–195. doi: 10.2478/s11687-006-0036-7. [DOI] [Google Scholar]
- 16.Skirnisson K, Jouet D, Ferte H, Nielsen OK. Occurrence of Mesocestoides canislagopodis (Rudolphi, 1810) Krabbe, 1865 in mammals and birds in Iceland and its molecular discrimination within the Mesocestoides species complex. Parasitol Res. 2016;115:2597–2607. doi: 10.1007/s00436-016-5006-5. [DOI] [PubMed] [Google Scholar]
- 17.Yanchev YI. Morphology, taxonomy and distribution of species of the genus Mesocestoides in Bulgaria. Khelmintologiya. 1986;21:45–65. [Google Scholar]
- 18.Wardle RA, Mcleod JA. The zoology of tapeworms. Minneapolis: University of Minnesota Press; 1952. [Google Scholar]
- 19.Zaleśny G, Hildebrand J. Molecular identification of Mesocestoides spp. from intermediate hosts (rodents) in central Europe (Poland) Parasitol Res. 2012;110:1055–1061. doi: 10.1007/s00436-011-2598-7. [DOI] [PubMed] [Google Scholar]
- 20.Karamon J, Dąbrowska J, Kochanowski M, Samorek-Pieróg M, Sroka J, Różycki M, et al. Prevalence of intestinal helminths of red foxes (Vulpes vulpes) in central Europe (Poland): a significant zoonotic threat. Parasit Vectors. 2018;11:436. doi: 10.1186/s13071-018-3021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kakietek D, Dwużnik D, Religa M, Mierzejewska EJ, Koczwarska J, Bajer A. Pasożyty jelitowe lisów z terenów województwa mazowieckiego, warmińsko-mazurskiego i kujawsko-pomorskiego. III Ogólnopolska Konferencja Doktorantów Nauk o Życiu BioOpen. Conference proceedings; 2017. p. 181.
- 22.Kołodziej-Sobocińska M, Yakovlev Y, Schmidt K, Hurníková Z, Ruczyńska I, Bednarski M, et al. Update of the helminth fauna in Eurasian lynx (Lynx lynx) in Poland. Parasitol Res. 2018;117:2613–2621. doi: 10.1007/s00436-018-5953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Littlewood DTJ, Olson PD. Small subunit rDNA and the platyhelminthes: signal, noise, conflict and compromise. In: Littlewood DTJ, Bray RA, editors. Interrelationships of the platyhelminthes. London: Taylor & Francis; 2001. pp. 262–278. [Google Scholar]
- 24.Von Nickisch-Rosenegk M, Silva-Gonzalez R, Lucius R. Modification of universal 12S rDNA primers for specific amplification of contaminated Taenia spp. (Cestoda) gDNA enabling phylogenetic studies. Parasitol Res. 1999;85:819–825. doi: 10.1007/s004360050638. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowles J, Blair D, McManus DP. A molecular phylogeny of the human schistosomes. Mol Phylogenet Evol. 1995;4:103–109. doi: 10.1006/mpev.1995.1011. [DOI] [PubMed] [Google Scholar]
- 27.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. Partition Finder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 32.Montalbano Di Filippo M, Meoli R, Cavallero S, Eleni C, De Liberato C, Berrilli F. Molecular identification of Mesocestoides sp. metacestodes in a captive gold-handed tamarin (Saguinus midas) Infect Genet Evol. 2018;65:399–405. doi: 10.1016/j.meegid.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Varcasia A, Sanna D, Casu M, Lahmar S, Dessi G, Pipia AP, et al. Species delimitation based on mt DNA genes suggests the occurrence of new species of Mesocestoides in the Mediterranean region. Parasit Vectors. 2018;11:619. doi: 10.1186/s13071-018-3185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanchev YI, Petrov IK. Mesocestoides melesi sp. n. (Cestoda, Mesocestoididae) in Meles meles L. from Bulgaria. Comput Rend Acad Bulg Sci. 1985;38:247–251. [Google Scholar]
- 35.Galimberti A, Romano DF, Genchi M, Paoloni D, Vercillo F, Bizzarri L, et al. Integrative taxonomy at work: DNA barcoding of taeniids harboured by wild and domestic cats. Mol Ecol Resour. 2012;12:403–413. doi: 10.1111/j.1755-0998.2011.03110.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Hu M, Jones A, Allsopp BA, Beveridge L, Schindler AR, et al. Characterization of Taenia madoquae and Taenia regis from carnivores in Kenya using genetic markers in nuclear and mitochondrial DNA, and their relationship with other selected taeniids. Mol Cell Probes. 2007;21:379–385. doi: 10.1016/j.mcp.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Hoeppli RJC. Mesocestoides corti, a new species of cestode from the mouse. J Parasitol. 1925;12:91–96. doi: 10.2307/3270771. [DOI] [Google Scholar]
- 38.Beaver PC. Mesocestoides corti: mouse type host, uncharacteristic or questionable? J Parasitol. 1989;75:815. doi: 10.2307/3283073. [DOI] [PubMed] [Google Scholar]
- 39.Etges FJ. The proliferative tetrathyridium of Mesocestoides vogae sp. n. (Cestoda) J Helm Soc Wash. 1991;58:181–185. [Google Scholar]
- 40.Specht D, Voge M. Asexual multiplication of Mesocestoides tetrathyridia in laboratory animals. J Parasitol. 1965;51:268–272. doi: 10.2307/3276097. [DOI] [PubMed] [Google Scholar]
- 41.Hrckova G, Miterpakova M, O’Connor A, Snabel V, Olson PD. Molecular and morphological circumscription of Mesocestoides tapeworms from red foxes (Vulpes vulpes) in central Europe. Parasitology. 2011;138:638–647. doi: 10.1017/S0031182011000047. [DOI] [PubMed] [Google Scholar]
- 42.Karamon J, Sroka J, Dąbrowska J, Bilska-Zając E, Zdybel J, Kochanowski M, et al. First report of Echinococcus multilocularis in cats in Poland: a monitoring study in cats and dogs from a rural area and animal shelter in a highly endemic region. Parasit Vectors. 2019;12:313. doi: 10.1186/s13071-019-3573-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Similarity (%) between selected 12S rDNA sequences of Mesocestoides generated in the present study and sequences of Mesocestoides spp. from the GenBank database. Table S2. Similarity (%) between selected 18S rDNA sequences of Mesocestoides generated in the present study and sequences of Mesocestoides spp. from the GenBank database. Table S3. Similarity (%) between selected cox1 sequences of Mesocestoides generated in the present study and sequences of Mesocestoides spp. from the GenBank database.
Additional file 2: Figure S1. Images of the larvae of M. melesi. Larvae from bank vole no. 029: free larva from peritoneal cavity (a-c) and liver cyst (d).
Additional file 3: Figure S2. Images of the adult M. melesi from the Eurasian badger no. 367. a Scolex with suckers. b Uterine proglottid, cirrus pouch visible. c Gravid proglottids with paruterine organ and cirrus pouch. d Scolex and gravid proglottid.
Additional file 4: Table S4. Comparison of the measurements of larval and adult M. melesi with M. litteratus and data from Yanchev and Petrov [34].
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files. Representative sequences are submitted to the GenBank database (accession numbers are provided in Table 1). Tapeworms from three Eurasian badgers, one Eurasian lynx, and one yellow-necked mouse were deposited in the scientific collection of the MRI PAS in Białowieża, Poland. A slide with adult tapeworms M. melesi has been deposited in the Natural History Museum, London, UK, under the accession number NHMUK 2019.9.23.1.