Abstract
PCV2 belongs to the genus Circovirus, family Circoviridae, who is recognized as the causative agents of postweaning multisystemic wasting syndrome. Since being found to China in 2000, it has caused serious damage to the pig industry. In this study, we downloaded 40 PCV2 genome‐wide sequences uploaded to GenBank from 2013 to 2018 in Shandong Province, including 23 uploaded by our laboratory. Construction of a genome‐wide evolution tree using MEGA V5.0 software. Phylogenetic tree analysis indicated that the genotype of PCV2 in Shandong Province was: three genotypes coexisted (2a, 2b, 2d); among them, PCV2d has become the main genotype in the province due to its number and spread range. Amino acid sequence analysis of different genotypes of ORF2 showed that specific amino acid sites exist in different genotypes, with the most significant range of 81–160; different genotypes of PCV2 can be distinguished at the molecular level. This study found that due to the increase in infections of the PCV2d genotype in recent years, it may replace PCV2b as the dominant base in Shandong.
Keywords: genetic evolution, genotype, ORFs, PCV2, phylogenetic analysis
In order to understand the genetic evolution of PCV2 in Shandong in recent years, we analyzed all PCV2 complete sequences collected from different farms in six regions in Shandong Province from 2013 to 2018. In this study, we also performed homologous comparison and amino acid site analysis on the ORF2‐encoded Cap protein sequences of 40 isolates. Some specific mutation sites may also serve as reference conditions for the genetic variation of the strain.

1. INTRODUCTION
Porcine circovirus (PCV) is the smallest animal virus known and can be divided into three types: the non‐pathogenic PCV1, the PCV2 and the new‐found PCV3 in 2016 (Palinski et al., 2016). Our laboratory first reported the occurrence of PCV3 in pigs without clinical infection signs in Shandong Province, eastern China (Zheng et al., 2018). PCV2 possesses a genome of 1766–1768 nucleotides (nt) in length, it is believed that there are five genotypes of PCV2a, PCV2b, PCV2c, PCV2d, and PCV2e in the world. However, PCV2c and PCV2e have not been found in Shandong Province so far. PCV2 genome contains 11 major open reading frames (ORFs). At present, more studies are ORF1, ORF2, ORF3, and ORF4 (Xiao, Halbur, & Opriessnig, 2015). The ORF2 gene which is considered to be the phylogenetic marker encodes the viral structural and virulence‐associated protein (Cap) involved in the host immune response (Fan et al., 2008), In past reports of our laboratory, through phylogenetic analysis revealed that PCV2b was the dominant genotype of Shandong Province before 2013 (Shi et al., 2015). According to reports from domestic and foreign countries in recent years, the genotypes of PCV2 are gradually becoming more complex and diversified. However, recent reports also suggest that an ongoing genotype shift occurs from PCV2b to PCV2d and that PCV2d appears widespread worldwide (Yang et al., 2018). In order to understand the genetic evolution of PCV2 in Shandong in recent years, we analysed all PCV2 complete sequences collected from different farms in six regions in Shandong Province from 2013 to 2018. In this study, we also performed homologous comparison and amino acid site analysis on the ORF2‐encoded Cap protein sequences of 40 isolates. Some specific mutation sites may also serve as reference conditions for the genetic variation of the strain.
2. MATERIAL AND METHODS
2.1. Samples
The 40 strains isolates by our laboratory were obtained from clinical samples (lymph nodes(8), spleen(6), liver(12), lung(11), and tonsil(3)) collected from commercial farms. After collected the clinical samples, the DNA were extracted according to the steps shown in the viral DNA extraction kit (BioFlux, BCC67MI), and stored at −20°C. The isolated PCV2 strains isolated in this study were obtained with ethical approval and written informed consent. All animal experimental procedures were performed in accordance with《The Regulations for the Administration of Affairs Concerning Experimental Animals》approved by institute of Animal Science and Veterinary Medicine, Shandong Academy of Agricultural Sciences.
2.2. Complete genome extraction and sequencing
To understand the genetic character of PCV2, primer F (5’‐GAACCGCGGGCTGGTGAACTTTTGAAAGT‐3’) and primer R (5’‐GCACCGCGGAAATTTCTGACAAACGTTACA‐3’) were used to amplify the whole 1.7 kb genome (Fenaux et al., 2002). The PCR reactions in a total volume of 20 μl contained 2 μl of extracted DNA, 1 μl of primer pairs, 10 μl 2 × Taq plus Master Mix II (TaKaRa) and 7 μl ddH2O. PCR amplification was initiated at a predenaturation stage of 95°C for 5 min, followed 30 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 30 s and extension at 72°C for 60 s. The amplified PCR product was then sent to Shenggong Biotechnology Co., Ltd. for sequencing. The whole genomes were sequenced three times each and submitted to the NCBI database.
2.3. Phylogenetic analysis
Sequences of PCV2 Shandong isolates were compared with other representative PCV2 genotype sequences in GenBank using MEGA V5.0 software. The homology between nucleotide sequences was analysed using MEGAlign software. The phylogenetic tree was calculated using the neighbour‐joining (NJ) method. Bootstrap values were calculated based on 1,000 repeats of the alignment. The phylogenetic tree was used to analyse the genotype evolution of PCV2 strains in Shandong Province from 2013 to 2018.
The amino acid homology of ORF2 of 40 strains was also analysed by Clustal W in MegAlign software. Find specific amino acid loci for different genotypes, analyze their relationship, and speculate whether this is a key factor leading to genotype evolution.
3. RESULTS
3.1. Phylogenetic analysis results of PCV2 isolates
Compared with three presentative PCV2 strains sequences, the phylogenetic analysis tree is shown in (Figure 1). It is concluded that 7 strains belong to PCV2a, 6 strains belong to PCV2b and 27 strains belong to PCV2d; from the number of infections, PCV2d has become the main genotype of Shandong at present. We also found from the phylogenetic tree that PCV2d not only highlights the advantages in quantity but also covers almost all areas of this sampling, covering all time periods from 2013 to 2018. On the other hand, PCV2a is mostly found in Yantai, Shandong. PCV2b was not detected in 2018, of course, this does not rule out the limitations of the sampling area (Table 1). Therefore, this has also been confirmed that the results of phylogenetic analysis indicate that PCV2d is currently the main epidemic strain of PCV2 in Shandong Province.
Figure 1.
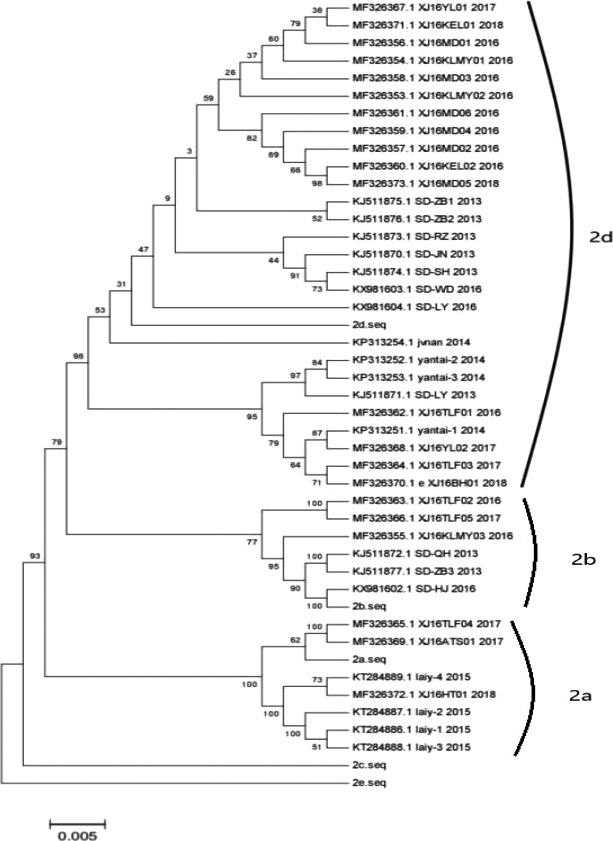
Complete genome nucleotide sequence phylogenetic tree of PCV2 (2a, 2b, 2d are a representative strain)
Table 1.
GenBank accession numbers, geographic origin, years, genome size and genotypes of shandong 40 strains PCV2 genomes sequenced in this study
| Sequence number | GenBank accession numbers | Geographic origin | Years | Genome size (bp) | Genotypes |
|---|---|---|---|---|---|
| 1 | KJ511870 a | JI NAN | 2013 | 1767 | 2d |
| 2 | KJ511871 a | YAN TAI | 2013 | 1767 | 2d |
| 3 | KJ511872 a | JI NAN | 2013 | 1767 | 2b |
| 4 | KJ511873 a | RI ZHAO | 2013 | 1767 | 2d |
| 5 | KJ511874 a | JI NAN | 2013 | 1767 | 2d |
| 6 | KJ511875 a | ZI BO | 2013 | 1767 | 2d |
| 7 | KJ511876 a | ZI BO | 2013 | 1767 | 2d |
| 8 | KJ511877 a | ZI BO | 2013 | 1767 | 2b |
| 9 | KP313251 | YAN TAI | 2014 | 1767 | 2d |
| 10 | KP313252 a | YAN TAI | 2014 | 1767 | 2d |
| 11 | KP313253 a | YAN TAI | 2014 | 1767 | 2d |
| 12 | KP313254 a | YAN TAI | 2014 | 1767 | 2d |
| 13 | KT284886 a | YAN TAI | 2015 | 1768 | 2a |
| 14 | KT284887 | YAN TAI | 2015 | 1768 | 2a |
| 15 | KT284888 | YAN TAI | 2015 | 1768 | 2a |
| 16 | KT284889 | YAN TAI | 2015 | 1768 | 2a |
| 17 | KX981602 a | BIN ZHOU | 2016 | 1767 | 2b |
| 18 | KX981603 a | BIN ZHOU | 2016 | 1767 | 2d |
| 19 | KX981604 a | YAN TAI | 2016 | 1767 | 2d |
| 20 | MF326353 | TAI AN | 2016 | 1767 | 2d |
| 21 | MF326354 | TAI AN | 2016 | 1767 | 2d |
| 22 | MF326355 | TAI AN | 2016 | 1767 | 2b |
| 23 | MF326356 | TAI AN | 2016 | 1767 | 2d |
| 24 | MF326357 | TAI AN | 2016 | 1767 | 2d |
| 25 | MF326358 | TAI AN | 2016 | 1767 | 2d |
| 26 | MF326359 | TAI AN | 2016 | 1767 | 2d |
| 27 | MF326360 | TAI AN | 2016 | 1767 | 2d |
| 28 | MF326361 | BIN ZHOU | 2016 | 1767 | 2d |
| 29 | MF326362 | BIN ZHOU | 2016 | 1766 | 2d |
| 30 | MF326363 a | BIN ZHOU | 2016 | 1767 | 2b |
| 31 | MF326364 a | BIN ZHOU | 2017 | 1767 | 2d |
| 32 | MF326365 a | BIN ZHOU | 2017 | 1768 | 2a |
| 33 | MF326366 a | JI NAN | 2017 | 1767 | 2b |
| 34 | MF326367 a | JI NAN | 2017 | 1767 | 2d |
| 35 | MF326368 a | JI NAN | 2017 | 1767 | 2d |
| 36 | MF326369 | YAN TAI | 2017 | 1768 | 2a |
| 37 | MF326370 | YAN TAI | 2018 | 1767 | 2d |
| 38 | MF326371 a | JI NAN | 2018 | 1767 | 2d |
| 39 | MF326372 a | JI NAN | 2018 | 1768 | 2a |
| 40 | MF326373 | RI ZHAO | 2018 | 1767 | 2d |
Represents the sequence collected and uploaded by our laboratory.
3.2. Sequence alignment and analysis of the amino acid sequence deduced from ORF2
Homology analysis of the amino acid sequences ORF2 of these 40 isolates used the MegAlign software in DNAstar. We found that homology of ORF2 was 85.3%–100% (Figure 2). Because the predecessors passed the sequence analysis of the PCV2 genome, the ORF2 gene has a high mutation rate, and there are many potential epitopes in the Cap protein and they can also mutated under immunological stress. We compared the 233 amino acid sequences encoded by ORF2 of 40 isolates with 3 different genotypes, it was found that there are specific mutation sites in the amino acid sequences of different genotypes. These specific mutation sites are concentrated in the amino acid sequence from 81 to 160. Among them, the specific site of PCV2d is 89L, 90T, 121T; PCV2b is 89R; PCV2a is 86T, 88K, 89I, 91I, 179P (Figure 3). According to these specific amino acid sites, different genotypes of PCV2 can be distinguished at the molecular level. Combined with the results of phylogenetic analysis, we can preliminarily conclude that the genotype epidemic of PCV2 in Shandong Province is characterized by multiple genotypes, and the dominant genotype changes from PCV2b to PCV2d. Mutations in amino acids on ORF2 may be one of the factors leading to the evolution of genotypes.
Figure 2.

Homology analysis of the amino acid sequences ORF2 of these 40 isolates used the MegAlign software in DNAstar (comparison of the ORF2 amino acid sequences showing that there was 85.3%–100% identity among the 40 isolates, the upper right triangle is the similarity, and the lower left triangle is the mutation rate)
Figure 3.

Analysis of amino acids 81–130 encoded by the ORF2 gene of PCV2 (different genotype amino acid‐specific mutation sites are indicated in the box)
4. DISCUSSION
In China, PCV2 infection has been described since 2000, PCV2 infection has been considered to be an important challenge to Chinese swine industry (Shuai et al., 2007). In order to understand the distribution and evolution of the major subtypes of PCV2 in our province in recent years, we collected 40 isolates for genome‐wide genetic evolution analysis and ORF2 sequence analysis. There are three genotypes of PCV2 in Shandong Province. Among them, PCV2d is considered to be the main dominant strain in Shandong Province because of its large quantity, wide coverage and longtime coverage. Two genotype changes in PCV2 occurred during the 20th century, including PCV2a being replaced by PCV2b in 2005 and PCV2b shifting to PCV2d in 2008. Since then, PCV2d has become a major genotype responsible for great losses in the swine industry worldwide, including the United States, South America, Europe, Korea and Thailand (Franzo, Cortey, Segalés, Hughes, & Drigo, 2016). There are similar phenomena in the prevalence of PCV2 strains in other provinces in China, there have been more cases of infection of the PCV2d genotype in pigs in Guangxi, PCV2d may replace PCV2b become the dominant genotype (Wang, Ge, Wang, Li, & Hu, 2013). At present, the genotypic shift was observed in Shandong too. Some studies have suggested that PCV2d is a mutant of PCV2b, also known as mPCV2b. It is a genotype that is produced after the large‐scale introduction of a viral vaccine against PCV2b. In China and South Korea, PCV2b was gradually replaced by PCV2d and PCV2d as a major genotype leads to Vaccine immunization failed, resulting in a significant increase in PCV2 infection rates (Xiao, 2012).
In conclusion, through phylogenetic analysis, combined with previous studies and predecessors, amino acid mutations encoded by ORF2 may be responsible for the emergence of PCV2d (Shen, 2012). The capsid protein encoded by the PCV2 ORF2 gene is the major structural protein of PCV2 and is an important target antigen that stimulates the body to produce an immune protective response and proved to be immunorelevant epitopes for virus type discrimination (Xiao et al., 2015). Studies have shown that mutations in the single amino acid of the PCV2 capsid protein may cause changes in the response phenotype of the PCV2 strain, changes in antibodies that would otherwise be neutralized by neutralizing antibodies (Guo et al., 2011). We compared the specific mutation sites of different genotypes and found that the partial‐specific mutation sites are located in the five epitope regions of ORF2. Whether the variation of these amino acids leads to the evolution between different genotypes? Will it cause changes in the antigenicity and virulence of the three genotype strains PCV2a, PCV2b, and PCV2d? These issues are for further study.
CONFLICT OF INTERESTS
The authors declare that there is no conflict of interest.
AUTHOR CONTRIBUTION
Jun Li and Shuo Wang conceived the project. Shuo Wang designed the experiments. Changxun Xin, Xiaoyan Wu, Jianli Shi and Zhe Peng performed most of the experiments. Yuwei Liu, Panpan Sun, Yan Wang, Mei Gao, Shaojian Xu and Fan Zhang contributed materials and participated in discussion. Shuo Wang wrote the manuscript. Jun Li supervised the work and edited the final version of the manuscript which was read and approved by all authors.
ETHICAL STATEMENT
We confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to and the appropriate ethical review committee approval has been received. The US National Research Council's guidelines for the Care and Use of Laboratory Animals were followed.
ACKNOWLEDGEMENTS
The study was partly supported by National Key R&D Program (2016YFD0500708), Shandong Province Modern Agricultural Industry Technology System (SDAIT‐08‐07), Doctor Fund of Shandong Province (ZR2017BC087), Shandong Natural Fund (ZR2017YL015 and ZR2017YL014), Taishan Scholars Project, Agricultural Science and Technology Innovation Project of Shandong Academy of Agricultural Sciences (CXGC2016B14), and the Youth Scientific Research Foundation of Shandong Academy of Agricultural Sciences (2016YQN53).
Wang S, Xin C, Wu X, et al. Genetic characterization of Porcine circovirus type 2 from 2013 to 2018 in Shandong Province, China. Vet Med Sci. 2020;6:76–81. 10.1002/vms3.196
Shuo Wang, Changxun Xin, and Xiaoyan Wu are contributed equally to this work.
REFERENCES
- Fan, H. , Xiao, S. , Tong, T. , Wang, S. , Xie, L. , Jiang, Y. , … Fang, L. (2008). Immunogenicity of porcine circovirus type 2 capsid protein targeting to different subcellular compartments. Molecular Immunology, 45(3), 653–660. 10.1016/j.molimm.2007.07.009 [DOI] [PubMed] [Google Scholar]
- Fenaux, M. , Halbur, P. G. , Haqshenas, G. , Royer, R. , Thomas, P. , Nawagitgul, P. , … Meng, X. J. (2002). Cloned genomic DNA of type 2 porcine circovirus is infectious when injected directly into the liver and lymph nodes of pigs: Characterization of clinical disease, virus distribution, and pathologic lesions. Journal of Virology, 76(2), 541 10.1128/JVI.76.2.541-551.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzo, G. , Cortey, M. , Segalés, J. , Hughes, J. , & Drigo, M. (2016). Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Molecular Phylogenetics and Evolution, 100, 269–280. 10.1016/j.ympev.2016.04.028 [DOI] [PubMed] [Google Scholar]
- Guo, L. , Lu, Y. , Wei, Y. , Huang, L. , Wu, H. , & Liu, C. (2011). Porcine circovirus genotype 2a (PCV2a) and genotype 2b (PCV2b) recombinant mutants showed significantly enhanced viral replication and altered antigenicity in vitro. Journal of General Virology, 419(2), 57–63. 10.1016/j.virol.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Palinski, R. , Piñeyro, P. , Shang, P. , Yuan, F. , Guo R., Fang Y., … Hause B. M.. (2016). A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. Journal of Virology, 91(1), pii: e01879-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H.‐G. , Halbur, P. G. , & Opriessnig, T. (2012). Prevalence and phylogenetic analysis of the current porcine circovirus 2 genotypes after implementation of widespread vaccination programmes in the usa. Journal of General Virology. Journal of General Virology, 93, 1345–1355. 10.1099/vir.0.039552-0 [DOI] [PubMed] [Google Scholar]
- Shi, J. , Xu, S. , Fu, F. , Cong, X. , Yuan, X. , Peng, Z. , … Wang, J. (2015). Phylogenetic analysis of 32 porcine circovirus type 2 isolates from Shandong, China. Archives of Virology, 160(2), 465–468. 10.1007/s00705-014-2300-3 [DOI] [PubMed] [Google Scholar]
- Shuai, J. , Wei, W. , Li, X. , Chen, N. , Zhang, Z. , Chen, X. , & Fang, W. (2007). Genetic characterization of porcine circovirus type 2 (PCV2) from pigs in high‐seroprevalence areas in southeastern China. Virus Genes, 35(3), 619–627. 10.1007/s11262-007-0121-0 [DOI] [PubMed] [Google Scholar]
- Wang, L. , Ge, C. , Wang, D. , Li, Y. , & Hu, J. (2013). The survey of porcine teschoviruses, porcine circovirus and porcine transmissible gastroenteritis virus infecting piglets in clinical specimens in China. Tropical Animal Health & Production, 45(5), 1087–1091. 10.1007/s11250-012-0329-4 [DOI] [PubMed] [Google Scholar]
- Xiao, C.‐T. , Halbur, P. G. , & Opriessnig, T. (2012). Complete genome sequence of a Novel Porcine Circovirus Type 2b variant present in cases of vaccine failures in the United States. Journal of Virology, 86(22), 12345–12496. 10.1128/JVI.02345-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, C. , Halbur, P. G. , & Opriessnig, T. (2015). Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. Journal of General Virology, 96(7), 1830–1841. 10.1099/vir.0.000100 [DOI] [PubMed] [Google Scholar]
- Yang, S. , Yin, S. , Shang, Y. , Liu, B. , Yuan, L. , Zafar Khan, M. U. , … Cai, J. (2018). Phylogenetic and genetic variation analyses of porcine circovirus type 2 isolated from China. Transboundary and Emerging Diseases, 65(2), e383–e392. 10.1111/tbed.12768 [DOI] [PubMed] [Google Scholar]
- Zheng, S. , Shi, J. , Wu, X. , Peng, Z. , Xin, C. , Zhang, L. , … Wang, J. (2018). Presence of Torque teno sus virus 1 and 2 in porcine circovirus 3‐positive pigs. Transboundary and Emerging Diseases, 65(2), 327–330. 10.1111/tbed.12792 [DOI] [PubMed] [Google Scholar]


