Abstract
This study aimed at determining the seroprevalence of foot‐and‐mouth disease (FMD) in domestic ruminants and at characterizing the virus strains circulating in four areas of Chad (East Batha, West Batha, Wadi Fira and West Ennedi). The study was carried out between October and November 2016. A total of 1,520 sera samples (928 cattle, 216 goats, 254 sheep and 122 dromedaries) were collected randomly for FMD serological analyses. Nine epithelial tissue samples were also collected from cattle showing clinical signs, for FMDV isolation and characterization. Serological results showed an overall NSP seroprevalence of 40% (375/928) in cattle in our sample (95% CrI [19–63]). However, seroprevalences of 84% (27/32), 78% (35/45) and 84% (21/25) were estimated in cattle over 5 years of age in East Batha, West Batha and Wadi Fira, respectively. In cattle under 1 year of age, 67% (18/27) seroprevalence was estimated in Wadi Fira, 64% (14/22) in East Batha and 59% (13/22) in West Batha. It was found that the high seroprevalences have been obtained in areas where pastures are shared by several different herds but also in farms where two to three species (bovine, caprine and ovine) are raised together. ELISA PrioCHECK®FMDV types O and A and in‐house solid phase competition ELISA serotyping results showed that the four O, A, SAT1 and SAT2 serotypes have circulated in Chad in 2016. However, the type SAT2 dominated with an overall seroprevalence of 43% (29/67) and was present in the four areas investigated. The phylogenetic analyses of the VP1 coding sequence allowed determining the serotype SAT2 topotype VII, close to viral strains found in Cameroon in 2015 with a similarity of 98.60%.
Keywords: Chad, foot‐and‐mouth disease virus, molecular characterization, seroprevalence
FMD virus serotyping results showed that four O, A, SAT1 and SAT2 serotypes circulated in Chad in 2015. Phylogenetic analyses of the coding sequence VP1 allowed to determine the serotype SAT2 topotype VII, close to the viral strains found in Cameroon in 2015 with a similarity of 98.60%.

1. INTRODUCTION
Foot‐and‐mouth disease (FMD) is a contagious disease of cloven‐hoofed animals, one of the most economically important diseases of livestock worldwide (Knight‐Jones & Rushton, 2013). It is caused by a virus of the genus Aphthovirus within the Picornaviridae family. It especially affects wild and domestic Artiodactyls, namely cattle, sheep, goats and pigs (Jamal & Belsham, 2013). FMD virus (FMDV) is a small virus consisting of a single‐stranded RNA genome of approximately 8,500 bases encoding for structural and non‐structural proteins. The virion is icosahedral, non‐enveloped with a positive‐sense single‐stranded RNA genome (Domingo et al., 1990; Thiry, Baranowski, & Domingo, 2001). There are seven distinct serotypes (O, A, C, SAT1, SAT2, SAT3 and Asia1). Each serotype has multiple subtypes because of the great antigenic variability resulting from genetic variation during FMDV replication (OIE, 2017).
FMD is enzootic in most of Africa, particularly in Central and West Africa (Couacy‐Hymann et al., 2006; Kouato, Souley, et al., 2018; Paton, Sumption, & Charleston, 2009; Rweyemamu et al., 2008). The incidence of FMD virus transmission is most likely related to animal mobility (Bertram, Bravo, et al., 2018; Bertram, Delgado, et al., 2018). In Chad, four serotypes have been identified: O, A, SAT1 and SAT2 (Gueme, 1998; Ouagal et al., 2018, 2017; Ouagal, Hendrikx, Saegerman, & Berkvens, 2010). The circulation of these serotypes was reported in countries neighbouring Chad, such as Sudan, Cameroon, Niger and Nigeria (Abu Elzein, 1983; Bertram, Bravo, et al., 2018; Bertram, Delgado, et al., 2018; Ehizibolo et al., 2017a, 2017b; Habiela, Alamin, Alamin, Raouf, & Ali, 2010; Habiela, Ferris, et al., 2010; Knowles, Bachanek‐Bankowska, et al., 2016; Kouato, Elliot, et al., 2018; Kouato, Souley, et al., 2018).
In Chad, despite the endemicity of the disease, few studies have been carried out on seroprevalence (Ouagal et al., 2018, 2017) and no studies on the molecular characterization of the FMDV have been yet published. That is why we decided to conduct a study to determine seroprevalence and characterize the serotypes of FMD viruses circulating in the area of the Pastoral Strengthening of Livestock in Chad (PREPAS) project. This project covers four potential areas livestock breeding in Chad (East Batha, West Batha, West Ennedi and Wadi Fira).
2. MATERIALS AND METHODS
2.1. Study area and sampling
The study area includes four livestock areas of Chad: East Batha, West Batha, Wadi Fira and West Ennedi (Figure 1). These areas are known for having favourable climatic conditions during the rainy season (July–October) for animal grazing, and therefore gathers most nomadic herds (cattle, small ruminants and dromedaries) from the sub‐region.
Figure 1.
Chad Departments within the region of PREPAS project (black rectangle)
A cross‐sectional survey for sample collection was conducted in October and November 2016. Chad's territorial administration is divided into regions that are divided into departments, whereas the latter are divided into sub‐prefectures. These sub‐prefectures are divided into cantons. Thus, the epidemiological unit selected for carrying out the surveys was the canton. On the basis of animal density, 20 cantons were sampled: six cantons in East Batha, six cantons in West Batha, four cantons in Wadi Fira and four cantons in West Ennedi. This number of 20 cantons was determined according to our material and human resources available and the duration of the investigation. Herds and animals were randomly sampled from a list of all animals in each herd. Indeed, in each herd, a list of all the animals was established by another team that was with us in the field but responsible for conducting a demographic study. In East Batha and West Batha, eight cattle herds, three goat and sheep herds, and one dromedary herd were sampled per canton. In Wadi Fira, five cattle herds, and three goat, sheep and camels herds were sampled. Finally, four goat and sheep herds, and five dromedary herds were sampled in West Ennedi. No cattle were sampled in West Ennedi because there are no cattle in this region.
A minimum of 11 sera per herd of cattle and 5 sera per herd of small ruminants and dromedaries were sampled to reach 95% chance to observe at least one positive test in the herd, assuming random sampling within herds with seroprevalence rates of 25% and 50%, respectively, in cattle and small ruminants. In total, 1,520 sera samples were collected for serological analyses (928 from cattle, 216 from goats, 254 from sheep and 122 from dromedaries). For virological testing, it was decided to sample all cattle showing FMD clinical signs. Nine epithelial tissue samples were collected from nine cattle showing clinical signs of disease. These sick cattle were not sampled in the serological study. Samples were sent according to the recommendations of the OIE (2008) to the FMD reference laboratory of Maisons‐Alfort (France), for confirmatory diagnosis and FMDV serotype characterization.
2.2. Laboratory analyses
Three serological tests (ELISA PrioCHECK® FMDV NSP, ELISA PrioCHECK® FMDV types O and A and ELISA internal solid phase competition (SPCE)) were performed at the EU/OIE/FAO reference laboratory for foot‐and‐mouth disease in Maisons‐Alfort (France). The tests ELISA PrioCHECK® FMDV (NSP, types O and A) were used according to the manufacturer's protocol. Indeed, the PrioCHECK® FMDV NSP ELISA was used to determine the overall seroprevalence of FMD. This test is not serotype specific. It detects the presence of antibodies induced by non‐structural proteins (in particular 3ABC protein) (Brocchi et al., 2006; Sorensen et al., 1998). The ELISA PrioCHECK® FMDV types O and A test were used to detect antibodies against FMDV O and A serotypes (Chénard, Miedema, Moonen, Schrijver, & Dekker, 2003; Relmy et al., 2017). The solid phase competition ELISA (SPCE) test was used for the detection of antibodies against FMDV serotypes SAT1, SAT2 and SAT3 (Li et al., 2012). The sensitivity (Se) and specificity (Sp) of these PrioCHECK® commercial tests were known, based on the validation data established by the FMD reference laboratory: ELISA PrioCHECK® FMDV NSP (Se: 86%–100% and Sp: 87%–99.5% data consistent with those reported by Brocchi et al., 2006 and Parida et al., 2007), ELISA PrioCHECK® FMDV type O (Se: 85.7%–98.8% and Sp: 99%–100%), or given by the supplier for the ELISA PrioCHECK® FMDV type A (Se: 90% and Sp: 99%). For the ELISA SPCE, the Se is 100% and the Sp is 99.41%–99.7% (Li et al., 2012). Serotyping tests were carried out only on sera from young animals under 1 year of age to determine the different serotypes of FMD virus which circulated in 2016.
For the detection of FMD virus, three tests were carried out: the real‐time RT‐PCR, multiplex RT‐PCR and conventional RT‐PCR. Indeed, real‐time RT‐PCR has been used for the detection of foot‐and‐mouth disease targeting the 3D and IRES regions of the viral genome (Laor, Torgersen, Yadin, & Becker, 1992; Reid, Ferris, Hutchings, Samuel, & Knowles, 2000). Multiplex RT‐PCR has been used for typing FMD virus (Callens & De Clercq, 1997; Giridharan, Hemadri, Tosh, Sanyal, & Bandyopadhyay, 2005; Gorna et al., 2016). However, this test does not allow the sequencing because the amplified part is very short (250 nt) which is insufficient for complete sequencing of the VP1 gene (Gorna et al., 2016). Finally, conventional RT‐PCR was used to amplify the complete gene encoding the VP1 protein (Relmy et al., 2017). Indeed, the classical RT‐PCR test was carried out using different primers specific to the four suspected FMD virus serotypes (O, A, SAT1 and SAT2). Thus, the following pairs of primers: O‐1C‐244F/EUR2B R; O‐1C‐272F/EUR2B R; O‐1C‐283F/EUR2B R; A‐1C‐562F/EUR2B R; A‐1C‐612F/EUR2B R; SAT1‐1C‐559F/SAT2B 208R; SAT2‐1C‐445F/SAT‐2B‐208R and SAT2‐P1‐1223F/SAT‐2B‐208R have been selected to amplify the genome region encoding the VP1 protein (Ayelet et al., 2009). DNA sequencing of the PCR products was carried out using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Life Technologies), according to the technical protocol described by Knowles, Wadsworth, Bachanek‐Bankowska, and King (2016).
2.3. Bioinformatic analysis of nucleotide sequences
The sequences obtained were assembled and verified using ContigExpress software (Vector NTI, Invitrogen). A complete sequence of the VP1 coding region was analysed and compared to the homologous genomic regions available in the NCBI GenBank database. Multiple sequence alignment and phylogenetic analyses were conducted using MEGA version 6 software (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013). FMDV sequences were aligned using the Muscle program with default parameters (Edgar, 2004). Phylogenetic analyses were conducted by Neighbour‐Joining (NJ) method (Saitou & Nei, 1987) with the Kimura two‐parameter model (Kimura, 1980). The confidence of the NJ tree was assessed by bootstrap analysis with 1,000 replicates.
2.4. Statistical analyses
Binomial regression was conducted using the package R‐INLA (Rue, Martino, & Chopin, 2009) of the R software (R Core Team, 2018) to estimate overall disease seroprevalence by species, age category and region. Since infection patterns are very different between species, species‐specific seroprevalence was estimated using separate models. However, one estimate was presented for small ruminants that is sheep and goats.
The epidemiological unit (EU) of the analysis was the group of animals of the same species and age category within a herd (thus, within a region as well). The EU was considered infected whenever at least one of the animals in the EU tested positive. The number of positive EUyij in region i and age category j for a given species has been modelled with a binomial likelihood, where nij is the number of sampled EU in region i and age group j.
The group‐prevalence pij has been modelled in a logit‐scale with a linear predictor with a region‐specific intercept βi and varying effects wij of the age category for each region:
so that within each region, a separate first‐order autoregressive model (AR1) has been used for the effect of the age category, all sharing the same marginal variance and autocorrelation parameter.
Given the small number of EU for some species (e.g. less than 10 for most age categories and regions of small ruminants), we fitted this model using Bayesian approach to reduce overfitting and improve the seroprevalence estimates. Finally, for the prior parameter of the scale, the approach of the penalized complexity prior was followed. This approach is defined in terms of the Kullback–Leibler distance from the model from a simpler model with zero variance. The penalty rate is determined by specifying a probability assessment such that P(σw > U) = α (Simpson, Rue, Riebler, Martins, & Sørbye, 2017). Thus, to make the calculation, U = 3 and α = 0.001 were set, which means that it is very unlikely that the scale parameter of the autoregressive effect was greater than 3.
3. RESULTS AND DISCUSSION
3.1. Seroprevalence results
Figure 2 shows the observed (black) and estimated (grey) seroprevalences of NSP antibodies at the group level by region and age category for the different animal species studied. Indeed, since in Chad, there has never been vaccination against foot‐and‐mouth disease, the antibodies found in animals were probably induced by wild viruses. Serological results show an overall NSP seroprevalence of 40% (375/928) in cattle in our sample, 95% credible interval (CrI) [19–63].
Figure 2.
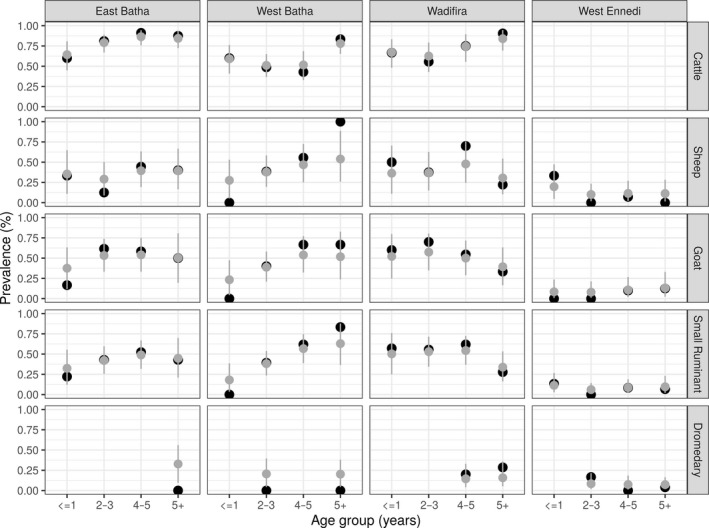
Estimated (grey) and observed (black) seroprevalence rates by group with 95% CrI
In cattle over 5 years of age, seroprevalence was estimated at 84% (27/32) (95% CrI: 72–93), 78% (35/45) (95% CrI: 65–89), 84% (21/25) (95% CrI: 69–95), respectively, in East Batha, West Batha and Wadi Fira. Too, seroprevalence estimates were made for cattle under 1 year of age: 67% (18/27) (95% CrI: 48–83) in Wadi Fira, 64% (16/25) (95% CrI: 45–81) in East Batha and 59% (13/22) (95% CrI: 41–76) in West Batha.
Furthermore, among small ruminants over 5 years of age, seroprevalences of 63% (15/24) (95% CrI: 36–88), 45% (13/29) (95% CrI: 21–70), 34% (15/44) (95% CrI: 16–53) and 10% (5/50) (95% CrI: 2–23) have been estimated in West Batha, East Batha, Wadi Fira and West Ennedi, respectively. Also, among young small ruminants under 1 year of age, seroprevalences were estimated in the different areas studied: 32% (9/28) (95% CrI: 12–55) in East Batha, 18% (8/45) (95% CrI: 4–39) in West Batha, 50% (14/28) (95% CrI: 25–76) in Wadi Fira and 11% (2/19) (95% CrI: 3–27) in West Ennedi.
Finally, seroprevalence has also been estimated among dromedaries over 5 years old in the different areas: 33% (5/15) (95% CrI: 10–56) in East Batha, 20% (2/10) (95% CrI: 4–38) in West Batha, 16% (6/38) (95% CrI: 5–36) in Wadi Fira, 7% (3/42) (95% CrI: 2–16) in West Ennedi. In dromedaries less than 1‐year old, seroprevalence was not determined due to lack of samples in this age group.
Based on the results of the seroprevalences estimates obtained, it was found that the three areas: West Batha, East Batha and Wadi Fira have high seroprevalence of FMD compared to the West Ennedi area. This can be explained by the fact that these three areas have high animal densities (cattle and small ruminants). In addition, these are transhumant areas where animal movements are uncontrolled. These uncontrolled movements may be one of the main sources of disease spread. Our results are consistent with previous results obtained by Ouagal and colleagues (Ouagal et al., 2010) in the same areas in Chad. On the other hand, the low seroprevalence rate obtained in Ennedi West could be explained by the fact that this area is arid, with a very low animal density and limited between herds contacts.
The low seroprevalence of dromedaries compared to other animal species (cattle, goats and sheep) is due to the fact that camels are less susceptible to FMD. Our results on the seroprevalence of FMD in dromedaries confirm the results obtained by Wungak and collaborators (2015) in Nigeria.
Seroprevalence rates by age group were found to be higher in older animals than in young animals under 1 year of age. These low seroprevalence rates in young animals (less than 1‐year old) may be due to low exposure of young animals to risk factors or to the fact that the virus has recently circulated at a low level. There is also the practice of herders to keep young animals separate from adult animals around the village, which may also explain the low seroprevalence rates. This may also be due to the fact that adult animals are repeatedly exposed and in close contact with other animals in pastures and water points. Our results are similar to those published by Wungak, Olugasa, Ishola, Lazarus, and Ularamu (2016). These authors determined seroprevalence by age group in Nigeria and also obtained higher seroprevalence in adults than in young animals.
Concerning the serotyping of the FMDV in Chad, serotyping tests were carried out on sera from young animals under 1 year of age (67/194) positive for the NSP ELISA test. The FMDV serotyping results have been broken down by geographical area and are presented in Table 1. These results show that the four serotypes O, A, SAT1 and SAT2 circulated well in Chad in 2016. The SAT2 type dominated with an overall seroprevalence of 43.3% and was present in all four areas. It is followed by serotype O (29.9%). This serotype O was present in West Batha and Wadi Fira. Then comes serotype A (22.4%) but it has only been detected in Wadi Fira. Finally, the SAT1 type (4.5%) was present in East Batha and West Batha.
Table 1.
Seroprevalence of FMD virus serotypes (ELISA PrioCHECK® types O and A and ELISA SPCE) in young animals by geographical area
| Areas | Number of young animals tested positive to NSP ELISA (age ≤ 1 year) | Percentage of positive animals to the different serotyping tests % (n) | |||
|---|---|---|---|---|---|
| Type SAT2 | Type O | Type SAT1 | Type A | ||
| West Batha | 13/49 | 61.5% (n = 8) | 30.8% (n = 4) | 7.7% (n = 1) | 0 |
| East Batha | 20/46 | 90% (n = 18) | 0 | 10% (n = 2) | 0 |
| West Ennedi | 2/17 | 100% (n = 2) | 0 | 0 | 0 |
| Wadi Fira | 32/82 | 3.1% (n = 1) | 50% (n = 16) | 0 | 46.9% (n = 15) |
| Total | 67/194 | 43.3% (n = 29) | 29.9% (n = 20) | 4.5% (n = 3) | 22.4% (n = 15) |
The presence of the four serotypes (A, O, SAT1 and SAT2) in the country could be due to the free cross‐border movement of animals in search of pastures and water points as well as uncontrolled trade in livestock between Chad, the Central African Republic, Sudan, Cameroon and Niger. Indeed, these four circulating serotypes (O, A, SAT1 and SAT2) have also been identified and not yet eradicated in countries bordering Chad (Bertram, Bravo, et al., 2018; Bertram, Delgado, et al., 2018; Habiela, Alamin, et al., 2010; Habiela, Ferris, et al., 2010; Kouato, Elliot, et al., 2018; Kouato, Souley, et al., 2018; Wungak et al., 2016).
3.2. Virological results
Virological analyses initially yielded five positives out of nine samples to the real‐time RT‐PCR test detecting both IRES and 3D targets of the viral genome. Then, the five real‐time RT‐PCR‐positive samples were tested by conventional multiplex RT‐PCR for virus typing. These typing results showed that only one out five was positive for the primer pair: SAT2‐P1‐1223F/SAT‐2B‐208R, confirming the presence of FMDV serotype SAT2. This serotype SAT2 sample was taken from the West Batha area. Several attempts to isolate and amplify the VP1 gene from the four negative samples were made but without success. This may be explained by the poor quality of these samples.
Finally, phylogenetic analyses of the sequence coding for VP1 of the SAT2 type obtained allowed determining the SAT2 topotype VII serotype, close to the virus strains of Cameroon 2015 with a similarity of 98.60% (Figure 3). This suggests the movements of infected animals between Chad and Cameroon. The resulting SAT2 sequence has been deposited in GenBank whose GenBank access number is BankIt2141564 Seq1 MH74858579.
Figure 3.
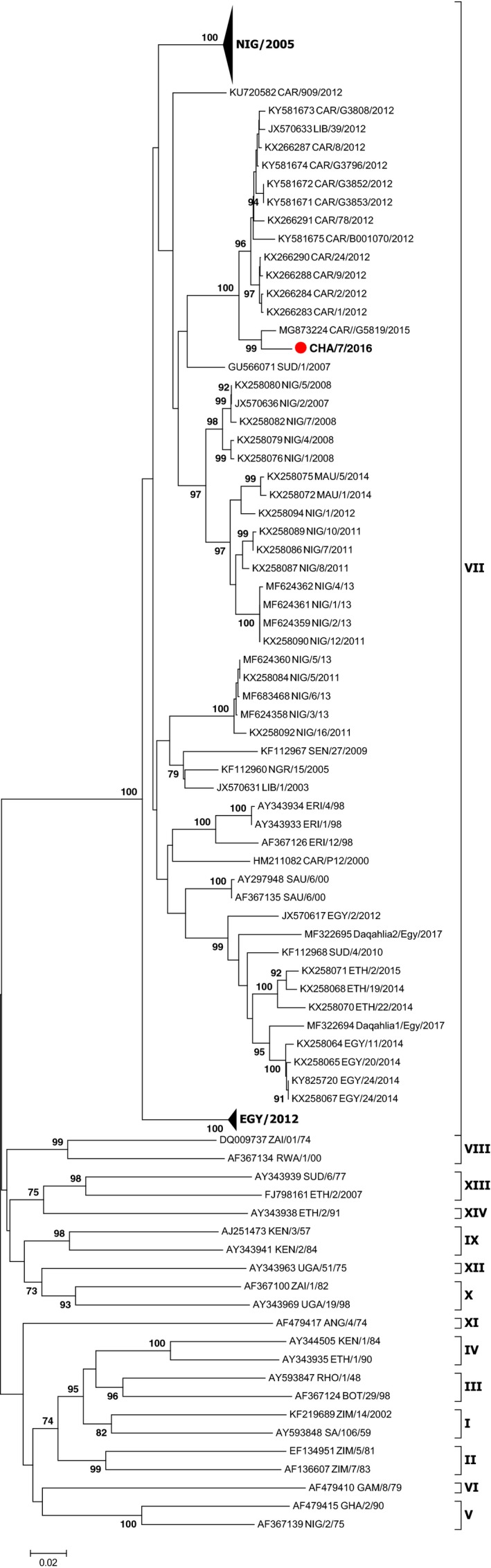
Phylogenetic tree of the FMD SAT2 virus sampled in Chad
4. CONCLUSION
This study showed that FMD is enzootic in the four study areas in Chad (East Batha, West Batha, Ennedi‐West and Wadi Fira) and that FMDV SAT2 serotype is circulating in all four areas. This is the first characterization of FMDV circulating in Chad. FMD endemicity may be maintained by primary foci scattered in pastoral areas where animal density is high. The high rate of seroprevalence of FMD and the potential for spread of the disease represent a major challenge for the control and eradication in Chad. Further studies in other area of Chad would provide more insight on the epidemiology of the disease at a national level.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
CONTRIBUTIONS
The authors were involved in the study design, analysis and interpretation of data. All authors contributed critically to the revision of the manuscript and had read and approved the final version.
ETHICS STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. The work described herein was approved by the Livestock Research Institute for Development in N'Djamena, the Veterinary Services Directorate (Chad). The disease described here occurred spontaneously in domestic animals without experimentation, inoculation or treatment of live animals.
ACKNOWLEDGEMENTS
The authors sincerely thank all the staff of the European Union/OIE/FAO reference laboratory for foot‐and‐mouth disease at ANSES in Maisons‐Alfort (France), the UMR ASTRE at CIRAD in Montpellier (France), the Livestock Research Institute for Development in N'Djamena (Chad) and the Veterinary Services Directorate (Chad) for their direct and indirect support.
Abdel‐Aziz AI, Romey A, Relmy A, et al. Seroprevalence and molecular characterization of foot‐and‐mouth disease virus in Chad. Vet Med Sci. 2020;6:114–121. 10.1002/vms3.206
Funding information
This study was co‐financed by ANSES/CIRAD as part of the doctoral thesis at AgroParisTech (France) of Mr Abdel‐Aziz.
REFERENCES
- Abu elzein, E. (1983). Foot and mouth disease in the Sudan. Revue Scientifique Et Technique De l'OIE, 2(1), 177–188. 10.20506/rst.2.1.106 [DOI] [PubMed] [Google Scholar]
- Ayelet, G. , Mahapatra, M. , Gelaye, E. , Egziabher, B. G. , Rufeal, T. , Sahle, M. , … Knowles, N. J. (2009). Genetic characterization of foot‐and‐mouth disease viruses, Ethiopia, 1981–2007. Emerging Infectious Diseases, 15(9), 1409–1417. 10.3201/eid1509.090091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram, M. R. , Bravo, C. , de Rueda, R. , Garabed, S. D. , Jumbo, M. M. , Pauszek, S. , … Arzt, J. (2018). Molecular epidemiology of foot‐and‐mouth disease virus in the context of transboundary animal movement in the far North Region of Cameroon. Frontiers in Veterinary Science, 5, 320 10.3389/fvets.2018.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram, M. R. , Delgado, A. , Pauszek, S. J. , Smoliga, G. R. , Brito, B. , Stenfeldt, C. , … Arzt, J. (2018). Effect of vaccination on cattle subclinically infected with foot‐and‐mouth disease virus in Cameroon. Preventive Veterinary Medicine, 155, 1–10. 10.1016/j.prevetmed.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Brocchi, E. , Bergmann, I. E. , Dekker, A. , Paton, D. J. , Sammin, D. J. , Greiner, M. , … De Clercq, K. (2006). Comparative evaluation of six ELISAs for the detection of antibodies to the non‐structural proteins of foot‐and‐mouth disease virus. Vaccine, 24(47–48), 6966–6979. 10.1016/j.vaccine.2006.04.050 [DOI] [PubMed] [Google Scholar]
- Callens, M. , & De Clercq, K. (1997). Differentiation of the seven serotypes of foot‐and‐mouth disease virus by reverse transcriptase polymerase chain reaction. Journal of Virological Methods, 67(1), 35–44. 10.1016/S0166-0934(97)00074-8 [DOI] [PubMed] [Google Scholar]
- Chénard, G. , Miedema, K. , Moonen, P. , Schrijver, R. S. , & Dekker, A. (2003). A solid‐phase blocking ELISA for detection of type O foot‐and‐mouth disease virus antibodies suitable for mass serology. Journal of Virological Methods, 107(1), 89–98. 10.1016/S0166-0934(02)00196-9 [DOI] [PubMed] [Google Scholar]
- Couacy‐Hymann, E. , Aplogan, G. L. , Sangaré, O. , Compaoré, Z. , Karimu, J. , Awoueme, K. A. , … Valarcher, J. F. (2006). Étude Rétrospective de La Fièvre Aphteuse En Afrique de l’Ouest de 1970 à 2003. Revue Scientifique Et Technique/Office International Des Epizooties, 25(3), 1013–1024. 10.20506/rst.25.3.1709 [DOI] [PubMed] [Google Scholar]
- Domingo, E. , Mateu, M. G. , Martínez, M. A. , Dopazo, J. , Moya, A. , & Sobrino, F. (1990). Genetic variability and antigenic diversity of foot‐and‐mouth disease virus In Kurstak R. G. M. E., Murphy F. A. & Regenmortel M. H. V. (Eds.), Virus variability, epidemiology and control (pp. 233–266). New York, NY: Plenum Publishing Corp. [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehizibolo, D. O. , Haegeman, A. , De Vleeschauwer, A. R. , Umoh, J. U. , Kazeem, H. M. , Okolocha, E. C. , … De Clercq, K. (2017a). Foot‐and‐mouth disease virus serotype SAT 1 in Cattle, Nigeria. Transboundary and Emerging Diseases, 64(3), 683–690. 10.1111/tbed.12629 [DOI] [PubMed] [Google Scholar]
- Ehizibolo, D. O. , Haegeman, A. , De Vleeschauwer, A. R. , Umoh, J. U. , Kazeem, H. M. , Okolocha, E. C. , … De Clercq, K. (2017b). Detection and molecular characterization of foot and mouth disease viruses from outbreaks in some states of Northern Nigeria 2013–2015. Transboundary and Emerging Diseases, 64(6), 1979–1990. 10.1111/tbed.12602 [DOI] [PubMed] [Google Scholar]
- Giridharan, P. , Hemadri, D. , Tosh, C. , Sanyal, A. , & Bandyopadhyay, S. K. (2005). Development and evaluation of a multiplex PCR for differentiation of foot‐and‐mouth disease virus strains Native to India. Journal of Virological Methods, 126(1–2), 1–11. 10.1016/j.jviromet.2005.01.015 [DOI] [PubMed] [Google Scholar]
- Gorna, K. , Relmy, A. , Romey, A. , Zientara, S. , Blaise‐Boisseau, S. , & Bakkali‐Kassimi, L. (2016). Establishment and validation of two duplex one‐step real‐time RT‐PCR Assays for diagnosis of foot‐and‐mouth disease. Journal of Virological Methods, 235, 168–175. 10.1016/j.jviromet.2016.03.020 [DOI] [PubMed] [Google Scholar]
- Gueme, A. (1998). “Contribution au sérotypage du virus aphteux dans quelques régions du Tchad”. Mémoire de fin d’études, Adjoint technique d’élevage à l’Ecole Nationale des Techniques d’Elevage, N’Djaména, Tchad. Bibliothèque de Laboratoire de Recherches Vétérinaires et Zootechnie (LRVZ)de. Farcha‐N’Djamena (Tchad). [Google Scholar]
- Habiela, M. , Alamin, M. A. G. , Raouf, Y. A. , & Ali, Y. H. (2010). Epizootiological study of foot and mouth disease in the sudan: the situation after two decades. Veterinarski Arhiv, 80(1), 11–26. [Google Scholar]
- Habiela, M. , Ferris, N. P. , Hutchings, G. H. , Wadsworth, J. , Reid, S. M. , Madi, M. , … Paton, D. J. (2010). Molecular characterization of foot‐and‐mouth disease viruses collected from Sudan. Transboundary and Emerging Diseases, 57(5), 305–314. 10.1111/j.1865-1682.2010.01151.x [DOI] [PubMed] [Google Scholar]
- Jamal, S. M. , & Belsham, G. J. (2013). Foot‐and‐mouth disease: past, present and future. Veterinary Research, 44(1), 116 10.1186/1297-9716-44-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16(2), 111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Knight‐Jones, T. J. D. , & Rushton, J. (2013). The economic impacts of foot and mouth disease—What are they, how big are they and where do they occur? Preventive Veterinary Medicine, 112(3), 161–173. 10.1016/j.prevetmed.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, N. J. , Bachanek‐Bankowska, K. , Wadsworth, J. , Mioulet, V. , Valdazo‐González, B. , Eldaghayes, I. M. , … King, D. P. (2016). Outbreaks of foot‐and‐mouth disease in Libya and Saudi Arabia during 2013 due to an exotic O/ME‐SA/Ind‐2001 lineage virus. Transboundary and Emerging Diseases, 63(5), e431–e435. 10.1111/tbed.12299 [DOI] [PubMed] [Google Scholar]
- Knowles, N. J. , Wadsworth, J. , Bachanek‐Bankowska, K. , & King, D. P. (2016). VP1 sequencing protocol for foot and mouth disease virus molecular epidemiology. Revue Scientifique Et Technique, 35(3), 741–755. 10.20506/rst.35.3.2565 [DOI] [PubMed] [Google Scholar]
- Kouato, B. S. , Elliot, F. M. King, D. P. , Hyera, J. , Knowles, N. J. , Ludi, A. B. , … Saegerman, C. (2018). Outbreak Investigations and Molecular Characterization of Foot-and-Mouth Disease Viruses Circulating in South-West Niger. Transboundary and Emerging Diseases, 65(1), 146–57. 10.1111/tbed.12642. [DOI] [PubMed] [Google Scholar]
- Kouato, B. , Souley, E. , Thys, V. , Renault, E. , Abatih, H. , Marichatou, S. I. , & Saegerman, C. (2018). Spatio‐temporal patterns of foot‐and‐mouth disease transmission in cattle between 2007 and 2015 and quantitative assessment of the economic impact of the disease in Niger. Transboundary and Emerging Diseases, 65(4), 1049–1066. 10.1111/tbed.12845 [DOI] [PubMed] [Google Scholar]
- Laor, O. , Torgersen, H. , Yadin, H. , & Becker, Y. (1992). detection of FMDV RNA amplified by the polymerase chain reaction (PCR). Journal of Virological Methods, 36(3), 197–207. 10.1016/0166-0934(92)90051-E [DOI] [PubMed] [Google Scholar]
- Li, Y. , Swabey, K. G. , Gibson, D. , Keel, P. J. , Hamblin, P. , Wilsden, G. , … Ferris, N. P. (2012). Evaluation of the solid phase competition ELISA for detecting antibodies against the six foot‐and‐mouth disease virus non‐O serotypes. Journal of Virological Methods, 183(2), 125–131. 10.1016/j.jviromet.2012.04.002 [DOI] [PubMed] [Google Scholar]
- OIE . (2008). “Manuel Des Tests de Diagnostic et Des Vaccins Pour Les Animaux Terrestres.” 2008. http://www.test.oie.int/fileadmin/Home/fr/Health_standards/tahm/Volume1_Manuel2008_fr.pdf. [Google Scholar]
- OIE . (2017). “Foot and Mouth Disease: Chapitre 2.1.8. Du Manuel Terrestre OIE”. 2017. http://www.oie.int/fileadmin/Home/fr/Health_standards/tahm/2.01.08_FMD.pdf.
- Ouagal, M. , Brocchi, E. , Grazioli, S. , Ade, B. Y. , Keith, S. , Kiram, D. , … Saegerman, C. (2018). Study on seroprevalence and serotyping of foot and mouth disease in Chad. Revue Scientifique Et Technique De l'OIE, 37(3), 937–947. 10.20506/37.3.2897 [DOI] [PubMed] [Google Scholar]
- Ouagal, M. , Brocchi, E. , Grazioli, S. , Ben Youssef, A. , Sumption, K. , Hendrikx, P. , … Saegerman, C. (2017). Evaluation de La Sensibilité Du Réseau d’épidémiosurveillance Des Maladies Animales Au Tchad Pour La Surveillance de La Fièvre Aphteuse. Épidémiologie Santé Animale, 72, 5–14. [Google Scholar]
- Ouagal, M. , Hendrikx, P. , Saegerman, C. , & Berkvens, D. (2010). Comparison between active and passive surveillance within the network of epidemiological surveillance of animal diseases in Chad. Acta Tropica, 116(2), 147–151. 10.1016/j.actatropica.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Parida, S. , Fleming, L. , Gibson, D. , Hamblin, P. A. , Grazioli, S. , Brocchi, E. , & Paton, D. J. (2007). Bovine serum panel for evaluating foot‐and‐mouth disease virus nonstructural protein antibody tests. Journal of Veterinary Diagnostic Investigation, 19(5), 539–544. 10.1177/104063870701900513 [DOI] [PubMed] [Google Scholar]
- Paton, D. J. , Sumption, K. J. , & Charleston, B. (2009). Options for control of foot‐and‐mouth disease: knowledge, capability and policy. Philosophical Transactions of the Royal Society of London B: Biological Sciences, 364(1530), 2657–2667. 10.1098/rstb.2009.0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2018). R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; https://www.r-project.org/. ISBN 3‐900051‐07‐0. [Google Scholar]
- Reid, S. M. , Ferris, N. P. , Hutchings, G. H. , Samuel, A. R. , & Knowles, N. J. (2000). Primary diagnosis of foot‐and‐mouth disease by reverse transcription polymerase chain reaction. Journal of Virological Methods, 89(1–2), 167–176. 10.1016/S0166-0934(00)00213-5 [DOI] [PubMed] [Google Scholar]
- Relmy, A. , Romey, A. , Gorna, K. , Blaise‐Boisseau, S. , Laloy, È. , Meenowa, D. , & Samoisy, K. (2017). Crise sanitaire dans l’Océan Indien: virus de la fièvre aphteuse aux Îles Maurice et Rodrigues en 2016. Épidémiologie Santé Animale, 71, 117–127. [Google Scholar]
- Rue, H. , Martino, S. , & Chopin, N. (2009). Approximate Bayesian inference for latent Gaussian models by using integrated nested laplace approximations. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 71(2), 319–392. 10.1111/j.1467-9868.2008.00700.x [DOI] [Google Scholar]
- Rweyemamu, M. , Roeder, P. , Mackay, D. , Sumption, K. , Brownlie, J. , Leforban, Y. , … Saraiva, V. (2008). Epidemiological patterns of foot‐and‐mouth disease worldwide. Transboundary and Emerging Diseases, 55(1), 57–72. 10.1111/j.1865-1682.2007.01013.x [DOI] [PubMed] [Google Scholar]
- Saitou, N. , & Nei, M. (1987). The neighbor‐joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution, 4(4), 406–425. [DOI] [PubMed] [Google Scholar]
- Simpson, D. , Rue, H. , Riebler, A. , Martins, T. G. , & Sørbye, S. H. (2017). Penalising model component complexity: a principled, practical approach to constructing priors. Statistical Science, 32(1), 1–28. 10.1214/16-STS576 [DOI] [Google Scholar]
- Sorensen, K. J. , Madsen, K. G. , Madsen, E. S. , Salt, J. S. , Nqindi, J. , & Mackay, D. K. J. (1998). Differentiation of infection from vaccination in foot‐and‐mouth disease by the detection of antibodies to the non‐structural proteins 3D, 3AB and 3ABC in ELISA using antigens expressed in baculovirus. Archives of Virology, 143(8), 1461–1476. 10.1007/s007050050390 [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Stecher, G. , Peterson, D. , Filipski, A. , & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12), 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry, E. , Baranowski, E. , & Domingo, E. (2001). Epidémiologie moléculaire de La Fièvre aphteuse. Epidémiology Santé Animals, 39, 59–67. [Google Scholar]
- Ularamu, H. G. , Wungak, Y. S. , Lazarus, D. D. , Woma, T. Y. , Ehizibolo, D. O. , Dogonyaro, B. B. , … Shamaki, D. (2015). Serological evidence of foot‐and‐mouth disease virus (FMDV) in camels (Camelus Dromedaries) in Nigeria. Journal of Veterinary Medicine and Animal Health, 7(7), 261–264. 10.5897/JVMAH2015.0393 [DOI] [Google Scholar]
- Wungak, Y. S. , Olugasa, B. O. , Ishola, O. O. , Lazarus, D. D. , & Ularamu, G. H. (2016). Foot‐and‐mouth disease (FMD) prevalence and exposure factors associated with seropositivity of cattle in North‐Central, Nigeria. African Journal of Biotechnology, 15(24), 1224–1232. 10.5897/AJB2016.15332 [DOI] [Google Scholar]


