Abstract
Resistant Acinetobacter baumannii isolates are not only known as opportunistic nosocomial bacteria but may also be regarded as emerging bacterial contaminants in foods of animal origins. The present investigation was done to assess the prevalence and antibiotic resistance pattern of A. baumannii isolated from different types of raw meat samples. One hundred and ninety‐four raw meat samples were collected and cultured for A. baumannii isolates. Culture‐positive bacteria were also approved using the loop‐mediated isothermal amplification (LAMP) technique. The disc diffusion method was used for antibiotic susceptibility testing. Out of 194 raw meat samples, 39 (20.10%) were positive for A. baumannii isolates. Ovine raw meat was the most commonly contaminated samples (32.14%). All of the culture‐positive A. baumannii isolates were also approved using the LAMP assay. A. baumannii isolates harboured the highest prevalence of resistance against gentamicin (87.17%), tetracycline (79.48%), erythromycin (74.35%), azithromycin (66.66%), ciprofloxacin (58.97%), trimethoprim/sulphamethoxazole (56.41%) and rifampin (51.28%). The lowest prevalence of resistance was found against imipenem (17.94%) and chloramphenicol (28.20%). Raw bovine, ovine, caprine, camel and poultry meat samples were considered as the important sources of isolates resistant to some of the categories of antimicrobials used to treat infections caused by A. baumannii. Further studies are required to find the exact role of resistant A. baumannii isolates in the dissemination of antibiotic resistance to human population.
Keywords: Acinetobacter baumannii, Antibiotic resistance, Iran, LAMP assay, Raw meat
Acinetobacter baumannii, LAMP‐PCR, Antibiotic resistance, Raw meat, Iran.
1. INTRODUCTION
The consumption of raw or undercooked meat has been associated with several outbreaks of bacterial food‐borne disease all around the world (Ghorbani, Gheisari, & Dehkordi, 2016; Momtaz, Davood Rahimian, & Safarpoor Dehkordi, 2013; Safarpoor Dehkordi, Gandomi, Akhondzadeh Basti, Misaghi, & Rahimi, 2017; Safarpoor Dehkordi, Haghighi, Momtaz, Rafsanjani, & Momeni, 2013; Safarpoor Dehkordi, Khamesipour, & Momeni, 2014; Safarpoor Dehkordi et al., 2012).
Acinetobacter species are saprophytic, ubiquitous and have emerged as an important nosocomial pathogen due to its ability for survival in the hospital environment on a wide range of dry and moist surfaces (Tavakol, Momtaz, Mohajeri, Shokoohizadeh, & Tajbakhsh, 2018). Human infections caused by Acinetobacter species include pneumonia, which is most often related to endotracheal tubes or tracheostomies, endocarditis, meningitis, skin and wound infections, peritonitis in patients receiving peritoneal dialysis, urinary tract infection (UTI) and bacteraemia (Tavakol et al., 2018). Acinetobacter spp., especially A. baumannii, are one of the newly emerged bacteria all‐around the world Tavakol et al., 2018). A. baumannii isolates have been rarely isolated from food products, especially meat (Tavakol et al., 2018), milk (Gurung et al., 2013) and vegetable (Berlau, Aucken, Houang, & Pitt, 1999). However, data about the clonality of A. baumannii from food are lacking, which precludes any speculation about the eventual exchange of A. baumannii clones between food and clinical settings (Berlau et al., 1999; Gurung et al., 2013; Tavakol et al., 2018).
A rapid and sensitive amplification platform for DNA called the loop‐mediated isothermal amplification (LAMP) assay has been developed to detect the A. baumannii isolates in recent years (Soo et al., 2013). It is based on autocycling strand displacement DNA synthesis in the presence of Bst DNA polymerase under isothermal conditions within 1 hr. The technique has been used widely in clinical diagnosis and detection of epidemic bacteria (Shahran, Dhingolia, Khatri, Singh Duban, & Kumar Gahlawat, 2014).
Owing to its remarkable ability to resist almost all available antimicrobial agents, A. baumannii infections are difficult to treat. Indeed, multidrug‐resistant or even pan‐drug‐resistant isolates are increasing alarmingly in the environment (Moradi, Hashemi, & Bahador, 2015; Xie, Zhang, Zhao, Peng, & Zheng, 2018).
Studies regarding the association of A. baumannii isolates with food‐borne illnesses are somewhat limited. Therefore, the present investigation was done to study the prevalence rate and phenotypic characterization of antibiotic resistance of the A. baumannii isolated from bovine, ovine, caprine, camel, chicken and turkey raw meat samples.
2. MATERIALS AND METHODS
2.1. Ethical consideration
The study was approved by the Ethical Council of Research of the Faculty of Basic Science, Islamic Azad University, Shahrekord, Iran. Verification of this research project and the licenses related to sampling process were approved by the Professor Hassan Momtaz (Approval Ref Number MCB 2017/1049).
2.2. Samples
From May to September 2017, a total of 194 various types of raw meat samples including bovine (n = 39), ovine (n = 28), caprine (n = 41), camel (n = 16), chicken (n = 52) and turkey (n = 18) meat samples were randomly collected from the butchers of different parts of the Isfahan province, Iran. All samples were collected from the femur muscle of animal species. The Isfahan province covers an area of approximately 107,027 km2 and is situated in the centre of Iran. All samples were taken from the femur muscle of animal species. A quantity of thirty grams of meat was collected from each animal. Samples were immediately transferred to the Microbiology Research Center of the Islamic Azad University of Shahrekord in cooler with ice packs. All meat samples showed normal physical characters including odour, colour and consolidation.
2.3. Bacterial isolation
Approximately 10 g of meat was homogenized in 90 ml of Luria‐Bertani broth in a stomacher (Stomacher 400 Circulator, Seward, Norfolk, UK) for 1 minutes and was incubated overnight at 37°C with agitation. From this overnight culture, 10 µl was streaked onto selective Chrom ID ESBL agar (bioMérieux, Marcy‐l’Étoile, France), which does not suppress the growth of A. baumannii (Tavakol et al., 2018), and was incubated at 37°C. All white colonies (presumptive A. baumannii) were transferred onto tryptic soy agar plates containing 5% sheep blood (BD, Franklin Lakes, NJ) and were incubated overnight at 37°C. Conventional biochemical methods, such as oxidase, citrate, urea urease, malonate consumption, oxidation and fermentation of sugars, motility and indole production, were used to identify A. baumannii. Additionally, the genus Acinetobacter was identified by Gram staining, cell and colony morphology, positive catalase test, negative oxidase test and absence of motility. Speciation of Acinetobacter was performed on the basis of glucose oxidation, gelatine liquefaction, beta haemolysis, growth at 37°C and 42°C, arginine hydrolysis and susceptibility to chloramphenicol. The colonies were identified using a matrix‐assisted laser desorption ionization time‐of‐flight mass spectrometer (Microflex LT, Bruker Daltonics, Bremen, Germany) (Espinal, Seifert, Dijkshoorn, Vila, & Roca, 2012).
2.4. Approval of bacterial isolates using the loop‐mediated isothermal amplification
Acinetobacter baumannii isolates were subcultured on TSB media (Merck, Darmstadt, Germany) and incubated for 48 hr at 37°C. Genomic DNA was then extracted from colonies using a DNA extraction kit (Thermo Fisher Scientific, St. Leon‐Rot, Germany). Procedure was done rendering to the manufacturer's guidelines. Purity (A260/A280) and concentration of extracted DNA were then checked (NanoDrop, Thermo Scientific). The truth of the DNA was assessed on a 2% agarose gel stained with DNA Safe Stain (0.5 μg/mL) (Cinnagen).
Table 1 represents the sequence of primers used for specific amplification of the ITS region of A. baumannii in the LAMP technique. A programmable DNA thermocycler (Eppendorf Mastercycler 5330, Eppendorf‐Nethel‐Hinz GmbH, Hamburg, Germany) was used in all LAMP assays. To optimize the reaction conditions of the primers for A. baumannii in the LAMP reaction, we observed various temperatures from 54°C to 72°C at 2°C intervals. To detect the rapidity of the LAMP assay, the LAMP products were analysed at intervals of 5 minutes using agarose gel electrophoresis (2% agarose gel electrophoresis) (Soo et al., 2013).
Table 1.
Sequence of primers used for specific amplification of the ITS region of A. baumannii using LAMP technique (Soo et al., 2013)
| Primer name | Type | Sequence (5′–3′) |
|---|---|---|
|
Ab‐ITS‐F3 Ab‐ITS‐B3 |
Forward outer Backward outer |
CGGTAATTAGTGTGATCTGAC CATTTCAGTTTAGAGCACTGT |
|
Ab‐ITS‐FIP Ab‐ITS‐BIP |
Forward inner Backward inner |
TTGCTTAACCTAAACTCTTGAGTGAGAAGACACATTAACTCATTAACAGA AGCAAATTAACTGAATCAAGCGTTTACTTAAGCACCGTACAGC |
|
Ab‐ITS‐LF Ab‐ITS‐LB |
Loop forward Loop backward |
AATTTATTTCAGACTCAATTTTGCCAA TGGTATGTGAATTTAGATTGAA |
2.5. Antibiotic resistance pattern
Antimicrobial resistance patterns of the A. baumannii isolates were studied using the simple disk diffusion technique. The Kirby‐Bauer technique was used for this purpose. The Mueller–Hinton agar (Merck, Darmstadt, Germany) medium was used for this purpose. The susceptibility of A. baumannii isolates were tested against several types of antibiotics with appropriate disks containing azithromycin (15 µg), erythromycin (15 µg), rifampin (5 µg), nitrofurantoin (300 µg), chloramphenicol (30 µg), imipenem (30 µg), ciprofloxacin (5 µg), tetracycline (30 µg), cephalotin (30 µg), trimethoprim/sulphamethoxazole(25 µg), gentamicin (10 µg) and streptomycin (10 µg) (produced by PadTan‐Teb, Tehran, Iran), according to the instruction of Clinical and Laboratory Standards Institute (CLSI, 2017). A. baumannii ATCC 19606 was used as a quality control organism in determination of antimicrobial susceptibility.
2.6. Statistical analysis
Statistical analysis was done using the SPSS 21.0 statistical software (SPSS Inc., Chicago, IL, USA). Chi‐square test and Fisher's exact two‐tailed test were used to assess any significant relationship between the prevalence of A. baumannii isolates and their antibiotic resistance pattern. A value of p < .05 was considered as statistically significant level.
3. RESULTS
The present investigation was done to assess the prevalence and antibiotic resistance pattern of A. baumannii isolated from different types of meat samples. Table 2 represents the prevalence of A. baumannii isolates in different types of raw meat samples. Out of 194 raw meat samples, 39 (20.10%) were positive for A. baumannii isolates. Ovine raw meat samples had the highest prevalence of A. baumannii isolates (32.14%), while turkey raw meat samples had the lowest (11.11%). All of the culture‐positive A. baumannii isolates were also approved using the LAMP assay (Figure 1). Statistically significant differences were seen between types of samples and prevalence of A. baumannii (p < .05).
Table 2.
Prevalence of A. baumannii isolates in different types of raw meat samples, the number of the positive isolates were approved by LAMP assay
| Types of raw meat | N. samples collected | N. samples positive in culture (%) |
|---|---|---|
| Bovine | 39 | 7 (17.94) |
| Ovine | 28 | 9 (32.14) |
| Caprine | 41 | 8 (19.51) |
| Camel | 16 | 2 (12.50) |
| Chicken | 52 | 11 (21.15) |
| Turkey | 18 | 2 (11.11) |
| Total | 194 | 39 (20.10) |
Figure 1.
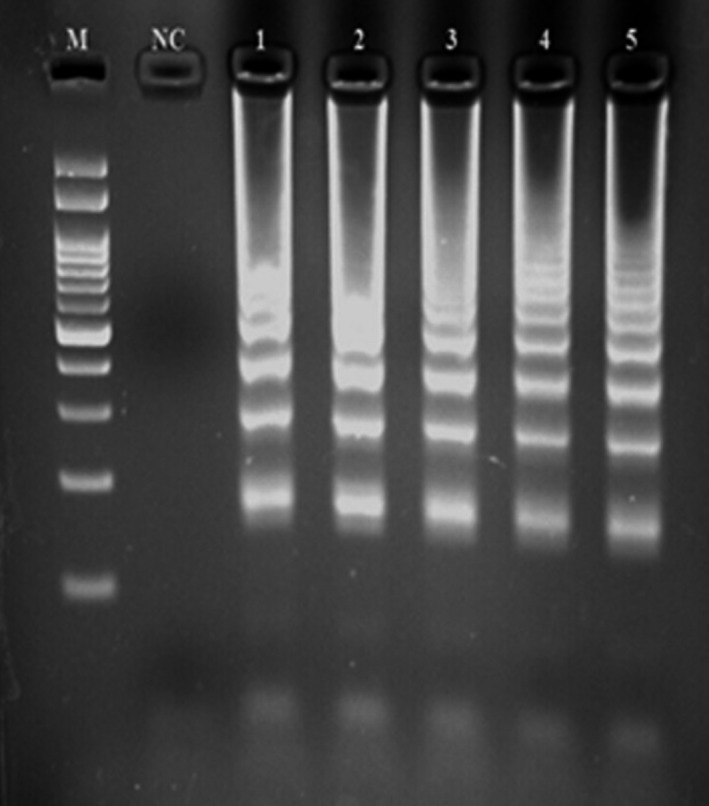
Loop‐mediated isothermal amplification‐polymerase chain reaction (LAMP‐PCR) gel electrophoresis for approval of Acinetobacter. baumannii isolates in meat samples. M: 100 bp ladder; NC: negative control; 1: positive control and 2–5: positive samples in the LAMP‐PCR reaction
Table 3 represents the antibiotic resistance pattern of A. baumannii isolated from different types of raw meat samples. A. baumannii isolates of the present research harboured the highest prevalence of resistance against gentamicin (87.17%), tetracycline (79.48%), erythromycin (74.35%), azithromycin (66.66%), ciprofloxacin (58.97%), trimethoprim/sulphamethoxazole (56.41%) and rifampin (51.28%) antibiotic agents. The prevalence of resistance against imipenem (17.94%) and chloramphenicol (28.20%) was lower than other tested antibiotic agents. Statistically significant differences were seen between types of samples and prevalence of antibiotic resistance in the A. baumannii isolates (p < .05).
Table 3.
Antibiotic resistance pattern of A. baumannii isolated from different types of raw meat samples
| Raw meat samples (N positive) | Antibiotic resistance pattern (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S10 | GM10 | SXT25 | CL30 | TE30 | CIP5 | IPM10 | C30 | F300 | AZM15 | RA5 | E15 | |
| Bovine (7) | 2 (28.57) | 6 (85.71) | 4 (57.14) | 2 (28.57) | 5 (71.42) | 4 (57.14) | 1 (14.28) | 1 (14.28) | 3 (42.85) | 4 (57.14) | 3 (42.85) | 5 (71.42) |
| Ovine (9) | 4 (44.44) | 8 (88.88) | 5 (55.55) | 5 (55.55) | 7 (77.77) | 5 (55.55) | 2 (22.22) | 3 (33.33) | 4 (44.44) | 6 (66.66) | 4 (44.44) | 7 (77.77) |
| Caprine (8) | 3 (37.50) | 7 (87.50) | 4 (50) | 3 (37.50) | 7 (87.50) | 5 (62.50) | 1 (12.50) | 2 (25) | 3 (37.50) | 5 (62.50) | 4 (50) | 6 (75) |
| Camel (2) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | — | — | 1 (50) | 1 (50) | 1 (50) | 1 (50) |
| Chicken (11) | 5 (45.45) | 11 (100) | 7 (63.63) | 6 (54.54) | 10 (90.90) | 7 (63.63) | 3 (27.27) | 4 (36.36) | 7 (63.63) | 9 (81.81) | 7 (63.63) | 9 (81.81) |
| Turkey (2) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) | — | 1 (50) | 1 (50) | 1 (50) | 1 (50) | 1 (50) |
| Total (39) | 16 (41.02) | 34 (87.17) | 22 (56.41) | 18 (46.15) | 31 (79.48) | 23 (58.97) | 7 (17.94) | 11 (28.20) | 19 (48.71) | 26 (66.66) | 20 (51.28) | 29 (74.35) |
Abbreviations: AZM15, Azithromycin; C30, Chloramphenicol; CIP5, Ciprofloxacin; CL30, Cephalothin; E15, Erythromycin; F300, Nitrofurantoin; GM10, Gentamicin; IPM10, Imipenem; RA5, Rifampin; S10, Streptomycin; SXT25, Trimethoprim/sulphamethoxazole; TE30, Tetracycline.
Figure 2 represents the distribution of multidrug resistant A. baumannii isolated from retail meat samples. We found that 89.74% of A. baumannii isolates had at least resistance against one antibiotic agents. The prevalence of resistance against two antibiotic agents was 79.48%, while the prevalence of resistance against more than eight antibiotic agents was 5.12%.
Figure 2.
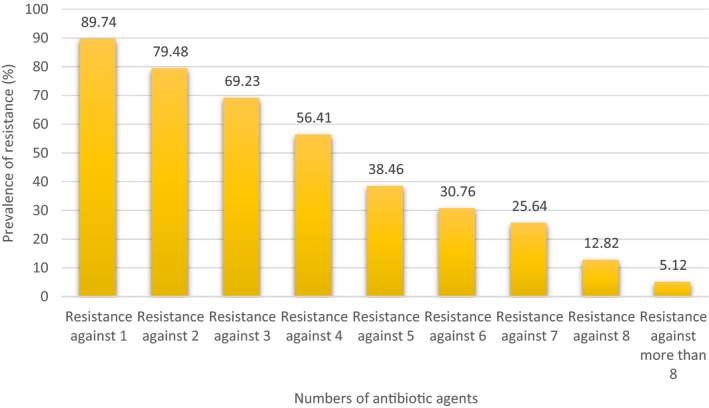
Distribution of multidrug resistant A. baumannii strains isolated from retail meat samples
4. DISCUSSION
The present research was done to study the prevalence rate and antibiotic resistance pattern of A. baumannii isolated from different types of raw meat samples. We found that 20.10% of all meat samples were positive for A. baumannii isolates. Isolates were also approved using the LAMP assay. Because LAMP recognizes the target by eight distinct sequences initially, it is expected to amplify the target sequence with high selectivity. LAMP is an isothermal approach to nucleic acid amplification (using a single temperature) and does not require the thermal cycling of PCR. This temperature requirement enables use of more portable or less expensive instruments, or even simple incubators or water baths. As it is not limited by a doubling‐by‐cycle amplification, LAMP generally produces more DNA than PCR in a more rapid incubation time. It is not uncommon to see ug yields in <15 minutes. Additionally, enzymes and conditions for LAMP provide a more robust and inhibitor‐tolerant amplification system, with detection of DNA or RNA species directly from a variety of crude sample preparations. Thus, the LAMP assay has been recommended for detection and approval of bacteria (Soo et al., 2013).
Rare studies have been conducted in the prevalence of A. baumannii isolates in raw meat samples. Carvalheira, Casquete, Silva, and Teixeira (2017) reported that the prevalence of A. baumannii isolates in chicken, turkey, beef and pork raw meat samples collected from Portugal were 42.85%, 42.85%, 0% and 14.28% respectively. Recent researches reported the prevalence rates of 75% in pork and beef samples collected from Hong Kong (Houang et al., 2001) and 28% in bovine meat samples collected from Lebanon (Rafei et al., 2015). These differences may be explained by the different methodologies used for the isolation and identification of A. baumannii isolates. A. baumannii was recently isolated by Lupo, Vogt, Seiffert, Endimiani, and Perreten (2014) from 62 out of 248 (25%) meat samples (chicken, turkey, veal, beef and pork). Nevertheless, A. baumannii was not detected in 27 meat samples (cow, chicken and pork) from retail supermarket chains in Edinburg (Hamouda, Findlay, Al Hassan, & Amyes, 2011; Zordan et al., 2011). It is possible that these results may reflect different levels of compliance with good manufacturing practices in different meat production chains.
Antibiotic resistance is one of the most important characters of the A. baumannii isolates. We found that A. baumannii isolates of our investigation exhibited considerable prevalence of resistance against gentamicin, tetracycline, erythromycin, azithromycin, ciprofloxacin, trimethoprim/sulphamethoxazole and rifampin antibiotic agents. Kiani, Momtaz, Serajian, and Tajbakhsh (2016) revealed the similar antibiotic resistance pattern of the A. baumannii isolated from different types of hospital infections. They showed that the prevalence of resistance against trimethoprim, tetracycline, amikacin, tobramycin, meropenem and imipenem were 50.74%, 89.55%, 5.97%, 7.46%, 2.98% and 4.47% respectively. Ahmad, Akhtar, Sheikh, Tipu, and Nasar (2018) reported that the prevalence of resistance of A. baumannii isolated from meat samples against ampicillin, ceftriaxone, imipenem, gentamicin, kanamycin, tetracycline, chloramphenicol, trimethoprim, sulfamethoxazole and norfloxacin were 100%, 20.80%, 33.30%, 16.60%, 54.10%, 79.10%, 66.60%, 100%, 8.30% and 16.60% respectively. Similar antibiotic resistance patterns of A. baumannii isolates was also reported from Iran (Moradi, Hashemi, & Bahador, 2015), Romania (Constantiniu, Romaniuc, Iancu, Filimon, & Taraşi, 2004), Turkey (Kulah et al., 2009), France (Kempf & Rolain, 2012) and Italy (Zarrilli, Pournaras, Giannouli, & Tsakris, 2013). Similar findings of antibiotic resistance were also reported in A. baumannii isolated from Iranian human clinical infections (Mirnejad, Mostofi, & Masjedian, 2013; Momtaz, Khamesipour, Tavakol, & Awosile, 2017; Tavakol et al., 2018).
The prevalence of antibiotic resistant bacteria in meat samples has been attributed, at least partially, to the extensive use of antimicrobials for treatment, prevention and control of diseases and finally for growth stimulate in food‐producing animals, since this enhances the antimicrobial selective pressure for strains present. Otherwise, using antibiotics for growth stimulation is allowed in Iran. The low prevalence rate of resistance against carbapenems, chloramphenicol and nitrofurantoin is due to the fact that these antibiotics are not allowed to treat food‐producing animals. However, their illegal prescription caused occurrence of significant antibiotic resistance against these antimicrobial agents especially in the poultry farms. High prescription of antibiotic agents in both medicine and veterinary caused significant increase in the levels of antibiotic resistance in some kinds of food‐borne bacteria (Nejat et al., 2015; Ranjbar, Yadollahi Farsani, & Safarpoor Dehkordi, 2018; Safarpoor Dehkordi, Akhondzadeh Basti, Gandomi, Misaghi, & Rahimi, 2018).
5. CONCLUSIONS
It was concluded that meat is also an important source of isolates resistant to some of the categories of antimicrobials used to treat infections caused by A. baumannii. Additional studies are required to assess the role of resistant isolates in the dissemination of resistance genes. The presence of clinically important species and multidrug‐resistant isolates in meat may be a threat to public health considering that meat may provide a vector for the spread of these opportunistic pathogen into both community and hospital settings environment. Poultry‐based A. baumannii isolates harboured the higher prevalence of resistance against all tested antibiotic agents. Therefore, higher levels of food‐related inspection and precision in antibiotic prescript should perform in poultry farms.
ETHICS STATEMENT
This study was done on meat samples collected from raw animal meats, so there have no ethical issue in this work.
PRACTICAL APPLICATION
Due to the high consumption rate of ruminants and poultry meat, they should have a high microbial quality. Foods with animal origins and especially meat are considered as a probable source of resistant Acinetobacter baumannii isolates. The incidence of contamination of meat by A. baumannii is fairly high, and nearly all of the isolates showed resistance against several types of antibiotics, so the risk of probable food‐borne diseases caused by A. baumannii in such products and also their high importance for transmission of resistance isolates to human population should not be neglected.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
The authors thank Dr. Abbas Farahani for his assistance in sample collection and LAMP assay analysis. This work was financially supported by the Islamic Azad University, Shahrekord Branch, Shahrekord, Iran.
Askari N, Momtaz H, Tajbakhsh E. Prevalence and phenotypic pattern of antibiotic resistance of Acinetobacter baumannii isolated from different types of raw meat samples in Isfahan, Iran. Vet Med Sci. 2020;6:147–153. 10.1002/vms3.199
Funding information
Hassan Momtaz received Research grants for Research at Islamic Azad University, Shahrekord Branch with Grant Number 97/1109.
REFERENCES
- Ahmad, A. , Akhtar, F. , Sheikh, A. A. , Tipu, M. Y. , & Nasar, M. (2018). Comparative antibiotic resistance profile of Acinetobacter species, isolated from fish, chicken and beef meat. Journal of Clinical Microbiology and Antimicrobials, 2(1), 1. [Google Scholar]
- Berlau, J. , Aucken, H. , Houang, E. , & Pitt, T. (1999). Isolation of Acinetobacter spp. including A. baumannii from vegetables: Implications for hospital‐acquired infections. Journal of Hospital Infection, 42(3), 201–204. [DOI] [PubMed] [Google Scholar]
- Carvalheira, A. , Casquete, R. , Silva, J. , & Teixeira, P. (2017). Prevalence and antimicrobial susceptibility of Acinetobacter spp. isolated from meat. International Journal of Food Microbiology, 243, 58–63. 10.1016/j.ijfoodmicro.2016.12.001 [DOI] [PubMed] [Google Scholar]
- CLSI (2017). Performance standards for antimicrobial susceptibility testing; twenty‐fifth informational supplement. CLSI document M100–S26. Wayne: Clinical and Laboratory Standards Institute. [Google Scholar]
- Constantiniu, S. , Romaniuc, A. , Iancu, L. S. , Filimon, R. , & Taraşi, I. (2004). Cultural and biochemical characteristics of Acinetobacter spp. strains isolated from hospital units. The Journal of Preventive Medicine, 12(3–4), 35–42. [Google Scholar]
- Espinal, P. , Seifert, H. , Dijkshoorn, L. , Vila, J. , & Roca, I. (2012). Rapid and accurate identification of genomic species from the Acinetobacter baumannii (Ab) group by MALDI‐TOF MS. Clinical Microbiology and Infection, 18(11), 1097–1103. 10.1111/j.1469-0691.2011.03696.x [DOI] [PubMed] [Google Scholar]
- Ghorbani, F. , Gheisari, E. , & Dehkordi, F. S. (2016). Genotyping of vacA alleles of Helicobacter pylori strains recovered from some Iranian food items. Tropical Journal of Pharmaceutical Research, 15(8), 1631–1636. [Google Scholar]
- Gurung, M. , Nam, H. , Tamang, M. , Chae, M. , Jang, G. , Jung, S. , & Lim, S. (2013). Prevalence and antimicrobial susceptibility of Acinetobacter from raw bulk tank milk in Korea. Journal of Dairy Science, 96(4), 1997–2002. 10.3168/jds.2012-5965 [DOI] [PubMed] [Google Scholar]
- Hamouda, A. , Findlay, J. , Al Hassan, L. , & Amyes, S. G. (2011). Epidemiology of Acinetobacter baumannii of animal origin. International Journal of Antimicrobial Agents, 38(4), 314–318. 10.1016/j.ijantimicag.2011.06.007 [DOI] [PubMed] [Google Scholar]
- Houang, E. T. , Chu, Y. , Leung, C. , Chu, K. , Berlau, J. , Ng, K. , & Cheng, A. (2001). Epidemiology and Infection Control Implications of Acinetobacter spp. Hong Kong. Journal of Clinical Microbiology, 39(1), 228–234. 10.1128/JCM.39.1.228-234.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf, M. , & Rolain, J.‐M. (2012). Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: Clinical impact and therapeutic options. International Journal of Antimicrobial Agents, 39(2), 105–114. 10.1016/j.ijantimicag.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Kiani, S. , Momtaz, H. , Serajian, A. A. , & Tajbakhsh, E. (2016). Detection of integrons in Acinetobacter baumannii strains isolated from the nosocomial infections of Ahvaz City and their relation with the resistance pattern. International Journal of Medical Laboratory, 3, 50–63. [Google Scholar]
- Kulah, C. , Aktas, E. , Comert, F. , Ozlu, N. , Akyar, I. , & Ankarali, H. (2009). Detecting imipenem resistance in Acinetobacter baumannii by automated systems (BD Phoenix, Microscan WalkAway, Vitek 2); high error rates with Microscan WalkAway. BMC Infectious Diseases, 9(1), 30 10.1186/1471-2334-9-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo, A. , Vogt, D. , Seiffert, S. N. , Endimiani, A. , & Perreten, V. (2014). Antibiotic resistance and phylogenetic characterization of Acinetobacter baumannii strains isolated from commercial raw meat in Switzerland. Journal of Food Protection, 77(11), 1976–1981. 10.4315/0362-028X.JFP-14-073 [DOI] [PubMed] [Google Scholar]
- Mirnejad, R. , Mostofi, S. , & Masjedian, F. (2013). Antibiotic resistance and carriage class 1 and 2 integrons in clinical isolates of Acinetobacter baumannii from Tehran, Iran. Asian Pacific Journal of Tropical Biomedicine, 3(2), 140 10.1016/S2221-1691(13)60038-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momtaz, H. , Davood Rahimian, M. , & Safarpoor Dehkordi, F. (2013). Identification and characterization of Yersinia enterocolitica isolated from raw chicken meat based on molecular and biological techniques. Journal of Applied Poultry Research, 22(1), 137–145. 10.3382/japr.2012-00549 [DOI] [Google Scholar]
- Momtaz, H. , Dehkordi, F. S. , Rahimi, E. , Asgarifar, A. , & Momeni, M. (2013). Virulence genes and antimicrobial resistance profiles of Staphylococcus aureus isolated from chicken meat in Isfahan province, Iran. Journal of Applied Poultry Research, 22(4), 913–921. 10.3382/japr.2012-00673 [DOI] [Google Scholar]
- Momtaz, H. , Khamesipour, F. , Tavakol, M. , & Awosile, B. (2017). Determination of antimicrobial resistance and resistant genes in Acinetobacter baumannii from human clinical samples. West Indian Medical Journal, 66(1), 56–64. [Google Scholar]
- Moradi, J. , Hashemi, F. B. , & Bahador, A. (2015). Antibiotic resistance of Acinetobacter baumannii in Iran: A systemic review of the published literature. Osong Public Health and Research Perspectives, 6(2), 79–86. 10.1016/j.phrp.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejat, S. , Momtaz, H. , Yadegari, M. , Nejat, S. , Safarpour Dehkordi, F. , & Khamesipour, F. (2015). Seasonal, geographical, age and breed distributions of equine viral arteritis in Iran. Kafkas University Veterinary Fakulty Dergisi, 21(1), 111–116. [Google Scholar]
- Rafei, R. , Hamze, M. , Pailhoriès, H. , Eveillard, M. , Marsollier, L. , Joly‐Guillou, M.‐L. , … Kempf, M. (2015). Extra‐human epidemiology of Acinetobacter baumannii in Lebanon. Applied and Environmental Microbiology, 81(7), 2359–2367. 10.1128/AEM.03824-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar, R. , Yadollahi Farsani, F. , & Safarpoor Dehkordi, F. (2018). Antimicrobial resistance and genotyping of vacA, cagA, and iceA alleles of the Helicobacter pylori strains isolated from traditional dairy products. Journal of Food Safety, 39(2), e12594. [Google Scholar]
- Safarpoor Dehkordi, F. , Akhondzadeh Basti, A. , Gandomi, H. , Misaghi, A. , & Rahimi, E. (2018). Pathogenic Staphylococcus aureus in hospital food samples; prevalence and antimicrobial resistance properties. Journal of Food Safety, 38(6), e12501. [Google Scholar]
- Safarpoor Dehkordi, F. , Barati, S. , Momtaz, H. , Hosseini Ahari, S. N. , & Nejat Dehkordi, S. (2013). Comparison of shedding, and antibiotic resistance properties of Listeria monocytogenes isolated from milk, feces, urine, and vaginal secretion of bovine, ovine, caprine, buffalo, and camel species in Iran. Jundishapur Journal of Microbiology, 6(3), 284–294. [Google Scholar]
- Safarpoor Dehkordi, F. , Gandomi, H. , Akhondzadeh Basti, A. , Misaghi, A. , & Rahimi, E. (2017). Phenotypic and genotypic characterization of antibiotic resistance of methicillin‐resistant Staphylococcus aureus isolated from hospital food. Antimicrobial Resistance & Infection Control, 6(1), 104 10.1186/s13756-017-0257-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safarpoor Dehkordi, F. , Haghighi, N. , Momtaz, H. , Rafsanjani, M. S. , & Momeni, M. (2013). Conventional vs real‐time PCR for detection of bovine herpes virus type 1 in aborted bovine, buffalo and camel foetuses. Bulgarian Journal of Veterinary Medicine, 16(2), 102–111. [Google Scholar]
- Safarpoor Dehkordi, F. , Khamesipour, F. , & Momeni, M. (2014). Brucella abortus and Brucella melitensis in Iranian bovine and buffalo semen samples: The first clinical trial on seasonal, Senile and geographical distribution using culture, Conventional and real‐time polymerase chain reaction assays. Kafkas Üniversitesi Veteriner Fakültesi Dergisi, 20(6), 821–828. [Google Scholar]
- Safarpoor Dehkordi, F. , Parsaei, P. , Saberian, S. , Moshkelani, S. , Hajshafiei, P. , Hoseini, S. , … Ghorbani, M. (2012). Prevalence study of Theileria annulata by comparison of four diagnostic techniques in southwest Iran. Bulgarian Journal of Veterinary Medicine, 15(2), 123–130. [Google Scholar]
- Shahran, P. , Dhingolia, S. , Khatri, P. , Singh Duban, J. , & Kumar Gahlawat, S. (2014). Loop‐mediated isothermal amplification (LAMP) based detection of bacteria: A review. African Journal of Biotechnology, 13(19), 1920–1928. 10.5897/AJB2013.13459 [DOI] [Google Scholar]
- Soo, P.‐C. , Tseng, C.‐C. , Ling, S.‐R. , Liou, M.‐L. , Liu, C.‐C. , Chao, H.‐J. , … Chang, K.‐C. (2013). Rapid and sensitive detection of Acinetobacter baumannii using loop‐mediated isothermal amplification. Journal of Microbiological Methods, 92(2), 197–200. 10.1016/j.mimet.2012.11.020 [DOI] [PubMed] [Google Scholar]
- Tavakol, M. , Momtaz, H. , Mohajeri, P. , Shokoohizadeh, L. , & Tajbakhsh, E. (2018). Genotyping and distribution of putative virulence factors and antibiotic resistance genes of Acinetobacter baumannii strains isolated from raw meat. Antimicrobial Resistance & Infection Control, 7(1), 120 10.1186/s13756-018-0405-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, R. , Zhang, X. D. , Zhao, Q. , Peng, B. , & Zheng, J. (2018). Analysis of global prevalence of antibiotic resistance in Acinetobacter baumannii infections disclosed a faster increase in OECD countries. Emerging Microbes & Infections, 7(1), 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrilli, R. , Pournaras, S. , Giannouli, M. , & Tsakris, A. (2013). Global evolution of multidrug‐resistant Acinetobacter baumannii clonal lineages. International Journal of Antimicrobial Agents, 41(1), 11–19. 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]
- Zordan, S. , Prenger‐Berninghoff, E. , Weiss, R. , van der Reijden, T. , van den Broek, P. , Baljer, G. , & Dijkshoorn, L. (2011). Multidrug‐resistant Acinetobacter baumannii in veterinary clinics, Germany. Emerging Infectious Diseases, 17(9), 1751–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]


