Abstract
The qRT-PCR method has been widely used to detect gene expression level in plants, helping to understand the molecular mechanisms. However, there are few researches which focus on the selection of the internal reference genes in Forsythia. To select the appropriate reference genes of Forsythia aimed at qRT-PCR normalization, twelve candidate reference genes were selected from our transcriptome data. Their expression was assessed by RT-PCR analysis in 47 Forsythia samples, including 12 species cultivars, different organs and tissues. GeNorm, NormFinder, and BestKeeper software were used to select the appropriate reference genes, AG and PSY were used to verify the accuracy of the outcome. The results showed that UKN1 was a stable reference gene in leaves of twelve Forsythia germplasms and in different developmental stages of fruits. MTP, ABCT + MTP, and ABCT + MTP + TIP were stable reference genes in different organs. ACT and SDH were stable reference genes in different flower tissues and different developmental stages of the flower buds. When Forsythia plants were stressed with PEG or ABA, SDH + UKN1 + G6PD was the stable reference gene group for qRT-PCR. The results provided the basis for investigating the physiological and biochemical processes of Forsythia related to medicinal and ornamental properties, and drought-resistance in the level of gene expression.
Electronic supplementary material
The online version of this article (10.1007/s12298-019-00731-y) contains supplementary material, which is available to authorized users.
Keywords: qRT-PCR, Forsythia, Reference genes, Flower, Fruit, Drought stress
Introduction
Forsythia belongs to Oleaceae, and is an important ornamental and medicinal plant (Ge et al. 2016; Rosati et al. 2007). The flowers of Forsythia are heterostylous, with the anther and stigma located at different heights (Long and Short) reciprocally (Sampson 1971). As an ornamental woody plant, Forsythia is widely found in the warm temperate zones of Asia and Europe, and plays an important role in the spring plant landscape (Rosati et al. 2007). The leaves and fruits of Forsythia are used most extensively for medical treatment and in functional diets (Guo et al. 2007; Piao et al. 2008). Forsythia is also listed as an herbal drug and food by the National Health Commission of the People’s Republic of China (Ge et al. 2016). The main components of Forsythia’s leaves and fruit, which including lignans, forsythoside A–E, and flavonoids, have anti-inflammatory, anti-infective, anti-oxidant, and anti-viral pharmacological influential effects on the human body (Piao et al. 2008). These components are valuable and have broad prospects in clinical applications and drug development. In recent years, research on the functional aspects of Forsythia mainly focus on the medicinal components of different species and varieties, the anabolism of medicinal components, and the critical period for the harvesting of medicinal parts (Satake et al. 2013, 2015; Piao et al. 2008). As for the ornamental characteristics, previous studies mainly focused on the molecular mechanisms of flower color (Rosati et al. 2003), leaf color (Wang et al. 2017), cold resistance (Flinn and Ashworth 1999; Mcnamara and Pellett 1993), and drought resistance (Zhao et al. 2007). With advancements in technology, molecular techniques determining the level of gene expression have been introduced into the research of Forsythia (Satake et al. 2013, 2015; Shiraishi et al. 2016). Gene expression analysis is an important tool to advance these studies.
At present, the methods used in detecting plant gene expression include northern blot, in situ hybridization, RT-PCR, and real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR). qRT-PCR is the most common method in current study, owing to its high sensitivity, specificity, accuracy, and high throughput (Gachon et al. 2004). Plant transcriptome sequence data and orthologous sequences of common housekeeping genes reported in model plant species were often used to screen for stable internal reference genes in non-model plants without the reference genome data (Li et al. 2017). However, the accuracy of the method is influenced by many factors, including plant materials, tissues and treatments, reverse transcription processes, and the amplification efficiency of primers. The optimal internal reference genes were varied among different species, different organs, and different developmental stages of specific species (Wang 2016; Hong 2016). Meanwhile, the use of improper reference genes for normalization in the relative quantification of gene expression could result in an incorrect conclusion (Gutierrez et al. 2010). Thus, for qRT-PCR analysis, it is important to screen for the optimal internal reference genes for different species and specific studies.
Appropriate reference gene selection has typically been reported for many plants used for herbal drugs and foods, such as Lilium (Xu et al. 2017), Citrus (Alves et al. 2018), Amygdalus (Kou et al. 2017). However, in Forsythia, genes selected from the Forsythia transcription data and housekeeping genes commonly used in model plant species were directly used as internal reference genes (Shiraishi et al. 2016). The application of these genes was limited in scientific research because they had not been standardized and tested systematically. There is still no report on the selection of an appropriate internal reference gene for Forsythia. In order to study the expression pattern and function of candidate genes in Forsythia, a selection of suitable internal reference genes is necessary. In our research, twelve candidate reference genes, often used as housekeeping genes or other stable reference genes in other plant species, were selected from our transcriptome data and evaluated by geNorm (Vandesompele et al. 2002), NormFinder (Andersen et al. 2004), and BestKeeper (Pfaffl et al. 2004) algorithms in order to select appropriate reference genes for qRT-PCR normalization in Forsythia. The target genes agamous (AG) and phytoene synthase (PSY) were used as gene of estimate to test the stability of the selected reference genes, which were involved in flower development (Hou et al. 2010) and carotenoid synthesis (Domonkos et al. 2013) respectively. The selected internal reference genes include UNK1, which has the best function for some stages in other plants (Chi et al. 2012). Therefore, it is possible that UKN1 performed as one of the stable reference gene in Forsythia. These selected internal reference genes could facilitate the study on gene expression during physiological and biochemical processes and play a significant role in the study of ornamental and medicinal molecular biology in Forsythia.
Materials and methods
Plant material
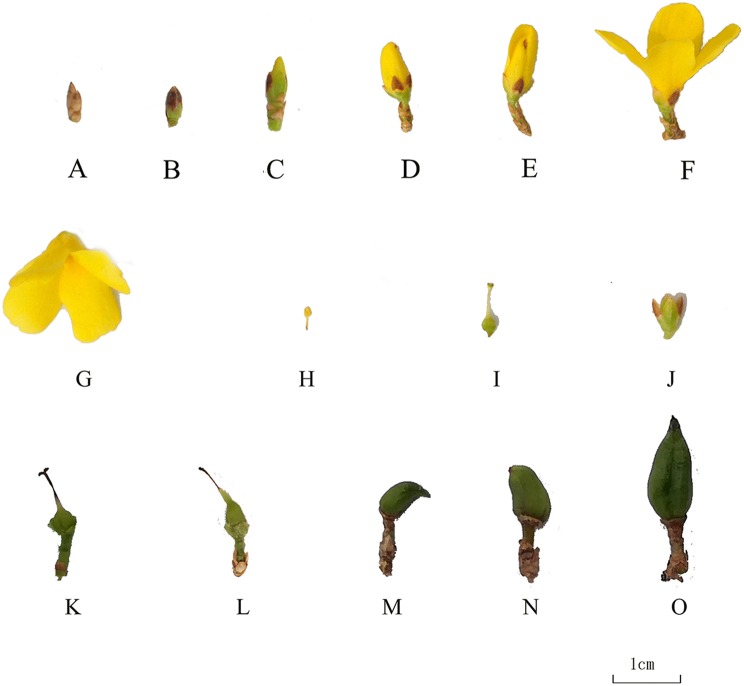
Forsythia seedlings were planted in July 2010 in the nursery of the National Engineering Research Center for Floriculture (NERCF) (40°02′ N, 115°50′ E), Beijing, China. The third fully expanded leaf of twelve Forsythia species and cultivars (Table 1) were collected on 20th July, 2018. Different plant organs of F. intermedia ‘Courtaneur’ were collected, including the root, stem, leaf, flower and fruit. During blossoming in the early spring of 2018, the flower buds of F. intermedia ‘Courtaneur’ were collected at different developmental stages and its fruits at different developmental stages were collected in autumn (Fig. 1). The development of the flower buds of F. intermedia ‘Courtaneur’ was divided into six stages, according to their shapes: grey bud (the buds are wrapped with grey flower scales) (Fig. 1a), green bud (the grey flower scales falls off, and the buds turn green) (Fig. 1b), start of yellow petals stage (half of the yellow petals are exposed) (Fig. 1c), fully yellow petals stage (the petals are fully exposed, but the flowers have not open) (Fig. 1d), start of blooming (the flowers start bloom) (Fig. 1e), and fully blooming (the flowers fully bloom) (Fig. 1f). Different parts of the mature flowers were then collected, (Fig. 1), including corolla (Fig. 1g), stamens (Fig. 1h), ovary and pistil (Fig. 1i), and sepals (Fig. 1j). The development of F. intermedia ‘Courtaneur’s fruits was divided into five stages: < 0.3 mm (Fig. 1k), 0.3–0.5 mm (Fig. 1l), 0.5–0.7 mm (Fig. 1m), 0.7–0.9 mm (Fig. 1n), and > 0.9 mm (Fig. 1o).
Table 1.
Information of 13 Forsythia species and cultivars used in our research
| Species | Cultivar | Style type | Code |
|---|---|---|---|
| Cultivars of F. intermedia | F. intermedia ‘Spectabilis’ | Sz | F1 |
| F. intermedia ‘Goldrausch’ | S | F2 | |
| F. intermedia ‘Lynwood’ | S | F3 | |
| F. intermedia ‘Courtalyn’ | S | F4 | |
| F. intermedia ‘Courtasol’ | S | F6 | |
| F. intermedia ‘Courtaneur’ | Ly | F7 | |
| Cultivars of F. koreana | F. koreana | S | Fk |
| F. koreana ‘Sawon Gold’ | S | FG | |
| F. viridissima var. koreana ‘Kumson’ | S | F8 | |
| Species | F. mandshurica | S | Fm |
| F. suspensa | L | Fp | |
| F. suspensa | S | Ft | |
| F.ovata | – | Fo |
zS means stigma shorter than anther
yL means stigma longer than anther
Fig. 1.
The different developmental stages of flower buds and fruits, and different tissues of mature flowers of F. ‘Courtaneur’
In June 2018, 1-year cutting plants of F. intermedia ‘Courtaneur’ were treated with abscisic acid (ABA) (100 mg/L) and polyethylene glycol (PEG 6000) (50 g/L) for 48 h, respectively, simulating drought stress (Gong et al. 2018). The concentrations of ABA and PEG 6000 were selected through concentration gradient treatment experiment, the plants were treated with aqueous solution containing different concentrations of ABA and PEG, then the semi-lethal concentration were sorted for further experiment. For each treatment fifty clones and their roots were soaked in the treatment solutions. After treatment, the plants were placed in the phytotron [temperature 25 °C, light intensity 300 µmol/m−2 s−1, 16 h/8 h (light/dark)] and the third pair of leaves were collected at 0 h, 2 h, 4 h, 6 h, 12 h, 24 h, 36 h and 48 h after treatment, respectively.
Three independent samples were taken at each stage or treatment.
Selection of candidate reference genes
Twelve candidate reference genes which have a stable expression (− 0.5 ≤ log2 ratio ≤ 0.5) in the mature leaves of Forsythia and are often used as housekeeping genes in model plant species were selected from our transcriptome data (SRA accession number PRJNA437859, Supplemental data Table S1). According to their CDS sequences, the primers were designed on the Web using the Primer Quest Tool (http://sg.idtdna.com/Primerquest/Home/Index) and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China), as shown in Table 2.
Table 2.
Reference genes and primers used for qRT-PCR in our research
| Gene | Forward primer sequence | Reverse primer sequence | Amplicon length (bp) | Tm (°C) | Amplification efficiency (%) | Correlation coefficient (R2) | Ct value |
|---|---|---|---|---|---|---|---|
| UKN1 | CAGACCAGCTTTGAGGAGTATC | GGCCAGAAACCAGTAGTCAATA | 92 | 60 | 98.0 | 0.976 | 21.00 ± 2.44 |
| CYP | TGCCAGAAGGACCCTTAAATC | TGGAGGAAGGGCTTGTAAATC | 83 | 60 | 115.5 | 0.989 | 23.46 ± 2.82 |
| TUB | ACATTCTCTGTGTTCCCATCTC | CCATACACTCATCGGCATTCT | 106 | 60 | 112.5 | 0.983 | 21.54 ± 2.32 |
| MTP | GGTAGGTCTGCTCTCTTATCC | GGAAACCCATTCCCGTACTT | 110 | 60 | 102.3 | 0.992 | 21.81 ± 2.24 |
| ACT | CGCCATCGAACTCTCCATAAA | TTGCAGAAGATGTCGGAAACTA | 90 | 60 | 81.0 | 0.960 | 19.64 ± 1.36 |
| GAPDH | TGAAGGGTGGTGCTAAGAAAG | GCATTGGAGACGATGTGTAGAT | 115 | 60 | 98.0 | 0.982 | 20.98 ± 2.88 |
| ABCT | GCTCCAGGGACTACAGAGATTA | AGGTTGAATGGGCACTATGG | 95 | 60 | 121.3 | 0.995 | 20.51 ± 3.73 |
| SDH | GAAATCGTCCCAACCTCTTACC | AGCTGTTCAGGACAAGGTTATG | 100 | 60 | 99.9 | 0.964 | 19.75 ± 2.21 |
| TIP | TGCATACGAGTGTCCCTTAATC | CCCAGTGGTTGGTTTCTTCT | 80 | 60 | 106.5 | 0.996 | 20.56 ± 2.10 |
| PTB | GAACTCCCAGCCTTCCTTATT | GTATACTGGGCTCCGTTGTATG | 84 | 60 | 104.0 | 0.996 | 21.73 ± 1.38 |
| G6PD | TGCTCGGGCTTGATAGAAAC | GAAGGACGCGGTGGATATTT | 119 | 60 | 91.2 | 0.989 | 21.28 ± 2.59 |
| F-Box | GCCAGCCCAGTAGACATAAC | TTCCAAAGTAAAGCCGACAAATC | 100 | 60 | 95.1 | 0.982 | 22.39 ± 2.61 |
Ct (Cycle threshold) means the number of cycles experienced when the fluorescent signal in each reaction tube reaches the set domain value
Ct values are mean ± SE (n = 3)
qRT-PCR analysis
The total cellular RNAs were extracted using the Trizol RNA reagent (Life Technologies, CA, USA) according to the manufacturer’s instructions and were stored at − 80 °C. RNA was assessed using 1% agarose gel electrophoresis and was quantified with a UV–visible spectrophotometer (UV-2550, Japan). The RNA samples with A260/A230 ratios between 1.7 and 2.0 were used for subsequent analysis and were used to synthesize the first strand of cDNA with the Perfect Real Time RT reagent kit (Takara, Japan) in a 20 µl reaction mixture according to the manufacturer’s protocol, once they were adjusted to the same concentration of RNA for each sample to 1 µg. The cDNA samples were diluted 1:5 with nuclease-free water prior to the qRT-PCR analysis.
qRT-PCR was performed using the PrimeScript™ RT reagent kit with the gDNA Eraser (perfect real time) (Code No. RR047, Takara, Japan), and was restricted to the guidelines as follows: the Mini Opticon Real-time PCR(Bio-Rad Laboratories Inc., USA) reaction mixture (25 µl) contained 12.5 µl of SYBR Premix Ex Taq (TaKaRa Bio Inc., Japan), 2 µl of 1:5 diluted cDNA, 0.5 µl of each primer (10 mM), and 9.5 µl of water. The reactions were incubated under the following cycling conditions: 30 s at 94 °C, 40 cycles of 94 °C for 5 s, Tm 60 °C for 30 s, and 72 °C for 30 s, with a single melt cycle from 65 to 94 °C in 5 s intervals.
The specificity of the primer pair was verified by the presence of a single peak in the melt curve analysis during the qRT-PCR process. Three independent biological replicates and three technical repetitions were performed for each of the quantitative PCR experiments. The threshold cycle (Ct) was measured automatically and the correlation coefficients (R2) together with the slope were calculated from the standard curve based on a fivefold series dilution of the cDNA templates. The corresponding qRT-PCR efficiencies (E) for each gene were determined from the given slope (Li et al. 2017).
Data analysis
GeNorm, NormFinder and BestKeeper algorithms were used to evaluate the stability of 12 candidate reference genes according to the Ct values, analyzing the stability of candidate reference genes comprehensively (Li et al. 2017).
Validation of reference genes
Agamous (AG) and phytoene synthase (PSY) were selected as control genes to confirm the reliability of the candidate reference genes, which are involved in flower development (Hou et al. 2010) and carotenoid synthesis (Domonkos et al. 2013). The stability in the expression of target genes AG and PSY was normalized using 5–8 genes that had been screened out. The qRT-PCR amplification conditions were the same as the conditions described above. AG was used to test the reliability of different developmental stages of flower buds and different parts of mature flowers, while PSY were used for other groups. The primer sequences of AG were as follows: Forward primer, 5′-TGAGCTCGCAGCATCTTTAC-3′, Reverse primer, 5′-GCTTGACCAGGATGTGGATATT-3′. The primer sequences of PSY were as follows: Forward primer, 5′-TCTCTCAGCAGACTCGTTATC-3′, Reverse primer, 5′-CAGGGATGTTGGAGAAGAGTA-3′.
Results
Candidate reference genes and PCR amplification
Twelve candidate reference genes traditionally often used as housekeeping genes or stable reference genes in other plant species were identified on the basis of NR annotation, including genes of unknown function (UKN1), Cyclophilin (CYP), Beta Tubulin (TUB), Metalloprotease (MTP), Actin (ACT), Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ATP-binding cassette transporter (ABCT), Succinate dehydrogenase (SDH), Tip41-like protein (TIP), Polypyrimidine tract-binding (PTB), 6-phosphogluconic dehydropenase (G6PD), and F-Box.
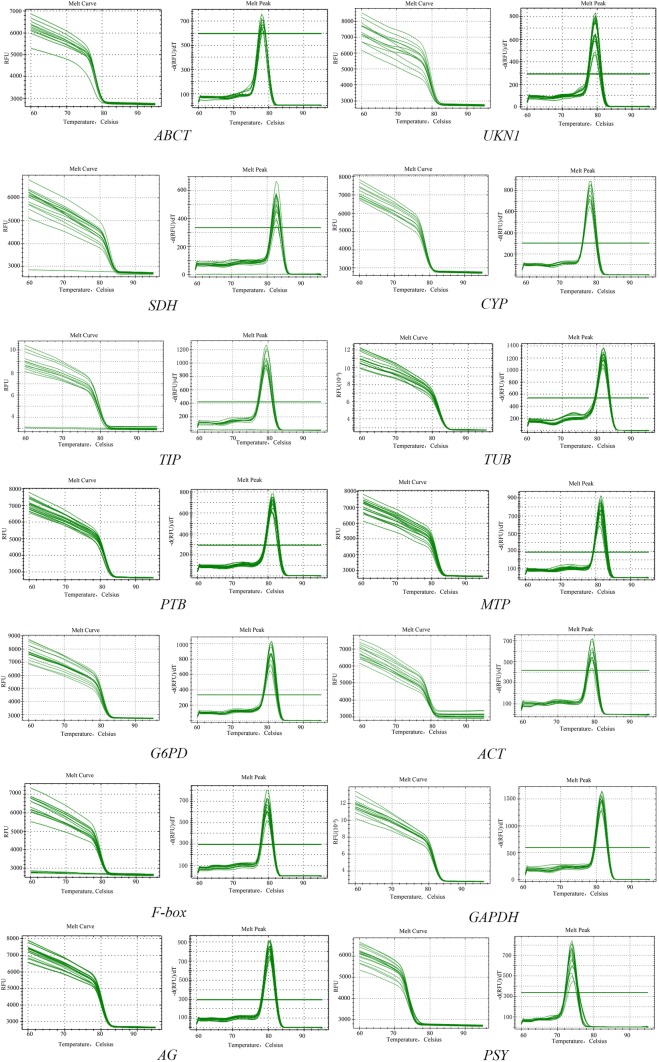
The amplification efficiencies of 12 candidate reference genes and 2 target genes ranged from 81.0 to 121.3%, and correlation coefficients ranged from 0.96 to 1 (Table 2). The single-peak melting curve showed that 14 primer pairs generated single fragments, indicating non-specific products amplified in RT-PCR reactions (Fig. 2).
Fig. 2.
Melting curves of the candidate reference genes and validate genes
Expression stability of candidate reference genes
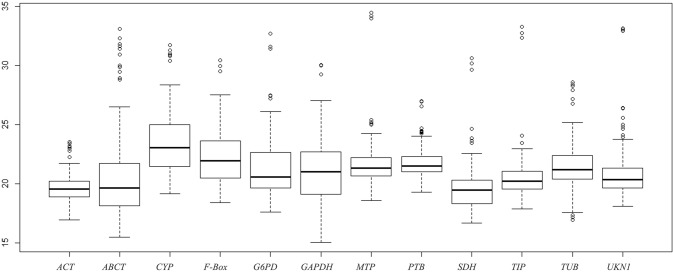
qRT-PCR analysis was used to detect the expression profiles of the 12 reference genes in different experimental materials, including the leaves of 12 Forsythia species and cultivars, different organs, different developmental stages of flower buds, tissues of flower parts, developmental stages of fruits, and the leaves of Forsythia plants treated with PEG and ABA. The results were shown in Fig. 3 and Table 2, which the mean CT values of the 12 reference genes ranged from 19.64 ± 1.36 (ACT) (mean ± SD) to 23.46 ± 2.82 (CYP). The expression of the 12 reference genes had a large fluctuation range and were not absolutely stable. Therefore, other calculation methods need to be introduced to compare the expression stability in different groups of samples, such as geNorm, NormFinder, and BestKeeper.
Fig. 3.
Mean Ct values of 12 candidate reference genes. (Note: The data were pooled from all samples. The line across the box are the medians, the boxes depicts the 25/75 percentiles, the dotted lines represent the 95% confidence intervals and the dots are outliers)
GeNorm analysis
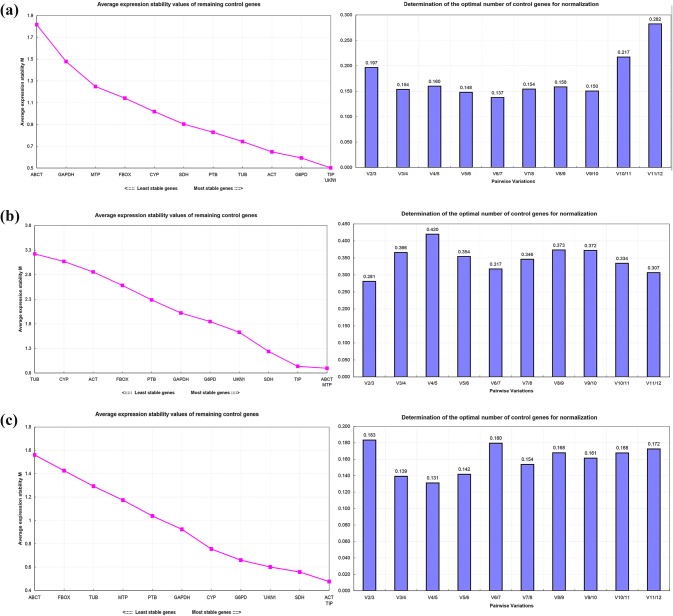
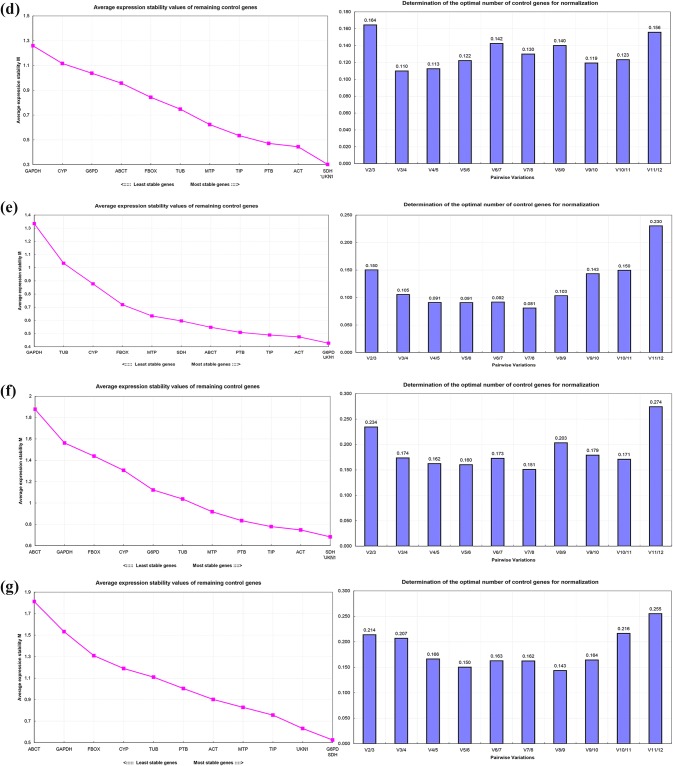
The average expression stability value (M) of reference genes was evaluated using the geNorm program. The M value reflects the average variability between the different genes indicating that the lower the M value, the more stable the gene expression. However, if the M value is greater than 1.5, the gene is not suitable as a reference gene. The optimal number of reference genes was estimated by the coefficient of variation (V) between two genes, such as Vn/Vn + 1 < 0.15, indicating that no additional reference genes are needed to be added. Otherwise, the n + 1 gene are needed to be introduced (n is the number of reference genes) for accurate normalization (Vandesompele et al. 2002).
As shown in Fig. 4a, for the group of different Forsythia species and cultivars, the top 2 stable expression genes were TIP and UKN1, while G6PD need to be added for normalization owing to the value of V2/V3 > 0.15; ABCT was the least stable gene because of the value of M > 1.5. ABCT and MTP were the most stable reference genes among the different organs of Forsythia, while TIP was added for normalization owing to the value of V2/V3 > 0.15; TUB, CYP, ACT, F-Box, PTB, GAPDH, G6PD and UKN1 were the least stable genes because of the value of M > 1.5 (Fig. 4b). ACT and TIP were the most stable reference genes in the different developmental stages of flower buds, while SDH was added for normalization owing to the value of V2/V3 > 0.15 (Fig. 4c). SDH and UKN1were the most stable reference genes in different tissue of flowers, while ACT was added for normalization owing to the value of V2/V3 > 0.15 (Fig. 4d). G6PD and UKN1 were the most stable and suitable reference genes for normalization in different developmental stages of fruits, owing to the value of V2/V3 = 0.15 (Fig. 4e). SDH and UKN1 were the most stable reference genes in mature leaves when plants were treated with ABA, while ACT was need to be added for normalization owing to the value of V2/V3 > 0.15; ABCT and GAPDH were the least stable genes because of the value of M > 1.5 (Fig. 4f). G6PD and SDH were the most stable reference genes in mature leaves when plants were treated by PEG, while UKN1 need to be added for normalization owing to the value of V2/V3 > 0.15, ABCT and GAPDH were the least stable genes because the value of M > 1.5 (Fig. 4g).
Fig. 4.
Expression stability, ranking and pairwise variation of 12 candidate reference genes by geNorm software. (a different species and cultivars; b different organs of Forsythia; c different developmental stages of Forsythia flowers; d different tissue of Forsythia lowers; e different developmental stages of Forsythia fruits; f leaves of Forsythia plants treated with ABA; g leaves of Forsythia plants treated with PEG)
The results showed that the most stable reference genes varied in leaves of different Forsythia species and cultivars, as well as in different organs, different developmental stages of flower buds, different flower tissues, different developmental stages of fruits, and in mature leaves of Forsythia plants stressed with PEG and ABA.
NormFinder analysis
Further verification of the stability of reference gene was performed using NormFinder. The smaller the stability value, the more suitable it is as reference gene (Andersen et al. 2004).
Results are shown in Table 3, and the gene selected in different groups by geNorm and NormFinder show a similarity, including the leaves of different Forsythia species and cultivars (TIP, UKN1 and G6PD), the different developmental stages of flower buds (ACT, TIP and SDH), and different part of flowers (SDH,UKN1 and ACT), as well as mature leaves of plants treated with ABA (SDH, UKN1 and ACT). However, the selected reference genes using NormFinder in different developmental stages of fruits (ACT, TIP and UKN1) and mature leaves of plants treated with PEG (G6PD, UKN1 and SDH) were different from the genes selected by the geNorm software. The most stable selected reference genes using NormFinder in different organs was GAPDH, which was quite different from the reference genes selected by the geNorm software.
Table 3.
Ranking of the reference gene and their expression stability values calculated by Normfinder
| Ranking order | Species and cultivars of Forsythia | Organs of Forsythia plants | Developmental stages of Forsythia flower buds | Forsythia flower plants | Developmental stages of Forsythia fruits | Leaves of Forsythia plants treated with ABA | Leaves of Forsythia plants treated with PEG | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | |
| 1 | TIP | 0.174 | GAPDH | 0.664 | TIP | 0.137 | SDH | 0.104 | ACT | 0.132 | ACT | 0.324 | G6PD | 0.036 |
| 2 | UKN1 | 0.290 | SDH | 1.039 | ACT | 0.220 | UKN1 | 0.139 | TIP | 0.140 | TIP | 0.491 | UKN1 | 0.132 |
| 3 | G6PD | 0.323 | G6PD | 1.109 | SDH | 0.220 | ACT | 0.212 | UKN1 | 0.143 | PTB | 0.533 | SDH | 0.182 |
| 4 | ACT | 0.410 | PTB | 1.182 | G6PD | 0.256 | PTB | 0.267 | G6PD | 0.148 | UKN1 | 0.537 | TIP | 0.546 |
| 5 | TUB | 0.497 | UKN1 | 1.357 | UKN1 | 0.430 | TIP | 0.388 | ABCT | 0.263 | G6PD | 0.553 | ACT | 0.593 |
| 6 | CYP | 0.566 | F-Box | 1.488 | CYP | 0.561 | TUB | 0.524 | PTB | 0.341 | SDH | 0.591 | MTP | 0.647 |
| 7 | SDH | 0.626 | TIP | 1.803 | GAPDH | 0.818 | MTP | 0.575 | MTP | 0.397 | TUB | 0.665 | TUB | 0.700 |
| 8 | PTB | 0.724 | ABCT | 1.822 | PTB | 0.926 | F-Box | 0.641 | SDH | 0.432 | MTP | 0.912 | CYP | 0.792 |
| 9 | F-Box | 0.996 | ACT | 2.139 | MTP | 0.980 | ABCT | 0.901 | F-Box | 0.663 | CYP | 1.071 | PTB | 0.941 |
| 10 | MTP | 1.017 | MTP | 2.214 | TUB | 1.111 | G6PD | 0.912 | TUB | 1.071 | F-Box | 1.212 | F-Box | 1.096 |
| 11 | GAPDH | 1.732 | CYP | 2.252 | F-Box | 1.321 | CYP | 0.932 | CYP | 1.147 | GAPDH | 1.355 | GAPDH | 1.738 |
| 12 | ABCT | 2.337 | TUB | 2.503 | ABCT | 1.417 | GAPDH | 1.283 | GAPDH | 1.909 | ABCT | 2.268 | ABCT | 2.108 |
Stability values was calculated by Normfinder and the smaller the stability value, the more suitable it is as reference gene
BestKeeper
The stability of the reference genes was evaluated by the standard deviation (SD) and coefficient of variance (CV) of the Ct values in the BestKeeper program. The genes with an SD value below 1.0 were considered to be stable and could be used for the gene expression normalization. The smaller the value was, the more stable the gene was considered to be (Pfaffl et al. 2004). The program can only analyze the stability of 10 candidate reference genes at most, therefore, the top ten candidate reference genes for each group were selected for BestKeeper analysis according to the stability calculated by GeNorm and NormFinder.
The results are shown in Table 4. UKN1, SDH, PTB, TIP, ACT, and G6PD were selected as suitable reference genes in the leaves of different Forsythia species and cultivars. Only the ACT gene was selected as a suitable reference gene in different organs. Only TIP and ACT were selected as suitable reference genes in the developmental stages of flower buds. ACT, PTB, TIP, MTP, ABCT and UKN1 were selected as suitable reference genes in different tissues of Forsythia flowers. G6PD, UKN1, ACT, TIP, MTP, PTB, ABCT and SDH were selected as suitable reference genes in different developmental stages of Forsythia fruits. Only TIP and MTP genes were selected as suitable reference genes in mature leaves when Forsythia plants were treated by PEG. However, there was no suitable reference gene in mature leaves when Forsythia plants were treated by ABA for gene expression normalization.
Table 4.
Ranking of the reference gene and their expression stability values calculated by Bestkeeper
| Ranking order | Species and cultivars of Forsythia | Organs of Forsythia plants | Developmental stages of Forsythia flower buds | Forsythia flower plants | Developmental stages of Forsythia fruits | Leaves of Forsythia plants treated with ABA | Leaves of Forsythia plants treated with PEG | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | |
| 1 | UKN1 | 0.74 | ACT | 0.55 | TIP | 0.79 | ACT | 0.69 | UKN1 | 0.18 | MTP | 1.08 | PTB | 0.95 |
| 2 | SDH | 0.75 | PTB | 1.48 | ACT | 0.98 | PTB | 0.71 | G6PD | 0.28 | TIP | 1.14 | MTP | 0.98 |
| 3 | PTB | 0.82 | F-Box | 1.86 | SDH | 1.05 | TIP | 0.72 | ACT | 0.34 | PTB | 1.28 | TIP | 1.15 |
| 4 | TIP | 0.82 | SDH | 3.15 | G6PD | 1.06 | MTP | 0.87 | TIP | 0.34 | ACT | 1.33 | UKN1 | 1.42 |
| 5 | ACT | 0.87 | GAPDH | 3.24 | PTB | 1.07 | ABCT | 0.87 | MTP | 0.36 | SDH | 1.38 | ACT | 1.61 |
| 6 | G6PD | 0.96 | TIP | 3.83 | UKN1 | 1.25 | UKN1 | 0.91 | PTB | 0.39 | UKN1 | 1.62 | SDH | 1.75 |
| 7 | MTP | 1.07 | UKN1 | 3.94 | CYP | 1.26 | SDH | 1.06 | ABCT | 0.44 | TUB | 1.89 | G6PD | 1.94 |
| 8 | TUB | 1.15 | ABCT | 3.96 | GAPDH | 1.37 | G6PD | 1.49 | SDH | 0.59 | G6PD | 2.16 | CYP | 2.1 |
| 9 | CYP | 1.28 | G6PD | 4.13 | MTP | 1.52 | TUB | 1.55 | F-Box | 1.05 | F-Box | 2.74 | TUB | 2.21 |
| 10 | F-Box | 1.51 | MTP | 4.13 | TUB | 1.62 | F-Box | 1.67 | CYP | 1.22 | CYP | 2.87 | F-Box | 2.64 |
Stability values was calculated by Bestkeeper and the genes value below 1.0 were considered to be stable and could be used for the gene expression normalization
In this study the stability of candidate reference genes was analyzed by the three software programs mentioned previously, and it was found that the most stable reference genes in the different developmental stages of the flower buds were TIP, ACT and SDH. The most stable reference genes in different developmental stages of the fruits were ACT, G6PD and UKN1. The results among three analysis methods were consistent.
The most stable reference genes selected from different Forsythia species and cultivars, different tissues of flowers, and mature leaves of Forsythia plants treated with PEG and ABA were slightly different when they were analyzed with different software.
Validation of the reference genes
In order to examine the reliability of the candidate reference genes for normalization, the stable and unstable reference genes recommended by the software analyses were used to normalize the expression trend of the test gene (AG or PSY gene) in different experimental materials. AG was chosen as the positive test gene in developmental stages and different tissues of Forsythia flowers, while PSY was chosen as the positive test gene in the other experimental materials.
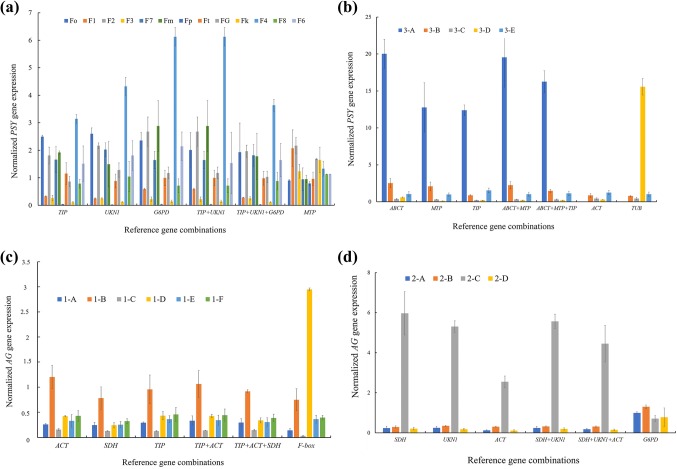
Candidate reference genes (TIP, UKN1, G6PD, TIP + UKN1, TIP + UKN1 + G6PD and MTP) were used for normalization in the mature leaves of different Forsythia species and cultivars, only UKN1 and TIP + UKN1 + G6PD were suitable as the reference genes for qRT-PCR normalization through the PSY transcripts expression trend (Fig. 5A).
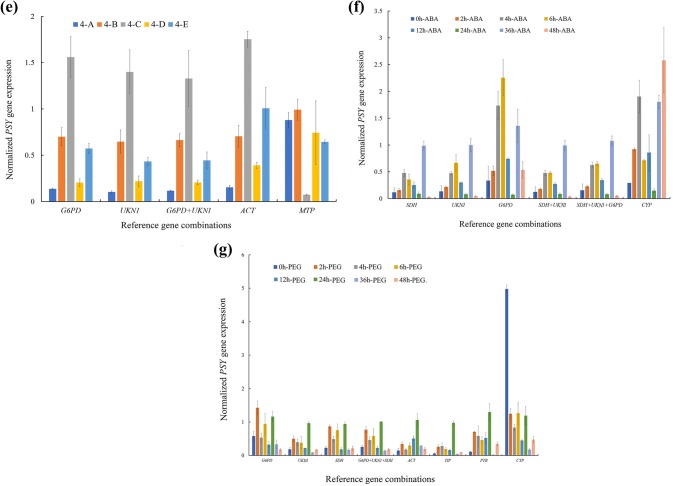
Fig. 5.
Test of the stability of reference genes. (a different Forsythia species and cultivars; b different organs of Forsythia; c different developmental stages of Forsythia flower buds; d different tissues of Forsythia flowers; e different developmental stages of Forsythia fruits; f leaves of Forsythia plants treated with ABA; g leaves of Forsythia plants treated with PEG)
When ABCT, MTP, TIP, ABCT + MTP, ABCT + MTP + TIP, ACT and TUB were used for normalization in the different organs of Forsythia, only MTP, ABCT+MTP and ABCT+MTP+TIP were found suitable as the reference genes for normalization through the PSY transcripts expression trend (Fig. 5b). The most stable (ACT, SDH, TIP, TIP + ACT, TIP + ACT + SDH) and least stable (F-box) reference genes in different developmental stages of Forsythia flower buds were used for normalization, and the AG transcripts expression trend were all generally consistent except when F-box was used as the reference gene (Fig. 5c). The most stable (SDH, UKN1, ACT, SDH + UKN1 and SDH + UKN1 + ACT) and least stable (G6PD) reference genes in different tissues of Forsythia flowers were used for normalization, and the AG transcripts expression trend were all consistent except when G6PD was used as the reference gene (Fig. 5d).When G6PD, UKN1, G6PD + UKN1, ACT and MTP were used for normalization in different developmental stages of Forsythia fruits, only G6PD, UKN1 and G6PD + UKN1 were found suitable as reference genes for normalization through the PSY transcripts expression trend (Fig. 5e).
SDH, UKN1, G6PD, SDH + UKN1, SDH + UKN1 + G6PD and CYP were used for normalization in the mature leaves of Forsythia when plants were treated by ABA. However, only SDH + UKN1 and SDH + UKN1 + G6PD were suitable as reference genes for normalization through the PSY transcripts expression trend (Fig. 5f). G6PD, UKV, SDH, G6PD+UKN1 + SDH, ACT, TIP, PTB and CYP were used for normalization in mature leaves when the plants were treated by PEG. Only SDH and G6PD + UKV + SDH were suitable as reference genes for normalization through the PSY transcripts expression trend (Fig. 5g).
In summary, the results suggested that the relative expression levels of the target gene are strongly affected by the choice of the reference genes.
Discussion
Proper reference genes for normalization in qRT-PCR could provide insight into the complex regulatory networks in plant science research (Gutierrez et al. 2010). The optimal reference genes selected from different species, different development stages, and different organs of the specific species usually vary (Wang 2016; Hong 2016). Forsythia is an important ornamental and medicinal plant, however, there is no report on the identification and validation of a suitable reference gene for gene expression study in qRT-PCR (Guo et al. 2007; Piao et al. 2008; Flinn and Ashworth 1999; Mcnamara and Pellett 1993; Rosati et al. 2003; Wang et al. 2017; Zhao et al.2007). In this study, the optimal reference genes for different Forsythia species and cultivars, different organs, different developmental stages, and different stress conditions were selected from 12 candidate genes, which provided a basis for studying the physiological and biochemical processes of Forsythia at the level of gene expression.
In order to select stable internal reference genes scientifically and accurately, diverse methods have been used to evaluate the candidate reference genes, such as geNorm, NormFinder and BestKeeper (Wang 2016; Hong 2016). Due to different calculation methods, the stability of these internal reference gene candidates may be different. In our study the stability of candidate reference genes was analyzed by the three software programs mentioned before, and it was found that the most stable reference genes in the different developmental stages were consistent among three analysis methods, while these genes selected from different Forsythia species and cultivars, different tissues of flowers, and mature leaves of Forsythia plants treated with PEG and ABA were slightly different among three anaylsis method, and the results from geNorm and NormFinder analysis were more similar. However, the stable reference genes selected from different organs differed greatly, and the stability of the reference genes need to be further tested. Therefore, it is necessary to conduct comprehensive analyses in order to ensure that the selected internal reference genes could correctly normalize the results of qPR-PCR. In this study, the results of geNorm and NormFinder analysis were similar, but they showed differences when compared with the results obtained from the BestKeeper analysis. For example, in the samples of different organs of Forsythia, ACT, PTB and F-box were selected by BestKeeper, and MTP, ABCT and TIP were selected by geNorm, and GAPDH, SDH and G6PD were selected by NormFinder. The stable reference genes selected by the three software analyses were examined by the PSY gene in different organs of Forsythia (Fig. 5b). We found that only MTP was suitable as internal reference gene, while TUB, F-box, ABCT, TIP and ACT were not suitable in different organs. And the same problem exists in citrus (Yan 2012). Therefore, it was necessary to use control genes to test the accuracy and validity of the selected reference genes. This study revealed that the results of the geNorm, NormFinder analysis were more accurate and reliable compared with the results of BestKeeper analysis.
In our study, the optimal internal reference genes selected from different species and cultivars, different organs, and different developmental stages of Forsythia varied (Table 5). UKN1 was the stable reference gene in different species and cultivars of Forsythia and different developmental stages of Forsythia fruits, which was consistent with the most stable reference genes in soybean (Hu et al. 2009). MTP, ABCT + MTP and ABCT + MTP + TIP were the stable reference genes in different organs of Forsythia. ACT and SDH were the stable reference genes in different developmental stages and different tissues of Forsythia flowers. SDH + UKN1 + G6PD was the stable reference gene combination when Forsythia was treated by PEG or ABA. While the stable internal reference genes of different organs, different developmental stages, and different stress treatment conditions were inconsistent with previous studies, such as those conducted with soybeans (Wang 2016), ash (Rivera-Vega et al. 2011), peach (Tong et al. 2009), and citrus (Yan 2012).
Table 5.
Expression stability ranking of the 12 candidate reference genes as calculated by software
| Materials | Group | Software | Ranking (stability of reference gene) (← most stable genes least stable genes →) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |||
| Forsythia | 12 species and cultivars | G* | UKV1 | TIP | G6PD | ACT | TUB | PTB | SDH | CYP | FBOX | MTP | GAPDH | ABCT |
| N* | TIP | UKV1 | G6PD | ACT | TUB | CYP | SDH | PTB | FBOX | MTP | GAPDH | ABCT | ||
| B* | UKV1 | SDH | TIP | PTB | ACT | G6PD | MTP | TUB | CYP | FBOX | / | / | ||
| F. ‘Courtaneur’ | Different organs | G* | MTP | ABCT | TIP | SDH | UKV1 | G6PD | GAPDH | PTB | FBOX | ACT | CYP | TUB |
| N* | GAPDH | SDH | G6PD | PTB | UKV1 | FBOX | TIP | ABCT | ACT | MTP | CYP | TUB | ||
| B* | ACT | PTB | FBOX | SDH | GAPDH | TIP | UKV1 | ABCT | G6PD | MTP | / | / | ||
| Different developmental stages of flower buds | G* | TIP | ACT | SDH | UKV1 | G6PD | CYP | GAPDH | PTB | MTP | TUB | FBOX | ABCT | |
| N* | TIP | ACT | SDH | G6PD | UKV1 | CYP | GAPDH | PTB | MTP | TUB | FBOX | ABCT | ||
| B* | TIP | ACT | SDH | G6PD | PTB | UKV1 | CYP | GAPDH | MTP | TUB | / | / | ||
| Different tissue of flowers | G* | UKV1 | SDH | ACT | PTB | TIP | MTP | TUB | FBOX | ABCT | G6PD | CYP | GAPDH | |
| N* | SDH | UKV1 | ACT | PTB | TIP | TUB | MTP | FBOX | ABCT | G6PD | CYP | GAPDH | ||
| B* | ACT | PTB | TIP | MTP | ABCT | VKV1 | SDH | G6PD | TUB | FBOX | / | / | ||
| Different developmental stages of fruit | G* | UKV1 | G6PD | ACT | TIP | PTB | ABCT | SDH | MTP | FBOX | CYP | TUB | GAPDH | |
| N* | ACT | TIP | UKV1 | G6PD | ABCT | PTB | MTP | SDH | FBOX | TUB | CYP | GAPDH | ||
| B* | UKV1 | G6PD | ACT | TIP | MTP | PTB | ABCT | SDH | CYP | FBOX | / | / | ||
| Stressed by ABA | G* | UKV1 | SDH | ACT | TIP | PTB | MTP | TUB | G6PD | CYP | FBOX | GAPDH | ABCT | |
| N* | ACT | TIP | PTB | UKV1 | G6PD | SDH | TUB | MTP | CYP | FBOX | GAPDH | ABCT | ||
| B* | MTP | TIP | PTB | ACT | SDH | UKV1 | TUB | G6PD | FBOX | CYP | / | / | ||
| Stressed by PEG | G* | SDH | G6PD | UKV1 | TIP | MTP | ACT | PTB | TUB | CYP | FBOX | GAPDH | ABCT | |
| N* | G6PD | UKV1 | SDH | TIP | ACT | MTP | TUB | CYP | PTB | FBOX | GAPDH | ABCT | ||
| B* | PTB | MTP | TIP | UKV1 | ACT | SDH | G6PD | CYP | TUB | FBOX | / | / | ||
G* means the gene stability calculated by GeNorm; N* means the gene stability calculated by NormFinder; B* means the gene stability calculated by BestKeeper
The results reveal that UKN1 was the stable reference gene in the mature leaves of twelve Forsythia species and cultivars. Moreover, the expression stability of 12 candidate reference genes was assessed by the GeNorm, NormFinder, and BestKeeper software programs in different taxonomic groups of Forsythia, including in different species, different cultivars of F. intermedia and F. koreana, and different style types (Supplemental data Table S2, S3, S4 and Fig. S1). We found that UKN1 was also the stable reference gene in the mature leaves of different taxonomic groups of Forsythia, but not the top ranked gene in these groups. TIP, SDH, G6PD were the most stable reference genes in different species and in different cultivars of F. intermedia and F. koreana. TIP was the most stable reference gene in the Forsythia species and cultivars with style type ‘S’ according to the result of software analysis, while stable reference gene recommended by the GeNorm, NormFinder and BestKeeper software programs in Forsythia species and cultivars with style type ‘L’ had a large discrepancy when calculated by different software.
As mentioned above, it is extremely important to select the appropriate internal reference genes for the specific species and under specific experimental conditions. Selection of the optimal internal reference gene in Forsythia involved a molecular mechanism with consideration for the genetic expression of the medicinal and ornamental traits desired.
In our study, it was also found that ABCT and TIP were not suitable as internal reference genes for qRT-PCR normalization in different organs of Forsythia. However, ABCT + MTP and ABCT + MTP + TIP were suitable as internal reference genes for qRT-PCR normalization. It can be seen that the normalization, with a combination of multiple reference genes, could provide more accurate quantification of the relative expression than with a single gene in qRT-PCR analysis (Peng et al. 2006).
In summary, 12 candidate reference genes were selected based on our transcriptome sequence data and previous studies and their expression stability was assessed and selected in different species and cultivars, organs and tissues of Forsythia. This study could provide a basis for investigating the physiological and biochemical processes of Forsythia related to growth and development, drought-resistance, ornamental characteristics, and medicinal properties at the level of gene expression.
Conclusion
In our study, we selected appropriate reference genes in different species, cultivars, organs, tissues and stress conditions of Forsythia for the first time, filling the gaps for suitable reference genes referenced during the study of the regulatory mechanisms of Forsythia on metabolite biosynthesis, biological processes, and molecular breeding. Through systematic analysis and research (Table 5 and Fig. 5), we found that UKN1 was the stable reference gene in mature leaves of twelve Forsythia species and cultivars and was also the stable reference gene in the developmental stages of the fruits. MTP, ABCT + MTP and ABCT + MTP + TIP were the stable reference genes in different Forsythia organs. ACT and SDH were the stable reference genes in different Forsythia flower tissues and developmental stages of the flower buds. SDH + UKN1 + G6PD was the stable reference gene combination in the leaves of Forsythia when plants were treated by PEG or ABA. Our study may provide a basis for investigating physiological and biochemical processes of Forsythia in order to obtain more accurate and reliable data for the levels of relative gene expression.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1 Expression stability, ranking and pairwise variation of 12 candidate reference genes in in different species, different cultivars of F. intermedia and F. koreana, and different style types by geNorm software. (A, Ranking in different Forsythia speciesl; B, Ranking in different cultivars of F. intermedia; C, Ranking in different cultivars of F.koreana; D, Ranking in species and cultivars with Style type ‘S’; E, Ranking in species and cultivars with Style type ‘L’) (TIFF 12317 kb)
Acknowledgements
The funding was supported by the World-Class Discipline Construction and Characteristic Development Guidance Funds for Beijing Forestry University (Grant No. 2019XKJS0323), Beijing Municipal Science and Technology Project (Grant No. Z181100002418006), Fundamental Research Fund for the Central University (Grant No. 2015ZCQ-YL-03), National Key Clinical Specialty Discipline Construction Program of China (CN), Hebei Provincial Department of science and Technology Project (Grant No. 18226316D), Key Technology Research and Development Program of Shandong (CN) (Grant No. HBCT2018060203).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alves MS, Al-Sadi AM, Carvalho CM. Selection of reference genes for quantitative PCR analysis in Citrus aurantifolia during phytoplasma infection. Trop Plant Pathol. 2018;43:402–412. doi: 10.1007/s40858-018-0224-2. [DOI] [Google Scholar]
- Andersen CK, Jensen JL, Qrntoft FT. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Chi X, Hu R, Yang Q, Zhang X, Pan L, Chen N, et al. Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol Genet Genom. 2012;287(2):167–176. doi: 10.1007/s00438-011-0665-5. [DOI] [PubMed] [Google Scholar]
- Domonkos I, Kis M, Gombos Z, Ughy B. Carotenoids, versatile components of oxygenic photosynthesis. Prog Lipid Res. 2013;52:539. doi: 10.1016/j.plipres.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Flinn CL, Ashworth EN. Supercooling in dormant flower buds of Forsythia, and the correlation between pistil size and bud hardiness. J Environ Hortic. 1999;17:57–62. [Google Scholar]
- Fu J, WangY Huang H, Zhang C, Dai S. Reference gene selection for RT-qPCR analysis of Chrysanthemum lavandulifolium during its flowering stages. Mol Breed. 2013;31:205–215. doi: 10.1007/s11032-012-9784-x. [DOI] [Google Scholar]
- Gachon C, Mingam A, Charrier B. Real-time PCR: What relevance to plant studies? J Exp Bot. 2004;55:1445–1454. doi: 10.1093/jxb/erh181. [DOI] [PubMed] [Google Scholar]
- Galpaz N, Wang Q, Menda N, Dani Z, Joseph H. Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 2008;53:717–730. doi: 10.1111/j.1365-313X.2007.03362.x. [DOI] [PubMed] [Google Scholar]
- Ge Y, Wang Y, Chen P, Wang Y, Hou C, Wu Y, Zhang M, Li L, Huo C, Shi Q, Gao H. Polyhydroxy triterpenoids and phenolic constituents from Forsythia suspensa (thunb.) vahl leaves. J Agric Food Chem. 2016;64:121–135. doi: 10.1021/acs.jafc.5b04509. [DOI] [PubMed] [Google Scholar]
- Gong L, Song JL, Gan XY, Liu X, Chen YC, Guo ZQ, Song YX. Correlation analysis of StNCED1 expression level and ABA content of Potato under simulated drought stress. J Plant Gene Resour. 2018;19:561–567. doi: 10.13430/j.cnki.jpgr.2018.03.023. [DOI] [Google Scholar]
- Guo H, Liu AH, Ye M, Yang M, Guo DA. Characterization of phenolic compounds in the fruits of Forsythia suspensa by high-performance liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:715–729. doi: 10.1002/rcm.2875. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J. 2010;6:609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- Hong Y (2016) Molecular mechanism of light-dependent anthocyanin biosynthesis in Chrysanthemum × morifolium [D]. Beijing Forestry University
- Hou JH, Gao ZH, Zhang Z, Chen SM, Ando T, Zhang JY, Wang XW. Isolation and characterization of an agamous, homologue pmag, from the japanese apricot (Prunus mume, sieb. et zucc.) Plant Mol Biol Rep. 2010;29:473–480. doi: 10.1007/s11105-010-0248-3. [DOI] [Google Scholar]
- Hu R, Fan C, Li H, Zhang Q, Fu YF. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol. 2009;10:93. doi: 10.1186/1471-2199-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou XY, Zhang L, Yang SZ, Li GH, Ye JL. Selection and validation of reference genes for quantitative RT-PCR analysis in peach fruit under different experimental conditions. Sci Hortic. 2017;225:195–203. doi: 10.1016/j.scienta.2017.07.004. [DOI] [Google Scholar]
- Li W, Zhang L, Zhang Y, Wang G, Song D, Zhang Y. Selection and validation of appropriate reference genes for quantitative real-time PCR normalization in staminate and perfect flowers of andromonoecious Taihangia rupestris. Front Plant Sci. 2017;8:729. doi: 10.3389/fpls.2017.00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcnamara S, Pellett H. Flower bud hardiness of Forsythia cultivars. J Environ Hortic. 1993;11:39–40. [Google Scholar]
- Peng F, James S, Niclas O, Reid KE, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006;6(1):27. doi: 10.1186/1471-2229-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper-excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Piao XL, Jang MH, Cui J, Piao X. Lignans from the fruits of Forsythia suspensa. Bioorg Med Chem Lett. 2008;18:1980–1984. doi: 10.1016/j.bmcl.2008.01.115. [DOI] [PubMed] [Google Scholar]
- Rivera-Vega L, Mamidala P, Koch JL, Mason ME, Mittapalli O. Evaluation of reference genes for expression studies in ash (Fraxinus spp.) Plant Mol Biol Rep. 2011;30:242–245. doi: 10.1007/s11105-011-0340-3. [DOI] [Google Scholar]
- Rosati C, Simoneau P, Treutter D. Engineering of flower color in Forsythia by expression of two independently transformed dihydroflavonol 4-reductase and anthocyanidin synthase genes of flavonoid pathway. Mol Breed. 2003;12:197–208. doi: 10.1023/a:1026364618719. [DOI] [Google Scholar]
- Rosati C, Cadic A, Duron M, Simoneau P. Forsythia. In: Pua EC, Davey MR, editors. Transgenic crops VI. Biotechnol agriculture and forestry. Berlin: Springer; 2007. pp. 299–318. [Google Scholar]
- Sampson DR. Mating group ratios in distylic Forsythia (Oleaceae) Can J Genet Cytol. 1971;13:368–371. doi: 10.1139/g71-057. [DOI] [Google Scholar]
- Satake H, Ono E, Murata J. Recent advances in the metabolic engineering of lignan biosynthesis pathways for the production of transgenic plant-based foods and supplements. J Agric Food Chem. 2013;61:11721–11729. doi: 10.1021/jf4007104. [DOI] [PubMed] [Google Scholar]
- Satake H, Koyama T, Bahabadi SE, Matsumoto E, Ono E, Murata J. Essences in metabolic engineering of lignan biosynthesis. Metabolites. 2015;5:270–290. doi: 10.3390/metabo5020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi A, Jun M, Erika M, Shin M, Eiichiro O, Honoo S. De novo transcriptomes of Forsythia koreana using a novel assembly method: insight into tissue- and species-specific expression of lignan biosynthesis-related gene. PLOS ONE. 2016;11:e0164805. doi: 10.1371/journal.pone.0164805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZG, Gao ZH, Wang F, Zhang Z, Zhou J. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol. 2009;10:71. doi: 10.1186/1471-2199-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De PK, Pattyn F, Bruce P, Nadine VR, Paepe AD, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genom Biol. 2002;3:1–11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X (2016) Selection of candidate reference genes for gene expression studies by RT-qPCR in soybean [D]. Shanxi Agricultural University
- Wang JY, Shen JS, Gu MM, Wang J, Cheng TR, Pan HT, Zhang QX. Leaf coloration and photosynthetic characteristics of hybrids between Forsythia ‘Courtaneur’ and Forsythia koreana ‘Suwon Gold’. HortScience. 2017;52:1661–1667. doi: 10.21273/HORTSCI12177-17. [DOI] [Google Scholar]
- Xu LF, Hua X, Yuwei C, Panpan Y, Yayan F, Yuchao T, Yuan SX, Ming J. Validation of reference genes for quantitative real-time PCR during bicolor tepal development in asiatic hybrid lilies (Lilium spp.) Front Plant Sci. 2017;8:669. doi: 10.3389/fpls.2017.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J. Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol Biol Rep. 2012;39:1831–1838. doi: 10.1007/s11033-011-0925-9. [DOI] [PubMed] [Google Scholar]
- Zhao P, Feng A, Ming T. Effects of arbuscular mycorrhiza fungi on drought resistance of Forsythia suspensa. Acta Bot Boreali Occident Sin. 2007;27:396–399. doi: 10.1016/S1872-2075(07)60055-7. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Expression stability, ranking and pairwise variation of 12 candidate reference genes in in different species, different cultivars of F. intermedia and F. koreana, and different style types by geNorm software. (A, Ranking in different Forsythia speciesl; B, Ranking in different cultivars of F. intermedia; C, Ranking in different cultivars of F.koreana; D, Ranking in species and cultivars with Style type ‘S’; E, Ranking in species and cultivars with Style type ‘L’) (TIFF 12317 kb)