Abstract
Background:
Amyloid fibrils are insoluble arranged aggregates of proteins that are fibrillar in structure and related to many diseases (at least 20 types of illnesses) and also create many pathologic conditions. Therefore understanding the circumstance of fibril formation is very important
Objectives:
This study aims to work on fibrillar structure formation of fibroin (as a model protein)
Material and Methods:
In this experimental study, fibroin was extracted from bombyx mori silk cocoon, and the concentration was obtained by Bradford method. The protein was incubated in a wide range of times (0 min to 7 days) in specific acidity and thermal conditions (pH=1.6, T=70 °C). The assays of UV-vis spectroscopy with congo red, field emission scanning electron microscopy, transmission electron microscopy, atomic force microscopy and circular dichroism spectroscopy were employed to monitor the fibrillation process.
Results:
Fibroin assemblies were formed upon the process of aggregation and fibril formation with a variety of morphology ranging from nanoparticles to elongated fibrils.
Conclusion:
The results showed progressive pathway of fibril formation
Keywords: Silk Fibroin; Circular Dichroism; Amyloid Fibrils; Microscopy, Electron, Scanning; Transmission Electron Microscopy; Fibroins
Introduction
Fibrous and globular proteins are the two groups of proteins contributing to the structural and functional purposes in biological entities, respectively [ 1 ]. Proper folding of proteins is significant in intercellular transportation, signals transmission and nervous impulses, cell adhesion, change in cytoskeleton, metabolic reactions and in general the proper functioning at cellular and intercellular levels [ 2 ]. On the other hand, protein aggregation is a biological phenomenon through which protein molecules have lost their normal structure generally as soluble and transformed to abnormal structures, and these abnormal molecules later have been connected to each other [ 3 ]. Protein aggregations are commonly categorized based on their microscopic shapes into two groups of regular aggregation (amyloid) and irregular aggregation (amorphous) [ 3 ]. Amyloid fibrils are insoluble fibrillar forms of proteins composed from peptide monomers [ 4 ]. In fact, many diseases are caused by protein structural changes and thus cellular disorders [ 2 ].
The general impression now is that all the proteins, under appropriate conditions, have the capacity to form amyloid fibers and this can be an intrinsic feature of proteins [ 5 ]. Nevertheless, the tendency of a protein to produce amyloid fibrils depends on several factors, including the polypeptide load and sequence as well as hydrophobicity and its secondary structure [ 6 ]. Mutations in a protein structure may also decrease or increase fibril production. Amyloid fibrils are also created when proteins are exposed to abnormal conditions such as temperature and pH [ 7 ]. Despite the differences in amino acid sequence and the primary structure of different proteins, they all gain the structure of beta sheets after the formation of fibrils.
The mechanism of amyloid fibers formation in vitro consists of two phases, including lag phase and growth phase. At lag phase, fibril formation is slow with no visible fibrils. The time required for this phase may range from a few hours or days to even a few weeks. This depends on several factors, including the structure of the protein, the ionic strength of the environment, temperature, pH and the redox potential. At growth phase with a higher speed, initiation and progression of the growth phase continues to a point which no other peptide is added to the end of a fibril, and finally stops without any further progression [ 5 , 8 , 9 ].
According to the extensive studies carried out on fibril formation, several models have been proposed to explain its mechanism, including the following [ 10 , 11 ]: i) nucleation conformational conversional model and ii) monomer-derived conversional model. Based on the first model, after the formation of a core, it grows in fibrous form to turn into long amyloid fiber. In the second model, a protein molecule changes from its original state and creates amyloid capable to induce amyloid features in other protein monomers. Finally, through the modification of protein structure, amyloid state is generated.
Fibril formation of proteins has attracted particular attention due to its significant role in the development of neurodegenerative and system amyloiosis diseases such as Alzheimer’s, Pick’s, Parkinson’s and type two diabetes [ 12 - 16 ]. Hence, the term “Amyloidosis” describes a particular type of disease associated with the formation of amyloid fibrils.
Under in vitro conditions, protein fibrils can be produced through the methods as following: i) mild denaturing when the primary protein is partially unfolded, ii) incubating at high temperatures and concentrations, iii) adding salt of high concentrations (salting out), iv) changing the solution pH, v) adding polar organic solvents, vi) adding heavy metal ions, and vii) adding nonionic polymers [ 17 - 20 ].
Fibril formation can be studied by the methods of fluorescence in thioflavin T upon protein bonding [ 21 ], UV-vis absorption of congo red upon binding into protein [ 22 - 23 ], scanning electron microscopy (SEM) [ 24 ], transmission electron microscopy (TEM) [ 25 ], atomic force microscopy (AFM) [ 9 , 26 ], cryo-electron microscopy [ 27 ], Fourier-transform infrared spectroscopy (FTIR) [ 28 ], X-ray diffraction (XRD) [ 29 ] and solid state nuclear magnetic resonance (NMR) [ 30 ].
On the other hand, stability and insolubility of amyloid fibrils prove their applications in different areas [ 31 - 37 ], while it also represents the intensity of their impact on pathogenicity processes [ 4 , 38 ]. However, the shape, size and positioning of the fibers should be controllable during their production. For example, this controlling leads to reducing cytotoxicity [ 39 ].
Fibroin is mainly derived from spider web and silkworm cocoons. Fibroin extracted from silkworm cocoon possesses features such as high biodegradation, absence of immune system stimulation, low toxicity, and high flexibility and mechanical strength [ 40 ]. Fibroin constitutes a light-chain, a heavy-chain and a sub-chain, and 85% of the molecular weight of this protein results from the heavy chain. Fibroin heavy chain contains 5263 amino acids mainly including glycine, alanine and serine [ 30 ]. Fibroin has applications in oxygen- and vapor-permeable films, enzyme immobilization, tissue engineering, artificial ligament production and drug delivery with a high adaptability with mammals’ cells [ 41 ].
In this study, fibroin was underwent conditions of fibril formation to evaluate the formation of amyloid and fibrillar structures.
Material and Methods
It is an experimental study. All materials and chemicals were purchased from Merck (Germany) or Sigma (USA) and used without further purification. Silkworm cocoon was supplied from a local store. In all the stages of the experiments, double distilled water was used.
To extract fibroin from silkworm cocoon, the cocoons of silkworms were firstly cut and rinsed three times with water. Then they were boiled four times with a solution of 0.2 mol L-1 sodium carbonate, rinsed again with water, and dried at 37 °C. The dried cocoons were transferred into a mixture of water, ethanol and calcium chloride in molar ratios of 8:2:1 at 90 °C for 6 hours to dissolve. The resulting solution was centrifuged at 5000 rpm for 20 minutes, and the supernatant was then dialyzed using a dialysis bag (MWCO 12 kDa) against water for three days, while the external water changed every 4 hours. Phosphate buffer saline 100 mmol L-1, pH=7.4 (PBS) was added into the remaining solution in the bag, passed through a 0.22 μm filter membrane and kept at 4 °C [ 31 ].
The concentration of fibroin solutions was determined by the Bradford test. A Bradford stock solution was prepared by mixing 10 mL ethanol 95%, 20 mL phosphoric acid 85% and 35 mg Coomassie blue G and stored at 4 °C in dark. Human serum albumin solutions of known concentrations were employed as standards, and a standard curve was constructed. To use the standard curve and the absorbance values of the fibroin solutions, the fibroin concentration was determined.
For fibroin fibrillation induction, fibroin solutions were reached pH=1.6 with hydraulic acid, and incubated at 70 °C at different time intervals.
Congo red binding test was performed by mixing 300 μL congo red stock solution of 20 mg mL-1 and 20 μL fibroin solutions. After 30 minutes, UV-vis absorbance values were measured.
A Rayleigh UV-2100 spectrophotometer (China) was employed to determine the concentration of protein and congo red absorption. Field-emission scanning electron microscopy (FESEM) was performed by a Zeiss, Sigma-IGMA/VP microscope (Germany) and transmission electron microscopy (TEM) was performed using a Zeiss, EM10C microscope (Germany). To prepare protein sample for FESEM, glass slides were rinsed with ethylene glycol and then double distilled water. The incubated protein solutions were stirred by a vortex for a few minutes and then put on the slide. The slides were sputtered with a gold layer as reported elsewhere [ 42 ], before observing under the microscope. Protein sample preparation for TEM was performed by sonication of protein solutions using a probe-ultrasound for 45 minutes. Then, a 2% uranyl acetate solution dissolved in water was utilized to stain the fibrils, and placed on the microscope grid.
Atomic force microscopy (AFM) was performed by JPK AG-Nano-Wizard II microscope at ambient conditions and noncontact mode as described elsewhere [ 43 ].
Circular dichroism (CD) spectra in the far-UV region were recorded using a J-810 Jasco spectropolarimeter with a 1 mm path cell at 25 °C, according to the procedures described elsewhere [ 44 ].
Results
The spontaneous amyloidogenesis of fibroin in PBS at pH=1.6 and 70 °C during different time intervals was recorded by time-lapse UV-vis spectra of bound congo red into the protein. Figure 1 shows the results obtained. Time intervals of fibroin incubation with a concentration of 180 μg mL-1 were 1, 2, 7.5, 10, 16, 28, 35, 45, 60, 75, 120 and 240 minutes, and 7, 15, 15, 20, 48 hours, and 4 and 7 days. Two phases of fibrillation called lag phase and growth phase are presented in Figure 2 schematically.
Figure 1.
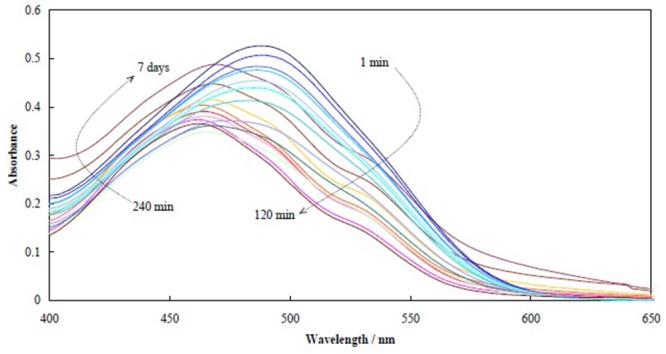
UV-vis spectra of bound congo red into fibroin for different time intervals of 1, 2, 7.5, 10, 16, 28, 35, 45, 60, 75, 120 and 240 min, and 7, 15, 15, 20, 48 h, and 4 and 7 days.
Figure 2.
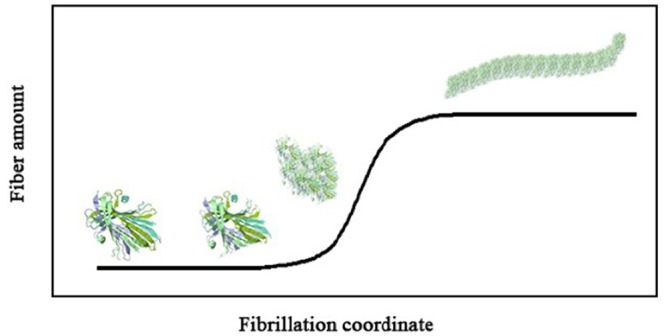
Illustration of fibrillation process of proteins.
The progression of fibroin fibrillation was followed at a concentration of 9 μg mL-1 and different selected time of incubation in PBS at pH=1.6 and 70 °C by FESEM. Figure 3 shows FESEM images of the resultant fibrillation products after incubation times of 30, 60, 90 and 240 minutes and 7 hours. Figures 3A and B show that after about 30 minutes, very small particles of ~18.0±3.5 nm are appeared in the protein solution due to post-nuclei formation. After 60 minutes, these small particles were aggregated and form larger ones (Figures 3C and D). It is clear from Figure 3D that these larger particles are collections of the smaller ones. Upon prolonging the fibrillation conditions to 90 minutes, a network-resembled of the particles, i.e. adhered particles, are observed as depicted in Figures 3E and F. These adhered particles would be the building blocks of the fibrils. A closer scrutiny of these images indicated that swollen fibroin particles tend to align within the surrounding area of the early fibrils. After 240 minutes, the fibrils were appeared with a texture of the fibrillation product (Figures 3G and H) with a variety of diameter and length. Upon the formation of sufficient amounts of nuclei, the fibrils’ elongation became the main dominated process and lead to the formation of a large number of fibrils accompanied by increasing in their topology complexity. These tenuous fibrils were transferred lastly into tick rod-like fibers upon 7 hours of incubation (Figure 3I). The fibroin fibrils had a thickness of 20-60 nm and a length of a few micrometers and could have different appearance even at equal formation conditions. The protein fibrils usually consist of 2-6 protofilaments having a beta sheets structure [ 4 ].
Figure 3.
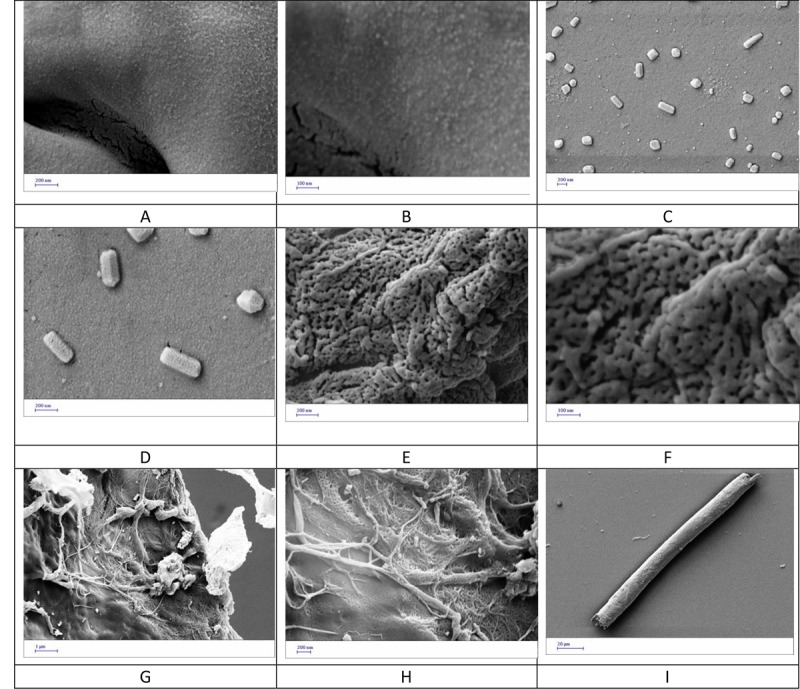
FESEM images from fibroin samples incubated under fibrillation conditions for different time intervals of 30 (A,B) , 60 (C,D) , 90 (E,F) and 240 (G,H) min and 7 h (I).
TEM images were recorded at elapsed fibrillation times of 30 (A), 90 (B) and 240 (C) minutes and 7 hours (D), and are shown in Figure 4. The initial nanoparticles formation in the lag phase was followed by small fibril creation, growing the small fibrils, and establishment of thick rod-like fibers. The images also confirmed that the final fibers were not condensed and seem to be hollow.
Figure 4.
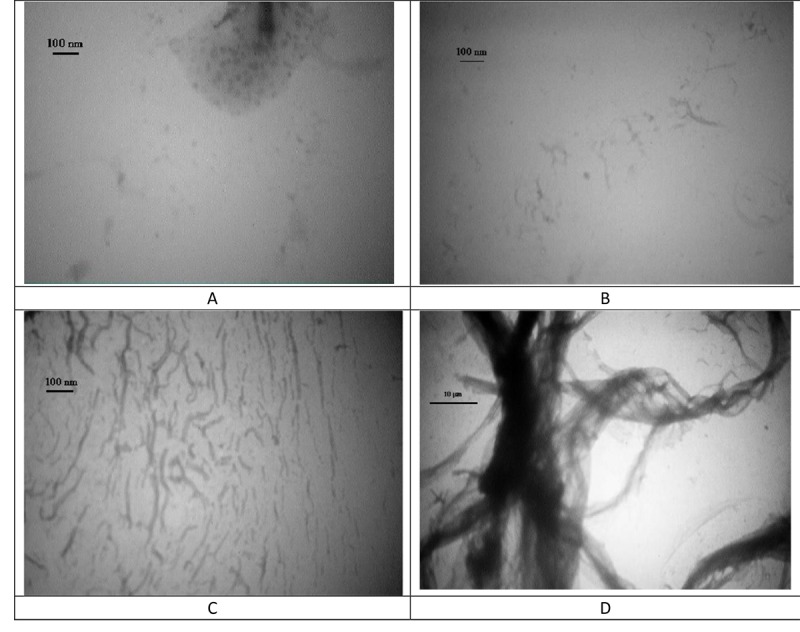
TEM images from fibroin samples incubated under fibrillation conditions for different time intervals of 30 (A), 90 (B) and 240 (C) min and 7h (D).
Figure 5 represents AFM images of the fibroin fibrils after incubation of the fibroin solution for 240 minutes (A) and 7 hours (B). The patterns of these figures are in line with those from FESEM and TEM images. At the shorter time (240 minutes), the fibrils were smoother and then achieve growth to provide thicker fibers. The fiber observed in Figure 5B was really comprised from thinner fibrils, and it seems that the thick fibers were formed by accumulating the thinner ones formed at shorter times.
Figure 5.
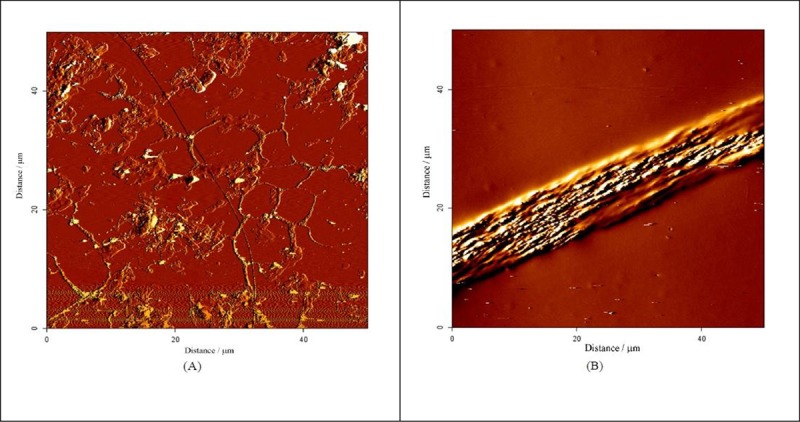
AFM images from fibroin samples incubated under fibrillation conditions for two time intervals of 240 min (A) and 7h (B).
Figure 6 shows far-UV CD spectra recorded for the native fibroin, and after its incubation under fibrillation conditions for 30, 60 and 120 minutes. In the spectra, upon increment in the fibrillation time, the ellipticity values at 197 nm increased. This indicated that in comparison with the native protein, incubation under fibrillation conditions lead to increasing the random coil conformations in the protein structure and eliminating the structural characteristic of the protein. This is the first step of the fibrillation.
Figure 6.
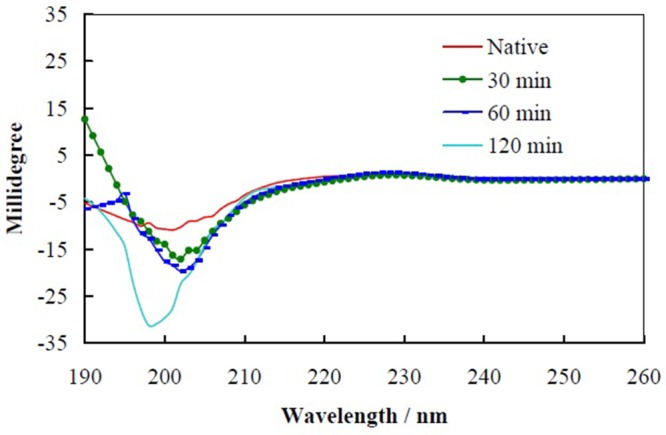
Far-UV CD spectra recorded for the native fibroin, and after its incubation under fibrillation conditions for 30, 60 and 120 min.
Discussion
Although proteins such as fibroin are not related to diseases, knowledge regarding their fibrillation is important for the basic studies in this field as well as applications in different areas of nanobiotechnology. Up to now, fibrillation of proteins which are not causing a disease such as casein [ 45 ], globulins [ 46 ] and lysozyme [ 47 , 48 ] have been studied. Protein fibril formation can be studied from two views of fibril production and structural changes upon and/or during the fibrillation process.
Binding to congo red occurs due to inherent core-sheet structural entity of the protein fibrils [ 49 , 50 ]. Congo red with a structure of diazo sulfonate has absorbance spectra with a maximum at 498 nm. However, upon congo red binding with peptides or proteins, either red or blue shift is occurred contingently upon protein structure and situation.
Congo red binding into fibroin under fibrillation conditions over time showed some distinct behavior. Form beginning of the fibrillation up to 120 minutes, the absorbance intensity decreased and the absorbance maxima had a blue shift. After this time (120 minutes) and up to the end of fibrillation time (7 days), the absorbance intensity increased and the absorbance maxima had a red shift. These two sets of spectra indicated two phases of fibrillation. The former was due to fibril nucleation [ 51 - 53 ] that is named as the lag phase; the absorbance intensity decreased due to accumulation of oligopeptides and nuclei formation [ 51 - 53 ]. These processes took about 2 hours, and after this time, the spectra patterns of congo red were related to fibril formation and growth of fibrils [ 49 ]. These results are well matched with previous studies [ 54 , 55 ].
The lag phase of aggregation is the initial feature of protein amyloidogenesis that is a thermodynamically unfavorable process of nucleation [ 56 , 57 ]. In this stage, tiny seeds of fibrils formed are followed by appearance of nuclei. Upon aggregation and growth of the nuclei, fibrils are generated. Previous studies have also shown that low PHs and/or high temperatures (without using any other solvent) induce fibrillation of proteins [ 9 , 58 ].
According to microscopic findings, lag phase and growth phase are obvious and by following these pictures from beginning to end, small particles are demonstrated at lag phase following by long fibrils; microns in length; at the end of the growing phase. In line with other studies [ 46 , 54 , 58 ], these two phases are shown. Process of fibrillation is in accordance with ionic strength and pH and temperature and amino acid composition of proteins [ 54 ]. Addition of some elements to the solution of protein can accelerate or decelerate the process of fibrillation; for instance it is shown that copper speeds up the fibrillation of HAS [ 55 ] and cysteine postpones this process in lysosome [ 47 ]. These items can be studied about fibroin to achieve a thermodynamically favorable method for production of fibrils.
Transmission and scanning electron microscopy indicates that in the primary stages of aggregation globular species are appeared subsequently converting to fibrils. In high temperature and in presence of acidic condition, these action happen in shorter period of time [ 54 ]. In our study, we have the same results too.
According to CD spectroscopic studies, α-helix structure is changed to β-sheet during fibrillation [ 54 , 56 ] and also these studies show that soluble proteins convert to protofilament, then they gather and make fibrils and at the last state, fibrils twist and form fibrils with large length and dimension [ 56 ].
In addition to studying the mechanism of producing fibrils, other studies are conducted to assess the possibility of controlling the dimension and length of fibrils. Concentration, temperature, pH and storage condition are factors influencing the process and length of final fibrils [ 58 ]. Higher concentration can limit the lag phase and also acidic content of the solution versus natural condition accelerate the speed of lag phase [ 57 ].
Studying model proteins such as fibroin can help us to have knowledge about fibrillation process to gain achievement in cure of diseases or achieve methods to have useful biological tools.
Conclusion
Fibroin assemblies were formed upon the process of aggregation and fibril formation with a variety of morphology ranging from nanoparticles to elongated fibrils. The solution pH and temperatures were key factors to form the fibroin fibrils and absence of any additional solvent to the protein content is very important. Growth of fibrils regarding to the incubation time shows the process of progressive fibril formation and presence of fibrils with different diameters and also lengths can be a proof for synergic effect of fibrillation process. If there are the more fibrillar content in a solution, there will be the more tendency to fibril formation.
Acknowledgement
This paper has been extracted from the M. Ahrami’s M.Sc. thesis supported by the Research Council of Shiraz University of Medical Sciences (94-7449).
Conflict of Interest: None
References
- 1. BioNinja [Internet] . Fibrous vs Globular Proteins. Available from: [http://ib.bioninja.com.au/standard-level/topic-2-molecular-biology/24-proteins/fibrous-vs-globular-protein.html. ]
- 2.Zerovnik E. Amyloid-fibril formation. Proposed mechanisms and relevance to conformational disease. Eur J Biochem. 2002;269:3362–71. doi: 10.1046/j.1432-1033.2002.03024.x. [DOI] [PubMed] [Google Scholar]
- 3.Lee C F. Self-assembly of protein amyloids: a competition between amorphous and ordered aggregation. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80:031922. doi: 10.1103/PhysRevE.80.031922. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson M R. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–60. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Morris A M, Watzky M A, Agar J N, Finke R G. Fitting neurological protein aggregation kinetic data via a 2-step, minimal/“Ockham’s Razor” model: The Finke− Watzky mechanism of nucleation followed by autocatalytic surface growth. Biochemistry. 2008;47:2413–27. doi: 10.1021/bi701899y. [DOI] [PubMed] [Google Scholar]
- 6.Rambaran R N, Serpell L C. Amyloid fibrils: abnormal protein assembly. Prion. 2008;2:112–7. doi: 10.4161/pri.2.3.7488. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohnishi S, Takano K. Amyloid fibrils from the viewpoint of protein folding. Cell Mol Life Sci. 2004;61:511–24. doi: 10.1007/s00018-003-3264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rochet J C, Lansbury Jr P T. Amyloid fibrillogenesis: themes and variations. Curr Opin Struct Biol. 2000;10:60–8. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 9.Jansen R, Dzwolak W, Winter R. Amyloidogenic self-assembly of insulin aggregates probed by high resolution atomic force microscopy. Biophys J. 2005;88:1344–53. doi: 10.1529/biophysj.104.048843. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy R M. Kinetics of amyloid formation and membrane interaction with amyloidogenic proteins. Biochim Biophys Acta. 2007;1768:1923–34. doi: 10.1016/j.bbamem.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Serio T R, Cashikar A G, Kowal A S, Sawicki G J, Moslehi J J, Serpell L, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–21. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 12.Toyama B H, Weissman J S. Amyloid structure: conformational diversity and consequences. Annu Rev Biochem. 2011;80:557–85. doi: 10.1146/annurev-biochem-090908-120656. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiti F, Dobson C M. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 14.Chiti F, Dobson C M. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 15.Lee C C, Sun Y, Huang H W. How type II diabetes-related islet amyloid polypeptide damages lipid bilayers. Biophys J. 2012;102:1059–68. doi: 10.1016/j.bpj.2012.01.039. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abedini A, Schmidt A M. Mechanisms of islet amyloidosis toxicity in type 2 diabetes. FEBS Lett. 2013;587:1119–27. doi: 10.1016/j.febslet.2013.01.017. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohnishi S, Takano K. Amyloid fibrils from the viewpoint of protein folding. Cell Mol Life Sci. 2004;61:511–24. doi: 10.1007/s00018-003-3264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sattarahmady N, Heli H, Moosavi-Movahedi A A, Karimian K. Deferiprone: structural and functional modulating agent of hemoglobin fructation. Mol Biol Rep. 2014;41:1723–9. doi: 10.1007/s11033-014-3021-0. [DOI] [PubMed] [Google Scholar]
- 19.Sattarahmady N, Heli H, Moosavi-Movahedi A A. Desferal as improving agent for hemoglobin fructation: structural and functional impacts. Protein J. 2012;31:651–5. doi: 10.1007/s10930-012-9444-3. [DOI] [PubMed] [Google Scholar]
- 20.Sattarahmady N, Khodagholi F, Moosavi-Movahedi A A, Heli H, Hakimelahi G H. Alginate as an antiglycating agent for human serum albumin. Int J Biol Macromol. 2007;41:180–4. doi: 10.1016/j.ijbiomac.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Whittingham J L, Scott D J, Chance K, Wilson A, Finch J, Brange J, et al. Insulin at pH 2: structural analysis of the conditions promoting insulin fibre formation. J Mol Biol. 2002;318:479–90. doi: 10.1016/S0022-2836(02)00021-9. [DOI] [PubMed] [Google Scholar]
- 22.Wang S S, Liu K N, Han T C. Amyloid fibrillation and cytotoxicity of insulin are inhibited by the amphiphilic surfactants. Biochim Biophys Acta. 2010;1802:519–30. doi: 10.1016/j.bbadis.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Divry P. Surles proprietes optiques de 1’amyloide. crsoc Bilge Biol. 1927;97:1808–10. [Google Scholar]
- 24.Takai E, Ohashi G, Ueki R, Yamada Y, Fujita J-I, Shiraki K. Scanning electron microscope imaging of amyloid fibrils. American Journal of Biochemistry & Biotechnology. 2014;10:31–39. doi: 10.3844/ajbbsp.2014.31.39. [DOI] [Google Scholar]
- 25.Cohen A S, Calkins E. Electron microscopic observations on a fibrous component in amyloid of diverse origins. Nature. 1959;183:1202–3. doi: 10.1038/1831202a0. [DOI] [PubMed] [Google Scholar]
- 26.Stine W B, Snyder S W, Ladror U S, Wade W S, Miller M F, Perun T J, et al. The nanometer-scale structure of amyloid-beta visualized by atomic force microscopy. J Protein Chem. 1996;15:193–203. doi: 10.1007/BF01887400. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Zhao C, Luo F, Liu Z, Gui X, Luo Z, et al. Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy. Cell Res. 2018;28:897–903. doi: 10.1038/s41422-018-0075-x. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zurdo J, Guijarro J I, Dobson C M. Preparation and characterization of purified amyloid fibrils. J Am Chem Soc. 2001;123:8141–2. doi: 10.1021/ja016229b. [DOI] [PubMed] [Google Scholar]
- 29.Eanes E D, Glenner G G. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem. 1968;16:673–7. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 30.Jiang T, Zhou P. Environment-Induced Silk Fibroin Conformation Based on the Magnetic Resonance Spectroscopy. On Biomimetics. 2011:357. doi: 10.5772/18479. [DOI] [Google Scholar]
- 31.Sattarahmady N, Vais R D, Heli H. Fibroin nanofibrils as an electrode material for electrical double-layer biosupercapacitor applications. Journal of Applied Electrochemistry. 2015;45:577–83. doi: 10.1007/s10800-015-0812-5. [DOI] [Google Scholar]
- 32.Pilkington S M, Roberts S J, Meade S J, Gerrard J A. Amyloid fibrils as a nanoscaffold for enzyme immobilization. Biotechnol Prog. 2010;26:93–100. doi: 10.1002/btpr.309. [DOI] [PubMed] [Google Scholar]
- 33.Domigan L J, Healy J P, Meade S J, Blaikie R J, Gerrard J A. Controlling the dimensions of amyloid fibrils: toward homogenous components for bionanotechnology. Biopolymers. 2012;97:123–33. doi: 10.1002/bip.21709. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L, Li N, Gao F, Hou L, Xu Z. Insulin amyloid fibrils: an excellent platform for controlled synthesis of ultrathin superlong platinum nanowires with high electrocatalytic activity. J Am Chem Soc. 2012;134:11326–9. doi: 10.1021/ja302959e. [DOI] [PubMed] [Google Scholar]
- 35.Andersson B V, Skoglund C, Uvdal K, Solin N. Preparation of amyloid-like fibrils containing magnetic iron oxide nanoparticles: effect of protein aggregation on proton relaxivity. Biochem Biophys Res Commun. 2012;419:682–6. doi: 10.1016/j.bbrc.2012.02.077. [DOI] [PubMed] [Google Scholar]
- 36.Li C, Adamcik J, Mezzenga R. Biodegradable nanocomposites of amyloid fibrils and graphene with shape-memory and enzyme-sensing properties. Nat Nanotechnol. 2012;7:421–7. doi: 10.1038/nnano.2012.62. [DOI] [PubMed] [Google Scholar]
- 37.Sattarahmady N, Moosavi-Movahedi A A, Habibi-Rezaei M, Ahmadian S, Saboury A A, Heli H, et al. Detergency effects of nanofibrillar amyloid formation on glycation of human serum albumin. Carbohydrate research. 2008;343(13):2229–34. doi: 10.1016/j.carres.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 38.Sipe J D, Cohen A S. Review: history of the amyloid fibril. J Struct Biol. 2000;130:88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 39.Domigan L J, Healy J P, Meade S J, Blaikie R J, Gerrard J A. Controlling the dimensions of amyloid fibrils: toward homogenous components for bionanotechnology. Biopolymers. 2012;97:123–33. doi: 10.1002/bip.21709. [DOI] [PubMed] [Google Scholar]
- 40.Hu Y, Zhang Q, You R, Wang L, Li M. The relationship between secondary structure and biodegradation behavior of silk fibroin scaffolds. Advances in Materials Science and Engineering. 2012;2012 doi: 10.1155/2012/185905. [DOI] [Google Scholar]
- 41.Motta A, Migliaresi C, Faccioni F, Torricelli P, Fini M, Giardino R. Fibroin hydrogels for biomedical applications: preparation, characterization and in vitro cell culture studies. J Biomater Sci Polym Ed. 2004;15:851–64. doi: 10.1163/1568562041271075. [DOI] [PubMed] [Google Scholar]
- 42.Rahi A, Sattarahmady N, Heli H. An ultrasensitive electrochemical genosensor for Brucella based on palladium nanoparticles. Anal Biochem. 2016;510:11–7. doi: 10.1016/j.ab.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Sattarahmady N, Heli H. A non-enzymatic amperometric sensor for glucose based on cobalt oxide nanoparticles. Journal of Experimental Nanoscience. 2012;7:529–46. [Google Scholar]
- 44.Mousavy S J, Riazi G H, Kamarei M, Aliakbarian H, Sattarahmady N, Sharifizadeh A, et al. Effects of mobile phone radiofrequency on the structure and function of the normal human hemoglobin. Int J Biol Macromol. 2009;44:278–85. doi: 10.1016/j.ijbiomac.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Pukala T L, Musgrave I F, Williams D M, Dehle F C, Carver J A. Gallic acid is the major component of grape seed extract that inhibits amyloid fibril formation. Bioorg Med Chem Lett. 2013;23:6336–40. doi: 10.1016/j.bmcl.2013.09.071. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Tang C-H. Heat-induced fibril assembly of vicilin at pH 2.0: Reaction kinetics, influence of ionic strength and protein concentration, and molecular mechanism. Food research international. 2013;51:621–32. doi: 10.1016/j.foodres.2012.12.049. [DOI] [Google Scholar]
- 47.Takai E, Uda K, Matsushita S, Shikiya Y, Yamada Y, Shiraki K, et al. Cysteine inhibits amyloid fibrillation of lysozyme and directs the formation of small worm-like aggregates through non-covalent interactions. Biotechnol Prog. 2014;30:470–8. doi: 10.1002/btpr.1866. [DOI] [PubMed] [Google Scholar]
- 48.Leung W H, Zou L, Lo W H, Chan P H. An Amyloid-Fibril-Based Colorimetric Nanosensor for Rapid and Sensitive Chromium (VI) Detection. ChemPlusChem. 2013;78:1440–5. doi: 10.1002/cplu.201300267. [DOI] [PubMed] [Google Scholar]
- 49.Klunk W E, Pettegrew J W, Abraham D J. Quantitative evaluation of congo red binding to amyloid-like proteins with a beta-pleated sheet conformation. J Histochem Cytochem. 1989;37:1273–81. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 50.Levine III H. Thioflavine T interaction with synthetic Alzheimer’s disease β-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993;2:404–10. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inouye H, Kirschner D A. Alzheimer’s beta-amyloid: insights into fibril formation and structure from Congo red binding. Subcell Biochem. 2005;38:203–24. doi: 10.1007/0-387-23226-5_10. [DOI] [PubMed] [Google Scholar]
- 52.Myers J K. Spectroscopic characterization of amyloid fibril formation by lysozyme. J Chem Educ. 2014;91:730–3. doi: 10.1021/ed400400x. [DOI] [Google Scholar]
- 53.Khurana R, Uversky V N, Nielsen L, Fink A L. Is Congo red an amyloid-specific dye? . J Biol Chem. 2001;276:22715–21. doi: 10.1074/jbc.M011499200. [DOI] [PubMed] [Google Scholar]
- 54.Juarez J, Taboada P, Mosquera V. Existence of different structural intermediates on the fibrillation pathway of human serum albumin. Biophys J. 2009;96:2353–70. doi: 10.1016/j.bpj.2008.12.3901. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pandey N K, Ghosh S, Dasgupta S. Fibrillation in human serum albumin is enhanced in the presence of copper(II) J Phys Chem B. 2010;114:10228–33. doi: 10.1021/jp103876p. [DOI] [PubMed] [Google Scholar]
- 56.Bouchard M, Zurdo J, Nettleton E J, Dobson C M, Robinson C V. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 2000;9:1960–7. doi: 10.1110/ps.9.10.1960. [ PMC Free Article ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nielsen L, Khurana R, Coats A, Frokjaer S, Brange J, Vyas S, et al. Effect of environmental factors on the kinetics of insulin fibril formation: elucidation of the molecular mechanism. Biochemistry. 2001;40:6036–46. doi: 10.1021/bi002555c. [DOI] [PubMed] [Google Scholar]
- 58.Domigan L J, Healy J P, Meade S J, Blaikie R J, Gerrard J A. Controlling the dimensions of amyloid fibrils: Toward homogenous components for bionanotechnology. Biopolymers. 2012;97:123–33. doi: 10.1002/bip.21709. [DOI] [PubMed] [Google Scholar]


