Abstract
Background
Trypanosomes are protozoan flagellates that cause human African trypanosomiasis (HAT) and African animal trypanosomiasis (AAT). HAT is caused by Trypanosoma brucei rhodesiense in East and Central Africa and T.b. gambiense in West Africa, whereas AAT is caused by a number of trypanosome species, including T. brucei brucei, T. evansi, T. vivax, T. congolense, T. godfreyi and T. simiae. The aim of this study was to establish if tsetse flies at Liwonde Wild Life Reserve (LWLR) are infected with these trypanosomes and thus pose a risk to both humans and animals within and surrounding the LWLR.
Methods
A total of 150 tsetse flies were caught. Of these, 82 remained alive after capture and were dissected such that the mid-gut could be examined microscopically for trypanosomes. DNA extractions were performed from both mid-guts and the 68 dead flies using a Qiagen Kit. Amplification techniques involved the Internal Transcriber Spacer 1 (ITS 1) conventional polymerase chain reaction (PCR) with primers designed to identify trypanosome species, and Repetitive Insertion Mobile Element — Loop Mediated Isothermal Amplification (RIME LAMP), a sequence specific to T. brucei.
Results
Analysis showed that 79/82 (96.3%) of the mid-guts examined microscopically were positive for trypanosomes and that 75/150 (50%) of the DNA extracts (from the mid-gut, and tsetse fly carcasses) were positive for T. brucei, as determined by the RIME LAMP method. ITS1 PCR further showed that 87/150 (58.0%) flies were positive for trypanosomes, of which 56/87 (64.4%) were T. brucei, 9/87 (10.3%) were T. vivax; 7/87 (8.1%) were T. simiae; 6/87 (6.9%) were T. congolense, and 6/87 (6.9%) were T. godfreyi. Ten samples had a mixture of infections.
Conclusion
Our analysis demonstrated a mixture of infections from trypanosome species in tsetse flies at LWLR, and that T. brucei, the species that causes HAT, was the most common. Our study successfully used molecular techniques to demonstrate the presence of T. b. rhodesiense at LWLR, a species that causes HAT in both East and Central Africa.
Keywords: Polymerase Chain Reaction, Tsetse Flies, African Trypanosomiasis, DNA, Southern Africa, Malawi
Introduction
Trypanosomes are protozoan flagellates responsible for two major diseases in Africa: human African trypanosomiasis (HAT), and animal African trypanosomiasis (AAT) or Nagana1. The three Trypanosoma brucei subspecies responsible for trypanosomiasis are T. brucei brucei, which is responsible for animal trypanosomiasis; T. brucei gambiense (T.b. gambiense, which causes HAT and is common in West Africa; and T. brucei rhodesiense (T.b. rhodesiense), which is common in East and Central Africa, and also causes HAT1,2. Despite the lack of appropriate molecular diagnostic tools, it is believed that T.b. rhodesiense is responsible for HAT in Malawi1.
HAT is endemic in West, East and Central regions of Africa. In Malawi, HAT is endemic to districts that have wildlife reserves and vast ranching areas, such as Rumphi, Kasungu and Nkhotakota1. In 2006, the prevalence of HAT in Nkhotakota and Rumphi was 6 and 16 cases per 100,000 people, respectively2.
Tsetse flies, of the Glossina genus, are the main vectors responsible for HAT2. In Malawi, the most predominant species is Glossina morsitans1,2. These tsetse flies are mostly localised to wildlife reserves where they depend on wild animals for their blood meal.
Little is known about the trypanosomes responsible for HAT and AAT at Liwonde Wild Life Reserve (LWLR), despite the clear presence of tsetse flies3. The range of trypanosome species infecting tsetse flies need to be accurately identified in order to gain a better understanding of the epidemiology of the disease. If left unattended, trypanosomiasis could have serious health implications for both humans and animals. The subsequent financial demands to cure HAT and the economic effect on livestock production as a result of AAT cannot be overemphasised4. It is therefore important to determine the prevalence of trypanosomes in tsetse flies in order to establish the risk posed to the people working at LWLR, and those in the surrounding villages, including their domestic animals. We sought to investigate the prevalence of trypanosomes in tsetse flies at LWLR using molecular techniques. Specifically, we aimed to determine the common species of tsetse flies, the trypanosome species infecting these tsetse flies and the prevalence of Trypanosoma brucei species responsible for HAT and Nagana.
Materials and methods
Study area
The study was conducted at LWLR, which is located in the Southern Region of Malawi, approximately 116 km away from the commercial city of Blantyre. This Wildlife Reserve harbours different types of wild animals since the Shire river (an outlet of Lake Malawi) passes through this reserve. The area has had reported cases of both HAT and AAT3.
Study population and sample size
Using a purposive sampling method to set traps in hotspot areas (near pathways for animals), we set Epsilon (Zimbabwe) and biconal (Challier-Laveissière, Burkina Faso) tsetse fly traps at Chinguni and Riverside camps, using liquid phenol and acetone as attractants5–7. The traps were set 50–100 m apart and the flies were harvested every 24hrs until the required sample size of 150 was reached. The flies, which were classified as either dead or alive, were then transported to the College of Medicine Molecular Biology Laboratories for species identification and processing.
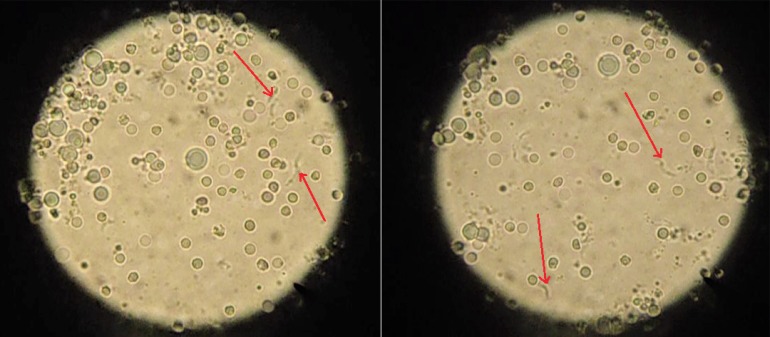
All flies were classified as either male or female by investigating the morphological features of the abdomen. We used standard identification methods for the tsetse flies. Male tsetse flies have a button-like structure called a hypopygium, while females have a slit. At the time of processing, 82 tsetse flies were still alive and were dissected; the mid-guts were removed and examined microscopically for the presence of trypanosomes (Figure 1). After microscopic examination, the mid-guts were stored in 400 µl of phosphate buffered saline (PBS). The abdomens of dead flies were extracted and kept in 400 µl of 99.9% ethanol in 1.5-ml microcentrifuge tubes. The mid-guts were homogenised and 200 µl was removed for DNA extraction. The abdomens from the 68 dead flies were dried in liquid nitrogen and crushed in 2 ml microcentrifuge tubes using clean pestles; 200 µl was also taken for DNA extraction.
Figure 1.
Brightfield microscopy images showing trypanosomes (arrows)
DNA extraction
DNA was extracted using the Qiagen DNA extraction kit (Qiagen, Germany). The starting material for the method was 200 µl of homogenate in a freshly labelled tube. A clean pipette was used to homogenise the mid-guts and crushed abdomens, to obtain a uniform mixture. Next, 20 µl of proteinase K was added, followed by 200 µl of lysis buffer, buffer AL; this was then mixed by vortexing. The mixture was incubated at 56°C for 1 h. Then, 200 µl of absolute ethanol was added and mixed thoroughly by vortexing. The mixture was pipetted into a DNeasy Mini Spin Column mounted in a 2-ml collecting tube and centrifuged at 6000 g (8000 rpm) for 1 min.
The flow-through and collecting tubes were discarded and the spin columns were placed in new 2-ml collection tubes. Then 500 µl of buffer AW1 was added to the spin columns and centrifuged for 1 min at 6000 g. The flow-through and collection tubes were discarded and the spin columns were placed in new 2-ml collection tubes. Next, we added 500 µl of buffer AW2 and centrifuged for 3 min at 12,000 g. The flow-through and collection tubes were discarded and the spin columns were placed onto sterile 1.5-ml microcentrifuge tubes. Next, 50 µl of buffer AE was added to each column, followed by incubation for 1 min at room temperature. Columns were then centrifuged at 6000 g for 1 min to collect the DNA. This last step was repeated in order to obtain sufficient DNA for all PCR reactions.
Molecular techniques
Two polymerase chain reaction (PCR) techniques were used to determine the presence of trypanosomes: Internal Transcriber Spacer region 1 (ITS 1) and RIME-LAMP PCR. ITS 1 region primers (Table 1) that were originally designed by Davilla in 2002 (unpublished) but approved by Njiru et al. (2005) were used to amplify the trypanosome genomes8. The ITS 1 region is highly conserved and different trypanosome species yield PCR amplicons of different sizes (Table 2). RIME-LAMP primers were designed by Njiru et al. and specifically target the T. brucei genome9.
Table 1.
List of primers used, their source, and expected product size
| Method | Primer | Sequence (5′–3′) | Amplicon size | Reference |
| ITS 1 PCR | Primer CF Primer BR |
CCGGAAGTTCACCGATATTG TTGCTGCGTTCTTCAACGAA |
Variable between species |
[8] |
| RIME LAMP PCR |
F3 B3 FIP BIP LF LB |
CTGTCCGGTGATGTGGAAC CGTGCCTTCGTGAGAGTTTC GGAATACAGCAGATGGGGCGAGGCCAATTGGCATCTTTGGGA AAGGGAGACTCTGCCACAGTCGTCAGCCATCACCGTAGAGC GCCTCCCACCCTGGACTC AGACCGATAGCATCTCAG |
179 bp | [9] |
Table 2.
ITS 1 polymerase chain reaction: species and expected product sizes
| Species | Expected product size |
Reference |
| T. brucei | 480 bp | [8] |
| T. congolense savannah | 700 bp | |
| T. congolense kilifi | 620 bp | |
| T. congolense forest | 710 bp | |
| T. simiae tsavo | 370 bp | |
| T. simiae | 400 bp | |
| T. godfreyi | 300 bp | |
| T. vivax | 250 bp |
Amplification reactions were carried out in 25 µl reaction mixtures for ITS 1 and RIME-LAMP. Thermocycling was carried out in a Techne Genius Thermo Cycler (Bibby Scientific, UK). The ITS 1 PCR 25 µl reaction tube contained 5 µl of 5× Phusion buffer, 0.5 µl of 10 mM dNTPs (Thermo Scientific, USA), 1.0 µl of each (forward and reverse) primer (Microsynth, Sweden), 0.25 µl of Phusion polymerase (Thermo Scientific, USA), 13.25 µl of nuclease free water and 5 µl of template DNA.
During the cycling, initial denaturation was carried out at 98°C for 10 s, followed by a second period of denaturation at 98°C for 30 s, annealing at 66°C for 30 s, and extension at 72°C for 30 s.
This sequence was repeated for 35 cycles, followed by final extension at 72°C for 10 min. The RIME-LAMP 25 µl reaction mixture contained 10 µl of lyophilised master mix containing dNTPs, Bsm polymerase, MgCl2, 5 µl of 5× reaction buffer (Thermo Scientific, USA), 1.0 µl of the outer forward primer (F3), 1.0 µl of the outer backward primer (B3), 1.0 µl of the forward inner primer (FIP), 1.0 µl of the backward inner primer (BIP), 1.0 µl of each of the loop forward and loop backward primers, LF & LB (Microsynth, Sweden), 2.0 µl of nuclease free water and 2.0 µl of template DNA. Amplification was performed in one step only at 62°C for 60 min and was terminated at 80°C for 5 min. Products were detected and visualised using Syber Green 1 dye (Life Technologies, USA). The detection of PCR products for ITS 1 was carried out using a 2% agarose gel (Thermo Scientific, USA) and stained with ethidium bromide (Sigma). Gels were visualised with a U-genius UV illuminator (Syngene, UK).
Results
Of the 150 tsetse flies caught, 32 were males and 138 were females. All of the tsetse flies caught were Glossina morsitans. Of the 150 flies caught, 82 were alive. These were dissected and the mid-guts were removed for microscopic analyses; 79/82 (96.3%) were positive for trypanosomes.
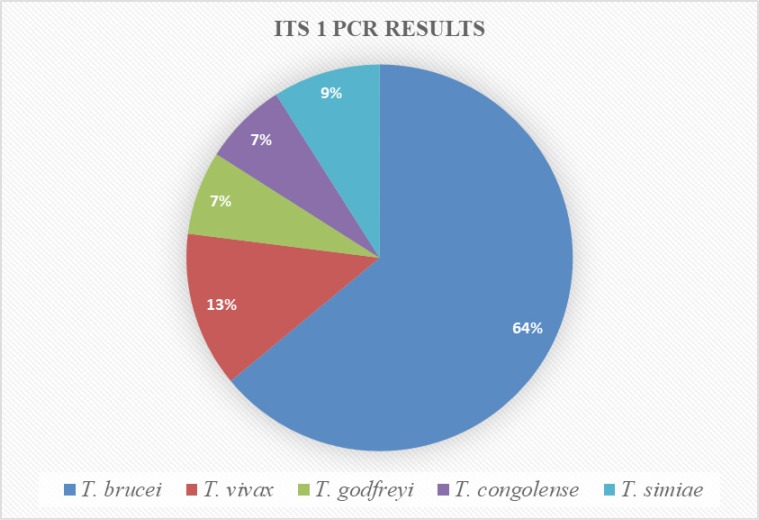
DNA analysis was performed on all of the 150 tsetse flies (using mid-guts from the dissected live flies as well as from the abdomens of dead flies). ITS 1 PCR analysis showed that 87/150 (58.0%) of the DNA extracts were positive for the presence of trypanosome DNA. Out of the 87 positives, 56 (64%) were T. brucei, 11 (13%) were T. vivax, 6 (7%) were T. godfreyi, 6 (7%) were T. congolense, and 8 (9%) were T. simiae (Figure 2). Ten samples had infections with more than one species of trypanosome (Figure 3). Out of the 150 samples, RIME-LAMP demonstrated that 75 (50.0%) were positive for trypanosomes (Figure 4).
Figure 2.
Trypanosome species identified by Internal Transcriber Spacer 1 polymerase chain reaction (ITS 1 PCR).
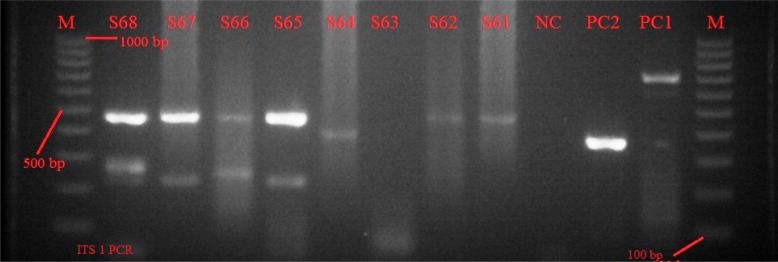
Figure 3.
Image of an agarose gel showing samples S61–S68: M is a 1000 bp marker; PC1 and PC2 were positive controls for T. congolense and T. simiae species; S61, S62, S64 are infections by T. brucei; S65 & S67 are mixed infections by T. brucei and T. vivax; S66 is a mixed infection by T. brucei and T. godfreyi; and S68 is a mixed infection by T. brucei, T. godfreyi and T. vivax.
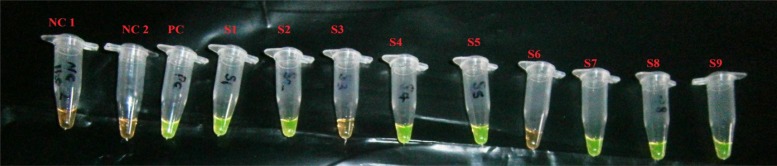
Figure 4.
RIME LAMP results: NC 1 is a negative control (nuclease free water); NC2 is a negative control (prepared master mix); PC is a positive control for a known T. brucei sample from Liwonde; S1, S2, S4, S5, S7–S9 are positives; and S3, S6 are negatives.
Discussion
This study has confirmed previous research findings which reported that most cases of HAT in Malawi are transmitted by Glossina morsitans tsetse fly species; this species is considered to be the most efficient transmitter of trypanosomes1. Microscopic analysis further established a very high infection rate (96.3%) in the tsetse flies at LWLR. This is by far the highest infection rate within the regions that are endemic with tsetse flies (Kenya, Tanzania, DRC, Congo Brazzaville, Malawi, Sudan and Uganda). The Xenomonitoring research consortium (2009–2013), which investigated infection rates caused by tsetse flies, established the average infection rate as 44.46% in Kasungu, 85.12% in Nkhotakota, and 94.46% in Liwonde, Malawi10. In the present study, microscopy identified a much higher infection rate than PCR. This was because microscopy identifies almost all trypanosome species. PCR does not detect all species, at least using the specific primers used in this study. This is consistent with previous research findings which reported a higher infection rate when determined by microscopy as compared with PCR4.
In most areas of Africa there are eleven different endemic pathogenic trypanosomes8. The present study identified five major pathogenic trypanosomes (Table 3); the most common was T. brucei (37.3%), as determined by ITS 1 PCR, and 50.0%, as determined by RIME LAMP. This is a very worrying finding because T. brucei is responsible for the disease, both in humans and animals. The other species identified were T. vivax, T. godfreyi, T. congolense, and T. simiae, which are solely for animal infection (African animal trypanosomiasis or Nagana), but are equally a cause for concern for our domestic animals.
Table 3.
Summary of results
| Method | Total number of samples |
Positives | Percentage of positives |
Negatives | |
| ITS PCR | 150 | 87 | 58.0% | 63 | |
| Specie | No. of positives |
||||
|
T. brucei T. godfreyi T. congolense T. simiae T. vivax |
56 6 6 8 11 |
64% 7% 7% 9% 13% |
|||
| Rime Lamp | 150 | 75 | 50% | 75 | |
Conclusion and recommendations
This study confirmed the existence of Glossina morsitans, the most effective transmitter of trypanosomes, in tsetse flies inhabiting LWLR. The study also established that there are mixed infections of different species of trypanosomes in the tsetse flies at LWLR. Five major species have been identified, including T. brucei, which was the most prevalent, and causes both human and animal disease. Our findings recommend that fly control strategies such as aerial spraying or insecticide impregnated target traps should be put in place at LWLR to reduce the levels of tsetse flies, or even eradicate them all together. Because we identified T. brucei, there is a significant concern for the agriculture sector because of the potential impact on animal infections. We therefore recommend the engagement of partners from agriculture (estates) to scale up campaigns for the eradication of tsetse flies by aerial sprays.
Further research is also needed on a large scale to determine the prevalence of trypanosomes in domesticated livestock and humans. A more specific study would be to determine the prevalence of Trypanosoma brucei rhodesiense in humans living close to wildlife and nature reserves.
Acknowledgements
We would like to acknowledge the College of Medicine Research Support Centre for funding this project. We also would like to acknowledge our Laboratory Technician, Mr Peter Nambala, for his technical assistance with laboratory work during the study. We acknowledge all the field workers who helped us with setting traps for the tsetse flies. Lastly, we wish to acknowledge the College of Medicine Biochemistry Department for giving us access to the Molecular Laboratory where we did all the laboratory work.
Authors' contributions (roles)
Symon F. Nayupe, Nelson V. Simwela and Peace Morrison Kamanga contributed in the study conception and design, acquisition of data, analysis and interpretation of data. They drafted the manuscript and made critical revision for intellectual content. John E. Chisi participated in the conception and design of the study and was the overall Principal Investigator for trypanosome projects and was the primary supervisor of this study. Edward Senga played a role in study conception and design and was also the secondary supervisor for the study. Janelisa Musaya was involved in manuscript drafting and critical revision for intellectual content. Emmanuel Maganga was the study Entomologist. He contributed in data acquisition and analysis and manuscript revision for intellectual content. All authors read and approved the final manuscript.
Funding
This study was funded by the College of Medicine Research Support Centre.
Availability of data and materials
Data supporting the findings of this study are available from the corresponding author, S.F., upon reasonable request.
Competing interests
The authors do not have an association that might pose competing interests (e.g., pharmaceutical stock ownership, consultancy, advisory board membership, relevant patents, or research funding).
References
- 1.Chisi J, Nkhoma A, Sternberg J. Presentation of trypanosomiasis in Nkhotakota. Malawi Med J. 2007;19(4):140–141. doi: 10.4314/mmj.v19i4.10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chisi JE, Muula AS, Ngwira B, Kabuluzi S. A retrospective study of human African trypanosomiasis in three Malawian districts. Tanzan J Health Res. 2011;13(1):62–68. doi: 10.4314/thrb.v13i1.61014. [DOI] [PubMed] [Google Scholar]
- 3.Bruce D, Hamerton AE, Watson DP, Bruce L. The trypanosome causing disease in man in Nyasaland: The Liwonde strain. Part I.- Morphology. Part II. -Susceptibility of animals. Roy Soc Proc B. 1914;88(601):97–111. [Google Scholar]
- 4.Morlais I, Grebaut P, Bodo JM, Djoha S, Cuny G, Herder S. Detection and identification of trypanosomes by polymerase chain reaction in wild tsetse flies in Cameroon. Acta Trop. 1998;70:109–117. doi: 10.1016/S0001-706X(98)00014-X. [DOI] [PubMed] [Google Scholar]
- 5.Auty H, Anderson N, Picozzi K, Lembo T, Mubanga J, Hoare R, et al. Trypanosome diversity in wildlife species from the Serengeti and Luangwa Valley ecosystems. PLoS Negl Trop Dis. 2012;6(10):e1828. doi: 10.1371/journal.pntd.0001828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohammed YO, Mohamed-Ahmed MM, Lubna TK, El Rayah IE. Detection of Trypanosoma brucei gambiense and T. b. rhodesiense in Glossina fuscipes fuscipes (Diptera: Glossinidae) and Stomoxys flies using the polymerase chain reaction (PCR) technique in southern Sudan. Afr J Biotechnol. 2010;9(38):6408–6412. [Google Scholar]
- 7.Nakayima J, Nakao R, Alhassan A, Mahama C, Afakye K, Sugimoto C. Molecular epidemiological studies on animal trypanosomiases in Ghana. Parasit Vectors. 2012;5:217. doi: 10.1186/1756-3305-5-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RCA, et al. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005;95:186–192. doi: 10.1007/s00436-004-1267-5. [DOI] [PubMed] [Google Scholar]
- 9.Njiru ZK, Mikoza A, Matovu E, Enyaru JCK, Ouma JO, Kibona SN, et al. African trypanosomiasis: sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int J Parasitol. 2008;38(5):589–599. doi: 10.1016/j.ijpara.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alibu VP, Enyaru JCK, Matovu E, Malele II, Chisi JE, Mbongo N, et al. Molecular xenomonitoring of trypanosomes in tsetse flies. J Parasitol Vector Biol. 2015;7(6):108–114. doi: 10.5897/JPVB2015.0188. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the findings of this study are available from the corresponding author, S.F., upon reasonable request.