Abstract
Mono(ADP-ribosylation) (MARylation) and poly(ADP-ribosylation) (PARylation) are posttranslational modifications found on multiple amino acids. There are 12 enzymatically active mono(ADP-ribose) polymerase (monoPARP) enzymes and 4 enzymatically active poly(ADP-ribose) polymerase (polyPARP) enzymes that use nicotinamide adenine dinucleotide (NAD+) as the ADP-ribose donating substrate to generate these modifications. While there are approved drugs and clinical trials ongoing for the enzymes that perform PARylation, MARylation is gaining recognition for its role in immune function, inflammation, and cancer. However, there is a lack of chemical probes to study the function of monoPARPs in cells and in vivo. An important first step to generating chemical probes for monoPARPs is to develop biochemical assays to enable hit finding, and determination of the potency and selectivity of inhibitors. Complicating the development of enzymatic assays is that it is poorly understood how monoPARPs engage their substrates. To overcome this, we have developed a family-wide approach to developing robust high-throughput monoPARP assays where the enzymes are immobilized and forced to self-modify using biotinylated-NAD+, which is detected using a dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA) readout. Herein we describe the development of assays for 12 monoPARPs and 3 polyPARPs and apply them to understand the potency and selectivity of a focused library of inhibitors across this family.
Keywords: poly(ADP-ribose) polymerase (PARP), mono(ADP-ribosylation) (MARylation), self-modification, nicotinamide adenine dinucleotide (NAD+), dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA)
Introduction
ADP-ribosylation has evolved as a posttranslational protein modification heavily involved in cellular stress response pathways and regulating host–pathogen interactions. The writers of the modification, poly(ADP-ribose) polymerase (PARP) enzymes are expressed throughout evolution. Human PARPs are responsible for the mono(ADP-ribosylation) (MARylation) and poly(ADP-ribosylation) (PARylation) of serine, aspartic acid, glutamic acid, lysine, tyrosine, and cysteine.1 The protein family is divided into two groups such that there are 12 mono(ADP-ribose) polymerases (monoPARPs) and 4 poly(ADP-ribose) polymerases (polyPARPs) and 1 inactive member (Suppl. Fig. S1). While monoPARPs and polyPARPs have similar active sites and all use nicotinamide adenine dinucleotide (NAD+) as the ADP-ribose donating substrate, the specificity for performing MARylation versus PARylation is correlated with a specific amino acid difference within the catalytic domains.2 PolyPARPs have a conserved H-Y-E motif in the active site, whereas most monoPARPs have a conserved H-Y motif and a leucine, isoleucine, or tyrosine in place of the glutamate with the exception of PARP3 and PARP4, which have the H-Y-E motif.
The polyPARPs have been extensively studied using small-molecule inhibitors; however, there is much less known about the monoPARPs, as evidenced by the number of publications for each PARP subfamily (Suppl. Fig. S2). There are limited reports of cell-active monoPARP inhibitors, and of those published, potencies and intra-PARP family selectivity are modest.3–10 Approved drugs that target PARP1 and PARP2 (niraparib and talazoparib) or PARP1, PARP2, and PARP3 (olaparib and rucaparib) are now in use for the treatment of a variety of cancers, and potent and selective inhibitors of PARP5a and PARP5b (tankyrases 1 and 2) have been reported to have antiproliferative activity in cancer cell lines and in vivo models.11 MonoPARPs have been reported to have important roles in immunology, inflammation, and cancer;12–14 however, these studies have relied almost exclusively on genetic perturbation techniques that often do not distinguish loss of catalytic activity from loss of the entire protein. Additionally, many monoPARPs are linked to RNA regulation,15 which can complicate the interpretation of results from siRNA knockdown and CRISPR knockout studies. To address these gaps, high-quality chemical probes are needed for the monoPARPs.
Typically, a family-wide approach to the generation of potent and selective chemical probes begins with the development of high-throughput enzyme activity or biophysical assays to enable rapid, high-fidelity rank-ordering of inhibitors. A major barrier to the development of enzyme activity assays for the monoPARPs is the gap in knowledge regarding what their substrates are and how they engage them, and at the time this work was initiated, there were no antibodies available that recognized MARylation. There have been limited reports describing inhibitor screening using enzyme assays, and in many cases high concentrations of enzyme have been required to generate sufficient signal to background to make the assays robust.8,16–19 Also, previous reports have used thermal shift assays (TSAs),20 surface plasmon resonance (SPR),20 DNA-encoded libraries (DELs)21 and small-molecule microarrays (SMMs)7 to identify and characterize monoPARP ligands; however, these techniques are either not quantitative (i.e., TSAs) or low-throughput (i.e., SPR), or require the development of expertise in complex chemistry (i.e., DELs and SMMs). Additionally, there has been a report of an AlphaScreen assay to identify inhibitors of the ADP-ribose binding macrodomains of PARP9, PARP14, and PARP15; however, this strategy was not shown to identify inhibitors of the catalytic activity of these proteins.22 While there have been efforts to identify substrates for monoPARPs,23–27 none of the substrates identified have been used to develop high-throughput inhibitor screening assays, and many of our attempts to develop assays using several of these substrates have not been successful. Additionally, there are no x-ray structures of monoPARPs bound to their substrates in the Protein Data Bank (PDB).
To enable the discovery of potent and selective monoPARP inhibitors, we have developed self-modification assays for nearly all of the PARPs. The enzymes are immobilized on Ni-NTA plates to bring them into close proximity and overcome the weak KM for self-modification, generating a low, yet detectable level of enzymatic activity in the absence of a true physiologically relevant substrate. The incorporation of a biotinylated ADP-ribose moiety derived from biotin-NAD+ is detected using a highly sensitive dissociation-enhanced lanthanide fluorescence immunoassay (DELFIA) readout capable of monitoring less than 1% enzymatic turnover. To calibrate the potency values generated in self-modification enzyme assays, we developed SPR assays for nearly all of the PARPs, and found the Kd values generated were similar to the potencies measured in the enzyme assays. Additionally, we employed a variation of this DELFIA assay format to validate a published report that ubiquitin is an ADP-ribosylation substrate of the PARP9/DTX3L complex.28 We used these assays to screen a focused library of PARP inhibitors against all PARP enzymes, generating a comprehensive dataset of potency and selectivity across the enzyme family that can be further used to develop chemical probes for understanding the biology of MARylation and its role in human disease.
Materials and Methods
Reagents
β-Nicotinamide-N6-(2-(6-(6-[biotinyl]aminohexanoyl)aminohexanoyl)aminoethyl)-adenine dinucleotide (Biotinylated-NAD+) was purchased from Biolog (Bremen, Germany). DELFIA Eu-N1 streptavidin, DELFIA assay buffer, diethylenetriamine pentaacetic acid (DTPA)-purified bovine serum albumin (BSA), DELFIA Enhancement Solution, and biotinylated hexahistidine peptide were purchased from PerkinElmer (Waltham, MA). Olaparib and PJ-34 were purchased from AdooQ Biosciences (Irvine, CA); rucaparib, niraparib, and veliparib were purchased from Selleckchem (Houston, TX); and RBN010860 and AZ1262949529 were synthesized. All other buffer reagents were purchased from Millipore-Sigma (Burlington, MA) at the highest level of purity possible. White polystyrene 384-well nickel-nitrilotriacetic acid (Ni-NTA)-coated microplates were custom-made by Thermo Fisher (Waltham, MA) and clear 384-well polypropylene microplates and white polystyrene high-binding 384-well microplates were purchased from Greiner (Monroe, NC). DNA oligomers were synthesized by Integrated DNA Technologies (Skokie, IL). 32P-NAD+ was purchased from American Radiolabeled Chemicals (St. Louis, MO). Histone H1, histone H2A, histone H2B, histone H3.1, and histone H4 were purchased from New England Biolabs (Ipswich, MA).
Protein Purification
PARP enzymes were purified to greater than 80% purity using an N-terminal hexahistidine (His6) tag. Construct, expression, and purification details can be found in Supplemental Table S1. The His6 tag was left on the protein following purification in order to capture the protein on the Ni-NTA plates in the assays. Constructs with Avi tags were biotinylated by BirA either in vitro using recombinant enzyme or in cellulo using bacterial cell lines expressing BirA and confirmed to have close to 100% modification via mass spectrometry. Protein purity was assessed on a Bioanalyzer 2100 (Agilent, Santa Clara, CA), and only proteins with >80% purity were used in assay development.
PARP9 and DTX3L were co-expressed in Sf9 insect cells on two separate pFastBacI plasmids. PARP9 (NM_001146102.1) was fused to an MBP tag with a TEV cleavage site between the MBP tag and PARP9. DTX3L (NM_138287.3) was fused to a His6 tag with a thrombin cleavage site between the His6 tag and DTX3L. The complex was purified on a nickel affinity column followed by an MBP column. The two proteins remained as a complex throughout the purification. Full-length proteins, UBE1 (NM_003334.3), UBE2D1 (NM_003338.4), and ubiquitin (NM_021009), were purified by affinity chromatography for their respective tags, followed by size exclusion chromatography. UBE2D1 and ubiquitin were both fused to a His6 tag, while UBE2D1 also had a TEV cleavage site between the tag and the gene sequence. UBE1 was fused to a Flag tag. UBE2D1 was treated with TEV protease and subjected to an additional round of purification on a nickel column to remove the His-tagged protease. Ubiquitin required one additional passage over a monoQ column following the size exclusion column to achieve high purity.
Equipment
Compounds were serially diluted on a Fluent (Tecan, Mannedorf, Switzerland) and spotted into white 384-well polystyrene Ni-NTA-coated microplates using a Mosquito (TTP Labtech, Melbourn, UK). During assay development, reagents were added to the microplates with multichannel pipets for some assay development steps; otherwise, they were added by Multidrop Combi (Thermo Fisher). During screening assays, all reagents were added by Multidrop Combi. Microplates were washed using an Elx-406 (Biotek, Winooski, VT) and read on an Envision plate reader (PerkinElmer) using a LANCE/DELFIA top mirror and a 340 nm TRF filter for excitation and 615 nm TRF filter for emission. SPR assays were developed on Biacore T200, Biacore 4000, and Biacore 8K systems (GE Healthcare Life Sciences, Marlborough, MA).
General Self-Modification Enzymatic Activity Assay Procedure
Reactions were performed in a 25 µL volume in 384-well white polystyrene Ni-NTA-coated microplates at 25 °C. Enzyme assay buffer was 20 mM HEPES (pH = 7.5), 100 mM NaCl, 2 mM DTT, 0.1% DTPA-purified BSA, and 0.002% Tween 20. Compounds were stored in 100% DMSO and 0.5 µL was dry-spotted into the microplates. Uninhibited control wells contained DMSO (final concentration [f.c.] = 2%) and fully inhibited control wells contained rucaparib, RBN010860, or AZ12629495 (f.c. = 200 µM), depending on the PARP being tested. His-tagged PARP enzymes were added in a 20 µL volume to the microplates and incubated for 30 min before the addition of 5 µL of biotinylated-NAD+ to initiate the reaction. The assays were ended while in the linear range of product versus time formation by the addition of 5 µL of NAD+ (f.c. = 2 mM) to outcompete the incorporation of biotinylated-NAD+. PARP1, PARP2, and PARP3 are activated by DNA;30,31 therefore, DNA oligomers were included in the reactions by addition to the biotinylated-NAD+ solution. The sequences of the DNA oligomers used for each PARP are listed in Supplemental Table S2. The details on concentrations of enzyme, biotinylated-NAD+, and activating DNA used, as well as reaction time for each PARP, are indicated in Table 1 . Note that 5 µM of unlabeled NAD+ is also added to the PARP2 reaction to stimulate the formation of poly(ADP-ribose). Quenched reactions were washed five times using 100 µL of Tris-buffered saline + Tween 20 (TBS-T), followed by the addition of 1:1000 DELFIA Eu-N1 streptavidin diluted in DELFIA assay buffer, and then incubated for 30 min at 25 °C to allow the streptavidin to bind to the incorporated biotin. Next, the reactions were washed five times with 100 µL of TBS-T, followed by the addition of 25 µL of DELFIA enhancement solution. Microplates were incubated 30 min; then the DELFIA signal was read on an Envision plate reader (excitation = 340 nm, emission = 615 nm).
Table 1.
| PARP Assay | Enzyme Concentration (µM) | Biotin-NAD+ Concentration (µM) | Activating DNA Concentration (µM) | Length of Assay (min) |
|---|---|---|---|---|
| PARP1 | 0.002 | 2 | 0.002 | 60 |
| PARP2 | 0.002 | 3 | 0.4 | 120 |
| PARP3 | 0.0025 | 2 | 0.1 | 120 |
| PARP4 | 0.075 | 2 | None | 180 |
| PARP5a | 0.01 | 3 (+10 µM unlabeled NAD+) | None | 120 |
| PARP6 | 0.003 | 3 | None | 180 |
| PARP7 | 0.075 | 2 | None | 240 |
| PARP8 | 0.05 | 3 | None | 180 |
| PARP9 | 0.008 | 3 | None | 180 |
| PARP10 | 0.015 | 3 | None | 180 |
| PARP11 | 0.008 | 3 | None | 180 |
| PARP12 | 0.015 | 3 | None | 180 |
| PARP14 | 0.05 | 3 | None | 180 |
| PARP15 | 0.001 | 1 | None | 1440 |
| PARP16 | 0.15 | 6 | None | 180 |
PARP5a Enzymatic Activity Assay Procedure
White polystyrene high-binding 384-well microplates were coated with 25 µL of 0.5 mg/mL histone H1 for 16 h at 4 °C, and then washed three times with 100 µL of TBS-T, followed by blocking with 100 µL of TBS Superblock for 1 h, and washed three times with 100 µL of TBS-T. Reactions were performed in a 25 µL volume in 384-well white polystyrene Ni-NTA-coated microplates at 25 °C. Enzyme assay buffer was 20 mM HEPES (pH = 7.5), 100 mM NaCl, 2 mM DTT, 0.1% DTPA-purified BSA, and 0.002% Tween 20. Compounds were stored in 100% DMSO and 0.5 µL was dry-spotted into the microplates. Uninhibited control wells contained DMSO (f.c. = 2%) and fully inhibited control wells contained rucaparib (f.c. = 200 µM). PARP5a (f.c. = 10 nM) was added in a 20 µL volume to the microplates and incubated with compound for 1 h before the addition of 5 µL of a mixture of biotinylated-NAD+ (f.c. = 3 µM) and NAD+ (f.c. = 10 µM) to initiate the reaction. The assay was ended after 2 h by the addition of 5 µL of NAD+ (f.c. = 2 mM) to outcompete the incorporation of biotinylated-NAD+. Quenched reactions were washed five times using 100 µL of TBS-T, followed by the addition of 1:1000 DELFIA Eu-N1 streptavidin diluted in DELFIA assay buffer, and then incubated for 30 min at 25 °C to allow the streptavidin to bind to the incorporated biotin. Next, the reactions were washed five times with 100 µL of TBS-T, followed by the addition of 25 µL of DELFIA enhancement solution. Microplates were incubated 30 min; then the DELFIA signal was read on an Envision plate reader (excitation = 340 nm, emission = 615 nm).
PARP9 Enzymatic Activity Assay Procedure
Reactions were performed in a 25 µL volume in clear 384-well polypropylene microplates at 25 °C. Enzyme assay buffer was 20 mM HEPES (pH = 7.5), 100 mM NaCl, 0.1% DTPA-purified BSA, and 0.002% Tween 20. Compounds were stored in 100% DMSO and 0.5 µL was dry-spotted into the microplates. Uninhibited control wells contained DMSO (f.c. = 2%) and fully inhibited control wells contained NAD+ (f.c. = 10 mM). PARP9/DTX3L (f.c. = 8 nM), UBE1 (f.c. = 50 nM), UBE2D1 (f.c. = 400 nM), His-tagged ubiquitin (f.c. = 1 µM), and biotinylated-NAD+ (f.c. = 3 µM) were added in a 20 µL volume and incubated with test compounds for 1 h before the addition of 2.5 µL of biotin-NAD+. The reaction was initiated by adding 2.5 µL of ATP (f.c. = 20 µM), and the reaction proceeded at 25 °C for 3 h before adding 25 µL of NAD+ (f.c. = 10 mM) to stop the incorporation of biotinylated-NAD+. Next, the stopped reactions were transferred from the clear 384-well polypropylene microplate to a 384-well white polystyrene Ni-NTA-coated microplate and incubated for 30 min to allow the His-tagged ubiquitin to bind. The microplate with captured His-tagged ubiquitin was washed five times using 100 µL of TBS-T, followed by the addition of 1:1000 DELFIA Eu-N1 streptavidin diluted in DELFIA assay buffer, and then incubated for 30 min at 25 °C to allow the streptavidin to bind to the incorporated biotin. Next, the reactions were washed five times with 100 µL of TBS-T, followed by the addition of 25 µL of DELFIA enhancement solution. Microplates were incubated 30 min; then the DELFIA signal was read on an Envision plate reader (excitation = 340 nm, emission = 615 nm).
DELFIA Metal-Chelation Counterscreen Assay Procedure
Reactions were performed in a 25 µL volume in 384-well white polystyrene Ni-NTA-coated microplates at 25 °C. The counterscreen assay buffer was 20 mM HEPES (pH = 7.5), 100 mM NaCl, 2 mM DTT, 0.1% DTPA-purified BSA, and 0.002% Tween 20. Compounds were stored in 100% DMSO and 0.5 µL was dry-spotted into the microplates. All control wells contained DMSO (f.c. = 2%). Biotinylated hexahistidine peptide (f.c. = 5 nM) was incubated with test compound in the counterscreen assay buffer for 60 min. Counterscreen assay buffer without peptide was added to fully inhibited control wells. The microplates were washed five times using 100 µL of TBS-T, followed by the addition of 1:1000 DELFIA Eu-N1 streptavidin diluted in the counterscreen assay buffer, and then incubated for 30 min at 25 °C to allow the streptavidin to bind to the incorporated biotin. Next, the reactions were washed five times with 100 µL of TBS-T, followed by the addition of 25 µL of DELFIA enhancement solution. Microplates were incubated for 30 min, and then the DELFIA signal was read on an Envision plate reader (excitation = 340 nm, emission = 615 nm).
Enzyme Inhibition Assay and Counterscreen Assay Data Analysis
Enzyme kinetics and parameters such as KM and kcat were determined using Prism software (GraphPad Software Inc., San Diego, CA) to analyze Michaelis–Menten fits of steady-state enzyme velocities. Screening data were processed using Dotmatics (Bishop’s Stortford, UK) and IC50 values and Hill slopes were generated using four-parameter fits. The quality and robustness of the assay was determined by analysis of the Z′ factor32 and performance of the 50% inhibition and IC50 control compounds.
General Surface Plasmon Resonance Procedure
The PARP SPR buffer was 50 mM HEPES (pH = 7.5), 100 mM NaCl, 1 mM TCEP, and 0.05% Tween 20, and assays were run at 25 °C. PARPs were captured either via N-terminal His6 tags on CM5 Ni-NTA sensor chips or via N-terminal biotinylated Avi tags on streptavidin SA sensor chips (GE Healthcare Life Sciences). The constructs used in the SPR assay are listed in Table 1 . Typically, 3000–6000 RU of protein was immobilized for each assay. Compounds were screened using a flow rate of 30 µL/min, using 60 s of association time, typically followed by observing dissociation for 90 s. For higher-affinity compounds, the dissociation window was increased, and in some instances compound binding was analyzed using single-cycle kinetics. Solvent correction was applied using a six-point curve from 1.5% to 2.75% DMSO injected before and after every 96 cycles. Data were normally fit to an equilibrium binding model to derive the binding constant KD, and when significant off-rates were observed, data were fit to a kinetic model to derive parameters for association and dissociation (ka and kd) and the binding constant Kd.
Results
Determining Conditions That Activate Enzymes to Perform ADP-Ribosylation
Several commercially available proteins and/or synthesized peptides corresponding to proteins reported to be substrates of PARP7,25,33,34 PARP14,35–37 and PARP1638 were tested as substrates under a variety of conditions. We looked for the incorporation of 32P-NAD+ or biotinylated-NAD+ using gel-, far-Western blot-, scintillation proximity counting-, or microplate-based readouts, but did not observe any signal above background under multiple reaction conditions. In contrast, we observed robust activity of PARP1 and PARP2 on purified histones, which are reported to be substrates for those enzymes39 (e.g., see Suppl. Fig. S3).
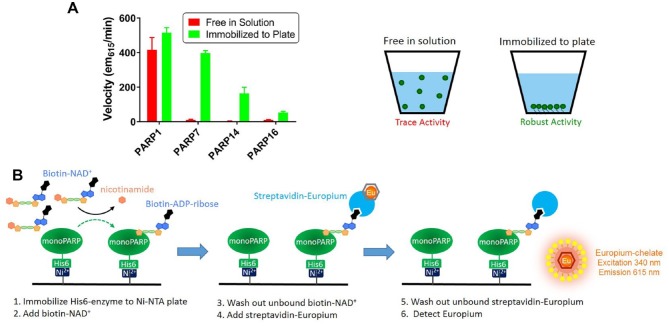
PARP enzymes are reported to self-ADP-ribosylate; however, in the literature high concentrations of proteins have been needed to observe this activity. In the work of Vyas et al.,2 self-ADP-ribosylation of nearly all monoPARP and polyPARP enzymes was observed on immunoprecipitated enzymes that were immobilized on beads and incubated with 32P-NAD+. To recapitulate this condition, recombinant purified His6-tagged PARP7, PARP14, and PARP16 enzymes were immobilized on Ni-NTA microplates, incubated with biotinylated-NAD+, and then an enzyme-linked immunosorbent assay (ELISA) readout was used to detect incorporation of biotinylated-MAR. The plates were washed prior to detection to remove the unincorporated biotinylated-NAD+ to avoid interfering with the streptavidin-based detection. Parallel reactions were run where the enzymes were incubated with biotinylated-NAD+, followed by Ni-NTA capture and ELISA detection. We observed robust activity for the monoPARPs only in conditions where the enzyme was immobilized before addition of the biotinylated-NAD+; however, PARP1 was active in both scenarios (Fig. 1). Using protein that was biotinylated with an Avi-tag, we empirically determined that the binding capacity of each well of the Ni-NTA microplate was approximately 5 pmol of protein, which corresponds to the ability to capture nearly all 2 µM of protein in a 25 µL reaction (data not shown). We compared streptavidin-coupled chemiluminescent, fluorescent, and DELFIA readouts of the biotin detection and selected DELFIA for all future studies as it gave the best combination of signal to background and reproducibility (data not shown).
Figure 1.
Immobilization of monoPARPs forces a self-modification enzymatic reaction. (A) Biotinylated-NAD+ was incubated with purified recombinant polyPARP and monoPARP enzymes that were either free in solution or immobilized to a Ni-NTA microplate. The free-in-solution reactions were transferred to a Ni-NTA microplate for capture; then all reactions were subjected to the detection of incorporation of biotinylated-ADP-ribose via the DELFIA assay protocol. The PARP1 enzyme showed robust activity in both conditions; however, the monoPARP enzymes PARP7, PARP14, and PARP16 only showed robust activity when they were immobilized prior to the reaction. (B) General procedure for performing and detecting self-ADP-ribosylation reactions on immobilized enzymes.
Development of Self-Modification Assays for Screening Inhibitors of the PARP Family
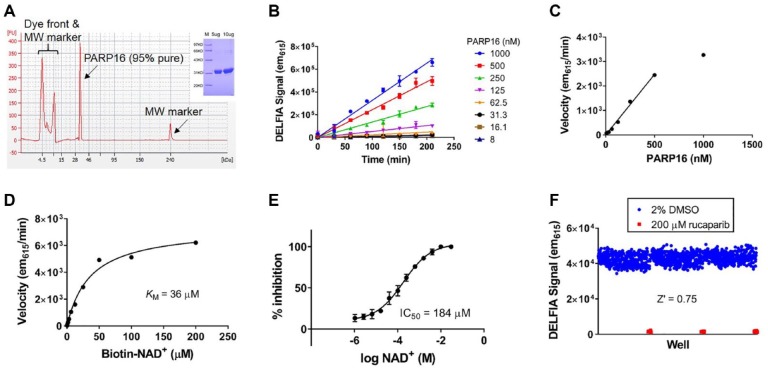
Based on the observation that immobilization led to forced self-ADP-ribosylation of PARP7, PARP14, and PARP16, we postulated that this strategy could be a family-wide solution to developing monoPARP enzyme assays without knowledge of their physiologically relevant substrates. We then subjected all reported enzymatically active monoPARP and polyPARP enzymes to assay development beginning with recombinant protein that was at least 80% pure (in most cases, purity exceeded 95%). Multiple assay development steps were performed to generate robust assays that would be sensitive to inhibitors of all mechanisms of inhibition. An example of this assay development process is demonstrated for PARP16 in Figure 2 and for the remaining PARPs in Supplemental Figures S4–S16. The biotinylated-NAD+ KMapp and IC50 values of unlabeled NAD+ under final assay conditions are tabulated in Supplemental Table S3. Assuming the IC50 of unlabeled NAD+ is an approximation for its KM, we observe that biotin-NAD+ has higher affinity for most PARP enzymes. PARP1, PARP2, and PARP3 are activated by DNA; therefore, specific DNA oligos were used in each case for assay development. The identities of the sequences are listed in Supplemental Table S2, and the amount of each oligo used in each assay is listed in Table 1 . PARP5a did not self-modify when immobilized to Ni-NTA microplates; however, similar to PARP1, we did observe activity of this enzyme on histones. We developed DELFIA screening assays for PARP5a on histones that were immobilized to high-binding microplates, and the workflow of washing and detection postenzymatic reaction was identical to that of the self-modification assays. To simplify the experimental setup and data analysis in the assay development and screening activities, the deposition of MARylation using biotinylated-NAD+ is shown in fluorescence units; however, in Supplemental Figure S17 we provide an example of a method that can be used to convert fluorescent signal to molarity using a standard curve of protein that is fully modified with a single biotin on a BirA tag.
Figure 2.
Assay development of PARP16 using the strategy of immobilizing enzyme to force self-modification. (A) A recombinant enzyme purified from Escherichia coli via a His6 tag was 91% pure as judged by capillary electrophoresis (main) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (inset). (B) The linearity of product formation versus time was evaluated at multiple enzyme concentrations. (C) The velocity versus enzyme concentration was plotted, and the linear range of this relationship goes up to 500 nM enzyme. (D) The KMapp for biotin-NAD+ was measured to be 36 µM. (E) Unlabeled NAD+ outcompetes biotin-NAD+ in the final assay conditions with an IC50 of 184 µM. (F) Uniformity experiments were performed using the final assay conditions, and the assay was robust and reproducible as judged by a Z′ of 0.75.
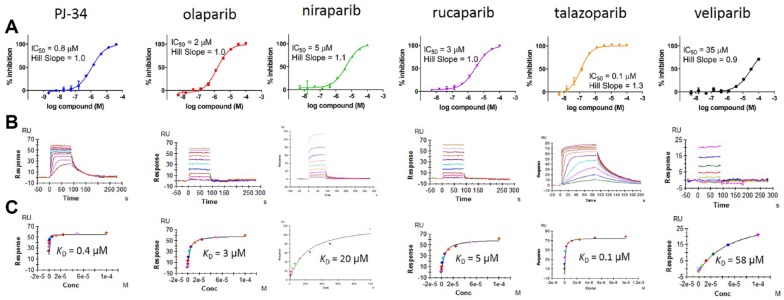
Enzyme assays were calibrated by determining the IC50 values of six key literature PARP1 inhibitors (structures shown in Suppl. Fig. S18) or a pan-monoPARP inhibitor (RBN010860; structure to be presented in a future publication) and comparing them against SPR assays developed for each corresponding PARP. An example of the IC50 curves, SPR sensorgrams, and the correlation between these assays for PARP16 is shown in Figure 3 , and for the remaining PARPs, the correlations between the enzyme and SPR assays are shown in Supplemental Figure S19. In all cases, the enzyme inhibition potency correlated well with the SPR binding affinity.
Figure 3.
Testing PARP1 literature inhibitors versus PARP16. (A) IC50 values were generated in the self-modification DELFIA assay. (B) SPR sensorgrams resulting from compound binding to PARP16 surfaces. (C) The binding affinity of compounds in SPR is derived from equilibrium fitting of the raw sensorgrams.
Development of an Assay Measuring the ADP-Ribosylation of Ubiquitin by the PARP9/DTX3L Complex
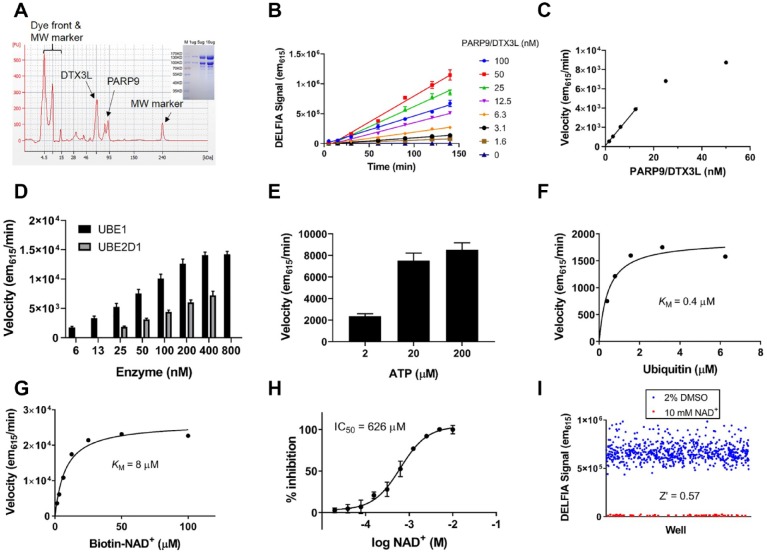
PARP9 is a macrodomain-containing PARP that heterodimerizes with DTX3L, an E3 ligase. Based on sequence alignments of its PARP domain, it has been predicted to be catalytically inactive.40 Empirical testing of this prediction via self-modification reactions has not shown PARP9 to possess enzymatic activity,2 and we have confirmed this ourselves (data not shown). However, a recent report was published showing that the PARP9/DTX3L complex MARylates the C-terminal carboxy terminus of ubiquitin when incubated with an E1 activating enzyme, an E2 conjugating enzyme, and ATP. We were able to confirm this activity using biotin-NAD+ to modify the ubiquitin and a far-Western blot readout to detect the incorporation of biotinylated-MAR (Suppl. Fig. S20A). We adapted this to a microplate-based assay using the same Ni-NTA microplates employed in the PARP self-modification assays. Enzymatic reactions were run free in solution and N-terminal His-tagged ubiquitin was subsequently captured on the Ni-NTA plates and all unbound biotinylated-NAD+ was removed by washing. The general scheme for the assay is shown in Supplemental Figure S21 and the screening assay development process for MARylation of ubiquitin by PARP9/DTX3L is shown in Figure 4 . We also confirmed that all components necessary to charge the E2 enzyme with ubiquitin are needed to see MARylation in the microplate-based assay format (Suppl. Fig. S20B).
Figure 4.
Assay development to follow ubiquitin MARylation by the PARP9/DTX3L complex. (A) Recombinant enzyme purified from Sf9 cells via a His6 tag was 80% pure as judged by capillary electrophoresis (main) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (inset). (B) Linearity of product formation versus time was evaluated at multiple enzyme concentrations. (C) Velocity versus enzyme concentration was plotted and the linear range of this relationship goes up to 13 nM enzyme. (D) Titration of the E1 (UBE1) and E2 (UBE2D1) enzymes was performed and 50 nM UBE1 and 400 nM UBE2D1 were selected for further assay development to maximize reaction velocity and conserve utilization of protein. (E) An ATP titration reveals that maximum reaction velocity is achieved at 20 µM ATP. (F) The KMapp for ubiquitin was measured to be 0.4 µM. (G) The KMapp for biotin-NAD+ was measured to be 8 µM. (H) Unlabeled NAD+ outcompetes biotin-NAD+ in the final assay conditions with an IC50 of 626 µM. (I) Uniformity experiments were performed using the final assay conditions and the assay was robust and reproducible as judged by a Z′ of 0.57.
Counterscreen Assay to Identify Compounds That Bind Nickel
Since the capture of His6-tagged PARP enzymes on Ni-NTA microplates is critical to observing enzymatic activity, compounds containing pharmacophores capable of binding to the nickel on the Ni-NTA microplates can lead to undesirable inhibition of the DELFIA signal. To quickly counterscreen compounds for this behavior, we developed a counterscreen assay examining the displacement of a biotinylated His6 peptide from the Ni-NTA microplates; the overall scheme for this assay is shown in Supplemental Figure S22.
Screening a Collection of PARP-Focused Compounds across All PARP Enzymes
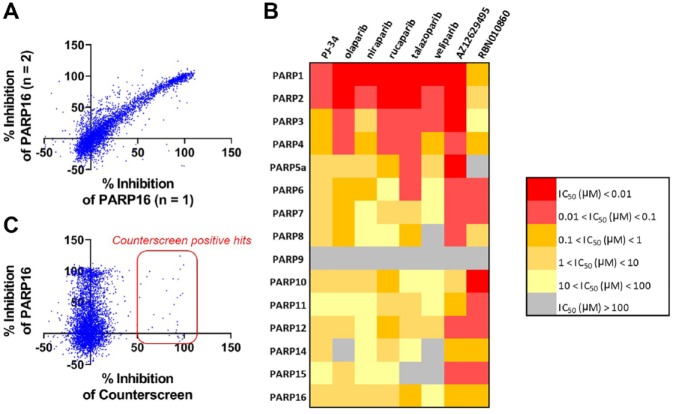
The PARP assays were used to generate potency and selectivity information across the entire enzyme family for a selection of literature PARP1 inhibitors and Ribon’s internal collection of >4000 PARP-focused inhibitors. Compounds were screened in duplicate using a single concentration screening paradigm, and selected compounds were further followed up with dose–response titrations to generate IC50 values. A correlation plot of n = 1 and n = 2 screening data for PARP16 (Fig. 5A) shows that this is a robust and reproducible assay suitable for inhibitor screening. An example of the type of potency and selectivity information that can be generated across the entire PARP family is shown for literature PARP1 inhibitors in Figure 5B , and the compounds flagged in the counterscreen are shown in Figure 5C . The structure–activity relationships across the entire PARP family will be detailed in future publications.
Figure 5.
Screening a library of PARP inhibitors against PARP16. (A) The PARP16 self-modification assay was used to screen a library of >4000 PARP-focused inhibitors at a concentration of 1 µM. Since the library is not a traditional diversity library and is enriched for PARP inhibitor templates, an arbitrary 50% inhibition cutoff was used, identifying 764 hits, for a hit rate of 19.1%. The correlation of percent inhibition between duplicate runs shows that the assay was robust in hit identification. (B) A heatmap depicting the potency of key literature PARP1 inhibitors (PJ-34, olaparib, niraparib, rucaparib, talazoparib, veliparib), a potent literature PARP inhibitor (AZ12629495), and a Ribon-designed pan-monoPARP inhibitor (RBN010860) across the entire PARP family. (C) A counterscreen to identify compounds that bind to nickel and prevent the immobilization of the PARP enzymes required to induce the forced self-modification reaction identifies 69 hits, for a hit rate of 0.9%.
Discussion
ADP-ribosylation is increasingly being recognized for playing a key role in stress response pathways in the context of immune function, inflammation, and cancer.12–14 The four known polyPARP enzymes have been extensively studied, and in the case of PARP1 and PARP2, Food and Drug Administration (FDA)-approved cancer drugs targeting those enzymes exist; however, much less is known about the 12 monoPARP enzymes. The lack of a clear understanding of how the monoPARP enzymes interact with their substrates has been a barrier to developing robust high-throughput assays that would enable a family-wide approach to generating chemical probes to study the function of these enzymes. We tested the ability of multiple monoPARPs to MARylate their published substrates by incubating recombinant proteins or synthetic peptides in the presence of 32P-NAD+ or biotinylated-NAD+. In most cases, we did not observe any detectable incorporation of MAR and determined that another paradigm to screening PARP enzymes was needed for this protein family.
PARP enzymes are noted to modify themselves with MAR; however, when we attempted to perform self-modification reactions using radioactive and ELISA-based formats, many PARPs showed little to no measurable activity and assay development was not possible. We discovered that when PARPs are immobilized via His6 tags onto Ni-NTA-coated microplates, we were able to observe robust self-modification that was detectable using a DELFIA readout. We hypothesize that immobilization of the PARPs on the microplate surface induces molecular crowding, which overcomes weak KM values for these interactions to generate detectable levels of self-modification. DELFIA assays are a variation of an ELISA that relies on lanthanide fluorescence to detect incorporation of a biotinylated ADP-ribose moiety. We compared DELFIA to other ELISA formats such as colorimetric, fluorometric, and chemiluminescence, and DELFIA gave the most reproducible data with the highest Z′ factors. Using this approach, we were able to develop robust self-modification assays for nearly all monoPARPs and polyPARPs. Ultimately, immobilization as a means to force self-modification may be a useful approach in the development of assays for targets where substrate information is lacking.
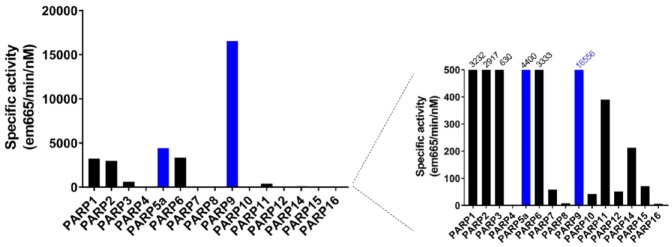
Notable exceptions to the paradigm of using self-modification reactions to generate a screening assay were observed for PARP5a and the PARP9/DTX3L complex. In the case of PARP5a, we found that the enzyme was able to PARylate immobilized histones, and we developed DELFIA screening assays with a similar detection workflow to the self-MARylation assays on histone-coated microplates. PARP9 and DTX3L, an E3 ligase, are located on chromosome 3q21 in a head-to-head orientation and are regulated by an IFN-γ responsive bidirectional promoter.41 Those proteins are always found in a heterodimer and we were not able to express and purify PARP9 without co-expressing DTX3L and purifying as a complex. Recently, it was shown that the PARP9/DTX3L complex MARylates the free C-terminal carboxylic acid on glycine 76 of ubiquitin only under conditions where the E2 enzyme is charged with ubiquitin.28 Since glycine 76 is normally used to conjugate ubiquitin to proteins, this modification precludes this ubiquitination in an NAD+-dependent manner. We were able to confirm these findings using a far-Western blot to visualize the ADP-ribosylation of ubiquitin and subsequently developed a microplate-based assay where the enzymatic reaction is run with all components free in solution followed by capture and detection of the ADP-ribosylated products. The PARP9/DTX3L reaction specific activity is several orders of magnitude larger than any self-modification reaction using immobilized PARP enzymes and underscores the need to perform further studies to identify monoPARP substrates (Fig. 6). This suggests that the in vitro catalytic efficiency of the monoPARP enzymes could be much greater if they are incubated with their natural substrates, and potentially in complex with other proteins that facilitate interaction with their substrates. There is precedent for PARP1 and PARP2 to require the accessory factor HPF1 to selectively PARylate serine residues in target proteins such as histones,42,43 and there may be a similar requirement by monoPARPs that is not recapitulated in our reductionist approach. Recent work has also shown that in vitro, 3′ and 5′ phosphates in nucleic acids can also be modified by PARP enzymes,44–47 and this may be another avenue to consider when developing PARP assays.
Figure 6.
Comparing specific activities of PARP enzymes under final assay conditions. The specific activity for each enzyme assay was calculated. The forced self-modification of immobilized enzymes (black bars) overall showed lowered specific activity than assays where an enzyme and its substrate were allowed to react freely in solution (blue bars).
Using assays generated for nearly the entire PARP family, we are able to rapidly and efficiently screen inhibitors in parallel to identify hits for each PARP as well as to build structure–activity relationships within the family. The ability of the assays to accurately assess inhibitor potency was validated by generating SPR assays for all PARPs and comparing IC50 values from the enzyme assays generated at KMapp or as close to KMapp as possible. The fact that SPR assays could be generated for nearly all PARPs tested also indicates the amenability of these proteins to biophysical approaches for hit identification, such as fragment screening. The strategies presented here are being used to generate potent and selective chemical probes for each family member to enable interrogation of the role of their catalytic activity in disease biology, and ultimately develop targeted therapeutics.
Supplemental Material
Supplemental material, Wigle_DELFIA_and_automodification_Supp_Info_Revised for Forced Self-Modification Assays as a Strategy to Screen MonoPARP Enzymes by Tim J. Wigle, W. David Church, Christina R. Majer, Kerren K. Swinger, Demet Aybar, Laurie B. Schenkel, Melissa M. Vasbinder, Arne Brendes, Claudia Beck, Martin Prahm, Dennis Wegener, Paul Chang and Kevin W. Kuntz in SLAS Discovery
Acknowledgments
We thank Mr. Phil Anwer for his efforts to obtain the custom Ni-NTA-coated microplates, Mr. Andrew Santospago for preparing compound plates, and Dr. Heike Keilhack and Dr. Victoria Richon for helpful discussions and critical review of this manuscript. Additionally, we are grateful to Ping Wei and her team at Viva Biotech and Hara Black and her team at Evotec SE for their contributions to protein production efforts.
Footnotes
Supplemental material is available online with this article.
Declaration of Conflicting Interests: The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: T.J.W., W.D.C., C.R.M., K.K.S., D.A., L.B.S., M.M.V., P.C., and K.W.K. are current or former employees and stockholders of Ribon Therapeutics Inc.; A.B., C.B., M.P., and D.W. are employees of Evotec SE.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Leslie Pedrioli D. M., Leutert M., Bilan V.; et al. Comprehensive ADP-Ribosylome Analysis Identifies Tyrosine as an ADP-Ribose Acceptor Site. EMBO Rep. 2018, 19, 45310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vyas S., Matic I., Uchima L.; et al. Family-Wide Analysis of Poly(ADP-Ribose) Polymerase Activity. Nat. Commun. 2014, 5, 4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindgren A. E., Karlberg T., Thorsell A. G.; et al. PARP Inhibitor with Selectivity toward ADP-Ribosyltransferase ARTD3/PARP3. ACS Chem. Biol. 2013, 8, 1698–1703. [DOI] [PubMed] [Google Scholar]
- 4. Johannes J. W., Almeida L., Daly K.; et al. Discovery of AZ0108, an Orally Bioavailable Phthalazinone PARP Inhibitor That Blocks Centrosome Clustering. Bioorg. Med. Chem. Lett. 2015, 25, 5743–5747. [DOI] [PubMed] [Google Scholar]
- 5. Venkannagari H., Verheugd P., Koivunen J.; et al. Small-Molecule Chemical Probe Rescues Cells from Mono-ADP-Ribosyltransferase ARTD10/PARP10-Induced Apoptosis and Sensitizes Cancer Cells to DNA Damage. Cell Chem. Biol. 2016, 23, 1251–1260. [DOI] [PubMed] [Google Scholar]
- 6. Carlile G. W., Robert R., Matthes E.; et al. Latonduine Analogs Restore F508del-Cystic Fibrosis Transmembrane Conductance Regulator Trafficking through the Modulation of Poly-ADP Ribose Polymerase 3 and Poly-ADP Ribose Polymerase 16 Activity. Mol. Pharmacol. 2016, 90, 65–79. [DOI] [PubMed] [Google Scholar]
- 7. Peng B., Thorsell A. G., Karlberg T.; et al. Small Molecule Microarray Based Discovery of PARP14 Inhibitors. Angew. Chem. Int. Ed. Engl. 2017, 56, 248–253. [DOI] [PubMed] [Google Scholar]
- 8. Yoneyama-Hirozane M., Matsumoto S. I., Toyoda Y.; et al. Identification of PARP14 Inhibitors Using Novel Methods for Detecting Auto-Ribosylation. Biochem. Biophys. Res. Commun. 2017, 486, 626–631. [DOI] [PubMed] [Google Scholar]
- 9. Morgan R. K., Kirby I. T., Vermehren-Schmaedick A.; et al. Rational Design of Cell-Active Inhibitors of PARP10. ACS Med. Chem. Lett. 2019, 10, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirby I. T., Kojic A., Arnold M. R.; et al. A Potent and Selective PARP11 Inhibitor Suggests Coupling between Cellular Localization and Catalytic Activity. Cell Chem. Biol. 2018, 25, 1547–1553e12. [DOI] [PubMed] [Google Scholar]
- 11. Haikarainen T., Krauss S., Lehtio L. Tankyrases: Structure, Function and Therapeutic Implications in Cancer. Curr. Pharm. Des. 2014, 20, 6472–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vyas S., Chang P. New PARP Targets for Cancer Therapy. Nat. Rev. Cancer 2014, 14, 502–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Butepage M., Eckei L., Verheugd P.; et al. Intracellular Mono-ADP-Ribosylation in Signaling and Disease. Cells 2015, 4, 569–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iwata H., Goettsch C., Sharma A.; et al. PARP9 and PARP14 Cross-Regulate Macrophage Activation via STAT1 ADP-Ribosylation. Nat. Commun. 2016, 7, 12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bock F. J., Todorova T. T., Chang P. RNA Regulation by Poly(ADP-Ribose) Polymerases. Mol. Cell 2015, 58, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Venkannagari H., Fallarero A., Feijs K. L.; et al. Activity-Based Assay for Human Mono-ADP-Ribosyltransferases ARTD7/PARP15 and ARTD10/PARP10 Aimed at Screening and Profiling Inhibitors. Eur. J. Pharm. Sci. 2013, 49, 148–156. [DOI] [PubMed] [Google Scholar]
- 17. Thorsell A. G., Ekblad T., Karlberg T.; et al. Structural Basis for Potency and Promiscuity in Poly(ADP-Ribose) Polymerase (PARP) and Tankyrase Inhibitors. J. Med. Chem. 2017, 60, 1262–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen J., Lam A. T., Zhang Y. A Macrodomain-Linked Immunosorbent Assay (MLISA) for Mono-ADP-Ribosyltransferases. Anal. Biochem. 2018, 543, 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ji M., Wang L., Xue N.; et al. The Development of a Biotinylated NAD(+)-Applied Human Poly(ADP-Ribose) Polymerase 3 (PARP3) Enzymatic Assay. SLAS Discov. 2018, 23, 545–553. [DOI] [PubMed] [Google Scholar]
- 20. Wahlberg E., Karlberg T., Kouznetsova E.; et al. Family-Wide Chemical Profiling and Structural Analysis of PARP and Tankyrase Inhibitors. Nat. Biotechnol. 2012, 30, 283–288. [DOI] [PubMed] [Google Scholar]
- 21. Yuen L. H., Dana S., Liu Y.; et al. A Focused DNA-Encoded Chemical Library for the Discovery of Inhibitors of NAD+-Dependent Enzymes. J. Am. Chem. Soc. 2019, 141, 5169–5181. [DOI] [PubMed] [Google Scholar]
- 22. Ekblad T., Verheugd P., Lindgren A. E.; et al. Identification of Poly(ADP-Ribose) Polymerase Macrodomain Inhibitors Using an AlphaScreen Protocol. SLAS Discov. 2018, 23, 353–362. [DOI] [PubMed] [Google Scholar]
- 23. Feijs K. L., Kleine H., Braczynski A.; et al. ARTD10 Substrate Identification on Protein Microarrays: Regulation of GSK3beta by Mono-ADP-Ribosylation. Cell Commun. Signal. 2013, 11, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carter-O’Connell I., Cohen M. S. Identifying Direct Protein Targets of Poly-ADP-Ribose Polymerases (PARPs) Using Engineered PARP Variants—Orthogonal Nicotinamide Adenine Dinucleotide (NAD+) Analog Pairs. Curr. Protoc. Chem. Biol. 2015, 7, 121–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bindesboll C., Tan S., Bott D.; et al. TCDD-Inducible Poly-ADP-Ribose Polymerase (TIPARP/PARP7) Mono-ADP-Ribosylates and Co-Activates Liver X Receptors. Biochem. J. 2016, 473, 899–910. [DOI] [PubMed] [Google Scholar]
- 26. Carter-O’Connell I., Jin H., Morgan R. K.; et al. Identifying Family-Member-Specific Targets of Mono-ARTDs by Using a Chemical Genetics Approach. Cell Rep. 2016, 14, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu A. Z., Abo R., Ren Y.; et al. Enabling Drug Discovery for the PARP Protein Family through the Detection of Mono-ADP-Ribosylation. Biochem. Pharmacol. 2019, 167, 97–106. [DOI] [PubMed] [Google Scholar]
- 28. Yang C. S., Jividen K., Spencer A.; et al. Ubiquitin Modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9. Mol. Cell 2017, 66, 503–516.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mikule K., Wang Z. Treatment of Cancer. WO 2016116602, July 28, 2016. [Google Scholar]
- 30. Langelier M. F., Riccio A. A., Pascal J. M. PARP-2 and PARP-3 Are Selectively Activated by 5′ Phosphorylated DNA Breaks through an Allosteric Regulatory Mechanism Shared with PARP-1. Nucleic Acids Res. 2014, 42, 7762–7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eustermann S., Wu W. F., Langelier M. F.; et al. Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1. Mol. Cell 2015, 60, 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang J. H., Chung T. D., Oldenburg K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen. 1999, 4, 67–73. [DOI] [PubMed] [Google Scholar]
- 33. Yamada T., Horimoto H., Kameyama T.; et al. Constitutive Aryl Hydrocarbon Receptor Signaling Constrains Type I Interferon-Mediated Antiviral Innate Defense. Nat. Immunol. 2016, 17, 687–694. [DOI] [PubMed] [Google Scholar]
- 34. Diani-Moore S., Zhang S., Ram P.; et al. Aryl Hydrocarbon Receptor Activation by Dioxin Targets Phosphoenolpyruvate Carboxykinase (PEPCK) for ADP-Ribosylation via 2,3,7,8-Tetrachlorodibenzo-p-Dioxin (TCDD)-Inducible Poly(ADP-Ribose) Polymerase (TiPARP). J. Biol. Chem. 2013, 288, 21514–21525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cho S. H., Goenka S., Henttinen T.; et al. PARP-14, a Member of the B Aggressive Lymphoma Family, Transduces Survival Signals in Primary B Cells. Blood 2009, 113, 2416–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Begitt A., Cavey J., Droescher M.; et al. On the Role of STAT1 and STAT6 ADP-Ribosylation in the Regulation of Macrophage Activation. Nat. Commun. 2018, 9, 2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carter-O’Connell I., Vermehren-Schmaedick A., Jin H.; et al. Combining Chemical Genetics with Proximity-Dependent Labeling Reveals Cellular Targets of Poly(ADP-Ribose) Poly-merase 14 (PARP14). ACS Chem. Biol. 2018, 13, 2841–2848. [DOI] [PubMed] [Google Scholar]
- 38. Jwa M., Chang P. PARP16 Is a Tail-Anchored Endoplasmic Reticulum Protein Required for the PERK- and IRE1alpha-Mediated Unfolded Protein Response. Nat. Cell Biol. 2012, 14, 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wacker D. A., Ruhl D. D., Balagamwala E. H.; et al. The DNA Binding and Catalytic Domains of Poly(ADP-Ribose) Polymerase 1 Cooperate in the Regulation of Chromatin Structure and Transcription. Mol. Cell. Biol. 2007, 27, 7475–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kleine H., Poreba E., Lesniewicz K.; et al. Substrate-Assisted Catalysis by PARP10 Limits Its Activity to Mono-ADP-Ribosylation. Mol. Cell 2008, 32, 57–69. [DOI] [PubMed] [Google Scholar]
- 41. Juszczynski P., Kutok J. L., Li C.; et al. BAL1 and BBAP Are Regulated by a Gamma Interferon-Responsive Bidirectional Promoter and Are Overexpressed in Diffuse Large B-Cell Lymphomas with a Prominent Inflammatory Infiltrate. Mol. Cell. Biol. 2006, 26, 5348–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bonfiglio J. J., Fontana P., Zhang Q.; et al. Serine ADP-Ribosylation Depends on HPF1. Mol. Cell 2017, 65, 932–940e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gibbs-Seymour I., Fontana P., Rack J. G. M.; et al. HPF1/C4orf27 Is a PARP-1-Interacting Protein That Regulates PARP-1 ADP-Ribosylation Activity. Mol. Cell 2016, 62, 432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Munnur D., Ahel I. Reversible Mono-ADP-Ribosylation of DNA Breaks. FEBS J. 2017, 284, 4002–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zarkovic G., Belousova E. A., Talhaoui I.; et al. Characterization of DNA ADP-Ribosyltransferase Activities of PARP2 and PARP3: New Insights into DNA ADP-Ribosylation. Nucleic Acids Res. 2018, 46, 2417–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Munnur D., Bartlett E., Mikolcevic P.; et al. Reversible ADP-Ribosylation of RNA. Nucleic Acids Res. 2019, 47, 5658–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Belousova E. A., Ishchenko A. C., Lavrik O. I. DNA Is a New Target of Parp3. Sci. Rep. 2018, 8, 4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Wigle_DELFIA_and_automodification_Supp_Info_Revised for Forced Self-Modification Assays as a Strategy to Screen MonoPARP Enzymes by Tim J. Wigle, W. David Church, Christina R. Majer, Kerren K. Swinger, Demet Aybar, Laurie B. Schenkel, Melissa M. Vasbinder, Arne Brendes, Claudia Beck, Martin Prahm, Dennis Wegener, Paul Chang and Kevin W. Kuntz in SLAS Discovery