Abstract
Background: Acute right heart failure (RHF) is the main cause of death in patients with acute pulmonary embolism and emergent pulmonary hypertension. However, the molecular mechanisms underpinning the acute RHF and the interactions between the right (RV) and left ventricles (LVs) under the diseased condition remain unknown. Methods and results: The Sprague Dawley male rats were randomly divided into the normal control, sham, and pulmonary artery banding (PAB) groups. One hour after the PAB operation, after measuring the haemodynamic and anatomical parameters, the free walls of RV and LV were harvested to detect the differential gene expression profiling by high-throughput RNA sequencing. The results showed that the PAB lead to 50–60% obstruction of the main pulmonary artery, which was accompanied by the significant elevation in the positive rate of rise in RV pressure and the maximum RV pressure as compared to the sham group. Moreover, compared with the counterparts in the sham group, the RV and LV in the PAB group exhibited 2057 differentially expressed genes (DEGs, 1159 upregulated and 898 downregulated) and 1196 DEGs (709 upregulated and 487 downregulated), respectively (DEG criteria: |log2 fold change| ≥1, q value ≤0.05). In comparison to the sham group, the enriched pathways in the PAB group include nuclear factor-κB signalling pathway, extracellular matrix–receptor interaction, and nucleotide oligomerization domain-like receptor signalling pathway. Conclusions: The PAB rat model exhibited the haemodynamic and gene expression changes in the RV that lead to acute RHF. Further, the acute RHF induced by pressure overload also caused gene expression changes in the LV, suggesting the molecular interactions between the RV and LV under the diseased condition.
Keywords: right heart failure, pulmonary artery banding, RNA-sequencing, pulmonary embolism, bioinformatics
Introduction
Acute right heart failure (RHF) is a complex syndrome with structural and functional cardiac abnormalities of the right ventricle (RV).1 RHF is a part of heart failure, whose mortality rate was 17% within 12 months of hospitalization.2 The aetiologies of acute RHF include acute pulmonary embolism (APE), sepsis, emergent pulmonary hypertension and cardiac surgery.1,3 APE, one of the most common cardiovascular diseases, can cause a sharp increase in the right ventricular afterload and rapidly progress to acute RHF.4,5 As a consequence, the acute RHF has become a significant cause of death in the APE patients.5 Moreover, mechanical ventilation after thoracotomy may also contribute to RHF by activating inflammatory factors.6,7
Several differences in anatomical structures and physiological functions exist between the RV and left ventricle (LV). More specifically, the myocardium of RV is thinner and more compliant than that of LV, hence RV is less tolerant to a sharp increase of afterload due to the weaker contractility. In contrast, LV tolerates high afterload and functions as the main driving force for systemic circulation owing to the thicker ventricle and greater contractility.8 These anatomical and functional characteristics indicate that RV is more resistant to the abrupt changes in volume than the changes in pressure, whereas LV is the opposite. However, the differential responses between LV and RV as well as the interactions between the two ventricles under the diseased conditions, in particular the condition of RHF, remain unknown.
RNA sequencing (RNA-seq), based on the high-throughput and deep-sequencing technology, has become a powerful tool to study gene expression profiling and molecular mechanisms at the transcript level.9 The RNA-seq has advantages over conventional approaches, it provides huge amount of information on gene expression and is free from temporal and spatial restrictions accompanied by the conventional gene expression analysis approaches, such as quantitative real-time PCR and gene chips. Moreover, it pinpoints the sequence variations to the resolution of a single nucleotide, including the substantial information on single nucleotide polymorphisms (SNPs).9
In this study, a rat model of acute RHF was established through a short-term pulmonary artery banding (PAB), following the measurement of morphological and haemodynamic parameters, the gene expression profiling of the free walls of RV and LV was detected by the high-throughput RNA-seq. The sequencing data were analysed by algorithms of bioinformatics, such as gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG), aiming to clarify the differential gene expression patterns between the RV and LV, to identify the molecular pathways mediating the interactions between the two ventricles under the acute RHF induced by the abrupt increase of afterload, and to find the potential therapeutic targets to this fatal disease.
Materials and methods
Animals
Fifteen four-week-old male Sprague Dawley rats were purchased from the Animal Core Facility of Nanjing Medical University. The rats were housed in a temperature- and humidity-conditioned facility (25 ± 1℃ with the relative humidity of 40–70%) and fed ad lib with food and water. The illumination was under 12 h light–dark cycles. All experimental procedures conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. The study was approved by the Ethical Committee of Gansu Provincial Hospital.
Establishment of the rat model of acute right heart failure
The animals were randomly divided into three groups: normal control group, sham group, and PAB group (n = 5 / group). In the sham and PAB groups, each rat was subjected to general anaesthesia by intraperitoneal administration of sodium pentobarbital according to the body weight (50 mg/kg). The RV catheterization and PAB were performed as previously described.10 Briefly, the rats were intubated with a 16-gauge Teflon tube and attached to a mechanical ventilator (Micro-Ventilator, UNO, Zevenaar, Netherlands) with the breathing frequency of 80 breaths / min, the pressures at 9/0 cmH2O, and the inspiratory/expiratory ratio of 1:1. The heads and limbs of the animals were fixed on an operating table. A middle thoracotomy was performed to access the RV. After the steady state was reached, the pressures of RV, including the positive and negative rate of rise in RV pressure (±dP/dT), minimum RV pressure (RVP min), maximum RV pressure (RVP max), as well as other physiological parameters, such as systolic blood pressure (SBP), diastolic blood pressure (DBP), and heart rate (HR) were recorded by a high-fidelity catheter tip transducer (Mikro-Tip SPR671, Millar Instruments, Houston, TX) over an interval of 10 s and averaged. Then the main pulmonary artery was carefully exposed, and its diameter measured by a Vernier caliper. A blunt needle with an appropriate gauge was selected based on the artery diameter and the percentage of the artery to be obstructed. The selected needle was placed in parallel to the main pulmonary artery. Then a vessel clip was applied, the cross section shape of the main pulmonary artery immediately turned from circular to elliptic, with the short shaft of the ellipse equal to the needle gauge. Thereby a similar degree of banding (50–60%) could be achieved among individual animals. In addition, two knots were made around the pulmonary artery and the vessel clip to secure the vascular banding. The rats in the sham group were subjected to the identical treatments except the vascular clip application.
After 1 h of the PAB, the ligation diameter, pulmonary artery radius, elliptic short half-shaft, elliptic long half-shaft, and the above-mentioned pressures were measured in the animals of the sham and PAB groups. Then, all the rats were sacrificed, the heart weight was measured. The RV from the three experimental groups (n = 3/group) were collected and processed for Masson’s trichrome staining as described elsewhere.11 Then the 12 free wall samples, including RV of normal control 1 (NC1R), LV of normal control 1 (NC1L), RV of normal control 2 (NC2R), LV of normal control 2 (NC2L), RV of sham 1 (S1R), LV of sham 1 (S1L), RV of sham 2 (S2R), LV of sham 2 (S2L), RV of PAB 1 (P1R), LV in PAB 1 (P1L), RV of PAB 2 (P2R), and LV in PAB 2 (P2L), were harvested and immediately frozen in liquid nitrogen and stored at −80℃.
RNA extraction and library construction
All samples were delivered through cold chain to Vazyme Biotech Co., Ltd for processing and RNA-seq. First, total RNA was extracted from all the samples using TRIzol (Thermo Fisher Scientific, Waltham, MA) following the company’s protocols. The extracted RNA was subjected to quality control assessments and the mRNA was enriched with mRNA capture beads. Then first-strand cDNA was synthesized using the mRNA as template and random hexamers as primers, the second strand of cDNA was subsequently generated using RNase H and DNA polymerase I. The double strand cDNA was purified by VAHTSTM DNA clean beads. The purified cDNA was repaired at both termini, added with poly-A tails and sequencing adaptors. The resulting cDNA fragments were then size-selected using VAHTSTM DNA Clean Beads and amplified by PCR. The purified PCR products constituted the libraries to be sequenced, which underwent another round of quality control examination on an Agilent 2100 Bioanalyzer. Finally, the libraries were sequenced using the Illumina HiSeq platform (Fig. 1a).12
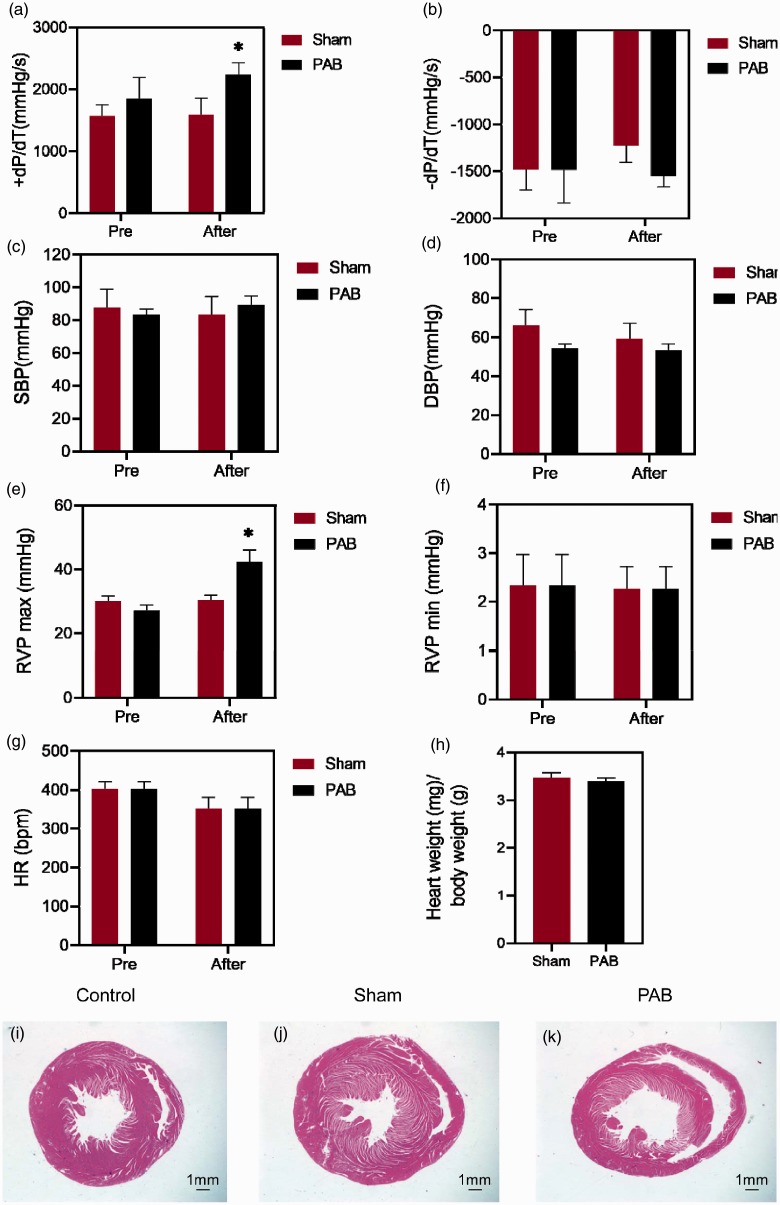
Fig. 1.
Functional characterizations of the PAB model. (a) +dP/dT; (b) −dP/dT; (c) SBP; (d) DBP; (e) RVP max; (f) RVP min; (g) HR; (h) ratio of heart weight over body weight in the sham and PAB groups. (i–k) Representative pictures of staining of the RV paraffin sections in the normal control, sham and PAB groups. Scale bar = 1.0 mm. PAB: pulmonary artery banding. *p < 0.05, sham vs. PAB. n = 5 / group.
Bioinformatics
The original sequencing data (raw reads) were pre-filtered to remove the reads with adapters, the reads containing more than 5% unidentifiable bases, and the reads comprising more than 50% bases with the mass value less than 10 (Q ≤ 10), whereby the high-quality sequencing data (clean reads) were obtained. Then the clean reads with a minimum 200-bp overlaps were aligned to the reference sequences, which subserved the second quality control of bioinformatics analysis. The results of this quality control depended upon the distribution and coverage of the clean reads on the reference sequences.
Subsequently, the analyses of differentially expressed genes (DEGs) and SNPs were performed. The abundance of each gene transcript was measured by the number of fragments per kilo bases per million reads, which was calculated by the software Cufflink v2.1.1. The genes whose transcript abundance in different groups met the criteria of |log2 fold change| ≥1 and q value ≤0.05 were deemed as DEGs between the two experimental groups. In addition, the GO enrichment, protein interaction network, and KEGG pathways were analysed.
Statistics
Statistical analyses were performed using Statistical Program for Social Sciences 20.0 (IBM SPSS Inc., New York, NY). All the data related to anatomical and haemodynamic parameters were expressed as mean ± SEM. The data were examined by D’Agostino and Pearson omnibus normality test to confirm normal distribution, after which by Levene test to confirm homogeneity of variance. Then the differences in the parameters between the normal control, sham, and PAB groups were analysed by one-way ANOVA followed by Tukey post hoc. A p value less than 0.05 was considered significant.
Results
Functional and morphological characterizations of the PAB model
One hour after PAB, the +dP/dT and RVP max were significantly elevated in the PAB group as compared to those in the sham group, respectively (Fig. 1a and e; both p < 0.05, sham after vs. PAB after). There was a trendy increase in the SBP in the PAB group in comparison to the sham group, but the difference had no statistical significance (Fig. 1c; p > 0.05, sham after vs. PAB after). The HR in both sham and PAB groups exhibited a reduced trend following the surgical treatments, although the changes were not significant (Fig. 1g; p > 0.05, sham after vs. PAB after). Other parameters, such as −dP/dT, RV min, DBP, and the ratio of heart weight over body weight did not exhibit significant changes during the short term of PAB (Fig. 1b, d, f, h; all p > 0.05, sham after vs. PAB after). There was no apparent difference in the morphology of the RV sections from the three experimental groups (Fig. 1i–k). On the other hand, the anatomical changes in pulmonary artery were also measured (Table 1). As excepted, the ligation diameter and ligation degree were drastically increased in the PAB group as compared to the sham group (p < 0.05, sham vs. PAB). In comparison to the sham group, the pulmonary artery radius, the ellipse long and short half shafts in the PAB group were insignificantly increased (all p > 0.05, sham vs. PAB). There was no significant difference in the body weight between the two groups either (p = 0.069, sham vs. PAB).
Table 1.
Morphometric parameters of sham and PAB groups.
| Morphometric parameters | Sham (n = 5) | PAB (n = 5) |
|---|---|---|
| Ligation diameter (mm) | 0.00 | 1.32 ± 0.02*** |
| Pulmonary artery radius (mm) | 2.03 ± 0.01 | 2.10 ± 0.03 |
| Elliptic short half-shaft (mm) | NA | 0.66 ± 0.01 |
| Elliptic long half-shaft (mm) | NA | 2.92 ± 0.05 |
| Ligation degree (%) | 0.00 ± 0.00 | 56.35 ± 0.48*** |
| Body weight (g) | 241.75 ± 5.38 | 244.00 ± 3.00 |
PAB: pulmonary artery banding; NA: not applicable.
p < 0.001.
General information on transcriptomes
The minus ratio of raw reads/clean reads was 99.20%, and the GC content was 47–49% in the transcriptomes of all experimental groups. In all the groups, the mean percentages of the bases with Q ≥ 20 and Q ≥ 30 were 97.83% and 95.04%, respectively, indicating the good quality of the sequencing data (Table 2). On average, 86.08% of the clean reads in all groups were distributed in exons; whereas 6.28% and 7.64% of them were located in the intronic and intergenic regions, respectively (Table 3).
Table 2.
Reads information in different samples.
| Samples | Raw reads | Clean reads | Ratio | Reads length | Bases | GC | Q20 | Q30 |
|---|---|---|---|---|---|---|---|---|
| C1L | 57865004 | 57480604 | 99.34% | 150 bp | 8622090600 | 47.79% | 97.84% | 95.06% |
| C1R | 53335642 | 53025064 | 99.42% | 150 bp | 7953759600 | 48.02% | 97.47% | 94.36% |
| S1L | 48589272 | 48250418 | 99.30% | 150 bp | 7237562700 | 48.23% | 97.85% | 95.09% |
| S1R | 48626830 | 48309532 | 99.35% | 150 bp | 7246429800 | 47.86% | 97.76% | 94.94% |
| P1L | 46231212 | 45922140 | 99.33% | 150 bp | 6888321000 | 48.13% | 98.00% | 95.36% |
| P1R | 73309370 | 72867478 | 99.40% | 150 bp | 10930121700 | 48.34% | 98.27% | 95.97% |
| C2L | 63745644 | 63270048 | 99.25% | 150 bp | 9490507200 | 48.62% | 97.69% | 94.77% |
| C2R | 50041970 | 49712312 | 99.34% | 150 bp | 7456846800 | 48.26% | 97.69% | 94.78% |
| S2L | 42284120 | 41983762 | 99.29% | 150 bp | 6297564300 | 48.51% | 97.78% | 94.92% |
| S2R | 45628056 | 45277990 | 99.23% | 150 bp | 6791698500 | 48.34% | 97.83% | 95.03% |
| P2L | 47019802 | 46645544 | 99.20% | 150 bp | 6996831600 | 48.36% | 97.84% | 95.05% |
| P2R | 51322682 | 50941748 | 99.26% | 150 bp | 7641262200 | 48.45% | 97.91% | 95.20% |
Ratio” is Clean reads as a percentage of Raw reads. “Bases” is the number of bases (product of Clean reads and Read length), which was used for high-quality sequencing. “GC” is GC content of high-quality sequencing data. “Q20” and “Q30” are percentages of the bases with mass value not less than 20 and 30, respectively.
Table 3.
Distribution of the reads in the genome.
| Regions (%) | C1L | C1R | S1L | S1R | P1L | P1R | C2L | C2R | S2L | S2R | P2L | P2R |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon | 86.71 | 85.17 | 87.41 | 83.27 | 88.89 | 85.06 | 88.84 | 84.19 | 88.52 | 83.41 | 88.19 | 83.26 |
| Intronic | 5.61 | 6.96 | 5.48 | 8.58 | 4.14 | 7.00 | 3.93 | 7.78 | 4.28 | 8.50 | 4.62 | 8.49 |
| intergenic | 7.68 | 7.87 | 7.11 | 8.15 | 6.98 | 7.94 | 7.24 | 8.03 | 7.20 | 8.09 | 7.20 | 8.24 |
C: normal control; S: sham; P: pulmonary artery banding; L: free wall of left ventricle; R: free wall of right ventricle.
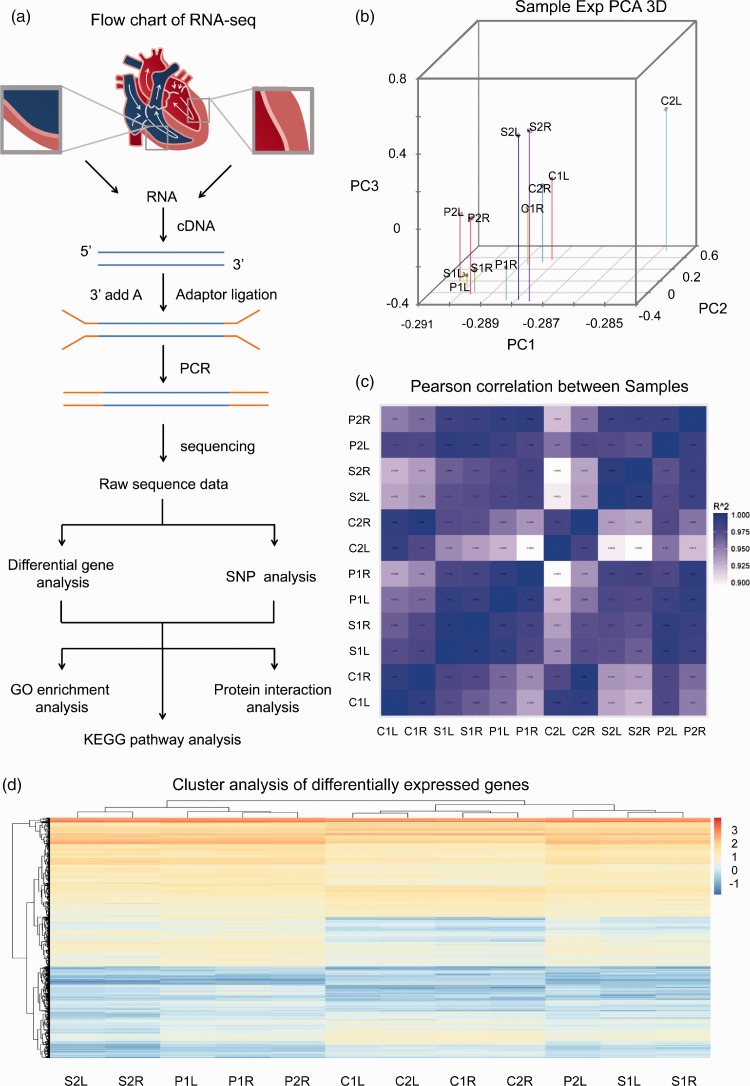
On the other hand, the principal component analysis showed that the variations were small between the two samples from the same group (Fig. 2b). Moreover, the correlation analysis showed that the correlation coefficients of the two samples from the same group were more than 0.9 (Fig. 2c). The results of these two analyses indicate the reliability of experimental performance and the rationality of sample selection in the current study. The heat map of cluster analysis showed the general information on DEGs in each group, both the sham and PAB treatment resulted in a trend of upregulation in gene expression as compared to the normal controls (Fig. 2d).
Fig. 2.
The RNA-seq analysis of the 12 samples from experimental groups. (a) Flow chart of RNA-seq. (b) The principal component analysis (PCA) 3D diagram of different genes in 12 samples. Wherein the position of each dot represents the value of the sample on each principal component, the different colours represent different samples. (c) RNA-seq correlation heat map among the 12 samples. The closer the correlation coefficient to 1 indicates the greater similarity between the expression patterns of the samples. (d) The heat map deciphering the cluster analysis of the DEGs between different experimental groups. C refers to normal control group; S refers to sham group; P refers to PAB group; R stands for the free walls of right ventricle; L stands for the free walls of left ventricle.
The differentially expressed genes among experimental groups
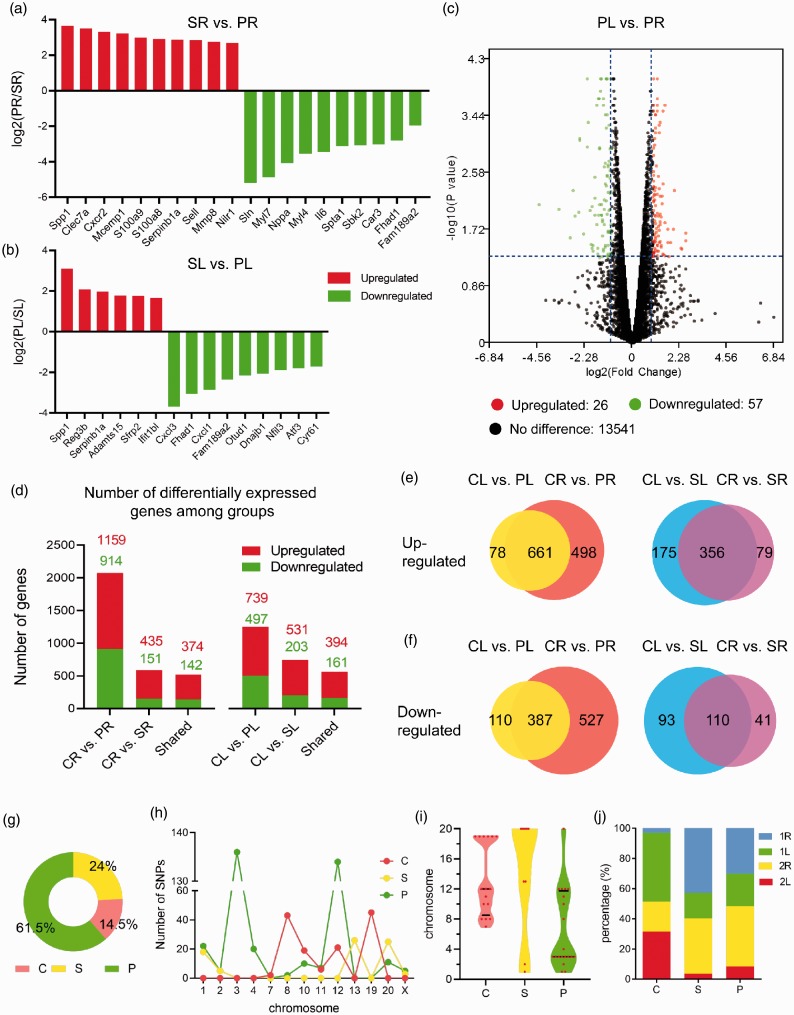
As for the RV samples, 30 genes were differentially expressed in the PAB group as compared to the sham group (Fig. 3a). Among these 30 genes, 20 were significantly upregulated, such as secreted phosphoprotein 1 (Spp1), C-type lectin domain family 7, member A (Clec7a), and C-X-C motif chemokine receptor 2 (Cxcr2); the other 10 were downregulated, such as sarcolipin (Sln), myosin light chain 7 (Myl7), and natriuretic peptide A (ANP) (Fig. 2a). Moreover, for the LV samples, 15 DEGs were identified in the PAB group as compared to the sham group. Six of these DEGs were upregulated and nine downregulated. The upregulated genes included Spp1, regenerating islet-derived 3β (Reg3b), Serpinb1a, and so forth; the downregulated ones were comprised of chemokine (C-X-C motif) ligand 3 (Cxcl3), forkhead-associated phosphopeptide binding domain 1 (Fhad1), chemokine (C-X-C motif) ligand 1 (Cxcl1) and so on (Fig. 3b).
Fig. 3.
The differentially expressed genes in three experimental groups and the distribution and analysis of single nucleotide polymorphisms. (a) The DEGs in the RV or LV of the PAB group as compared to the corresponding ventricles of the sham group. (b) Volcano plot of the DEGs in the LV of PAB group as compared to the RV of the same group. The y-axis represents −log10 (p value), and x-axis log2(fold change). (c) The number of DEGs in the RV (left) or LV (right) of the sham group and PAB group when compared to the normal control group. (d) The number of DEGs in both ventricles of the PAB group (left) and sham group (right) as compared to the normal controls. (e) The number of upregulated genes in RV and LV when PAB and sham groups compared with normal controls. (f) The number of downregulated genes in RV and LV when PAB and sham groups compared with normal controls. (g) The percentages of SNPs in the experimental groups. (h) The chromosomal distribution of the SNP numbers in different experimental groups. (i) The violin plot illustrated the chromosomal distribution of the SNPs in different experimental groups. (j) The percentage of SNPs in the ventricle samples of each experimental group.
When focusing on the gene expression differences between RV and LV in the PAB group, 83 genes were differentially expressed, of which 26 (31%) were upregulated and 57 (69%) downregulated (Fig. 3c).
Moreover, as compared to the RV in the normal control group, 1159 genes in the RV of the PAB group were upregulated and 914 genes were downregulated. For the LV, 739 genes were upregulated and 497 genes downregulated in the PAB group when compared to the normal controls (Fig. 3d). In addition, the upregulated and downregulated genes were also found in the RV and LV samples, respectively, from the sham group when compared to the normal control group (Fig. 3d).
In the PAB group of rats, greater changes in gene expression were observed in the RV than in the LV. In particular, when compared to the corresponding normal control group, the expression of 498 genes in the RV of the PAB group was significantly upregulated; by contrast, only 78 genes in the LV of the same group exhibited the significant upregulation (Fig. 3e). This finding was also true for the downregulated genes in the PAB group (Fig. 3f). However, this was not the case for the sham group, the DEGs were predominantly identified in the LV, where the number of the DEGs was two times that in the RV for both upregulated and downregulated genes (Fig. 3e and f).
The single nucleotide polymorphisms among experimental groups
The SNPs play a critical role in the pathogenesis of diseases.13 The SNPs were detected at the highest frequency in the PAB group (61.5%), next to which was the sham group (24.0%; Fig. 3g). Furthermore, the substantial increase in the number of the SNPs was detected on chromosome 8 and 19 in the normal control group, on chromosome 13 and 20 in the sham group, and on chromosome 3 and 12 in the PAB group (Fig. 3h and i). More interestingly, the SNP frequency in the RV was higher than that in the LV for both the sham and PAB groups (Fig. 3j). However, some SNPs were detected only in the LV. For instance, at position 93796941 of chromosome 2, five SNPs were only found in the LV of the sham group. Likewise, the SNPs at positions 17343932/17343936/17343940 of chromosome 12 were merely observed in the LV of the PAB group (Table 4). Moreover, some SNPs specifically occurred in one group. For instance, at the position 52667195 and 52667197 of chromosome 13, 13 SNPs were found only in the sham group; whereas at the position 176807057 on chromosome 4, 20 SNPs were observed only in the PAB group (Table 4).
Table 4.
The number of SNPs in three experimental groups.
| Chr | Position | Ref | Alt | NC | Sham | PAB |
|---|---|---|---|---|---|---|
| 1 | 1.71 E + 08 | C | T | 0 | 0 | 14 |
| 1 | 2.28 E + 08 | A | G | 0 | 18 | 8 |
| 2 | 93796941 | C | T | 0 | 5 | 5 |
| 3 | 11249139 | A | T | 0 | 0 | 18 |
| 3 | 11249141 | A | T | 0 | 0 | 18 |
| 3 | 11249143 | A | T | 0 | 0 | 18 |
| 3 | 11249145 | A | T | 0 | 0 | 18 |
| 3 | 46802179 | C | A | 0 | 0 | 16 |
| 3 | 46802181 | G | A | 0 | 0 | 16 |
| 3 | 46802182 | A | G | 0 | 0 | 16 |
| 3 | 46802185 | G | A | 0 | 0 | 16 |
| 4 | 1.77 E + 08 | C | G | 0 | 0 | 20 |
| 7 | 1.19 E + 08 | A | G | 2 | 0 | 0 |
| 8 | 33435904 | A | G | 0 | 0 | 2 |
| 8 | 52764002 | A | C | 15 | 0 | 0 |
| 8 | 52764005 | C | A | 15 | 0 | 0 |
| 8 | 52764007 | C | A | 13 | 0 | 0 |
| 10 | 55679059 | G | A | 15 | 0 | 10 |
| 10 | 61178850 | C | A | 4 | 0 | 0 |
| 11 | 30372675 | T | C | 0 | 0 | 7 |
| 11 | 34610817 | T | C | 6 | 0 | 0 |
| 12 | 17343932 | G | A | 0 | 0 | 25 |
| 12 | 17343936 | G | A | 7 | 0 | 35 |
| 12 | 17343940 | G | A | 7 | 0 | 37 |
| 12 | 17343944 | G | A | 7 | 0 | 37 |
| 13 | 52667195 | A | C | 0 | 13 | 0 |
| 13 | 52667197 | T | C | 0 | 13 | 0 |
| 19 | 24741561 | G | A | 7 | 0 | 0 |
| 19 | 24741563 | G | A | 7 | 0 | 0 |
| 19 | 24741565 | G | A | 7 | 0 | 0 |
| 19 | 24741567 | G | A | 8 | 0 | 0 |
| 19 | 24741569 | G | A | 8 | 0 | 0 |
| 19 | 24741571 | G | A | 8 | 0 | 0 |
| 20 | 21882498 | C | T | 0 | 0 | 11 |
| 20 | 3395701 | C | T | 0 | 5 | 0 |
| 20 | 3395706 | T | C | 0 | 5 | 0 |
| 20 | 3395707 | C | T | 0 | 5 | 0 |
| 20 | 3395711 | C | G | 0 | 5 | 0 |
| 20 | 3395715 | T | C | 0 | 5 | 0 |
| X | 37572079 | T | G | 0 | 8 | 0 |
Chr: chromosome; Ref: reference sequence; Alt: alternative genotype; NC: normal control.
The KEGG pathway and protein–protein network analysis
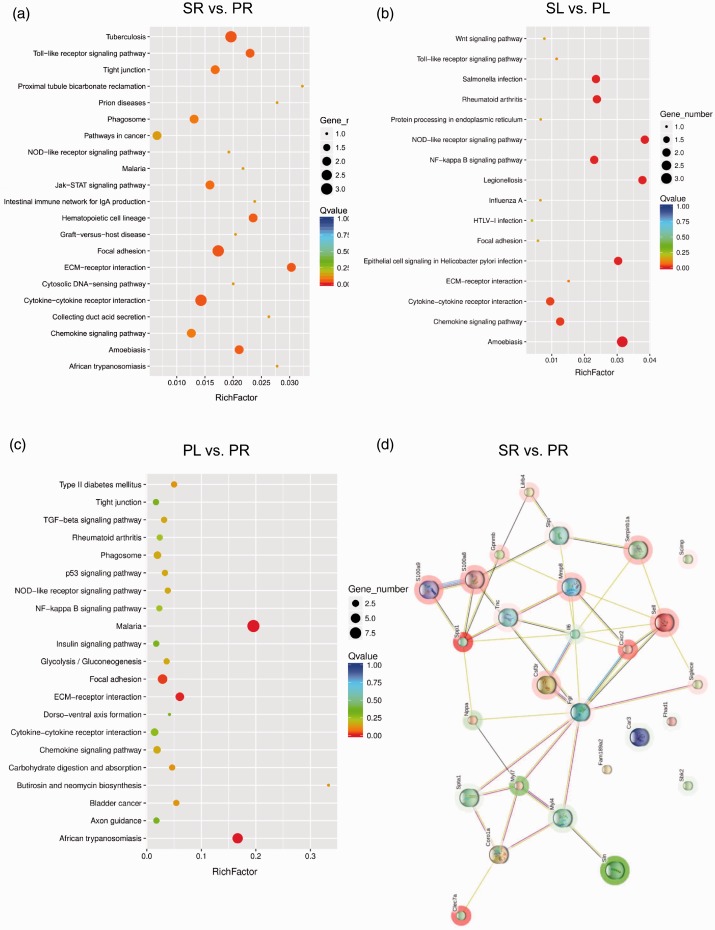
By KEGG analysis, the top 20 enriched pathways in RV or LV among the sham, PAB, and normal control groups were shown in Fig. 3 and Supplementary Fig. 1. In specifics, as compared to RV of the sham group, the DEGs in the PAB counterparts were enriched in focal adhesion and cytokine–cytokine receptor interaction pathways (Fig. 4a). The focal adhesion pathway includes tenascin C (TnC), Spp1, and Myl7 genes; whereas the cytokine–cytokine receptor interaction pathway comprises colony stimulating factor 3 receptor, Cxcr2 and interleukin-6 (IL-6) genes. In the case of LV, the most enriched four pathways in the PAB group were nucleotide oligomerization domain-like receptor (NLR) signalling pathway, nuclear factor-κB (NF-κB) signalling pathway, cytokine–cytokine receptor interaction, and chemokine signalling pathway (Fig. 4b). It is of note that Cxcl1 and DnaJ heat shock protein family member B1 were enriched in all of the four pathways, implicating the importance of these two genes in the pathological changes of LV under acute RHF. On the other hand, within the PAB group, when the gene expression profiling was compared between the two ventricles, the enriched pathways in the RV include extracellular matrix (ECM)–receptor interaction, focal adhesion, metabolism, and phagosome pathways (Fig. 4c). The ECM–receptor interaction pathway contains Spp1, cartilage oligomeric matrix protein, thrombospondin 4 and Tnc genes. On the other hand, when compared with the normal control group, the enriched pathways in the RV of the sham group were cytokine–cytokine receptor interaction, JAK-STAT signalling pathway and cell adhesion molecules. While in comparison to the LV of normal controls, the enriched pathways in the LV of the sham group were cytokine–cytokine receptor interaction, JAK-STAT signalling pathway, phagosome and NF-κB signalling pathway (Supplementary Fig. 1a and b).
Fig. 4.
Bubble diagram of top 20 KEGG pathway analysis and protein–protein network analysis. (a–c) The y-axis represents different pathways; the x-axis indicates richfactor. The richfactor refers to the ratio of the number of DEGs enriched in the pathway to the number of pathway genes annotated. The larger the richfactor, the greater the enrichment degree. The size of the bubble is proportional to the number of EDGs in the pathway; the colour corresponds to the q value, indicating the enrichment statistical significance. (d) The protein–protein network between the RVs in the PAB and the sham groups analysed by STRING algorithm. The red halo around the gene indicates upregulation, the green halo downregulation. KEGG: Kyoto Encyclopedia of Genes and Genomes pathway.
Moreover, the protein–protein network of RV in the PAB group as compared to the sham group had 27 protein nodes and 44 interactions (9 expected interactions). Among these proteins, TNC, SPP1, S100A8, S100A9, colony stimulating factor 3 receptor, CXCR2, matrix metallopeptidase 8, and selectin L were indicated to have strong interactions. The expression of these proteins was upregulated, whereas the expression of Myl4 and Myl7 was downregulated (Fig. 4d). On the other hand, within the PAB group, when the protein–protein network of RV was compared to that of LV, there were 74 nodes and 114 interactions, 77 of which were expected. The proteins with more interactive evidence include eph receptor A3, MYL4, potassium voltage-gated channel interacting protein 2, MYL7, phosphoglycerate mutase 2, C-C motif chemokine ligand 2 (Supplementary Fig. 2).
The GO analysis of DEGs
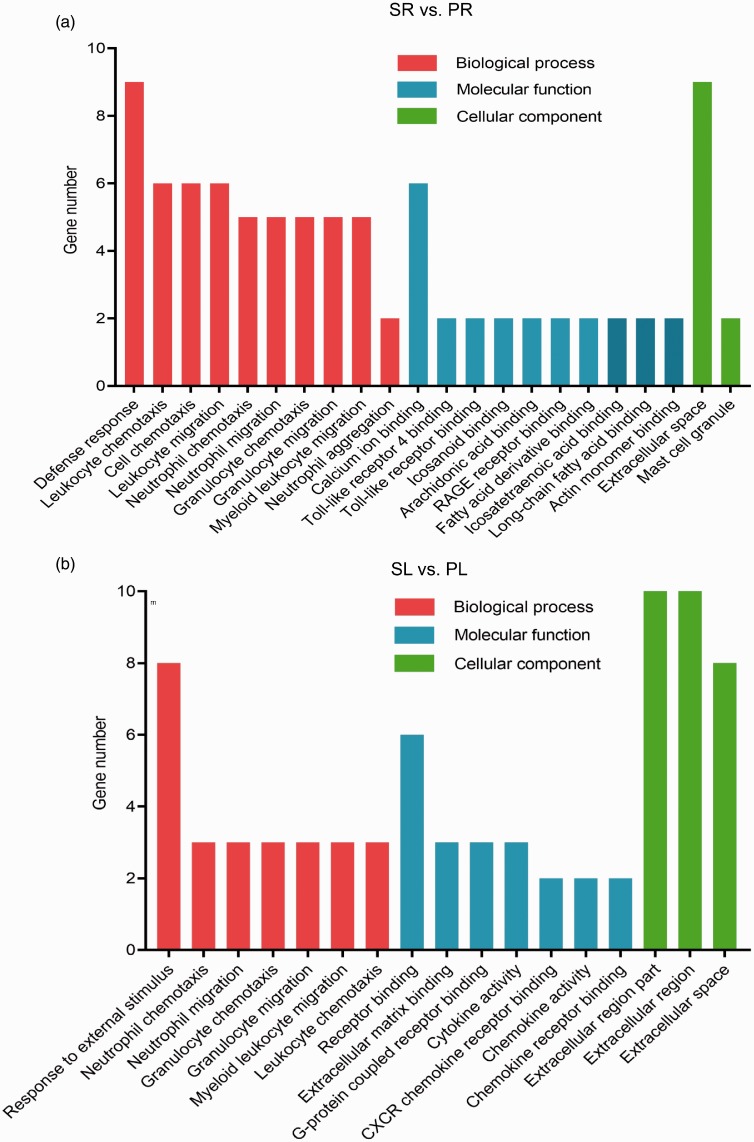
The results of GO analysis showed that the functions of the DEGs were enriched in three parts: cellular component, molecular function, and biological process (Fig. 5). For example, when compared to the RV in the sham group, the ontology nodes of the RV in the PAB group were ranked as the following: defence response (biologic process), such as Cxcr2, Spp1, IL-6 and S100a8 genes; calcium ion binding (molecular function), including natriuretic peptide A, IL-6, Spp1, matrix metallopeptidase 8 genes and ect; and extracellular space (cellular component), such as Tnc, S100a8, S100a9 and Spp1 genes (Fig. 5a).
Fig. 5.
The GO enrichment analysis of the LV and RV in the PAB group as compared to the sham group. GO analysis indicates the enrichment of the DEGs in the LV (a) and RV (b) of the PAB group in comparison to the sham group (p value ≤ 0.05). GO: gene ontology.
As for the LV, when compared to the sham group, the top three ontology nodes in the PAB group include response to external stimulus (biologic process), receptor binding (molecular function), and cellular component (most pathways related to extracellular region; Fig. 5b). The DEGs enriched in external stimulus pathway were Cxcl1, Cxcl3, Spp1 and Reg3b; those enriched in receptor binding included Cxcl1, Cxcl3, Spp1 and secreted frizzled-related protein genes; the DEGs in extracellular region were secreted frizzled-related protein, Cxcl1, Cxcl3 and Spp1 genes.
Discussion
APE may be caused by a deep vein thrombus that enters the pulmonary artery along the systemic circulation and occludes the pulmonary artery. The massive or submassive occlusion leads to an abrupt elevation in pulmonary artery pressure, subsequently a sharp increase in RV afterload, resulting in decompensation in right ventricular function.14 In the meanwhile, the hypoxia caused by the APE further deteriorates the functions of RV.15 Eventually acute RHF occurs. Therefore, the acute RHF is a severe complication and an important cause of mortality of APE.16 Therefore, it is particularly important to study the molecular mechanisms of the acute RHF caused by APE. Unfortunately, there is currently no perfect animal model of the APE-induced acute RHF. The rat model of pulmonary hypertension induced by monocrotaline has been used to study RHF. However, this model exhibits serious pulmonary interstitial fibrosis apart from pulmonary hypertension, hence its phenotypes are not merely caused by pressure overload.17 In this study, we sought to investigate the molecular mechanisms underpinning the acute RHF caused only by the increase of afterload. Therefore, a rat model of PAB was adopted, because no pathological changes have been observed in the pulmonary parenchyma of this rat model and its phenotypes of acute RHF are predominantly, if not completely, caused by simple increase of afterload.4 In the current study, also in the previous study,10 we clipped the main pulmonary artery to induce a rapid increase in RV afterload and acute RHF. Afterwards, the high-throughput RNA-seq combined with bioinformatics analyses were employed to analyse gene expression profiling of the experimental groups.
In general, the ontology and haemodynamics of RV are different from those of LV. During early development, both RV and interventricular septum are derived from the second field cardiac progenitor cells, whereas LV stems from the first heart field cardiac progenitor cells.18,19 However, functionally speaking, the interventricular septum is similar to LV.19 Therefore, only free walls of the ventricles were harvested for RNA-seq analysis in this study. On the other hand, previous studies showed that the two ventricles may interact with each other when the afterload of RV underwent an abrupt increase.8 Indeed, when the pulmonary artery was acutely obstructed and the RV afterload was suddenly elevated, RV and LV contract asynchronously. That is to say, when the LV diastolic phase begins, the RV is still in the systolic phase as a result of delayed contraction caused by substantially enhanced afterload. Thus the RV shifts the interventricular septum towards the LV, altering the geometry of LV. Consequently, the LV preload was decreased and the LV filling was reduced due to the obstructed pulmonary circulation. These two pathologies act additively or synergistically to impair the systemic circulation.8,20 Therefore, acute RHF may contribute to LV damage. Accordingly, in the current study, although the number of DEGs in the LV was less than that in the RV, we did detect certain gene expression changes in the LV in PAB group as compared to the normal controls, indicating the possible deleterious influence of the RV dysfunctions on the LV under the acute RHF.
A previous study demonstrated that inflammation and fibrosis were critical for RHF progression.21 Consistently, KEGG in the current study revealed that the DEGs induced by PAB were enriched in the pathways such as ECM–receptor interaction, Toll-like receptor (TLR) signalling pathway, NLR and NF-κB pathways. TLR signalling and NF-κB pathways are typical inflammatory pathways.22 The TLRs can promote inflammation and cell apoptosis by activating NF-κB and MAPK pathways, hence may contribute to heart failure.23 Moreover, the NLR signalling pathway stimulates the formation of inflammasome, increases the production of and activates the downstream cytokines, IL-1β and IL-18.24 Furthermore, a recent study showed that ECM–receptor interactions and focal adhesion molecules can mediate cardiac fibrosis induced by PAB.25 Thus, the PAB-induced RHF may be associated with inflammation and fibrosis, the signalling pathways revealed by this study could provide hints on the potential targets to the RHF induced by acute obstruction of pulmonary circulation.
We also noticed that treatment of thoracotomy without PAB can elicit gene expression changes when compared to the normal control group. Research has proved that the inflammatory response is a critical contributor to the thoracotomic complications, such as multiple organ failure.6,7 Therefore, the involvement of the DEGs of the sham group in the inflammation caused by thoracotomy warrants further investigation.
There are some limitations in the current study. First, the number of animals in each experimental group for RNA-seq may not be enough for biological repeats. However, due to the special method used to establish the PAB model, the consistent 50–60% of obstruction in the main pulmonary artery could be achieved among individual experimental animals, therefore, the variability caused by the PAB treatment should be substantially reduced. In fact, the results of the principal component analysis and correlation analysis have demonstrated that the variations between the two samples from the same group are small. Therefore, the current RNA-seq results might be able to shed lights on the gene expression trends in the RV and LV under the acute RHF and the possible therapeutic targets to such a catastrophic disease. Further, it would be more convincing and scientifically rigid to validate the expression patterns of important and representative genes among the experimental groups through a conventional method, such as quantitative real-time PCR.
Taken together, this study demonstrated that the PAB caused the anatomical, haemodynamic, and gene expression changes in the RV that lead to acute RHF in rats. The novel finding of this study is that the acute RHF induced by pressure overload can cause gene expression changes in the LV, suggesting the molecular interactions between RV and LV under the diseased condition. Moreover, the gene expression profiling of both heart ventricles in an acute RHF model may provide hints for a potential therapeutic target to this devastating disease.
Supplemental Material
Supplemental material, PUL879396 Supplemental Material1 for RNA-sequencing analysis of gene expression in a rat model of acute right heart failure by Yunshan Cao, Yahong Li, Mianmian Wu, Jiyang Song, Min Zhang, Yichao Duan, Kaiyu Jiang, Xing Zhou and Yan Zhang in Pulmonary Circulation
Supplemental material, PUL879396 Supplemental Material2 for RNA-sequencing analysis of gene expression in a rat model of acute right heart failure by Yunshan Cao, Yahong Li, Mianmian Wu, Jiyang Song, Min Zhang, Yichao Duan, Kaiyu Jiang, Xing Zhou and Yan Zhang in Pulmonary Circulation
Conflict of interest
The author(s) declare that there is no conflict of interest.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: the National Natural Science Foundation of China (81460072, 81770300, 81860059); CAS “Light of West China” Program; the Natural Science Foundation of Gansu Province (17JR5RA057, 18JR3RA060); International Joint Research Program of Gansu Province (18YF1WA046); Innovation and entrepreneurship project of Lanzhou City (2018-RC-78).
Ethical approval
Approved by Gansu Provincial Hospital.
Authors’ contributions
Yunshan Cao designed the study. Yunshan Cao, Yahong Li and Mianmian Wu performed the experiments. Jiyang Song did pulmonary artery banding for rats. Min Zhang wrote the drafts. Yichao Duan and Kaiyu Jiang made the figures. Xing Zhou and Yan Zhang designed the study and revised the manuscript.
ORCID iD
Yunshan Cao https://orcid.org/0000-0001-8463-1093
Supplemental material
Supplemental material for this article is available online.
References
- 1.Gorter TM, van Veldhuisen DJ, Bauersachs J, et al. Right heart dysfunction and failure in heart failure with preserved ejection fraction: mechanisms and management. Position statement on behalf of the Heart Failure Association of the European Society of Cardiology. Eur J Heart Fail 2018; 20: 16–37. [DOI] [PubMed] [Google Scholar]
- 2.Ponikowski P, Voors AA, Anker SD, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC)Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J 2016; 37: 2129–2200. [DOI] [PubMed] [Google Scholar]
- 3.Simon MA. Assessment and treatment of right ventricular failure. Nat Rev Cardiol 2013; 10: 204–218. [DOI] [PubMed] [Google Scholar]
- 4.Guihaire J, Bogaard HJ, Flecher E, et al. Experimental models of right heart failure: a window for translational research in pulmonary hypertension. Semin Respir Crit Care Med 2013; 34: 689–699. [DOI] [PubMed] [Google Scholar]
- 5.Cildag MB, Gok M, Karaman CZ. Pulmonary artery obstruction index and right ventricular dysfunction signs in initial and follow up pulmonary computed tomography angiography in acute pulmonary embolism. J Clin Diagn Res 2017; 11: TC21–TC25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paparella D, Yau TM, Young E. Cardiopulmonary bypass induced inflammation: pathophysiology and treatment. An update. Eur J Cardiothorac Surg 2002; 21: 232–244. [DOI] [PubMed] [Google Scholar]
- 7.Zupancich E, Paparella D, Turani F, et al. Mechanical ventilation affects inflammatory mediators in patients undergoing cardiopulmonary bypass for cardiac surgery: a randomized clinical trial. J Thorac Cardiovasc Surg 2005; 130: 378–383. [DOI] [PubMed] [Google Scholar]
- 8.Haddad F, Hunt SA, Rosenthal DN, et al. Right ventricular function in cardiovascular disease, part I: Anatomy, physiology, aging, and functional assessment of the right ventricle. Circulation 2008; 117: 1436–1448. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y, Song J, Shen S, et al. Trimedazidine alleviates pulmonary artery banding-induced acute right heart dysfunction and activates PRAS40 in rats. Oncotarget 2017; 8: 92064–92078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bae HK, Lee H, Kim KC, et al. The effect of sildenafil on right ventricular remodeling in a rat model of monocrotaline-induced right ventricular failure. Korean J Pediatr 2016; 59: 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Morley M, Brandimarto J, et al. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics 2015; 105: 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tebbutt SJ, James A, Pare PD. Single-nucleotide polymorphisms and lung disease: clinical implications. Chest 2007; 131: 1216–1223. [DOI] [PubMed] [Google Scholar]
- 14.Bryce YC, Perez-Johnston R, Bryce EB, et al. Pathophysiology of right ventricular failure in acute pulmonary embolism and chronic thromboembolic pulmonary hypertension: a pictorial essay for the interventional radiologist. Insights Imaging 2019; 10: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthews JC, McLaughlin V. Acute right ventricular failure in the setting of acute pulmonary embolism or chronic pulmonary hypertension: a detailed review of the pathophysiology, diagnosis, and management. Curr Cardiol Rev 2008; 4: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konstam MA, Kiernan MS, Bernstein D, et al. Evaluation and management of right-sided heart failure: a scientific statement from the American Heart Association. Circulation 2018; 137: e578–e622. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Wang Y, Zhang J, et al. Galectin-3 contributes to vascular fibrosis in monocrotaline-induced pulmonary arterial hypertension rat model. J Biochem Mol Toxicol 2017; 31: e21879. [DOI] [PubMed] [Google Scholar]
- 18.Reddy S, Bernstein D. Molecular mechanisms of right ventricular failure. Circulation 2015; 132: 1734–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 2005; 6: 826–835. [DOI] [PubMed] [Google Scholar]
- 20.Greyson CR. Pathophysiology of right ventricular failure. Crit Care Med 2008; 36: S57–S65. [DOI] [PubMed] [Google Scholar]
- 21.Voelkel NF, Gomez-Arroyo J, Abbate A, et al. Mechanisms of right heart failure – a work in progress and a plea for failure prevention. Pulm Circ 2013; 3: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor CT, Doherty G, Fallon PG, et al. Hypoxia-dependent regulation of inflammatory pathways in immune cells. J Clin Invest 2016; 126: 3716–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, Feng Z. The role of toll-like receptor signaling in the progression of heart failure. Mediators Inflamm 2018; 2018: 9874109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukata M, Vamadevan AS, Abreu MT. Toll-like receptors (TLRs) and Nod-like receptors (NLRs) in inflammatory disorders. Semin Immunol 2009; 21: 242–253. [DOI] [PubMed] [Google Scholar]
- 25.Buttner P, Ueberham L, Shoemaker MB, et al. Identification of central regulators of calcium signaling and ECM–receptor interaction genetically associated with the progression and recurrence of atrial fibrillation. Front Genet 2018; 9: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, PUL879396 Supplemental Material1 for RNA-sequencing analysis of gene expression in a rat model of acute right heart failure by Yunshan Cao, Yahong Li, Mianmian Wu, Jiyang Song, Min Zhang, Yichao Duan, Kaiyu Jiang, Xing Zhou and Yan Zhang in Pulmonary Circulation
Supplemental material, PUL879396 Supplemental Material2 for RNA-sequencing analysis of gene expression in a rat model of acute right heart failure by Yunshan Cao, Yahong Li, Mianmian Wu, Jiyang Song, Min Zhang, Yichao Duan, Kaiyu Jiang, Xing Zhou and Yan Zhang in Pulmonary Circulation