Abstract
Background & Aims
Bacterial flagellin is an important antigen in inflammatory bowel disease, but the role of flagellin-specific CD4+ T cells in disease pathogenesis remains unclear. Also unknown is how changes in intestinal microbiome intersect with those in microbiota-specific CD4+ T cells. We aimed to quantify and characterize flagellin-specific CD4+ T cells in Crohn’s disease (CD) and ulcerative colitis (UC) patients and study their relationship with intestinal microbiome diversity.
Methods
Blood was collected from 3 cohorts that included CD patients, UC patients, and healthy controls. Flow cytometry analyzed CD4+ T cells specific for Lachnospiraceae-derived A4-Fla2 and Escherichia coli H18 FliC flagellins, or control vaccine antigens. Serum antiflagellin IgG and IgA antibodies were detected by enzyme-linked immunosorbent assay and stool samples were collected and subjected to 16S ribosomal DNA sequencing.
Results
Compared with healthy controls, CD and UC patients had lower frequencies of vaccine-antigen–specific CD4+ T cells and, as a proportion of vaccine-specific cells, higher frequencies of flagellin-specific CD4+ T cells. The proportion of flagellin-specific CD4+ T cells that were CXCR3negCCR4+CCR6+ Th17 cells was reduced in CD and UC patients, with increased proportions of CD39+, PD-1+, and integrin β7+ cells. Microbiome analysis showed differentially abundant bacterial species in patient groups that correlated with immune responses to flagellin.
Conclusions
Both CD and UC patients have relative increases in the proportion of circulating Fla2-specific CD4+ T cells, which may be associated with changes in the intestinal microbiome. Evidence that the phenotype of these cells strongly correlate with disease severity provides insight into the potential roles of flagellin-specific CD4+ T cells in inflammatory bowel disease.
Keywords: CD4+ T Cells, Crohn’s Disease, Ulcerative Colitis, Microbiome
Abbreviations used in this paper: BSA, bovine serum albumin; CD, Crohn’s disease; IBD, inflammatory bowel disease; mAb, monoclonal antibody; rDNA, ribosomal DNA; SEB, Staphylococcal enterotoxin B; TBST, Tris-buffered saline containing 0.05% Tween-20; Th, helper T cell; TLR, Toll-like receptor; TNFα, tumor necrosis factor-α; UC, ulcerative colitis
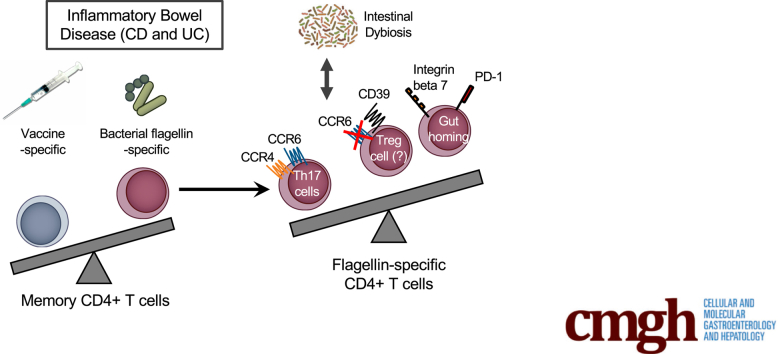
Graphical abstract
Summary.
Analyses of memory immune responses in ulcerative colitis and Crohn’s disease patients have shown reduced vaccine-specific responses and proportionally increased flagellin-specific responses compared with healthy controls. Flagellin-specific T cells may be a useful biomarker and provide insight into inflammatory bowel disease pathogenesis.
Inflammatory bowel disease (IBD), which includes Crohn's disease (CD) and ulcerative colitis (UC), is characterized by chronic relapsing intestinal inflammation mediated by a variety of dysfunctional immune responses. Serologic testing has shown that CD patients often develop antibodies to antigens derived from microbes such as Escherichia coli, Pseudomonas fluorescens, and Saccharomyces cerevisiae,1 with these antibody responses associated with disease severity.2 In addition, T-cell responses to intestinal commensal bacteria, such as segmented filamentous bacteria and Clostridiales family members, also have been associated with CD, leading to the concept that abnormal immunity to microbial antigens is an important aspect of IBD disease pathogenesis.3
An antigen commonly expressed by commensal and pathogenic bacteria is flagellin, the major structural protein of bacterial flagella. Flagellin interacts with the pattern-recognition receptors Toll-like receptor 5 (TLR5) and nucleotide-binding oligomerization domain-like receptor (NLR) family caspase activation and recruitment domain (CARD) domain containing 4, leading to production of proinflammatory cytokines and chemokines.4 However, unlike most other TLR agonists, flagellin is also a protein containing many antigenic epitopes, and indeed CD4+ T cells specific for bacterial flagellin are found in both mice and human beings. In mice, CD4+ T cells specific for CBir1, a flagellin derived from a commensal Lachnospiraceae strain, cause colitis when transferred to immunodeficient recipients.5 In human beings, flagellin-specific CD4+ T cells are found in circulation, and in intestinal tissues of IBD subjects are skewed toward a T helper (Th) cell 17/17.1 cell phenotype.6 In terms of flagellin-specific antibodies, approximately 50% of CD patients have circulating anti-CBir1 IgG and IgA antibodies, the presence of which is associated with small-bowel, internal-perforating, and fibrostenotic disease phenotypes.7,8
Another key feature of IBD is intestinal dysbiosis, characterized by altered microbial communities,9 loss of bacterial diversity, and an enrichment of potential pathogens.3,10 Although there are physiological sequelae of intestinal dysbiosis,11,12 whether dysbiosis is a cause or an effect of intestinal inflammation in IBD is unknown.9 Moreover, whether changes in the intestinal microbiome of IBD patients might interface with existing antimicrobial immune responses and/or drive their de novo development remains an unexplored question.
Here, we aimed to investigate how the frequency and phenotype of flagellin-specific CD4+ T cells in CD and UC patients changes with treatment and relates to disease severity and progression. We also investigated the relationship of CD4+ T cells with anti-flagellin antibodies, as well as with changes in the fecal microbiome composition.
Results
Identification of Circulating Flagellin-Specific CD4+ T Cells in IBD Patients
To measure circulating flagellin-specific CD4+ T cells, we used a whole-blood assay, hereafter referred to as the OX40 assay, which detects antigen-specific CD4+ T cells by measuring antigen-stimulated co-expression of CD25 and OX40 (CD134).13 CD25 and OX40, which are co-expressed at very low levels on circulating CD4+ T cells, are up-regulated after T cell receptor-mediated activation, enabling detection of rare, antigen-specific, memory CD4+ T cells.13 Major advantages of this approach are that HLA-restricted T-cell epitopes do not need to be known and, because cell quantification does not rely on the ability of cells to produce cytokines or divide, this assay accurately enumerates the entire population of antigen-specific CD4+ T cells.14
We first collected a cross-sectional cohort (cohort 1) of subjects with CD (n = 55) or UC (n = 9) (Table 1) to determine if the OX40 assay was sufficiently sensitive to detect circulating flagellin-specific CD4+ T cells. We tested responses to Lachnospiraceae-derived A4-Fla2 (hereafter Fla2) and E coli H18 FliC (hereafter FliC), because these species encode flagellins that are representative of their respective bacterial families and are known to be immunogenic.15 Of note, we previously found Fla2 to be a poor TLR5 agonist.16 We detected FliC- and Fla2-specific CD4+ T cells in both CD and UC patients (Figure 1A and B), and there were no differences between the percentages of CD4+ T cells specific for FliC or Fla2 in either group (Figure 1B).
Table 1.
Cohort 1 Characteristics
| ID | Age, y | Sex | Years since diagnosis | Type of disease | Type of treatment | Disease location/extent |
|---|---|---|---|---|---|---|
| YUL001 | 25 | F | 15 | CD | Mesalamine, Prednisone, 6-MP | Sigmoid, rectum/luminal |
| YUL002 | 20 | M | 1 | CD | Mesalamine | UGI, large intestine/luminal |
| YUL003 | 19 | M | 7 | CD | 6-MP | NA/luminal |
| YUL006 | 21 | F | 6 | CD | MTX | Descending colon/perianal |
| YUL007 | 23 | M | 12 | CD | Mesalamine, budesonide | Ileum, cecum/stricturing |
| YUL008 | 19 | F | 10 | CD | MTX, infliximab | UGI, cecum, transverse and descending colon, sigmoid/luminal |
| YUL009 | 18 | M | 6 | CD | Mesalamine | Ileum, cecum/luminal, perianal |
| YUL010 | 44 | F | 29 | CD | Budesonide, adalimumab | Ileum, cecum, colon/penetrating, perianal |
| YUL012 | 20 | M | 4 | CD | Azathioprine | UGI, ileum, cecum/perianal |
| YUL014 | 18 | M | 1 | CD | 6-MP | Ileum, cecum, ascending colon/penetrating |
| YUL016 | 42 | F | 18 | CD | Infliximab | Ileum, cecum, ascending colon, sigmoid/penetrating, perianal |
| YUL018 | 21 | M | 4 | CD | Prednisone, MTX, infliximab | UGI, cecum, transverse and descending colon, sigmoid, rectum/luminal |
| YUL021 | 22 | M | 7 | CD | None | Large intestine/luminal |
| YUL029 | 41 | M | NA | CD | None | Ileum, cecum, ascending colon/luminal |
| YUL030 | 46 | M | 28 | CD | 6-MP | Ileum, cecum, sigmoid/perianal |
| YUL031 | 25 | M | 11 | CD | Mesalamine, budesonide, 6-MP | Ileum/luminal |
| YUL032 | 25 | F | 23 | CD | Azathioprine | Colon, ascending and transverse colon, sigmoid, rectum/luminal |
| YUL033 | 21 | F | 5 | CD | Infliximab | UGI, ileum, cecum, ascending colon/perianal |
| YUL034 | 27 | F | 10 | CD | 6-MP, adalimumab | UGI, ileum, large intestine/stricturing |
| YUL036 | 35 | F | 2 | CD | Mesalamine | Cecum, rectum/luminal |
| YUL037 | 24 | F | 8 | CD | Mesalamine, azathioprine | Ileum/luminal |
| YUL038 | 24 | F | 7 | CD | Azathioprine | Ileum/penetrating |
| YUL039 | 20 | F | 2 | CD | Budesonide, 6-MP | Ileum, cecum, sigmoid, rectum/luminal |
| YUL040 | 18 | M | NA | CD | 6-MP | NA/stricturing |
| YUL042 | 21 | M | 10 | CD | Infliximab | UGI, ileum, cecum, colon/luminal |
| YUL043 | 25 | F | 8 | CD | Mesalamine, 6-MP | Jejunum, ileum, ascending colon/luminal |
| YUL044 | 20 | F | 18 | CD | Adalimumab | UGI, ileum, cecum, colon, sigmoid/luminal, perianal |
| YUL045 | 18 | M | 12 | CD | Mesalamine, 6-MP, adalimumab, infliximab | Descending colon, sigmoid, rectum/luminal |
| YUL047 | 31 | M | 14 | CD | None | Ileum, cecum/penetrating, perianal |
| YUL048 | 25 | M | 12 | CD | Azathioprine | Ileum/penetrating |
| YUL049 | 19 | F | 9 | CD | MTX, adalimumab | UGI, ileum, transverse and descending colon, sigmoid, rectum/luminal, perianal |
| YUL050 | 18 | F | 13 | CD | Mesalamine | NA |
| YUL051 | 23 | M | 1 | CD | Mesalamine | Ileum, cecum, sigmoid, rectum/luminal |
| YUL052 | 32 | M | NA | CD | Prednisone, 6-MP | Ileum, rectum/luminal |
| YUL054 | 18 | F | 1 | CD | MTX | Ascending and descending colon, sigmoid, rectum/luminal |
| YUL056 | 26 | M | 13 | CD | Sulfasalazine | Descending colon, sigmoid, rectum/luminal |
| YUL059 | 23 | F | 5 | CD | Azathioprine | UGI, ileum, descending colon, sigmoid/luminal, perianal |
| YUL060 | 26 | F | 24 | CD | Prednisone, azathioprine | Cecum, ascending and transverse colon, sigmoid, rectum/luminal |
| YUL062 | 24 | M | 17 | CD | Infliximab | Ileum, cecum, ascending, transverse and descending colon, sigmoid, rectum/luminal |
| YUL063 | 29 | M | 1 | CD | None | Jejunum, sigmoid/luminal |
| YUL064 | 18 | F | NA | CD | 6-MP | UGI, ileum, cecum, ascending, transverse and descending colon, sigmoid, rectum/stricturing |
| YUL065 | 19 | M | 8 | CD | MTX, infliximab | UGI, ileum, rectum/penetrating |
| YUL066 | 32 | F | 3 | CD | Azathioprine | Jejunum, ileum/luminal |
| YUL067 | 20 | F | 18 | CD | Ustekinumab | UGI, ileum, cecum, ascending, transverse and descending colon, sigmoid, rectum/stricturing |
| YUL070 | 25 | F | 11 | CD | Infliximab | UGI, cecum, ascending, transverse and descending colon, sigmoid, rectum/luminal |
| YUL073 | 24 | M | 10 | CD | Azathioprine | Ileum, cecum/luminal |
| YUL074 | 21 | F | 6 | CD | None | UGI, ileum, cecum, ascending, transverse and descending colon, sigmoid/luminal |
| YUL075 | 22 | F | 6 | CD | Infliximab | UGI, ileum, cecum, ascending colon, sigmoid, rectum/luminal, perianal |
| YUL076 | 27 | F | 25 | CD | Azathioprine | Cecum, ascending and transverse colon, sigmoid, rectum/luminal |
| YUL083 | 18 | M | NA | CD | Infliximab | NA/luminal |
| YUL086 | 25 | M | 11 | CD | Azathioprine | Ileum, cecum/luminal |
| YUL087 | 37 | F | 22 | CD | Adalimumab | Ileum, cecum, ascending, transverse and descending colon, sigmoid, rectum/penetrating, perianal |
| YUL088 | 35 | F | NA | CD | NA | NA/luminal |
| YUL089 | NA | NA | NA | CD | NA | NA |
| YUL090 | NA | NA | NA | CD | NA | NA |
| YUL020 | 23 | M | 7 | UC | Mesalamine | Large intestine/luminal |
| YUL035 | 21 | F | 4 | UC | Mesalamine, azathioprine | Transverse and descending colon, sigmoid, rectum/NA |
| YUL041 | 22 | F | 12 | UC | Mesalamine | Sigmoid, rectum/NA |
| YUL055 | 20 | F | 3 | UC | Azathioprine | Ascending and descending colon, sigmoid, rectum |
| YUL057 | 33 | F | 4 | UC | Azathioprine, adalimumab | Transverse and descending colon, sigmoid, rectum |
| YUL058 | 41 | M | 3 | UC | Mesalamine, budesonide, 6-MP | Sigmoid, rectum |
| YUL061 | 21 | M | 4 | UC | MTX, infliximab | Cecum, ascending, transverse and descending colon, sigmoid, rectum |
6-MP, mercaptopurine; MTX, methotrexate; NA, data not available; UGI, upper gastrointestinal tract.
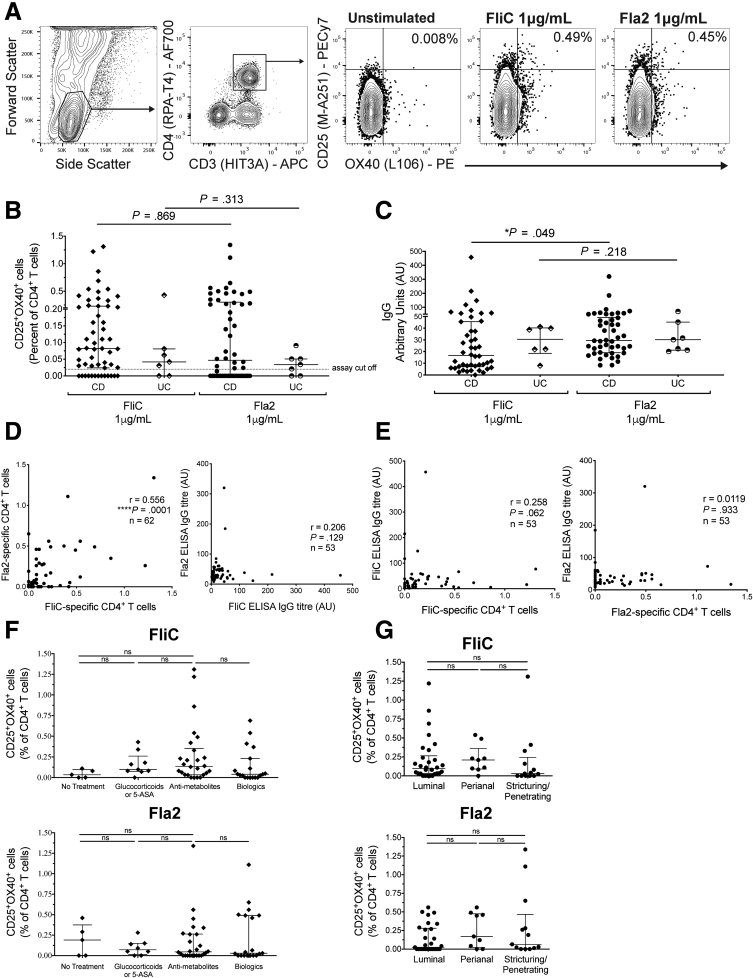
Figure 1.
Flagellin-specific CD4+T cells can be detected in IBD patients’ peripheral blood. (A) Gating strategy and representative flow cytometry data for cohort 1. (B) The percentage of CD25+OX40+ cells within CD4+ T cells after stimulation of whole blood with FliC or Fla2 for 44 hours (CD, n = 55; UC, n = 7). (C) Anti-FliC and anti-Fla2 IgG levels in plasma measured using an enzyme-linked immunosorbent assay (ELISA); concentrations are in arbitrary units based on high-titer pooled plasma standards. (D) Correlations between FliC– and Fla2–specific CD4+ T cells and IgG levels and (E) correlations between FliC- and Fla2-specific CD4+ T cells and IgG levels. FliC- and Fla2-specific CD4+ T-cell responses were compared between CD patients based on (F) current treatments (untreated, n = 5; glucocorticoids or mesalamine, (n = 9 for FliC and n = 8 for Fla2); Fla2, antimetabolites n = 26 and biologics n = 19) and (G) clinical phenotype of disease: perianal (n = 5), luminal (n = 20), or stricturing/penetrating (n = 9). Statistical analyses used (B and C) Mann–Whitney U tests, (D and E) calculated Spearman Rho (r), and (F and G) Kruskal–Wallis tests. APC, allophycocyanin; PECy7, phycoerythrin-cyanine 7.
Similar to previous reports examining antibody responses to CBir and FlaX flagella,7,8,17 we detected anti-Fla2 and anti-FliC IgG in plasma from both CD and UC subjects, with paired analysis showing significantly higher levels of anti-Fla2 IgG compared with anti-FliC IgG in CD, but not UC, patients (Figure 1C). Correlation analyses showed that the proportions of Fla2- or FliC-specific CD4+ T cells were significantly positively correlated with each other, indicating that individuals with a strong CD4+ T-cell response to 1 flagellin antigen were likely to have a similarly strong response to the second antigen, although this was not observed for IgG levels (Figure 1D). There was also no correlation between the proportion of flagellin-specific CD4+ T cells and anti-flagellin IgG levels, indicating that antibody levels cannot be used to predict the proportion of circulating flagellin-specific CD4+ T cells (Figure 1E). Proportions of flagellin-specific CD4+ T cells did not correlate with current medication (classified as follows: untreated, glucocorticoids or mesalamine only, glucocorticoids/mesalamine and an antimetabolite, or a biologic with or without any other medication), and/or disease location or severity (perianal, luminal, or stricturing) (Figure 1F and G). These data validate the use of the OX40 assay to measure flagellin-specific CD4+ T cells and confirm a previous report that circulating flagellin-specific CD4+ T cells are detectable in CD patients.6
Changes in Flagellin- and Vaccine-Specific CD4+ T Cells in CD Patients After Anti–Tumor Necrosis Factor-α Treatment
Although anti–tumor necrosis factor-α (TNFα) therapies increasingly are becoming standard of care for CD patients, the precise effects on immune responses, particularly memory responses, remains unknown. To assess the impact of anti-TNFα treatment on the proportion of circulating flagellin-specific CD4+ T cells, we enrolled 14 CD patients into a longitudinal cohort (cohort 2) and collected blood before commencing anti-TNFα therapy (week 0) and at 8 and 24 weeks (6 months) after therapy. The majority of patients reported symptom improvement after commencing anti-TNFα treatment, as measured by their Harvey–Bradshaw Index score (Table 2). For this cohort, we incorporated an additional control for the OX40 assay, which was stimulation with the pentavalent vaccine Pediacel (Sanofi Pasteur, Lyon, France). This vaccine is part of Canadian provincial schedules and thus the majority of subjects should have memory CD4+ T cells specific for 1 or more antigens in this mixture.
Table 2.
Cohort 2 Characteristics
| ID | Age, y | Sex | Years since diagnosis | Type of disease | Type of treatment before commencing anti-TNFα therapy | Harvey–Bradshaw Index |
||
|---|---|---|---|---|---|---|---|---|
| Week 0 | Week 8 | Week 24 | ||||||
| YVR001 | 28 | F | 12 | CD | Azathioprine, mesalamine | 10 | NA | 5 |
| YVR002 | 43 | F | 27 | CD | Budesonide | 7 | NA | NA |
| YVR003 | 55 | F | 7 | CD | None | NA | NA | NA |
| YVR004 | 55 | F | 25 | CD | Azathioprine | 0 | 1 | 1 |
| YVR005 | 27 | F | 2 | CD | Azathioprine, budesonide | 6 | 2 | 3 |
| YVR006 | 33 | M | 17 | CD | MTX | 1 | 5 | 3 |
| YVR007 | 22 | M | 7 | CD | None | 2 | 4 | 4 |
| YVR008 | 74 | F | 45 | CD | Prednisone | 2 | 6 | 1 |
| YVR009 | 60 | F | 20 | CD | Mesalamine | 5 | 1 | 2 |
| YVR010 | 54 | M | 19 | CD | Azathioprine | 9 | 5 | 0 |
| YVR011 | 35 | M | 7 | CD | MTX | 6 | 2 | 1 |
| YVR012 | 26 | M | 11 | CD | Azathioprine | 11 | 3 | 10 |
| YVR013 | 60 | F | 40 | CD | None | 3 | 0 | 0 |
| YVR014 | NA | NA | NA | CD | NA | 0 | 3 | 1 |
MTX, methotrexate; NA, data not available.
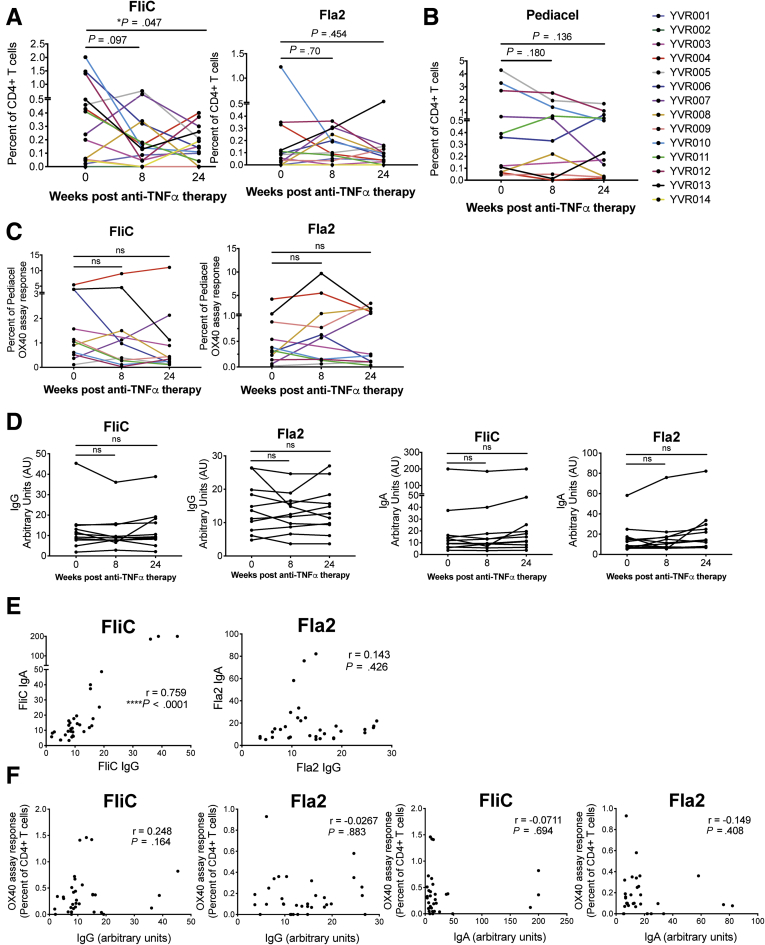
We first observed a reduction in circulating FliC-specific, but not Fla2-specific, CD4+ T cells after 6 months of anti-TNFα therapy (Figure 2A). Although not significant when analyzed cumulatively, some individuals also had reduced responses to Pediacel after anti-TNFα therapy (Figure 2B), raising the possibility that longitudinal comparisons of antigen-specific responses may be affected by nonspecific immune suppression as a result of therapy and/or disease progression. Because exposure to the antigens contained in the Pediacel vaccine should have occurred equally among the patient groups (either through vaccination or natural infection), the CD4+ T-cell memory responses to these pooled antigens provided a means to normalize the flagellin-specific responses by accounting for overall reduced frequencies in the CD4+ T-cell memory pool in IBD patients. We therefore re-analyzed the FliC- and Fla2-specific responses as a proportion of Pediacel responses, showing no significant changes from before to after anti-TNFα therapy (Figure 2C). Similarly, amounts of anti-FliC and anti-Fla2 IgG and IgA did not change significantly after anti-TNFα therapy (Figure 2D).
Figure 2.
Flagellin-specific immune responses in CD patients before and after anti-TNFα therapy. OX40 assays were performed for cohort 2 (n = 14 CD patients), before anti-TNFα treatment, and at 8 and 24 weeks after treatment. For each individual, data are shown for (A) the proportion of FliC- and Fla2-specific CD4+ T cells (n = 14 at weeks 0 and 8, n = 13 at week 24) and (B) the proportion of Pediacel-specific CD4+ T cells (n = 12 at weeks 0 and 24 and n = 11 at week 8). (C) FliC- and Fla2-specific CD4+ T cells are shown as a percentage of Pediacel-specific CD4+ T cells (n = 12 at weeks 0 and 24 and n = 11 at week 8). (D) Anti-FliC and anti-Fla2 IgG and IgA levels were measured by enzyme-linked immunosorbent assay at each time point (n = 12). Statistical analysis used Wilcoxon signed-rank tests. Spearman Rho (r) correlations between anti-FliC and anti-Fla2 (E) IgG and IgA levels from all time points (n = 11, each at 3 time points); and (F) OX40 assay responses and IgG and IgA levels.
Correlation analyses between amounts of IgA and IgG showed a strong positive correlation for anti-FliC, but not anti-Fla2, antibodies (Figure 2E). Similar to cohort 1, we did not find any correlation between FliC- or Fla2-specific CD4+ T-cell responses and IgG or IgA levels (Figure 2F). These data suggest that anti-TNFα treatment may cause a generalized suppression of memory CD4+ T-cell responses in some CD subjects, but that it does not affect amounts of antiflagellin IgG or IgA over a 24-week time period.
IBD Patients Have Altered Proportions of Vaccine- and Flagellin-Specific CD4+ T Cells
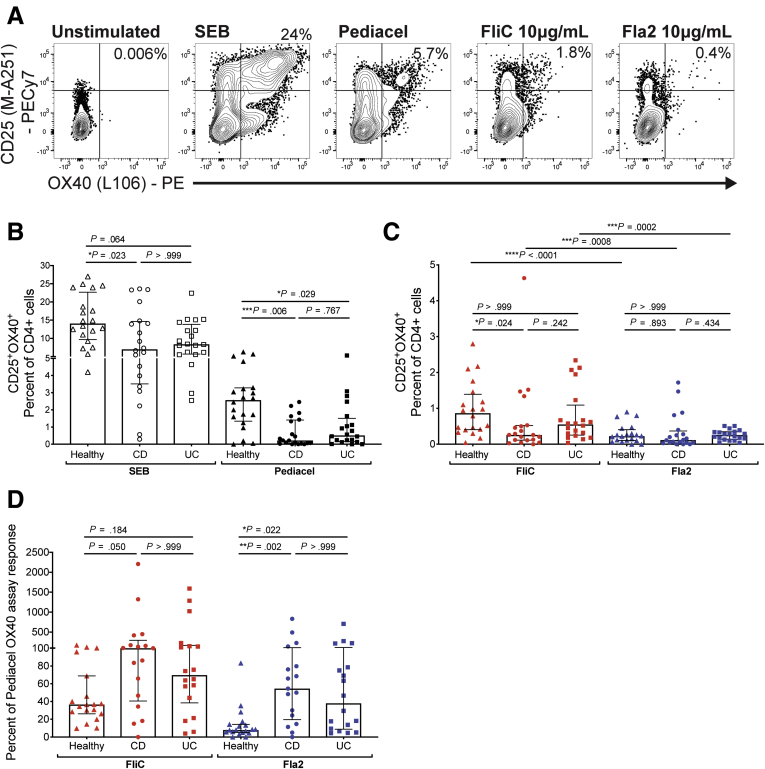
We next asked if the proportions and phenotype of flagellin-specific memory CD4+ T cells might be different between CD or UC subjects and healthy controls. To test this possibility, we enrolled a new cohort (cohort 3, with n = 20 each of CD, UC, and age- and sex-matched healthy controls) (Table 3). In anticipation of possible effects of IBD on responses to the Pediacel positive control, we also incorporated an additional positive control: Staphylococcal enterotoxin B (SEB), a superantigen that activates T cells by cross-linking TCR and major histocompatibility complex II molecules18 (Figure 3A). As expected, SEB stimulation up-regulated expression of OX40 and CD25 in all individuals, but in comparison with healthy, age-matched controls, CD patients had significantly reduced proportions of SEB-stimulated CD4+ T cells. Moreover, both CD and UC patients had reduced proportions of Pediacel-specific CD4+ T cells (Figure 3B), confirming the trend observed in cohort 2.
Table 3.
Cohort 3 Characteristics
| ID | Age, y | Sex | Years since diagnosis | Type of disease | Type of treatment | Harvey–Bradshaw Index |
|---|---|---|---|---|---|---|
| FL001 | 37 | F | 4 | UC | MTX, mesalamine | 14 |
| FL002 | 62 | F | 22 | UC | Azathioprine, infliximab | 3 |
| FL003 | 33 | M | 3 | CD | Ustekinumab | 0 |
| FL004 | 70 | F | 30 | CD | None | 7 |
| FL005 | 27 | M | 7 | CD | Adalimumab | 1 |
| FL006 | 35 | F | 2 | CD | Infliximab | 14 |
| FL007 | 30 | M | 2 | UC | Azathioprine, golimumab | 2 |
| FL008 | 46 | M | 16 | CD | Azathioprine, infliximab | 2 |
| FL009 | 48 | M | 28 | CD | Ustekinumab | 2 |
| FL010 | 74 | F | 10 | UC | Mesalamine | NA |
| FL011 | 26 | F | 11 | UC | Azathioprine, vedolizumab | 21 |
| FL012 | 44 | F | 1 | UC | Adalimumab | 6 |
| FL013 | 64 | M | 11 | UC | Mesalamine | 2 |
| FL014 | 48 | F | 25 | UC | Adalimumab | 2 |
| FL015 | 40 | M | 16 | CD | None | 2 |
| FL016 | 78 | F | 3 | UC | Vedolizumab | 2 |
| FL017 | 29 | M | 13 | CD | Infliximab | 2 |
| FL018 | 51 | M | 7 | CD | Prednisone, vedolizumab | 2 |
| FL019 | 48 | M | 7 | CD | Vedolizumab | 8 |
| FL020 | 28 | M | 12 | CD | Infliximab | 3 |
| FL021 | 35 | M | 7 | UC | Infliximab | 0 |
| FL022 | 40 | M | 19 | CD | Infliximab | 4 |
| FL023 | 44 | M | 27 | CD | Vedolizumab | 19 |
| FL024 | 43 | F | 15 | UC | Vedolizumab | 13 |
| FL025 | 28 | F | 8 | CD | Vedolizumab | 9 |
| FL026 | 39 | F | 18 | CD | None | 15 |
| FL027 | 38 | M | <1 | UC | Prednisone, infliximab | 7 |
| FL028 | 32 | F | 11 | CD | Hydrocortisone, infliximab | 3 |
| FL029 | 37 | M | 10 | CD | Infliximab | 1 |
| FL030 | 39 | M | 11 | CD | Infliximab | 8 |
| FL031 | 29 | M | 12 | CD | Infliximab | 6 |
| FL032 | 49 | M | 36 | CD | Infliximab | 4 |
| FL033 | 36 | F | 4 | UC | Infliximab | 1 |
| FL034 | 44 | F | 15 | UC | Azathioprine, vedolizumab, mesalamine | 4 |
| FL035 | 27 | F | 4 | UC | Vedolizumab | 3 |
| FL036 | 35 | M | 13 | UC | Infliximab | 1 |
| FL037 | 35 | F | <1 | UC | Infliximab | 13 |
| FL038 | 28 | M | 10 | UC | Infliximab | 0 |
| FL039 | 25 | F | 4 | UC | Vedolizumab | 2 |
| FL040 | 34 | F | 7 | UC | Infliximab | 3 |
| FL041 | 28 | M | – | Healthy | – | – |
| FL042 | 28 | M | – | Healthy | – | – |
| FL043 | 24 | M | – | Healthy | – | – |
| FL044 | 27 | M | – | Healthy | – | – |
| FL045 | 30 | F | – | Healthy | – | – |
| FL046 | 48 | F | – | Healthy | – | – |
| FL047 | 46 | F | – | Healthy | – | – |
| FL048 | 59 | M | – | Healthy | – | – |
| FL049 | 48 | F | – | Healthy | – | – |
| FL050 | 33 | F | – | Healthy | – | – |
| FL051 | 36 | M | – | Healthy | – | – |
| FL052 | 38 | M | – | Healthy | – | – |
| FL053 | 79 | M | – | Healthy | – | – |
| FL054 | 38 | F | – | Healthy | – | – |
| FL055 | 32 | M | – | Healthy | – | – |
| FL056 | 34 | F | – | Healthy | – | – |
| FL057 | 80 | M | – | Healthy | – | – |
| FL058 | 44 | F | – | Healthy | – | – |
| FL059 | 44 | M | – | Healthy | – | – |
| FL060 | 41 | F | – | Healthy | – | – |
MTX, methotrexate; NA, data not available.
Figure 3.
Flagellin-specific CD4+T cells are enriched proportionally in IBD patients. (A) Gating strategy and representative flow cytometry data for cohort 3 for all antigens tested (n = 20 each for healthy controls, CD patients, and UC patients). The percentage of CD25+OX40+ cells within CD4+ T cells after stimulation of whole blood for 44 hours with (B) SEB or Pediacel, or (C) FliC or Fla2 antigens. (D) Anti-FliC and anti-Fla2 for subjects with detectable (>0.02% of CD4+ T cells) Pediacel-specific CD4+ T cells (n = 19 healthy, n = 17 CD, n = 18 UC), the proportions of FliC- and Fla2-specific T cells were analyzed as a proportion of Pediacel-specific CD4+ T cells within each individual (Kruskal–Wallis test). PE, phycoerythrin.
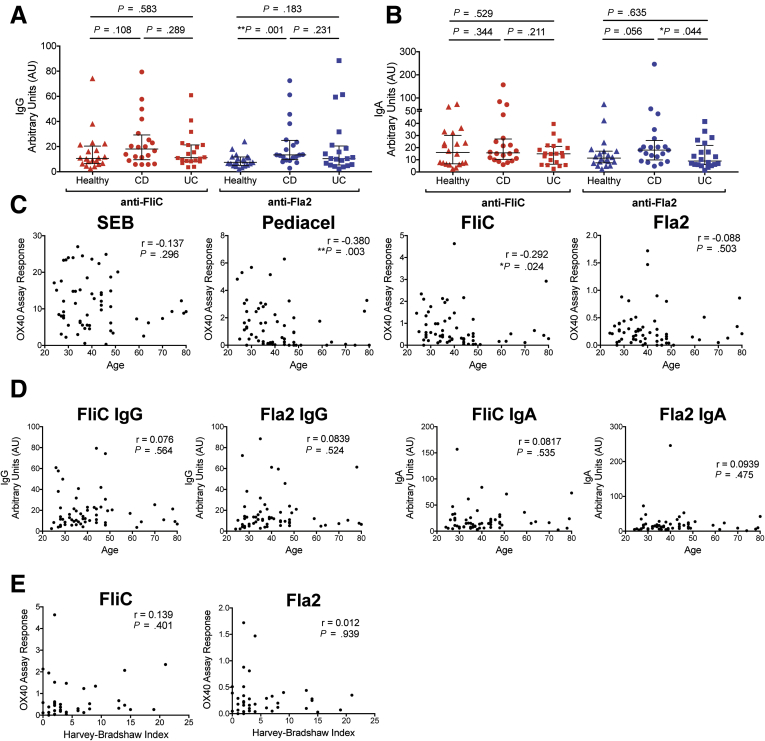
We measured the proportions of FliC- or Fla2-specific CD4+ T cells, finding no differences between the groups for Fla2, and a small, but significant, decrease in FliC-specific CD4+ T cells in CD patients (Figure 3C). However, we reasoned that proportions of disease-relevant, antigen-specific CD4+ T cells could not be compared directly between healthy control and IBD patients without correcting for the overall diminished proportion of CD4+ T cells specific for disease-irrelevant antigens. We therefore re-analyzed the proportion of FliC- or Fla2-specific CD4+ T cells as a proportion of the Pediacel response. This analysis showed that both CD and UC patients had significantly increased responses to Fla2 compared with healthy controls, with CD patients also having increased FliC responses (Figure 3D).
Correlation of Flagellin-Specific T- and B-Cell Responses With Age and Disease Severity
Similar to previous studies of CBir1 responses,7,8 compared with healthy controls, anti-Fla2 IgG, but not anti-FliC IgG, was increased in CD patients, but not in UC patients (Figure 4A). In contrast, for IgA, amounts of anti-FliC did not differ, but anti-Fla2 IgA was decreased significantly in UC compared with CD, with a trend of higher levels in CD compared with controls (Figure 4B). We next investigated the possible basis for diminished proportions of SEB-stimulated and Pediacel-specific CD4+ T cells in IBD patients. As expected, there was an inverse correlation between the proportions of Pediacel-specific CD4+ T cells, but not of SEB-stimulated CD4+ T cells, and age. Proportions of FliC-specific, but not Fla2-specific, CD4+ T cells also diminished with age, with no age-associated changes observed for anti-FliC or anti-Fla2 IgG or IgA levels (Figure 4C and D). However, because the healthy controls were age- and sex-matched, the reduced frequency of vaccine-antigen–specific CD4+ T cells in IBD patients was not attributed to age-related changes in immune functions. We also did not observe any consistent effect of treatment (the majority of this cohort was on a biologic treatment) (Table 3) or correlation between Harvey–Bradshaw Index score at the time of the blood draw and the proportion of FliC- or Fla2-specific CD4+ T cells (Figure 4E).
Figure 4.
Age and disease severity correlations with flagellin-specific immune responses. (A) IgG levels and (B) IgA levels (n = 20 for all groups) were measured by enzyme-linked immunosorbent assay. Correlation analyses of combined healthy controls, CD patients, and UC patients (n = 60, cohort 3) between age at time of blood collection and (C) OX40 responses to SEB, Pediacel, FliC, and Fla2, and (D) levels of anti-FliC and anti-Fla2 IgG and IgA. (E) Correlation analyses of CD and UC patients from cohort 3 (n = 40) between Harvey–Bradshaw Index (reported at time of blood collection) and OX40 responses to FliC and Fla2. Correlation analyses were calculated using Spearman rho (r).
Phenotype of Circulating Flagellin-Specific CD4+ T Cells in CD and UC
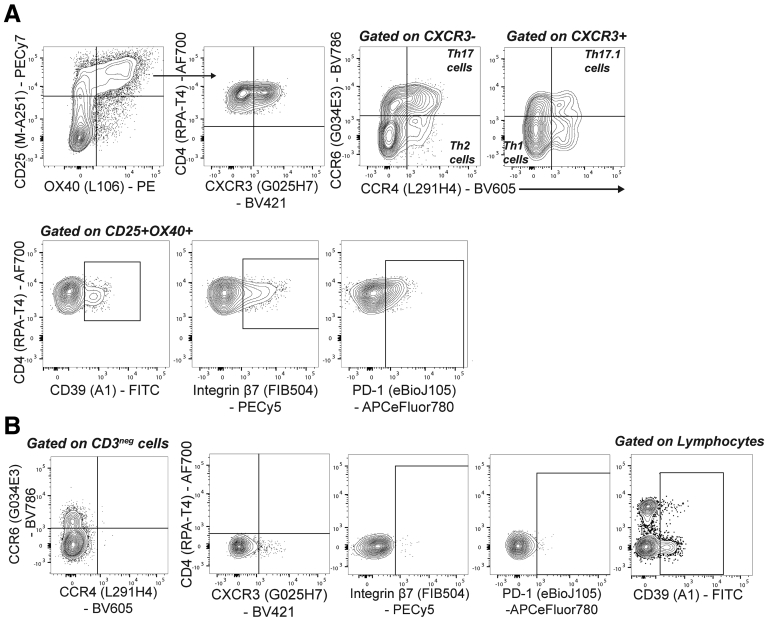
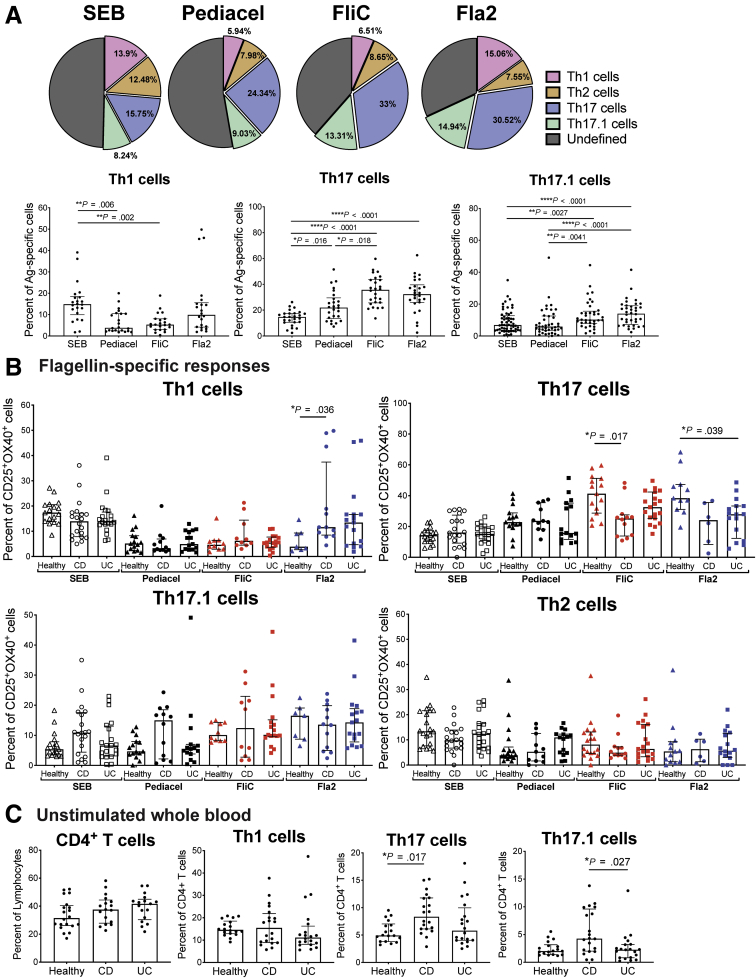
Compared with healthy controls, in CD patients, CD4+ T cells specific for FlaX- or A4-Fla2 flagellin, or E coli YidX protein, are biased toward a Th17/Th17.1 cell phenotype.6 To confirm these data, and extend them to CD patients before anti-TNFα therapy and UC patients, we performed a detailed phenotypic analysis of flagellin-specific CD4+ T cells. Specifically, within the antigen-specific CD25+OX40+ cells, we quantified proportions of Th17 (CXCR3−CCR4+CCR6+), Th17.1 (CXCR3+CCR6+CCR4+), Th1 (CXCR3+CCR4−CCR6−), and Th2 (CXCR3−CCR4+CCR6−) cells (gating in Figure 5).19,20 Compared with responses to control antigens, FliC- and Fla2-specific CD4+ T cells in the cohort as a whole were enriched for Th17 and Th17.1, but not Th1 cells (Figure 6A, the n used for each analysis is in Table 5). Surprisingly, comparisons between healthy controls and subjects with IBD showed that FliC-specific Th17 cells were decreased and Fla2-specific Th1 cells were increased in CD, and that Fla2-specific Th17 cells were decreased in UC patients (Figure 6B). Although, it should be noted that there was significant heterogeneity in the Fla2-specific Th1 cell frequencies in the CD group, so this observation requires confirmation in a larger cohort.
Figure 5.
Gating strategy for phenotypic analysis of flagellin-specific CD4+T cells. (A) Gating strategy for phenotype analysis of antigen-specific CD4+ T cells in cohort 3. (B) Gates for CCR4, CCR6, CXCR3, gut homing marker integrin β7, and the co-inhibitory marker PD-1 were set using CD3neg cells. Gates for CD39 (to identify a T regulatory (Treg)-enriched cell population) were set using lymphocytes. APC, allophycocyanin; FITC, fluorescein isothiocyanate; PECy7, phycoerythrin-cyanine 7.
Figure 6.
IBD patients have reduced proportions of flagellin-specific CD4+T cells with a Th17 cell surface phenotype. (A) Data from all subjects in cohort 3 (n = 20 each of healthy controls, CD patients, and UC patients) were combined to assess differences in Th1, Th17, and Th17.1 cell proportions within CD4+ T-cell responses to SEB, Pediacel, FliC, and Fla2 antigens (pie charts show mean responses). (B) Data in panel A were analyzed to determine differences between healthy controls (Healthy), CD patients, and UC patients for the proportions of Th1, Th2, Th17, and Th17.1 cells within all antigen-specific CD4+ T-cell responses measured. (C) Unstimulated whole blood for cohort 3 (n = 20 each for healthy controls, CD patients, and UC patients) was analyzed for the frequency of CD4+ T cells (percentage of lymphocytes, n = 20 each) and Th1, Th17, and Th17.1 cells (percentage of CD4+ cells). Statistical tests used were as follows: (A) Friedman test and (B and C) Kruskal–Wallis test; the n for analysis of each parameter not specified here is provided in Table 5. Ag, antigen.
Table 5.
List of n Used for Phenotype Analyses
| Antigen | Th1 cells |
Th2 cells |
Th17 cells |
CD39+ |
PD-1+ |
Integrin β7+PD-1+ |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HC | CD | UC | HC | CD | UC | HC | CC | UC | HC | CD | UC | HC | CD | UC | HC | CD | UC | |
| SEB | 17 | 20 | 20 | 20 | 20 | 20 | 17 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 20 | 18 | 20 |
| Pediacel | 15 | 12 | 15 | 18 | 11 | 15 | 15 | 11 | 15 | 18 | 12 | 16 | 18 | 12 | 16 | 18 | 8 | 14 |
| FliC | 10 | 10 | 17 | 18 | 12 | 19 | 15 | 12 | 19 | 19 | 15 | 19 | 19 | 15 | 19 | 15 | 9 | 13 |
| Fla2 | 7 | 12 | 17 | 13 | 6 | 16 | 11 | 6 | 16 | 15 | 12 | 18 | 15 | 12 | 18 | 9 | 7 | 9 |
| Unstimulated | 18 | 20 | 20 | 18 | 20 | 20 | 18 | 20 | 20 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
HC, healthy control; NA, not assessed.
To determine if the reduced proportions of FliC- and Fla2-specific Th17 cells were owing to reduced numbers of these cells within circulating CD4+ T cells of IBD patients, cell frequencies were measured in unstimulated whole blood. We observed that CD patients had higher frequencies of circulating Th17 cells compared with healthy controls, and higher frequencies of Th17.1 cells compared with UC patients, with no differences in Th1 cells (Figure 6C). Thus, the reduced proportions of FliC-/Fla2-specific Th17 cells in IBD patients is not driven by a systemic loss of differentiation toward these phenotypes. Overall, FliC- and Fla2-specific CD4+ T cells were skewed toward a Th17 cell phenotype. Evidence that this phenotype also was present in healthy controls and diminished in CD patients suggests that Th17-biased CD4+ T-cell responses to flagellin antigens may be a feature of a healthy immune system.
Novel Subsets of Flagellin-Specific CD4+ T Cells Discriminate IBD Patients From Healthy Controls and Associate With Disease Severity
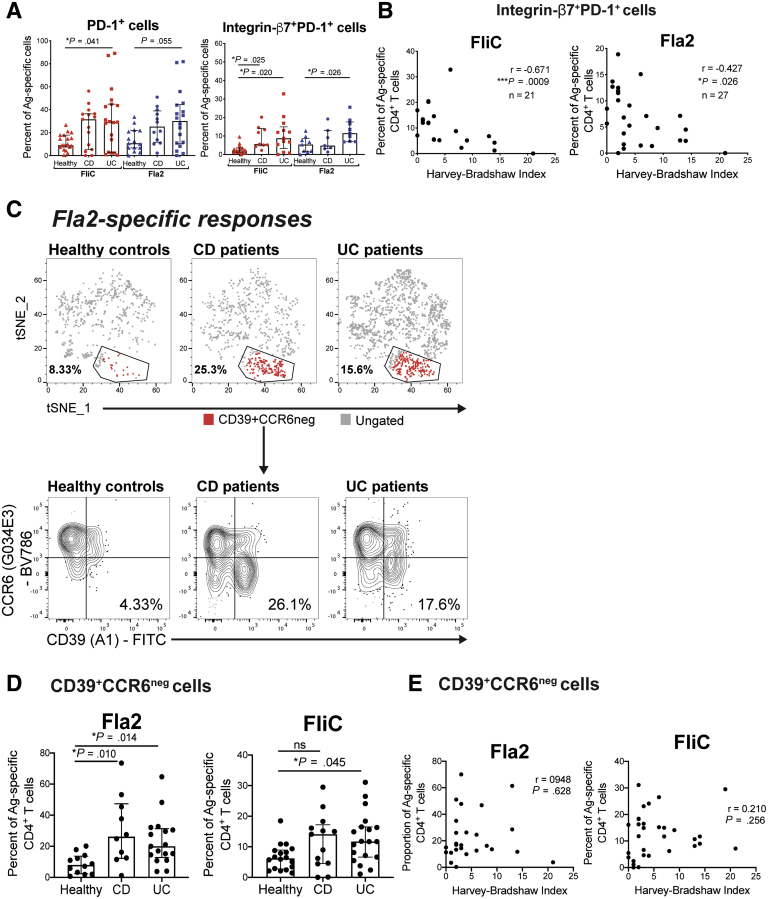
We next asked whether there were differences in the proportions of flagellin-specific CD4+ T cells that expressed the gut homing integrin β7, a target of the licensed IBD drug vedolizumab,21 and/or the co-inhibitory marker PD-1, which has a role in mucosal intestinal tolerance22 (gating in Figure 5). Both CD and UC patients had increased proportions of PD-1+ and integrin β7+PD-1+ FliC- or Fla2-specific CD4+ T cells (Figure 7A). There were no differences in expression of 2 other co-inhibitory markers: T cell immunoreceptor with immunoglobulin and immunoreceptor tyrosine-based inhibition motif domains (TIGIT) and CD226. This integrin β7+PD-1+ cell population may be a useful biomarker for disease severity because there was a significant inverse correlation with the Harvey–Bradshaw Index score (Figure 7B).
Figure 7.
Flagellin-specific gut homing and regulatory CD4+T-cell populations are enriched in IBD patients. (A) For cohort 3, differences were assessed between groups for the proportions of PD-1+ cells and integrin β7+PD-1+ cells within FliC- or Fla2-specific CD4+ T-cell responses. (B) Correlation analysis between the Harvey–Bradshaw Index and the proportion of FliC-specific (n = 21) and Fla2-specific (n = 16) CD4+ T cells that were integrin β7+PD-1+. (C) tSNE plots of concatenated Fla2-specific responses, with a gated CD39+CCD6neg cell population identified by FlowSOM clustering analysis. The CD39+CCD6neg cell population gate and frequency are shown for concatenated Fla2-specific responses. (D) Proportions of CD39+CCD6neg cells within Fla2-specific (n = 12 HC, n = 10 CD, and n = 17 UC) and FliC-specific (n = 18 HC, n = 13 CD, and n = 19 UC) responses. (E) Spearman rho correlation between frequency of Fla2-specific (n = 27) or FliC-specific (n = 32) CD4+ T cells and the Harvey–Bradshaw Index. The n for analysis of each parameter in panels A and D is shown in Table 5. (A, C, D, and F) The Kruskal–Wallis test was used. Ag, antigen; FITC, fluorescein isothiocyanate.
Because we found that Fla2-specific, but not FliC-specific, responses were enriched significantly in both CD and UC patients, we asked whether there may be novel subpopulations within these antigen-specific T cells that could discriminate between patients and controls. Unsupervised dimensionality reduction and clustering analysis of Fla2-specific CD4+ T cells showed a population of CD39+CCR6neg cells that was more prevalent in IBD patients compared with controls (Figure 7C). This observation was confirmed through traditional hierarchical gating analysis, with CD39+CCR6neg cells found to be increased significantly within Fla2-specific, but not FliC-specific, CD4+ T cells in CD and UC patients compared with controls (Figure 7D). The frequency of this population, however, did not correlate with disease severity (Figure 7E).
Gut Microbiota Diversity Is Reduced in IBD Patients and Correlates With Flagellin-Specific T Cells
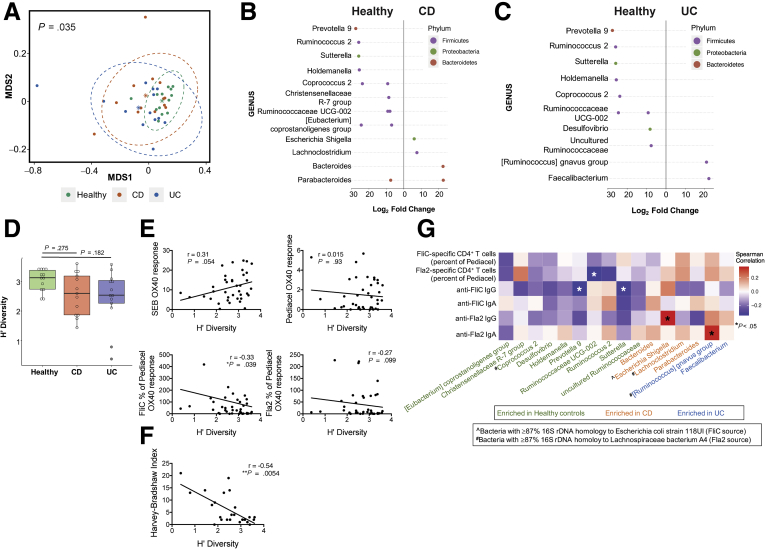
The fecal microbiome of IBD patients is reduced in diversity compared with healthy controls, with a distinct microbial signature described for CD patients.3 How these changes in the gut microbiota might correlate with anticommensal immunity is unknown. To investigate if there was a relationship between flagellin-specific adaptive immunity and gut microbiota composition, we performed 16S ribosomal DNA (rDNA) sequencing on stool samples from a subset of subjects in cohort 3. Unsupervised clustering on the basis of β-diversity showed that healthy individuals clustered together with significant differences in the dispersion patterns between healthy controls, CD patients, and UC patients (P = .035) (Figure 8A). At the genus level, several bacteria were differentially abundant between healthy controls and CD (Figure 8B) or UC patients (Figure 8C), confirming previous reports.3,23 Although there were no statistically significant differences in the Shannon’s diversity (H’) indices between groups, there was a trend toward lower diversity in both CD and UC groups compared with healthy controls (Figure 8D). Correlation analyses with the Shannon’s diversity (H’) index showed significant inverse correlations with both FliC-specific CD4+ T cells and the Harvey–Bradshaw Index (Figure 8E and F).
Figure 8.
Correlation of fecal microbiome composition with antiflagellin immune responses. (A) Nonmetric multidimensional scaling (MDS1 and MDS2) ordination plot from analysis of β-diversity within 16S rDNA samples for healthy controls, CD patients, and UC patients in cohort 3 (n = 13 each). Adonis multivariate analysis identified that both UC and CD patient groups clustered significantly distinctly from healthy controls (P = .035). Bacteria are shown that were significantly differently abundant in (B) CD patients and (C) UC patients compared with healthy controls. (B and C) Multiple dots indicate detection of >1 sequence for the bacterial genus. (D) Comparison of Shannon’s diversity (H’) index between groups (Kruskal–Wallis test). Spearman (r) correlation between Shannon’s diversity (H’) index and (E) immune responses in patients and healthy controls (n = 39) and (F) Harvey–Bradshaw index for UC and CD patients (n = 26). (G) Heat map showing Spearman correlation analyses between the relative abundance of differentially expressed bacteria and antiflagellin immune responses.
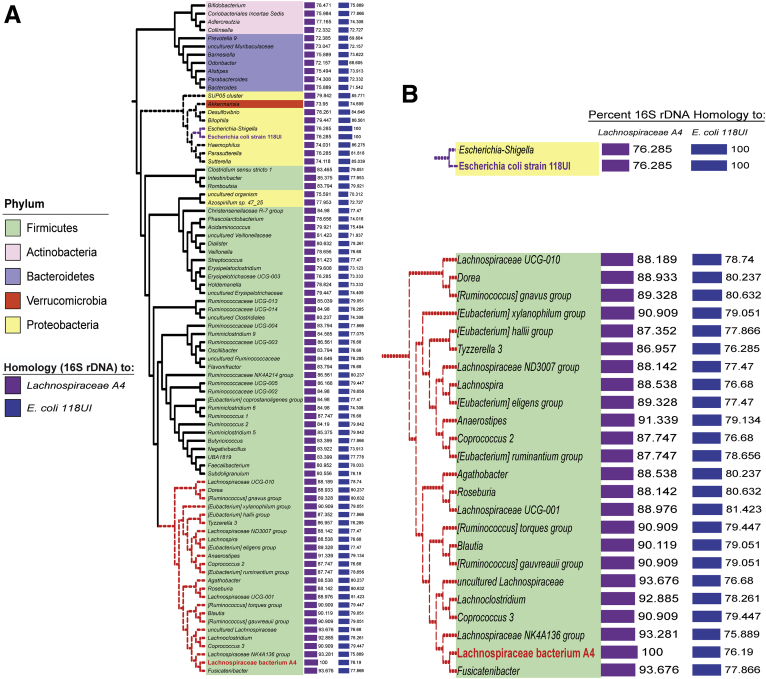
To identify bacterial species likely to produce similar flagellin antigens to FliC and Fla2, we used National Center for Biotechnology Information basic local alignment search tool (BLAST) to identify bacteria with more than 87% homology by 16s rDNA to the species that are the source of FliC (E coli 118UI) and Fla2 (Lachnospiraceae A4) (Figure 9). We then performed correlation analyses between bacteria identified as differentially abundant in CD and UC patients compared with healthy controls and FliC-/Fla2-specific T- and B-cell responses. We found significant positive correlations between the relative abundance of bacteria more prevalent in IBD patients (Escherichia/Shigella and [Ruminococcus] gnavus groups) and concentrations of anti-Fla2 IgG and anti-Fla2 IgA, respectively. Reciprocally, there were inverse correlations between the abundance of bacteria more prevalent in healthy controls (Prevotella 9, Ruminococcaceae UCG-002, and Sutterella) and the proportion of FliC- or Fla2-specific CD4+ T cells (Figure 8G). These data indicate that the increased abundance of CD-/UC-associated bacteria in the fecal microbiome, some of which are highly homologous to the bacterial strains producing FliC and Fla2, occurs in parallel with increased antiflagellin T- and B-cell responses.
Figure 9.
Phylogenetic clustering of bacterial species based on FliC/Fla2 16S rDNA homology. Phylogenetic tree of bacteria closely related by 16S rDNA sequence to E coli strain 118UI, the source of FliC, and Lachnospiraceae bacterium A4, the source of Fla2. Enlarged portions of the tree are shown for bacteria with >87% homology to these strains.
Discussion
We report here the quantification and phenotype of ex vivo flagellin-specific CD4+ T cells in peripheral blood from both CD and UC patients, including a longitudinal assessment before and after anti-TNFα therapy in CD patients. Our data show that, compared with healthy controls, both CD and UC patients have a proportional increase in circulating flagellin-specific CD4+ T cells, but that these cells are altered phenotypically with a decrease in Th17 cells and an increase in integrin β7+PD-1+ cells and CD39+CCR6neg cells. In combination with evidence for an association between changes in antiflagellin immunity and dysbiosis, these data suggest that altered immunity to flagellated bacteria may contribute to IBD pathogenesis.
An advantage of the OX40 assay is that it detects antigen-specific CD4+ T cells independently from their ability to produce cytokines and/or proliferate. By using this assay, we found that compared with age- and sex-matched healthy controls, both CD and UC patients (the majority of whom were being treated with a biologic therapy) had reduced frequencies of superantigen- and vaccine-specific CD4+ T cells. Our data on vaccine-specific immunity are consistent with a previous study that showed that IBD patients had reduced levels of antidiphtheria or antipertussis antibodies, particularly in subjects receiving anti-TNFα therapy,24 and that IBD patients had increased susceptibility to vaccine-preventable illnesses.25 In contrast, another study found that CD patients and healthy controls had similar levels of tetanus toxoid–specific CD4+ T-cell responses,6 but this was determined by measuring proliferation, which can underestimate cellular frequencies. The fact that IBD patients have reduced T-cell immunity to vaccine antigens suggests that the disease and/or treatments affect long-lived, T-cell memory and indicate that future studies of disease-relevant, antigen-specific immunity in subjects with IBD should include appropriate disease-irrelevant antigens as controls.
We found that flagellin-specific CD4+ T cells were detectable in the peripheral blood of healthy individuals,6 and that both CD and UC patients had a significant increase in the frequency of Fla2-specific, and a similar trend for FliC-specific, CD4+ T cells. Similar to previous reports,26,27 CD patients, but not UC patients, also had significantly increased levels of anti-Fla2 IgG and a trend toward higher anti-Fla2 IgA levels (P = .056), but there were no differences between CD or UC patients or controls for the amounts of anti-FliC IgG or IgA. We also found that anti-Fla2 IgA levels were significantly higher in CD patients compared with UC patients; however, this may be owing to the fact that there were more males in the CD vs UC group (75% vs 30%), because males have higher IgA levels compared with females.28 Overall, although CD4+ T cells are important to generate high-affinity, class-switched antibodies, the lack of correlation between the amounts of circulating antiflagellin antibodies and flagellin-specific T cells shows that for the antigens tested, antibody levels cannot be used to infer relative frequencies of CD4+ T cells.
Phenotypic analysis of circulating flagellin-specific CD4+ T cells confirmed the previous observation that Fla2-specific cells in CD patients are biased toward a Th17 cell phenotype.6 Here, we extended these observations to UC patients, CD patients before to commencing anti-TNFα therapy, and for responses to FliC antigen. Interestingly, and in contrast to the previous study,6 we found that compared with healthy controls the Th17 cell proportion of flagellin-specific CD4+ T cells was reduced in CD and UC patients. This difference may be owing to the fact that Calderon-Gomez et al6 defined Th17 cells on the basis of cytokine production after in vitro expansion, whereas we quantified them on the basis of ex vivo characteristic chemokine-receptor expression. Our data showing reduced Th17 cells are supported by the observation that monocyte-derived dendritic cells from CD patients have an impaired ability to generate Th17 responses from memory cells. Future work should investigate differences in the cytokine secretion profiles of the ex vivo FliC- and Fla2-specific cells between IBD patients and healthy controls.
Interestingly, we found UC and, to a lesser extent, CD patients had increased expression of PD-1 on FliC- and Fla2-specific CD4+ cells, particularly on those that also were integrin β7+. Moreover, the significant inverse correlation between the Harvey–Bradshaw Index and the proportion of integrin β7+PD-1+ cells within FliC- or Fla2-specific CD4+ T cells suggests that as disease worsens these cells are no longer in circulation, possibly owing to increased migration to the intestinal tissue. An alternate explanation could be that the phenotype of flagellin-specific cells may influence disease severity, with co-expression of integrin β7 and PD-1 resulting in improved symptoms. Indeed, commensal–specific CD4+ T cells have been reported to be present in intestinal tissue from IBD patients.29 These observations were not driven by different frequencies of integrin β7+ cells between patients and controls, with healthy controls’ unstimulated CD4+ T cells actually containing higher proportions of gut-homing cells. PD-1 has been described as a marker of cellular exhaustion, although this has been largely from studies of CD8+ T cells in the setting of chronic viral infection.30 Evidence for PD-1 marking exhausted CD4+ T cells is limited, with more evidence pointing toward its expression being indicative of increased cell activation and/or of suppressive capacity.31 Therefore, these PD-1+ gut-homing, flagellin-specific cells could represent an activated, suppressive cell subset. The presence of integrin β7+PD-1+ flagellin-specific CD4+ T cells in healthy controls could be a contributing factor to why some cancer patients receiving anti–PD-1 therapy develop IBD.32
We also identified a novel population of CD39+CCR6neg Fla2-/FliC-specific CD4+ T cells that was enriched in both CD and UC patients. Because expression of CD39 identifies a T regulatory cell–enriched population of antigen-specific cells in this assay,33 CD39+CCR6neg cells may represent an IBD-relevant subpopulation of T regulatory cells that may have an impaired ability to migrate to sites of Th17 cell–driven inflammation.34 Analysis of fecal microbiome samples in our study showed that compared with healthy controls, CD and UC patients had distinct β-diversity patterns and a reduced Shannon’s Diversity index that correlated inversely with the Harvey–Bradshaw Index.3 Interestingly, the relative abundance of bacteria that were enriched in CD and UC subjects correlated positively with amounts of anti-Fla2 antibodies. Similarly, a loss of bacterial species that were associated with a healthy microbiome correlated with reduced levels of anti-FliC IgG and Fla2-specific CD4+ T cells. These data suggest that changes in the intestinal microbiome might be driving changes in anticommensal adaptive immune responses, which in turn may be shaping the microbial communities.
In conclusion, flagellin-specific CD4+ T cells are part of the normal immune repertoire in healthy individuals, and are increased proportionally in both CD and UC patients. Comparisons of T- and B-cell responses to FliC vs Fla2 showed that immunity to Fla2, rather than FliC, more accurately discriminates between healthy individuals and CD and UC patients. Notably, neither the proportion nor phenotype of flagellin-specific CD4+ T cells, nor amounts of circulating flagellin-specific IgG or IgA, were different between CD and UC patients, indicating that dysregulated antiflagellin immunity likely is relevant to both diseases. Evidence that changes in adaptive immunity were correlated with intestinal dysbiosis suggest that there could be a previously unappreciated link between alterations in the microbiome and the development and/or amplification of inappropriate immunity to bacteria.
Materials and Methods
Subjects
Human peripheral blood was obtained following protocols approved by Clinical Research Ethics Boards of the University of British Columbia (H09-02401, H09-01238, and H18-02553) and the McGill University Institutional Review Board (A06-M65-07A). IBD patients were recruited at the McGill University Health Centre (cohort 1) or the Vancouver Gastrointestinal Research Institute (cohorts 2 and 3); age- and sex-matched healthy controls were recruited from the British Columbia Children’s Hospital Research Institute. Exclusion criteria for all cohorts were human immunodeficiency virus infection, hematologic malignancy, immunodeficiency state unrelated to IBD, or hemorrhagic disorder. Patient characteristics and disease history were collected at enrolment and are detailed in Table 1, Table 2, Table 3. Disease activity was measured using the Harvey–Bradshaw Index, with the scores interpreted as follows: < 5 indicated disease remission, 5–7 indicated mild disease, 8–16 indicated moderate disease, and >16 indicated severe disease. Investigators were blinded to disease categorization until data analyses were completed. All authors had access to the study data and reviewed and approved the final manuscript.
Sample Collection and Processing
Peripheral blood (9 mL) was collected in sodium heparin vacutainers (Becton-Dickinson, Franklin Lakes, NJ) and transported at ambient temperature. For cohort 1, blood was processed within 30 hours of collection, and for cohorts 2 and 3, blood was processed within 8 hours of collection. An aliquot of whole blood was tested immediately in OX40 assays (described later), and the remaining blood was separated into plasma and peripheral blood mononuclear cells for storage at -80°C or in liquid nitrogen, respectively. Stool samples were collected into stool nucleic acid collection and preservation tubes (Norgen Biotek Corp, Thorold, ON, Canada), shipped at room temperature, and stored at -80°C.
Reagents
SEB was from Sigma-Aldrich (St. Louis, MO). Pediacel, a pentavalent vaccine containing components of pertussis vaccine, diphtheria and tetanus toxoids, inactivated poliomyelitis vaccine, and Haemophilus influenzae B conjugate vaccine, was from Sanofi Pasteur Ltd (Lyon, France). Recombinant FliC from enteroaggregative E coli 042 serotype O44:H18 (GenBank accession: AF194946) and A4-Fla2 (termed Fla2) from Lachnospiraceae family bacteria A4 (GenBank accession: DQ789126) were generated as previously described,35 with an additional step to remove endotoxins by passing protein preparations through a polymyxin B agarose column (P1411; Sigma-Aldrich) at least 10 times. Purified FliC and Fla2 were tested with Pyrotell lysate (GS003, Cape Cod, Inc, Falmouth, MA) to ensure endotoxin levels were <0.03 U/mL. Aliquots of all antigens were stored at -80°C.
Identification of Antigen-Specific T Cells Using the OX40 Assay
The OX40 assay was performed as previously described.13,36 Briefly, whole blood was diluted 1:1 with Iscove's modified Dulbecco's medium (Thermo Fisher Scientific, Waltham, MA) and 200 μL of blood/media mix was placed into each well in a 48-well tissue culture plate (Becton-Dickinson). Assay wells were either left unstimulated or incubated with SEB (1 μg/mL), Pediacel (1/40 dilution), FliC or Fla2 antigens were added at 1 μg/mL for cohort 1 and at 10 μg/mL for cohorts 2 and 3. After 44 hours at 37°C (5% CO2), cells were stained with the relevant monoclonal antibody (mAb) panels in 5mL polystyrene round-bottom tubes (Becton-Dickinson) for 15 minutes at room temperature followed by incubation with 500 μL Optilyse C (Beckman Coulter, Brea, CA) for 10 minutes to lyse red blood cells. When more than 2 Brilliant Violet conjugates were used in a panel, 50 μL of Brilliant Buffer (Becton-Dickinson) was added to the mAb master mix. Tubes were washed with 2 mL phosphate-buffered saline, cells were pelleted by centrifugation at 400 × g for 5 minutes, supernatant was discarded, and cells were resuspended in 200 μL phosphate-buffered saline. A 4-laser LSRII FortessaX20 flow cytometer (Becton-Dickinson) was used for data acquisition, using application settings established for each mAb panel. OX40 assay mAb panels are listed in Table 4 for cohorts 1, 2, and 3. A positive OX40+CD25+ response was defined as being >0.02% of CD4+ T cells (calculated as means ± 3 SD of unstimulated wells) and consisting of at least 20 cells. Responses not meeting these criteria were assigned a value of zero.
Table 4.
OX40 mAb Analysis Panels
| Specificity | Conjugation | Clone | Amount/test | Manufacturer | Catalogue number |
|---|---|---|---|---|---|
| OX40 mAb analysis panel for cohort 1 | |||||
| CD3 | APC | HIT3A | 4 μL | Becton-Dickinson | 555342 |
| CD4 | FITC | OKT4 | 4 μL | eBioscience San Diego, CA | 11-0048-42 |
| CD25 | PECy7 | M-A251 | 2 μL | Becton-Dickinson | 557741 |
| CD134(OX40) | PE | L106 | 10 μL | Becton-Dickinson | 340420 |
| CD14 | APCeFluor780 | 61D3 | 2 μL | eBioscience | 47-0149-42 |
| OX40 mAb analysis panel for cohort 2 | |||||
| CD3 | Brilliant Violet 785 | OKT3 | 0.5 μL | BioLegend San Diego, CA | 317330 |
| CD4 | Alexa Fluor 700 | RPA-T4 | 1 μL | Becton-Dickinson | 557922 |
| CD25 | PECy7 | M-A251 | 5 μL | Becton-Dickinson | 557741 |
| CD134(OX40) | PE | L106 | 20 μL | Becton-Dickinson | 340420 |
| CD39 | FITC | A1 | 2 μL | BioLegend | 328206 |
| CD14 | APCeFluor780 | 61D3 | 2 μL | eBioscience | 47-0149-42 |
| Integrin β7 | PECy5 | FIB504 | 2 μL | Becton-Dickinson | 551059 |
| CCR7 | PerCPCy5.5 | G043H7 | 2 μL | BioLegend | 353220 |
| CXCR3 | Brilliant Violet 451 | G025H7 | 2 μL | BioLegend | 353716 |
| CCR6 | APC | G034E3 | 2 μL | BioLegend | 353415 |
| CCR4 | Brilliant Violet 605 | L291H4 | 1 μL | BioLegend | 359418 |
| OX40 mAb analysis panel for cohort 3 | |||||
| CD3 | Brilliant Violet 510 | SK7 | 0.5 μL | BioLegend | 344828 |
| CD4 | Alexa Fluor 700 | RPA-T4 | 1 μL | Becton-Dickinson | 557922 |
| CD25 | PECy7 | M-A251 | 5 μL | Becton-Dickinson | 557741 |
| CD134(OX40) | PE | L106 | 20 μL | Becton-Dickinson | 340420 |
| CD39 | FITC | A1 | 2 μL | BioLegend | 328206 |
| PD-1 | APCeFluor780 | eBioJ105 | 2 μL | eBioscience | 47-2799 |
| Integrin β7 | PECy5 | FIB504 | 2 μL | Becton-Dickinson | 551059 |
| TIGIT | Alexa Fluor 647 | A15153G | 1 μL | BioLegend | 372724 |
| CXCR3 | Brilliant Violet 451 | G025H7 | 2 μL | BioLegend | 353716 |
| CCR4 | Brilliant Violet 605 | L291H4 | 1 μL | BioLegend | 359418 |
| CD226 | Brilliant Violet 711 | DX11 | 2 μL | Becton-Dickinson | 564796 |
| CCR6 | Brilliant Violet 785 | G034E3 | 2 μL | BioLegend | 353422 |
Flow Cytometry Data Analysis
Analysis was performed using FlowJo, LLC software (v10.5; Becton-Dickinson). For cohort 3, unsupervised analysis of antigen-specific CD4+ T-cell populations used the DownSample v3.0.0 plugin to generate populations of 100 cells (populations <100 cells were excluded), which then were concatenated (combining data from healthy controls, CD patients, and UC patients). tSNE v2.037 and FlowSOM v1.538 were run on these files as plugins in FlowJo with input parameters of CCR4, CCR6, CXCR3, CD39, PD-1, integrin β7, CD226, and TIGIT. The healthy controls, CD patients, and UC patients then were gated from keywords in the parent file and exported as separate files to enable comparative data visualization with embedded tSNE and FlowSOM analysis parameters. All plugins were downloaded from www.flowjo.com/exchange (FlowJo, LLC, Ashland, OR).
Enzyme-Linked Immunosorbent Assay
Quantification of antiflagellin IgG and IgA levels in plasma was performed by enzyme-linked immunosorbent assay. Maxisorp 96-well plates (Nunc, Roskilde, Denmark) were coated with 1 μg/mL FliC or Fla2 in 0.1 mol/L carbonate buffer (33.5 mmol/L Na2CO3, 0.1 mol/L NaHCO3, pH 9.6) and incubated overnight at 4°C. Plates were blocked with 3% bovine serum albumin (Sigma-Aldrich) in Tris-buffered saline containing 0.05% Tween-20 (3% BSA/TBST) for 2 hours at room temperature on a rapid shaker. All washes were performed with Tris-buffered saline/0.1% Tween-20. Plates were washed and incubated for 1 hour at room temperature with subject plasma diluted 1:100 in 3% BSA/TBST. Standards consisted of pooled high-titer plasma from the study cohort used at one-half serial dilutions from 1/25 to 1/1600. Samples and standards were assayed in triplicate. Plates were washed and incubated with goat anti-human IgG or IgA conjugated to horseradish peroxidase (109-035-088, 109-035-011; Jackson ImmunoResearch, West Grove, PA) at 1:5000 in 3% BSA/TBST for 1 hour at room temperature. Plates were developed with tetramethylbenzidine (Becton Dickinson) for 10–15 minutes at room temperature and the reaction was stopped by addition of 1 N HCl. The optical density of each well was analyzed at 450 nm on an iMark (BioRad, Hercules, CA) plate reader and analyzed using 5-parameter logistic curve fitting in Prism v7 (GraphPad Software, Inc, San Diego, CA).
DNA Extraction and Sequencing
DNA was isolated from stool samples using a modified DNAeasy Powersoil protocol (12888; Qiagen, Hilden, Germany). Briefly, 200 uL of bead solution was replaced with 200 uL of phenol:chloroform:isoamyl alcohol (P2069; Sigma-Aldrich, St. Louis, MO) before adding 25 mg of homogenized stool. Mechanical and chemical lysis was performed according to the manufacturers’ protocol. Then, 650 μL stool lysate was combined with 600 μL of 98% ethanol and 600 μL solution C4, vortexed, and loaded onto a silica membrane. The membrane was washed with 600 μL of 98% ethanol, 500 μL solution C5, and DNA eluted with 60 μL of solution C6 and stored at -80°C. DNA was quantified using the Qubit fluorometer (Q32857; Invitrogen, Carlsbad, CA) using the double-stranded DNA BR assay kit (Q32853; Invitrogen) and submitted to Microbiome Insights (Vancouver, British Columbia, Canada) for library preparation and Illumina Miseq sequencing of the V4 region on the 16s rRNA gene.
16s rDNA Sequence Analysis
FASTQ files were loaded into QIIME2 version 2018.1139 for preprocessing, quality filtering, taxonomic assignment, and phylogenetic analysis. Reads were truncated at 245 bp (forward) and 97 bp (reverse) to remove low-quality bases. The DADA240 pipeline was applied to truncated reads and taxonomy was assigned using the Naive Bayes pretrained SILVA taxonomic data set (version 13241). A phylogenetic tree was constructed using the QIIME2 plug-in, align-to-tree-mafft-fasttree pipeline with default settings. Sequence variants were removed if they appeared in <10% of the samples, or in >10% of the nontemplate controls. Cyanobacteria sequences were removed from analysis because they shared more homology to 16s rDNA of plants than to bacteria. We used National Center for Biotechnology Information basic local alignment search tool (BLAST) to compare our sequence data against 16S rDNA sequences from the species that are the source of FliC (E coli 118UI; GenBank: CP032515.1) and Fla2 (Lachnospiraceae A4; GenBank: DQ789118.1) to identify bacteria with >85% homology.
Statistics
Unless stated otherwise, statistical analyses of 3 or more groups used Kruskal–Wallis 1-way analysis of variance, or a Friedman 1-way analysis of variance if samples were paired, with the Dunn multiple comparison post-test. Comparisons between 2 groups used the Mann–Whitney U test or, if samples were paired, a Wilcoxon signed-rank test. Correlation analyses calculated Spearman rho (r). P values were considered significant when <.05. Prism v8 (GraphPad Software, Inc) was used for all statistical analyses. Unless stated otherwise, error bars represent medians ± interquartile range. For DNA analyses, the phyloseq R package (v1.26.142) was used for statistical analysis. Unweighted Unifrac43 was used to determine β-diversity, adonis244 was used to test statistical differences of sample composition, and Deseq245 was used to identify differentially abundant amplicon sequence variants.
Acknowledgments
The authors wish to thank Mr Matthew Suzuki at the Vancouver Gastrointestinal Research Institute for patient recruitment and sample collection; and Drs Nabila Seddiki, John J. Zaunders, and Anthony D. Kelleher for their helpful discussions on using the OX40 assay for this project.
Footnotes
Author contributions Laura Cook designed the experiments, acquired, analyzed, and interpreted data, and wrote the manuscript; Daniel J. Lisko performed all microbiome experiments and analyzed data; May Q. Wong, Rosa V. Garcia, and Megan E. Himmel designed the experiments and acquired and analyzed data; Ernest G. Seidman and Brian Bressler contributed to the study design, patient recruitment, and critical revision of the manuscript; and Megan K. Levings and Theodore S. Steiner obtained funding and contributed to the study concept, design, supervision, and critical revision of the manuscript.
Conflicts of interest These authors disclose the following: Megan K. Levings has received research funding from Bristol Myers Squibb, Takeda, CRISPR Therapeutics, and Sangamo, Inc, for work not related to this study; Theodore S. Steiner has received research funding from Merck, Rebiotix, Seres, NuBiyota, Actelion, Sanofi Pasteur, and Pfizer for work not related to this study; and Ernest G. Seidman has received research funding from AbbVie, Gilead, and Janssen for work not related to this study. The remaining authors disclose no conflicts.
Funding Supported by the Broad Medical Research Program at the Crohn's & Colitis Foundation of America (IBD-0326 to M.K.L and T.S.S); a Canadian Institutes of Health Research team grant in Immunoregulation and Inflammatory Bowel Disease (E.G.S, M.K.L., and T.S.S.); a Canada Research Chair in Immune Mediated Gastrointestinal Disorders and the B Kaufman McGill Chair in Inflammatory Bowel Disease (E.G.S.); a Frederick Banting and Charles Best Canada Graduate Scholarship (M.E.H.); and the British Columbia Children’s Hospital Research Institute Bertram Hoffmeister Postdoctoral Fellowship (L.C.).
References
- 1.Landers C.J., Cohavy O., Misra R., Yang H., Lin Y.C., Braun J., Targan S.R. Selected loss of tolerance evidenced by Crohn's disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–699. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- 2.Amre D.K., Lu S.E., Costea F., Seidman E.G. Utility of serological markers in predicting the early occurrence of complications and surgery in pediatric Crohn's disease patients. Am J Gastroenterol. 2006;101:645–652. doi: 10.1111/j.1572-0241.2006.00468.x. [DOI] [PubMed] [Google Scholar]
- 3.Pascal V., Pozuelo M., Borruel N., Casellas F., Campos D., Santiago A., Martinez X., Varela E., Sarrabayrouse G., Machiels K., Vermeire S., Sokol H., Guarner F., Manichanh C. A microbial signature for Crohn's disease. Gut. 2017;66:813–822. doi: 10.1136/gutjnl-2016-313235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Means T.K., Hayashi F., Smith K.D., Aderem A., Luster A.D. The Toll-like receptor 5 stimulus bacterial flagellin induces maturation and chemokine production in human dendritic cells. J Immunol. 2003;170:5165–5175. doi: 10.4049/jimmunol.170.10.5165. [DOI] [PubMed] [Google Scholar]
- 5.Feng T., Wang L., Schoeb T.R., Elson C.O., Cong Y. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med. 2010;207:1321–1332. doi: 10.1084/jem.20092253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calderon-Gomez E., Bassolas-Molina H., Mora-Buch R., Dotti I., Planell N., Esteller M., Gallego M., Marti M., Garcia-Martin C., Martinez-Torro C., Ordas I., Singh S., Panes J., Benitez-Ribas D., Salas A. Commensal-specific CD4(+) cells from patients with Crohn's disease have a T-helper 17 inflammatory profile. Gastroenterology. 2016;151:489–500 e3. doi: 10.1053/j.gastro.2016.05.050. [DOI] [PubMed] [Google Scholar]
- 7.Targan S.R., Landers C.J., Yang H., Lodes M.J., Cong Y., Papadakis K.A., Vasiliauskas E., Elson C.O., Hershberg R.M. Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn's disease. Gastroenterology. 2005;128:2020–2028. doi: 10.1053/j.gastro.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 8.Lodes M.J., Cong Y., Elson C.O., Mohamath R., Landers C.J., Targan S.R., Fort M., Hershberg R.M. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ni J., Wu G.D., Albenberg L., Tomov V.T. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14:573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M., Sun K., Wu Y., Yang Y., Tso P., Wu Z. Interactions between intestinal microbiota and host immune response in inflammatory bowel disease. Front Immunol. 2017;8:942. doi: 10.3389/fimmu.2017.00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., Eeckhaut V., Ballet V., Claes K., Van Immerseel F., Verbeke K., Ferrante M., Verhaegen J., Rutgeerts P., Vermeire S. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 12.Lin L., Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017;18:2. doi: 10.1186/s12865-016-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaunders J.J., Munier M.L., Seddiki N., Pett S., Ip S., Bailey M., Xu Y., Brown K., Dyer W.B., Kim M., de Rose R., Kent S.J., Jiang L., Breit S.N., Emery S., Cunningham A.L., Cooper D.A., Kelleher A.D. High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40) J Immunol. 2009;183:2827–2836. doi: 10.4049/jimmunol.0803548. [DOI] [PubMed] [Google Scholar]
- 14.Reiss S., Baxter A.E., Cirelli K.M., Dan J.M., Morou A., Daigneault A., Brassard N., Silvestri G., Routy J.P., Havenar-Daughton C., Crotty S., Kaufmann D.E. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS One. 2017;12 doi: 10.1371/journal.pone.0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duck L.W., Walter M.R., Novak J., Kelly D., Tomasi M., Cong Y., Elson C.O. Isolation of flagellated bacteria implicated in Crohn's disease. Inflamm Bowel Dis. 2007;13:1191–1201. doi: 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- 16.Steiner T., Ivison S., Wang C., Elson C. P-0163: the A4-Fla2 flagellin, a dominant antigen in Crohn's disease, is a poor TLR5 agonist. Inflamm Bowel Dis. 2009;15(Suppl 2) S55-S. [Google Scholar]
- 17.Sitaraman S.V., Klapproth J.M., Moore D.A., 3rd, Landers C., Targan S., Williams I.R., Gewirtz A.T. Elevated flagellin-specific immunoglobulins in Crohn's disease. Am J Physiol Gastrointest Liver Physiol. 2005;288:G403–G406. doi: 10.1152/ajpgi.00357.2004. [DOI] [PubMed] [Google Scholar]
- 18.Rodstrom K.E., Elbing K., Lindkvist-Petersson K. Structure of the superantigen staphylococcal enterotoxin B in complex with TCR and peptide-MHC demonstrates absence of TCR-peptide contacts. J Immunol. 2014;193:1998–2004. doi: 10.4049/jimmunol.1401268. [DOI] [PubMed] [Google Scholar]
- 19.Acosta-Rodriguez E.V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 20.Rivino L., Messi M., Jarrossay D., Lanzavecchia A., Sallusto F., Geginat J. Chemokine receptor expression identifies Pre-T helper (Th)1, Pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. J Exp Med. 2004;200:725–735. doi: 10.1084/jem.20040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb C.A., O'Byrne S., Keir M.E., Butcher E.C. Gut-selective integrin-targeted therapies for inflammatory bowel disease. J Crohns Colitis. 2018;12(Suppl 2):S653–S668. doi: 10.1093/ecco-jcc/jjy060. [DOI] [PubMed] [Google Scholar]
- 22.Pinchuk I.V., Saada J.I., Beswick E.J., Boya G., Qiu S.M., Mifflin R.C., Raju G.S., Reyes V.E., Powell D.W. PD-1 ligand expression by human colonic myofibroblasts/fibroblasts regulates CD4+ T-cell activity. Gastroenterology. 2008;135:1228–1237. doi: 10.1053/j.gastro.2008.07.016. 37 e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joossens M., Huys G., Cnockaert M., De Preter V., Verbeke K., Rutgeerts P., Vandamme P., Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 24.Caldera F., Saha S., Wald A., Garmoe C.A., McCrone S., Megna B., Ley D., Reichelderfer M., Hayney M.S. Lower sustained diphtheria and pertussis antibody concentrations in inflammatory bowel disease patients. Dig Dis Sci. 2018;63:1532–1540. doi: 10.1007/s10620-018-5043-2. [DOI] [PubMed] [Google Scholar]
- 25.Melmed G.Y., Ippoliti A.F., Papadakis K.A., Tran T.T., Birt J.L., Lee S.K., Frenck R.W., Targan S.R., Vasiliauskas E.A. Patients with inflammatory bowel disease are at risk for vaccine-preventable illnesses. Am J Gastroenterol. 2006;101:1834–1840. doi: 10.1111/j.1572-0241.2006.00646.x. [DOI] [PubMed] [Google Scholar]
- 26.Schoepfer A.M., Schaffer T., Mueller S., Flogerzi B., Vassella E., Seibold-Schmid B., Seibold F. Phenotypic associations of Crohn's disease with antibodies to flagellins A4-Fla2 and Fla-X, ASCA, p-ANCA, PAB, and NOD2 mutations in a Swiss cohort. Inflamm Bowel Dis. 2009;15:1358–1367. doi: 10.1002/ibd.20892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoepfer A.M., Schaffer T., Seibold-Schmid B., Muller S., Seibold F. Antibodies to flagellin indicate reactivity to bacterial antigens in IBS patients. Neurogastroenterol Motil. 2008;20:1110–1118. doi: 10.1111/j.1365-2982.2008.01166.x. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez-Quintela A., Alende R., Gude F., Campos J., Rey J., Meijide L.M., Fernandez-Merino C., Vidal C. Serum levels of immunoglobulins (IgG, IgA, IgM) in a general adult population and their relationship with alcohol consumption, smoking and common metabolic abnormalities. Clin Exp Immunol. 2008;151:42–50. doi: 10.1111/j.1365-2249.2007.03545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegazy A.N., West N.R., Stubbington M.J.T., Wendt E., Suijker K.I.M., Datsi A., This S., Danne C., Campion S., Duncan S.H. Owens BMJ, Uhlig HH, McMichael A, Oxford IBD Cohort Investigators, Bergthaler A, Teichmann SA, Keshav S, Powrie F. Circulating and tissue-resident CD4(+) T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology. 2017;153:1320––1337 e16. doi: 10.1053/j.gastro.2017.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulpa D.A., Lawani M., Cooper A., Peretz Y., Ahlers J., Sekaly R.P. PD-1 coinhibitory signals: the link between pathogenesis and protection. Semin Immunol. 2013;25:219–227. doi: 10.1016/j.smim.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keir M.E., Butte M.J., Freeman G.J., Sharpe A.H. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coutzac C., Adam J., Soularue E., Collins M., Racine A., Mussini C., Boselli L., Kamsukom N., Mateus C., Charrier M., Cassard L., Planchard D., Ribrag V., Fizazi K., Loriot Y., Lepage P., Scoazec J.Y., Robert C., Carbonnel F., Chaput N. Colon immune-related adverse events: anti-CTLA-4 and anti-PD-1 blockade induce distinct immunopathological entities. J Crohns Colitis. 2017;11:1238–1246. doi: 10.1093/ecco-jcc/jjx081. [DOI] [PubMed] [Google Scholar]
- 33.Seddiki N., Cook L., Hsu D.C., Phetsouphanh C., Brown K., Xu Y., Kerr S.J., Cooper D.A., Munier C.M., Pett S., Ananworanich J., Zaunders J., Kelleher A.D. Human antigen-specific CD4(+) CD25(+) CD134(+) CD39(+) T cells are enriched for regulatory T cells and comprise a substantial proportion of recall responses. Eur J Immunol. 2014;44:1644–1661. doi: 10.1002/eji.201344102. [DOI] [PubMed] [Google Scholar]
- 34.Chaudhry A., Rudra D., Treuting P., Samstein R.M., Liang Y., Kas A., Rudensky A.Y. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steiner T.S., Nataro J.P., Poteet-Smith C.E., Smith J.A., Guerrant R.L. Enteroaggregative Escherichia coli expresses a novel flagellin that causes IL-8 release from intestinal epithelial cells. J Clin Invest. 2000;105:1769–1777. doi: 10.1172/JCI8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cook L., Munier C.M.L., Seddiki N., van Bockel D., Ontiveros N., Hardy M.Y., Gillies J.K., Levings M.K., Reid H.H., Petersen J., Rossjohn J., Anderson R.P., Zaunders J.J., Tye-Din J.A., Kelleher A.D. Circulating gluten-specific FOXP3(+)CD39(+) regulatory T cells have impaired suppressive function in patients with celiac disease. J Allergy Clin Immunol. 2017;140:1592–1603 e8. doi: 10.1016/j.jaci.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 37.van der Maaten L., Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 38.Van Gassen S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., Saeys Y. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 39.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glockner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O'Hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Szoecs E., Wagner H. Vegan: community ecology package. https://CRAN.R-Project.org/Package=Vegan Available from: 2019. Accessed February 20, 2019.
- 45.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]