Abstract
Background
DEL, the weakest D variant, is mistyped as D-negative by routine serological assays. Transfusion of red blood cells expressing the DEL phenotype has the potential to elicit anti-D alloimmunization in D-negative recipients. The goal of this study was to recommend DEL typing strategies for serologically D-negative Asian donors.
Methods
RhCE phenotyping and the adsorption-elution test were performed on 674 serologically D-negative samples. RHD genotyping using real-time polymerase chain reaction and melting curve analysis were also undertaken to identify DEL alleles. Costs and turnaround time of RhCE phenotyping, the adsorption-elution test, and RHD genotyping were estimated.
Results
Sensitivity and specificity of the adsorption-elution test for serologically D-negative samples were 94.9% (93/98) and 91.5% (527/576), respectively. C+ phenotypes were detected in all 98 samples with DEL alleles. Despite comparable costs, RHD genotyping was more accurate and rapid than the adsorption-elution test.
Conclusions
Two practical DEL typing strategies using RhCE phenotyping as an initial screening method were recommended for serologically D-negative Asian donors. Compared with DEL typing using RHD genotyping, serological DEL typing using adsorption-elution test is predicted to increase the incidence of anti-D alloimmunization and decrease the D-negative donor pool without having any cost-competitiveness but can be used in laboratories where molecular methods are not applicable.
Keywords: Adsorption-elution test, DEL, RHD genotyping, D-negative donors
Introduction
Among the non-ABO blood group antigens, the D antigen is the most immunogenic, and as little as 0.1–1 mL of D-positive red blood cells (RBCs) can induce anti-D formation in D-negative recipients [1]. Maternal anti-D is transported across the placenta during gestation, capable of causing hemolytic disease of the fetus and newborn (HDFN) [2]. Clinical presentation of HDFN due to anti-D varies from asymptomatic mild anemia to hydrops fetalis, potentially leading to intrauterine death [3]. DEL, the weakest D variant, is defined by trace amounts of D antigen serologically detectable only via the adsorption-elution test. Without routine DEL screening, blood donors with the DEL phenotype are misplaced in the D-negative donor pool [4]. Anti-D alloimmunization cases by missed DEL units have been reported, albeit rarely [5, 6, 7, 8, 9, 10, 11, 12].
Accurately identifying DEL is particularly important in Asians, as the frequency of DEL among serologically D-negative donors is 1:5–1:8 for Asians compared to 1:350–1:2,000 for Europeans [13]. At present, 44 DEL alleles are listed by the International Society of Blood Transfusion (ISBT) Working Party on Red Cell Immunogenetics and Blood Group Terminology [14]. The RHD (c.1227G>A) allele, termed RHD*01EL.01 by the ISBT, is the most prevalent DEL allele in Asians, including Koreans [15, 16, 17, 18]. In addition, only the RHD (c.1222T>C) allele, termed RHD*01EL.10 by the ISBT, has been observed in Koreans [15, 18]. On the other hand, the most common DEL alleles in Europeans are RHD(c.885G>T) and RHD(c.486 + 1G>A), termed as RHD*11 and RHD*01EL.08 by the ISBT, respectively [19, 20]. Hence, the spectrum of DEL alleles in each ethnic group should be considered to design an optimal genotyping assay [21].
Several genotyping strategies to identify DEL alleles in serologically D-negative Koreans have been proposed [15, 18, 22, 23, 24]. Furthermore, RhCE phenotypes can be utilized for DEL typing strategies, as DEL is strongly associated with C+ phenotypes in many populations [13, 15, 18, 22, 25]. As mentioned above, the adsorption-elution test has the capacity to detect the DEL phenotype directly and can thus be used for routine DEL screening, particularly when molecular methods are not available. To release safer RBC units at low incremental cost, the benefits and costs of various DEL typing strategies should be comprehensively evaluated. Crottet et al. [26]addressed the cost-efficiency of routine RHD polymerase chain reaction (PCR) for donors typed as D-negative by direct agglutination compared with conventional weak D testing using the indirect antiglobulin test (IAT). However, no study has investigated the cost-efficiency of various DEL typing strategies for serologically D-negative donors.
The objective of this study was to compare RhCE phenotyping, the adsorption-elution test, and RHD genotyping, and to recommend practical DEL typing strategies for serologically D-negative Asian donors.
Material and Methods
Blood Samples and Serological Methods
In total, 674 serologically D-negative Koreans were included in this study between June 2008 and November 2017. Serologically D-negative was defined according to the typing results of the IAT, using both the tube technique and gel microcolumn assay with at least 5 anti-D reagents. The 6 anti-D reagents used were: BIOSCOT anti-D IgM/IgG blend (Merck Millipore, Darmstadt, Germany), anti-D (Anti-RH0) BioClone (Ortho Clinical Diagnostics, Raritan, NJ, USA), anti-D (RH1) TOTEM (Diagast, Loos, France), anti-D (RH1) IgG (Diagast), Combi anti-D Mono-Type (Medion Grifols Diagnostics, Düdingen, Switzerland), and NovaClone Anti-D IgM/IgG monoclonal blend (Immucor, Norcross, GA, USA). Since August 2015, Combi anti-D Mono-Type has been unavailable, so the IAT was performed using the remaining 5 anti-D reagents. All serologically D-negative samples were tested for RhCE phenotypes using the tube technique with anti-C, anti-c, anti-E, and anti-e reagents (Diagast).
Adsorption-Elution Test
In total, 674 serologically D-negative samples were tested for the DEL phenotype by means of the adsorption-elution test. A 500-μL aliquot of washed RBCs was incubated for 1 h at 37°C with an equal volume of BIOSCOT anti-D IgM/IgG blend. The cells were washed 8 times with 1× Dulbecco's PBS, and the elution procedure was performed using the rapid acid elution kit (DiaCidel, Bio-Rad, Cressier, Switzerland). The eluate and final washed supernatant were used for the IAT against D-positive (CCDee and ccDEE) and D-negative (ccdee) reagent RBCs. Next, 25 μL of eluate (or final washed supernatant) and 50 μL of 0.8% reagent RBCs suspended in low-ionic-strength saline (LISS) were added to the LISS/Coombs card (Bio-Rad). After a 15-min incubation at 37°C, the card was centrifuged at 1,030 rpm for 10 min. When an agglutination reaction occurred (≥1+), the sample was interpreted as serological DEL.
RHD Genotyping
RHD genotyping was performed on all 674 serologically D-negative samples according to the method described by Hong et al. [18]. Real-time PCR targeting the 3′-untranslated region of exon 10 was conducted. For analysis of exon 9, real-time PCR was monitored with fluorescently labeled hybridization probes, followed by postamplification melting curve analysis. The donor probe was designed in such a way that the 2 DEL alleles, RHD (c.1222T>C) and RHD (c.1227G>A), could be distinguished from the wild-type allele by melting-curve analysis. Melting curves of RHD (c.1222T>C) and RHD (c.1227G>A) peak at 62.62 and 61.95°C, respectively. Negative results for both exons 9 and 10 were regarded as the RHD deletion. Samples testing negative for exon 9 and positive for exon 10 were considered to harbor the RHD-CE-D hybrid. The RHD deletion and the RHD-CE-D hybrid are classified here as RHD null alleles, encoding the D-negative phenotype. The term “D-negative” refers to the complete absence of the D antigen. Positive results for both exons 9 and 10 were interpreted as D variants or DEL alleles. To exclude DEL alleles testing negative for exon 9 and positive for exon 10, further molecular characterization using Partial D-TYPE (BAG Health Care, Lich, Germany) was performed on 9 samples which had been shown to harbor the RHD-CE-D hybrid on real-time RHD PCR but the DEL phenotype by adsorption-elution test.
Estimate of Costs and Turnaround Time
Cost estimates for RhCE phenotyping, the adsorption-elution test, and RHD genotyping included the cost of the reagents required to perform the assays, such as monoclonal antibodies and PCR primer/probe sets, as well as the cost of labor. The cost of purchasing and maintaining the PCR equipment was excluded from the analysis. Cost, turnaround time (TAT), and hands-on time were calculated based on the assumption that 6 samples were run simultaneously for each test. Labor costs were computed by multiplying hands-on time by the average hourly wage of USD 15. TAT was the time interval between sample receipt and reporting of the results.
Results
RHD Genotypes in Serologically D-Negative Samples
Our RHD genotyping assay revealed underlying genetic causes in 674 serologically D-negative samples. The intact RHD gene was discovered in 98 samples (14.5%), composed of 96 RHD (c.1227G>A) samples (14.2%) and 2 RHD (c.1222T>C) samples (0.3%) (Table 1).
Table 1.
Results of adsorption-elution test and RHD genotyping in 674 serologically D-negative samples
| DEL alleles | RHD deletion | RHD-CED hybrid | Total | |
|---|---|---|---|---|
| Adsorption-elution (+) | 93* | 40 | 9 | 142 |
| Adsorption-elution (–) | 5 | 492 | 35 | 532 |
| Total | 98 | 532 | 44 | 674 |
Two samples harbored the RHD (c.1222T>C) allele.
Accuracy of the Adsorption-Elution Test in Serologically D-Negative Samples
Among the 98 samples with DEL alleles, 93 (94.9%) were identified as serological DEL by the adsorption-elution test. Among the 576 samples with RHD null alleles, 527 (91.5%) tested negative for the DEL phenotype by adsorption-elution test (Table 1). In all 9 samples with the RHD-CE-D hybrid but serological DEL, RHD*D-CE(2–9)-D was detected through further investigation using PCR with sequence-specific primers.
Distribution of RhCE Phenotypes according to the RHD Genotype
The RhCE phenotypes of 674 serologically D-negative samples are described in Table 2. C+ phenotypes were found in 178 (26.4%) samples. C+ phenotypes were detected in all 98 samples with DEL alleles. In addition, the RhCE phenotypes of 5 samples testing negative for the DEL phenotype but positive for RHD (c.1227G>A) consisted of 1 CCee, 3 Ccee, and 1 CcEe.
Table 2.
Distribution of RhCE phenotypes in 674 serologically D-negative samples with DEL alleles, RHD deletion, and RHD-CE-D hybrid
| CCee | CCEe | Ccee | CcEe | ccee | ccEe | ccEE | Total | |
|---|---|---|---|---|---|---|---|---|
| DEL alleles | 10 | 1 | 85 | 2 | 98 | |||
| RHD deletion | 1 | 34 | 3 | 422 | 64 | 8 | 532 | |
| RHD-CE-D hybrid | 1 | 40 | 1 | 1 | 1 | 44 | ||
| Total | 12 | 1 | 159 | 6 | 423 | 65 | 8 | 674 |
Comparison of RhCE Phenotyping, the Adsorption-Elution Test, and RHD Genotyping
Cost, TAT, and hands-on time were highest for the adsorption-elution test and lowest for RhCE phenotyping. The advantages and disadvantages of RhCE phenotyping, the adsorption-elution test, and RHD genotyping are summarized in Table 3.
Table 3.
Comparison of RhCE phenotyping, adsorption-elution test, and RHD genotyping
| RhCE phenotyping | Adsorption-elution test | RHD genotyping | |
|---|---|---|---|
| Cost1 TAT1 Hands-on time1 |
USD 34.5 (USD 5.8/sample) 30 min 30 min (5 min/sample) |
USD 120.0 (USD 20.0/sample) 4.5 h 3.5 h (35 min/sample) |
USD 106.5 (USD 17.8/sample) 2.5 h 1.5 h (15 min/sample) |
| Advantages | Rapid, inexpensive, and no special instruments required | No special instruments required, phenotypic characterization of novel alleles | Relatively rapid and an accurate (reference method) |
| Disadvantages | Only used to exclude D-negative samples with RHD null alleles | Technically demanding, laborious, and time-consuming | Usually unable to detect all DEL alleles High initial costs for instruments |
Cost, TAT, and hands-on time were estimated, assuming that 6 samples were tested simultaneously.
Establishing Practical DEL Typing Strategies for Serologically D-Negative Asian Donors
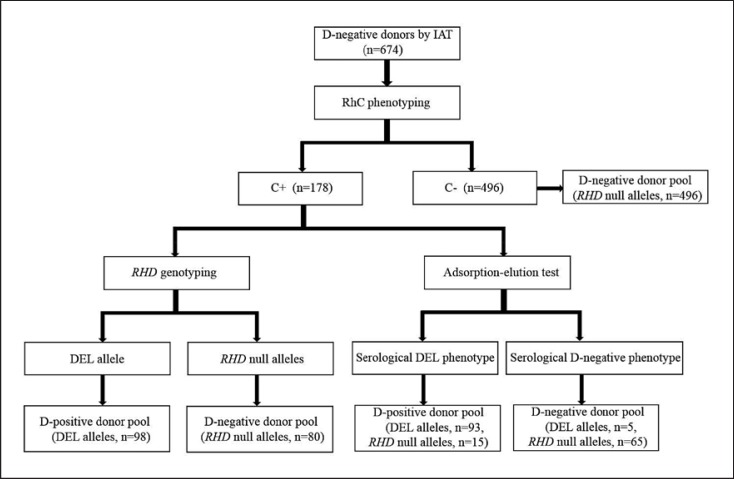
Based on the results of RhCE phenotyping, the adsorption-elution test, and RHD genotyping for 674 serologically D-negative samples, 2 practical DEL typing strategies were designed for serologically D-negative Asian donors (Fig. 1). These strategies employed RhCE phenotyping as an initial screening method to exclude serologically D-negative donors with RHD null alleles.
Fig. 1.
Two practical DEL typing strategies for serologically D-negative Asian donors. Using our data, aninitial screening method using RhCE phenotyping was predicted to exclude only donors with RHD null alleles. Serologically D-negative donors with C+ phenotypes were tested by RHD genotyping or the adsorption-elution test. Using RHD genotyping, the D-negative donor pool was predicted to contain only donors with RHD null alleles. Using the adsorption-elution test, 0.7% (5/674) of serologically D-negative donors were expected to have the potential to cause anti-D alloimmunization.
The first strategy was to genotype all serologically D-negative donors with C+ phenotypes. Using this strategy, D-negative recipients were predicted to receive blood only from donors with RHD null alleles, indicating virtually no risk of anti-D alloimmunization. A total of 576 serologically D-negative donors with RHD null alleles were included in the D-negative donor pool. The total cost of this DEL typing strategy was the sum of the cost of RhCE phenotyping of 674 serologically D-negative donors and RHD genotyping of 178 serologically D-negative donors with C+ phenotypes. On average, USD 10.5 were expected to be spent for DEL typing of each serologically D-negative donor.
The second DEL typing strategy was to detect the DEL phenotype of serologically D-negative donors with C+ phenotypes by means of the adsorption-elution test. Following this strategy, 5 donors with DEL alleles mistakenly included in the D-negative donor pool were predicted to have the potential to induce anti-D alloimmunization. A total of 561 serologically D-negative donors with RHD null alleles were included in the D-negative donor pool. The total cost of this DEL typing strategy was the sum of the cost of RhCE phenotyping of 674 serologically D-negative donors and the adsorption-elution test of 178 serologically D-negative donors with C+ phenotypes. On average, USD 11.1 were expected to be spent for DEL typing of each serologically D-negative donor.
Discussion
In many countries, including Korea, DEL blood donors are typed as D-negative because DEL typing methods are not currently used in routine practice. RHD genotyping is the current gold standard for detecting DEL [21] and is already used for routine donor screening in some countries [13, 19, 20, 26, 27]. However, the adsorption-elution test may not be applicable for routine donor screening, as it is laborious and time-consuming [19]. This test is prone to technical errors and differences in protocols and reagents, such as monoclonal anti-D and washing buffers [21]. The accuracy of this test for serologically D-negative samples has not been discussed sufficiently.
In this study, the sensitivity of the adsorption-elution test for serologically D-negative samples was 94.9% (93/98). The test may have been insufficiently sensitive to detect 5 samples with RHD (c.1227G>A). We also did not use reagent RBCs treated with papain capable of detecting very low levels of anti-D reagents in eluates. At the Blood Transfusion Service in Berne, papain-treated RBCs are used for the adsorption-elution test [26]. Further studies are needed to evaluate the sensitivity of papain-treated RBCs. In addition, C in trans with the DEL allele may have suppressed D antigen density to such low levels that the adsorption-elution test could not detect it [19]. In this study, only 1 false-negative sample had C in trans with the DEL allele; the other 4 samples could not be explained by the suppressive effects of C in trans. Lastly, the failure to detect DEL by adsorption-elution test may have been due to the low sensitivity of the anti-D reagents used, or technical errors. Future studies should evaluate various anti-D reagents used for the adsorption-elution test.
The specificity of the adsorption-elution test for serologically D-negative samples was 91.5% (527/576) in this study. Testing D variants other than DEL by adsorption-elution test is 1 of the causes of false-positive reactions. We minimized this risk by using at least 5 different anti-D reagents in the IAT. Consequently, no D variants other than DEL were discovered among 142 samples testing positive by adsorption-elution test. Furthermore, insufficient washing of adsorbed RBCs may have been a source of false-positive reactions, although the last washed supernatant was used for the IAT against D-positive reagent RBCs. Further studies are necessary to evaluate the impact of various washing buffers on the specificity of the adsorption-elution test. Finally, false-positive results may have arisen from unexplained technical errors.
One potential pitfall of our genotyping method targeting only exons 9 and 10 is that DEL alleles testing negative for exon 9 and positive for exon 10 are mistyped as RHD null alleles. DEL alleles such as RHD*-CE(4–9)-D (designated as RHD*01EL.44 by the ISBT) and RHD exon 9 deletion (no designation assigned by the ISBT) were previously reported [28, 29]. However, only RHD*D-CE(2–9)-D (designated as RHD*01N.03 by the ISBT), the second-most prevalent RHD null allele in Korea [15], was identified in 9 samples that had the RHD-CE-D hybrid but serological DEL. The most likely explanation for this is false-positive reactions of adsorption-elution test. Moreover, samples testing negative for bothexons 9 and 10 are very unlikely to harbor DEL alleles because the 2 possible RHD alleles are the RHD deletion and RHD*CE(1)-D(6)-CE(7–10), termed RHD*01N.01 and RHD*01N.42 by the ISBT, respectively. Likewise, positive reactions of the adsorption-elution test in 40 samples typed as the RHD deletion were believed to be false-positive. We conclude that there are few, if any, DEL alleles other than RHD (c.1222T>C) and RHD (c.1227G>A) in Korea. Designing a RHD genotyping assay to detect additional DEL alleles may thus not be necessary.
Our study shows that all serologically D-negative donors with C– are predicted to have RHD null alleles. Given that RhCE phenotyping is a quick and inexpensive method, DEL typing for serologically D-negative donors with C+ is more cost-efficient than for all serologically D-negative donors. Our data suggest that an initial screening method using RhCE phenotyping is expected to reduce the number of candidates for DEL typing, using the more expensive and time-consuming adsorption-elution test or RHD genotyping, to 26.4% (178/674). The cost-efficiency of RhCE phenotyping as an initial screening method varies among populations depending on the frequency of C+ phenotypes of serologically D-negative donors.
Carrying almost no risk of anti-D alloimmunization, the cost of DEL typing strategy using RHD genotyping (USD 10.5) was slightly lower than that of serological DEL typing by means of the adsorption-elution test (USD 11.1). As our cost estimates did not include the costs of introducing and maintaining the instruments used for RHD genotyping, the actual difference between the 2 strategies may have been negligible. Pooled testing is capable of driving down genotyping costs significantly but is only applicable in populations where DEL donors are not frequently observed among serologically D-negative donors [13].
The cost saved by implementing DEL typing strategy is of particular significance in women of childbearing potential, as managing anti-D alloimmunized pregnancy requires fetal surveillance and therapeutic procedures, such as amniocentesis, intrauterine transfusion, and phototherapy [30]. Health care prices vary significantly across countries, and hence, each country needs to assess its own cost of managing anti-D alloimmunized pregnancies. In addition, the immunogenicity of DEL units needs to be further evaluated to estimate the cost saved by implementing a DEL typing strategy. Although the immunogenicity of DEL units is generally deemed to be low, to date, no study has reliably assessed it. In a follow-up of 13 DEL units, only 1 recipient developed anti-D in Denmark; however, the alloimmunization event might have been triggered by the concurrent transfusion of D-positive platelets [12]. Despite the relatively high prevalence of DEL, only a few anti-D alloimmunization events have been reported in Korea [10, 11].
In summary, we recommend 2 practical DEL typing strategies for serologically D-negative Asian donors. Our study shows that RhCE phenotyping is a cost-efficient option as an initial screening method. Compared with DEL typing using RHD genotyping, serological DEL typing using the adsorption-elution test is predicted to increase the risk of anti-D alloimmunization and reduce the D-negative donor pool without having any cost-competitiveness, and it can be used in laboratories where molecular methods are not applicable. Although our study included only serologically D-negative Koreans, our DEL typing strategies can be utilized as guidance for other ethnic groups with comparable DEL spectra and price levels.
Statement of Ethics
This study was approved by the Institutional Review Board of Seoul National University Bundang Hospital, Seongnam, South Korea.
Disclosure Statement
The authors declare that they have no conflicts of interest with regard to this paper.
References
- 1.Basu S, Kaur R, Kaur G. Hemolytic disease of the fetus and newborn: current trends and perspectives. Asian J Transfus Sci. 2011 Jan;5((1)):3–7. doi: 10.4103/0973-6247.75963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eder AF. Update on HDFN: new information on long-standing controversies. Immunohematology. 2006;22((4)):188–95. [PubMed] [Google Scholar]
- 3.Urbaniak SJ, Greiss MA. RhD haemolytic disease of the fetus and the newborn. Blood Rev. 2000 Mar;14((1)):44–61. doi: 10.1054/blre.1999.0123. [DOI] [PubMed] [Google Scholar]
- 4.Scott SA, Nagl L, Tilley L, Liew YW, Condon J, Flower R, et al. The RHD(1227G[{GT}]A) DEL-associated allele is the most prevalent DEL allele in Australian D- blood donors with C+ and/or E+ phenotypes. Transfusion. 2014 Nov;54((11)):2931–40. doi: 10.1111/trf.12701. [DOI] [PubMed] [Google Scholar]
- 5.Yasuda H, Ohto H, Sakuma S, Ishikawa Y. Secondary anti-D immunization by Del red blood cells. Transfusion. 2005 Oct;45((10)):1581–4. doi: 10.1111/j.1537-2995.2005.00579.x. [DOI] [PubMed] [Google Scholar]
- 6.Wagner T, Körmöczi GF, Buchta C, Vadon M, Lanzer G, Mayr WR, et al. Anti-D immunization by DEL red blood cells. Transfusion. 2005 Apr;45((4)):520–6. doi: 10.1111/j.0041-1132.2005.04256.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Li L, Tsai SL, Lin K. Anti-D immunization by Del red blood cells in Taiwan: two case reports. Transfusion. 2006;46(Suppl.):129A. [Google Scholar]
- 8.Shao CP, Wang BY, Ye SH, Zhang WL, Xu H, Zhuang NB, et al. DEL RBC transfusion should be avoided in particular blood recipient in East Asia due to allosensitization and ineffectiveness. J Zhejiang Univ Sci B. 2012 Nov;13((11)):913–8. doi: 10.1631/jzus.B1100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang QP, Dong GT, Wang XD, Gu J, Li Z, Sun AY, et al. An investigation of secondary anti-D immunisation among phenotypically RhD-negative individuals in the Chinese population. Blood Transfus. 2014 Apr;12((2)):238–43. doi: 10.2450/2013.0184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim KH, Kim KE, Woo KS, Han JY, Kim JM, Park KU. Primary anti-D immunization by DEL red blood cells. Korean J Lab Med. 2009 Aug;29((4)):361–5. doi: 10.3343/kjlm.2009.29.4.361. [DOI] [PubMed] [Google Scholar]
- 11.Yang HS, Lee MY, Park TS, Cho SY, Lee HJ, Lim G, et al. Primary anti-D alloimmunization induced by “Asian type” RHD (c.1227G[{GT}]A) DEL red cell transfusion. Ann Lab Med. 2015 Sep;35((5)):554–6. doi: 10.3343/alm.2015.35.5.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krog GR, Clausen FB, Berkowicz A, Jørgensen L, Rieneck K, Nielsen LK, et al. Is current serologic RhD typing of blood donors sufficient for avoiding immunization of recipients? Transfusion. 2011 Nov;51((11)):2278–85. doi: 10.1111/j.1537-2995.2011.03156.x. [DOI] [PubMed] [Google Scholar]
- 13.Wagner FF. RHD PCR of D-negative blood donors. Transfus Med Hemother. 2013 Jun;40((3)):172–81. doi: 10.1159/000351604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Society of Blood Transfusion Red Cell Immunogenetics and Blood Group Terminology. http://www.isbtweb.org/working-parties/red-cell-immunogenetics-and-blood-group-terminology/
- 15.Kim JY, Kim SY, Kim CA, Yon GS, Park SS. Molecular characterization of D- Korean persons: development of a diagnostic strategy. Transfusion. 2005 Mar;45((3)):345–52. doi: 10.1111/j.1537-2995.2005.04311.x. [DOI] [PubMed] [Google Scholar]
- 16.Shao CP, Maas JH, Su YQ, Köhler M, Legler TJ. Molecular background of RhD-positive, D-negative, D(el) and weak D phenotypes in Chinese. Vox Sang. 2002 Aug;83((2)):156–61. doi: 10.1046/j.1423-0410.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen JC, Lin TM, Chen YL, Wang YH, Jin YT, Yue CT. RHD 1227A is an important genetic marker for RhD(el) individuals. Am J Clin Pathol. 2004 Aug;122((2)):193–8. doi: 10.1309/3XMF-2NV5-707T-JE7X. [DOI] [PubMed] [Google Scholar]
- 18.Hong YJ, Kim H, Song EY, Cho D, Park KU, Han KS. Development of Real-time PCR and Melting Curve Analysis for the Rapid Detection of DEL with RHD (c.1222T[{GT}]C) or RHD (c.1227G[{GT}]A) Clin Lab. 2016 Oct;62((10)):1995–2000. doi: 10.7754/Clin.Lab.2016.160320. [DOI] [PubMed] [Google Scholar]
- 19.Polin H, Danzer M, Gaszner W, Broda D, St-Louis M, Pröll J, et al. Identification of RHD alleles with the potential of anti-D immunization among seemingly D- blood donors in Upper Austria. Transfusion. 2009 Apr;49((4)):676–81. doi: 10.1111/j.1537-2995.2008.02046.x. [DOI] [PubMed] [Google Scholar]
- 20.Flegel WA, von Zabern I, Wagner FF. Six years' experience performing RHD genotyping to confirm D- red blood cell units in Germany for preventing anti-D immunizations. Transfusion. 2009 Mar;49((3)):465–71. doi: 10.1111/j.1537-2995.2008.01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nuchnoi P, Thongbus J, Srisarin A, Kerdpin U, Prachayasittikul V. Clinical and laboratory update on the DEL variant. Lab Med. 2014;45((4)):285–90. doi: 10.1309/LMTUZ00O7VFTGCEB. [DOI] [PubMed] [Google Scholar]
- 22.Seo MH, Won EJ, Hong YJ, Chun S, Kwon JR, Choi YS, et al. An effective diagnostic strategy for accurate detection of RhD variants including Asian DEL type in apparently RhD-negative blood donors in Korea. Vox Sang. 2016 Nov;111((4)):425–30. doi: 10.1111/vox.12450. [DOI] [PubMed] [Google Scholar]
- 23.Luettringhaus TA, Cho D, Ryang DW, Flegel WA. An easy RHD genotyping strategy for D- East Asian persons applied to Korean blood donors. Transfusion. 2006 Dec;46((12)):2128–37. doi: 10.1111/j.1537-2995.2006.01042.x. [DOI] [PubMed] [Google Scholar]
- 24.Kim B, Lee ST, Kim S, Choi JR, Kim HO. Application of Multiplex Ligation-Dependent Probe Amplification Assay for Genotyping Major Blood Group Systems Including DEL Variants in the D-Negative Korean Population. Ann Lab Med. 2018 Jan;38((1)):32–8. doi: 10.3343/alm.2018.38.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srijinda S, Suwanasophon C, Visawapoka U, Pongsavee M. RhC phenotyping, adsorption/elution test, and SSP-PCR: the combined test for D-elute phenotype screening in Thai RhD-negative blood donors. ISRN Hematol. 2012;2012:358316. doi: 10.5402/2012/358316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crottet SL, Henny C, Meyer S, Still F, Stolz M, Gottschalk J, et al. Implementation of a mandatory donor RHD screening in Switzerland. Transfus Apheresis Sci. 2014 Apr;50((2)):169–74. doi: 10.1016/j.transci.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Mota M, Dezan M, Valgueiro MC, Sakashita AM, Kutner JM, Castilho L. RHD allelic identification among D-Brazilian blood donors as a routine test using pools of DNA. J Clin Lab Anal. 2012 Feb;26((2)):104–8. doi: 10.1002/jcla.21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Hou L, Guo ZH, Ye LY, Yue DQ, Zhu ZY. Molecular basis of the RHD gene in blood donors with DEL phenotypes in Shanghai. Vox Sang. 2009 Aug;97((2)):139–46. doi: 10.1111/j.1423-0410.2009.01181.x. [DOI] [PubMed] [Google Scholar]
- 29.Lopez GH, Turner RM, McGowan EC, Schoeman EM, Scott SA, O'Brien H, et al. A DEL phenotype attributed to RHD Exon 9 sequence deletion: slipped-strand mispairing and blood group polymorphisms. Transfusion. 2018 Mar;58((3)):685–91. doi: 10.1111/trf.14439. [DOI] [PubMed] [Google Scholar]
- 30.Chilcott J, Tappenden P, Lloyd Jones M, Wight J, Forman K, Wray J, et al. The economics of routine antenatal anti-D prophylaxis for pregnant women who are rhesus negative. BJOG. 2004 Sep;111((9)):903–7. doi: 10.1111/j.1471-0528.2004.00226.x. [DOI] [PubMed] [Google Scholar]